Abstract
The bacterial CRISPR/Cas9 system for genome editing has greatly expanded the toolbox for mammalian genetics, enabling the rapid generation of isogenic cell lines and mice with modified alleles. Here, we describe a pooled, loss-of-function genetic screening approach suitable for both positive and negative selection that uses a genome-scale lentiviral single guide RNA (sgRNA) library. sgRNA expression cassettes were stably integrated into the genome, which enabled a complex mutant pool to be tracked by massively parallel sequencing. We used a library containing 73,000 sgRNAs to generate knockout collections and performed screens in two human cell lines. A screen for resistance to the nucleotide analog 6-thioguanine identified all expected members of the DNA mismatch repair pathway, while another for the DNA topoisomerase II (TOP2A) poison etoposide identified TOP2A, as expected, and also cyclin-dependent kinase 6, CDK6. A negative selection screen for essential genes identified numerous gene sets corresponding to fundamental processes. Finally, we show that sgRNA efficiency is associated with specific sequence motifs, enabling the prediction of more effective sgRNAs. Collectively, these results establish Cas9/sgRNA screens as a powerful tool for systematic genetic analysis in mammalian cells.
A critical need in biology is the ability to efficiently identify the set of genes underlying a cellular process. In microorganisms, powerful methods allow systematic loss-of-function genetic screening (1, 2). In mammalian cells, however, current screening methods fall short – primarily because of the difficulty of inactivating both copies of a gene in a diploid mammalian cell. Insertional mutagenesis screens in cell lines that are near-haploid or carry Blm mutations, that cause frequent somatic crossing-over, have proven powerful but are not applicable to most cell lines and suffer from integration biases of the insertion vectors (3, 4). The primary solution has been to target mRNAs with RNA interference (RNAi) (5–9). However, this approach is also imperfect as it only partially suppresses target gene levels and can have off-target effects on other mRNAs – resulting in false negative and false positive results (10–12). Thus, there remains an unmet need for an efficient, large-scale, loss of function screening method in mammalian cells
Recently, the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) pathway, which functions as an adaptive immune system in bacteria (13), has been co-opted to engineer mammalian genomes in an efficient manner (14–16). In this two-component system, a single guide RNA (sgRNA) directs the Cas9 nuclease to cause double-stranded cleavage of matching target DNA sequences (17). In contrast to previous genome-editing techniques, such as zinc-finger nucleases and TALENs, the target specificity of CRISPR/Cas9 is dictated by a 20-base pair sequence at the 5′-end of the sgRNA, allowing for much greater ease of construction of knockout reagents. Mutant cells lines and mice bearing multiple modified alleles can be generated with this technology (18, 19).
We set out to explore the feasibility of using the CRISPR/Cas9 system to perform large-scale, loss-of-function screens in mammalian cells. The idea was to use a pool of sgRNA-expressing lentivirus to generate a library of knockout cells that could be screened under both positive and negative selection. Each sgRNA would serve as a distinct DNA barcode that can be used to count the number of cells carrying it using high-throughput sequencing (Fig. 1A). Pooled screening requires that single-copy sgRNA integrants are sufficient to induce efficient cleavage of both copies of a targeted locus. This contrasts with the high expression of sgRNAs achieved by transfection that is typically used to engineer a specific genomic change using the CRISPR/Cas9 system.
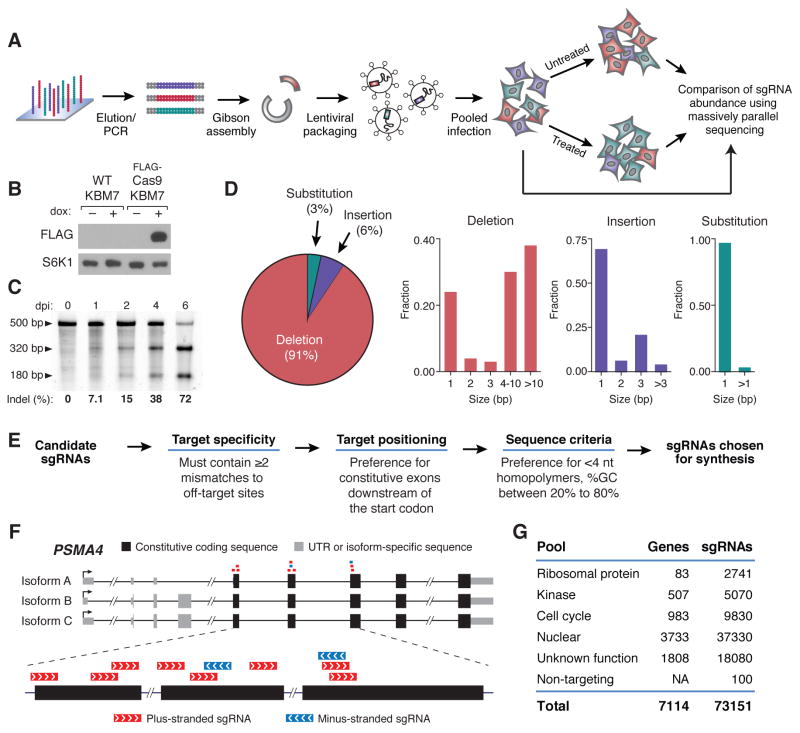
Fig. 1. A pooled approach for genetic screening in mammalian cells using a lentiviral CRISPR/Cas9 system.
(A) Outline of sgRNA library construction and genetic screening strategy (B) Immunoblot analysis of wild-type KBM7 cells and KBM7 cells transduced with a doxycycline inducible FLAG-Cas9 construct upon doxycycline induction. S6K1 was used as a loading control. (C) Sufficiency of single copy sgRNAs to induce genomic cleavage. Cas9-expressing KBM7 cells were transduced with AAVS1-targeting sgRNA lentivirus at low MOI. The SURVEYOR mutation detection assay was performed on cells at the indicated days post-infection (dpi). Briefly, mutations resulting from cleavage of the AAVS1 locus were detected through PCR amplification of a 500-bp amplicon flanking the target sequence, re-annealing of the PCR product and selective digestion of mismatched heteroduplex fragments. (D) Characterization of mutations induced by CRISPR/Cas9 as analyzed by high-throughput sequencing. (E) sgRNA library design pipeline. (F) Example of sgRNAs designed for PSMA4. sgRNAs targeting constitutive exonic coding sequences nearest to the start codon were chosen for construction. (G) Composition of genome-scale sgRNA library.
We first tested the concept in the near-haploid, human KBM7 CML cell line, by creating a clonal derivative expressing the Cas9 nuclease (with a FLAG-tag at its N-terminus) under a doxycycline-inducible promoter (Fig. 1B). Transduction of these cells at low multiplicity of infection (MOI) with a lentivirus expressing a sgRNA targeting the endogenous AAVS1 locus revealed substantial cleavage at the AAVS1 locus 48 hours after infection (Fig. 1C). Moreover, because the sgRNA was stably expressed, genomic cleavage continued to increase over the course of the experiment. Deep sequencing of the locus revealed that repair of Cas9-induced double-strand breaks resulted in small deletions (<20 bp) in the target sequence, with tiny insertions or substitutions (<3 bp) occurring at a lower frequency (Fig. 1D). The vast majority of the lesions, occurring in a protein-coding region, would be predicted to give rise to a nonfunctional protein product, indicating that CRISPR/Cas9 is an efficient means of generating loss-of-function alleles.
We also analyzed off-target activity of CRISPR/Cas9. Although the specificity of CRISPR/Cas9 has been extensively characterized in transfection-based settings (20–22), we wanted to examine its off-target behavior in our system, where Cas9 and a single guide RNA targeting AAVS1 (sgAAVS1) were stably expressed for two weeks. We compared the level of cleavage observed at the target locus (97%) to levels at 13 potential off-target cleavage sites in the genome (defined as sites differing by up to 3 bp from sgAAVS1). Minimal cleavage (<2.5%) was observed at all sites with one exception, which was the only site that had perfect complementarity in the ‘seed’ region (terminal 8 bp). On average, sgRNAs have ~2.2 such sites in the genome, almost always (as in this case) occurring in non-coding DNA and thus less likely to affect gene function (Note S1).
To test the ability to simultaneously screen tens of thousands of sgRNAs, we designed a sgRNA library with 73,151 members, consisting of multiple sgRNAs targeting 7,114 genes and 100 non-targeting controls (Methods, Fig. 1E, Table S1). sgRNAs were designed against constitutive coding exons near the beginning of each gene and filtered for potential off-target effects based on sequence similarity to the rest of the human genome (Fig. 1, F and G). The library included 10 sgRNAs for each of 7033 genes and all possible sgRNAs for each of the 84 genes encoding ribosomal proteins (Fig. 1H). To assess the effective representation of our microarray synthesized library, we sequenced sgRNA barcodes from KBM7 cells 24 hours after infection with the entire lentiviral pool and were able to detect the overwhelming majority (>99%) of our sgRNAs, with high uniformity across constructs (only 6-fold increase in abundance between the 10th and 90th percentiles) (Fig. S2A).
As an initial test of our approach, we screened the library for genes that function in DNA mismatch repair (MMR). In the presence of the nucleotide analog 6-thioguanine (6-TG), MMR-proficient cells are unable to repair 6-TG-induced lesions and arrest at the G2-M cell-cycle checkpoint, while MMR-defective cells do not recognize the lesions and continue to divide (23). We infected Cas9-KBM7 cells with the entire sgRNA library, cultured the cells in a concentration of 6-TG that is lethal to wild-type KBM7 cells, and sequenced the sgRNA barcodes in the final population. sgRNAs targeting the genes encoding the four components of the MMR pathway (MSH2, MSH6, MLH1 and PMS2) (24) were dramatically enriched in the 6-TG-treated cells. At least four independent sgRNAs for each gene showed very strong enrichment and barcodes corresponding to these genes made up >30% of all barcodes (Fig. 2, A and B). Strikingly, each of the twenty most abundant sgRNAs targeted one of these four genes. The fact that few of the other 73,000 sgRNAs scored highly in this assay suggests a low frequency of off-target effects.
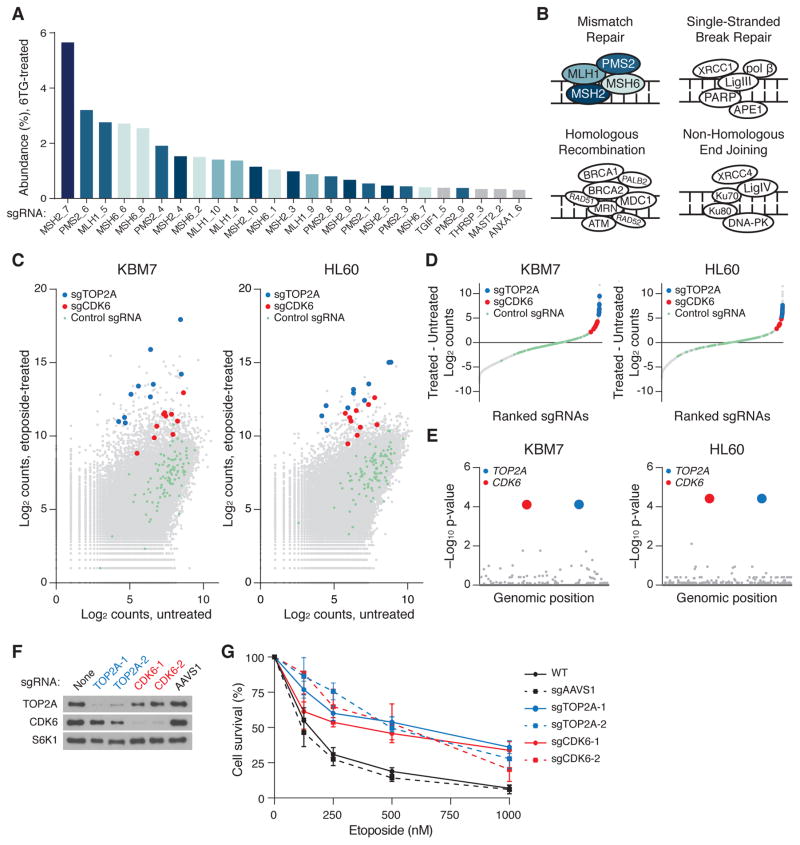
Fig. 2. Resistance screens using CRISPR/Cas9.
(A) Raw abundance (%) of sgRNA barcodes after 12 days of selection with 6-thioguanine (6-TG). (B) Mismatch repair (MMR) deficiency confers resistance to 6-TG. Diagram depicts cellular DNA repair processes. Only sgRNAs targeting components of the DNA MMR pathway were enriched. Diagram modified and adapted from (32). (C) Primary etoposide screening data. The count for a sgRNA is defined as the number of reads that perfectly match the sgRNA target sequence. (D) sgRNAs from both screens were ranked by their differential abundance between the treated versus untreated populations. For clarity, sgRNAs with no change in abundance are omitted. (E) Gene hit identification by comparing differential abundances of all sgRNAs targeting a gene to differential abundances of non-targeting sgRNAs in a one-sided Kolmogorov-Smirnov test. p-values are corrected for multiple hypothesis testing. (F) Immunoblot analysis of WT and sgRNA-modified HL60 cells 1 week after infection. S6K1 was used as a loading control. (G) Viability, as measured by cellular ATP concentration, of WT and sgRNA-modified HL60 cells at indicated etoposide concentrations. Error bars denote standard deviation (n=3).
We next addressed the challenge of loss of function screening in diploid cells, which require bi-allelic inactivation of a target gene. We therefore generated an inducible Cas9 derivative of the HL60 pseudo-diploid human leukemic cell line. In both HL60 and KBM7 cells, we screened for genes whose loss conferred resistance to etoposide, a chemotherapeutic agent that poisons DNA topoisomerase IIA (TOP2A). To identify hit genes, we calculated the difference in abundance between the treated and untreated populations for each sgRNA, calculated a score for each gene by using a Kolmogorov-Smirnov test to compare the sgRNAs targeting the gene against the non-targeting control sgRNAs, and corrected for multiple hypothesis testing (Fig. 2, C to E, Table S2). Identical genes were detected in both screens, with significance levels exceeding all other genes by more than 100-fold. As expected, loss of TOP2A itself conferred strong protection to etoposide (25). The screen also revealed a role for CDK6, a G1 cyclin-dependent kinase, in mediating etoposide-induced cytotoxicity. Notably, every one of the 20 sgRNAs in the library targeting TOP2A or CDK6 was strongly enriched (>90th percentile) in both screens, indicating that the effective coverage of our libraries is very high. We generated isogenic HL60 cell lines with individual sgRNAs against TOP2A and CDK6 and, consistent with the screen results, these lines were much more resistant to etoposide than parental or sgAAVS1-modified HL60 cells (Fig. 2, F and G). Thus, our Cas9/sgRNA system enables large-scale positive selection loss-of-function screens.
To identify genes required for cellular proliferation we screened for genes whose loss conferred a selective disadvantage on cells. Such a screen requires accurate identification of sgRNAs that are depleted from the final cell population. Importantly, a sgRNA will show depletion only if cleavage of the target gene occurs in the majority of cells carrying the construct.
As an initial test, we screened KBM7 cells with a small library containing sgRNAs targeting the BCR and ABL1 genes (Table S3). The survival of KBM7 cells depends on the fusion protein produced by the BCR-ABL translocation (26). As expected, depletion was seen only for sgRNAs targeting the exons of BCR and ABL1 that encode the fusion protein, but not for those targeting the other exons of BCR and ABL1 (Fig. 3A).
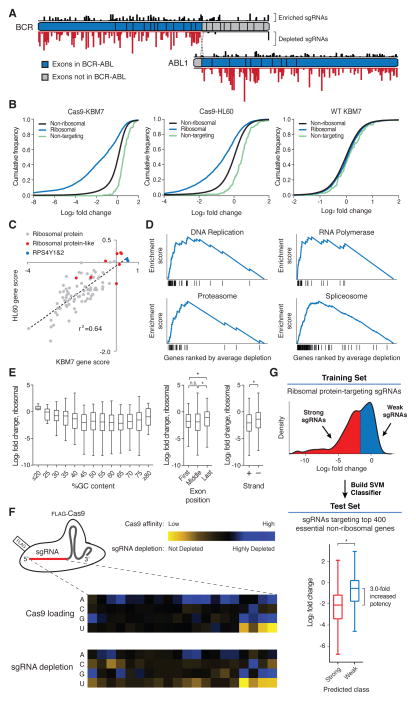
Fig. 3. Negative selection screens using CRISPR/Cas9 reveal rules governing sgRNA efficacy.
(A) Selective depletion of sgRNAs targeting exons of BCR and ABL1 present in the fusion protein. Individual sgRNAs are plotted according to their target sequence position along each gene and the height of each bar indicates the level of depletion observed. Boxes indicate individual exons. (B) Cas9-dependent depletion of sgRNAs targeting ribosomal proteins. Cumulative distribution function plots of log2 fold changes in sgRNA abundance before and after twelve cell doublings in Cas9-KBM7, Cas9-HL60 and WT-KBM7 cells. (C) Requirement of similar sets of ribosomal protein genes for proliferation in the HL60 and KBM7 cells. Gene scores are defined as the median log2 fold change of all sgRNAs targeting a gene. (D) Depleted sgRNAs target genes involved in fundamental biological processes. Gene Set Enrichment Analysis was performed on genes ranked by their combined depletion scores from screens in HL60 and KBM7 cells. Vertical lines underneath the x-axis denote members of the gene set analyzed. (E) Features influencing sgRNA efficacy. Depletion (log2 fold change) of sgRNAs targeting ribosomal protein genes was used as an indicator of sgRNA efficacy. Correlation between log2 fold changes and spacer %GC content (left), exon position targeted (middle) and strand targeted (right) are depicted. (*p<0.05) (F) sgRNA target sequence preferences for Cas9 loading and cleavage efficiency. Position-specific nucleotide preferences for Cas9 loading are determined by counting sgRNAs bound to Cas9 normalized to the number of corresponding genomic integrations. Heatmaps depict sequence-dependent variation in Cas9 loading (top) and ribosomal protein gene-targeting sgRNA depletion (bottom). The color scale represents the median value (of Cas9 affinity or log2 fold-change) for all sgRNAs with the specified nucleotide at the specified position. (G) sgRNA efficacy prediction. Ribosomal protein gene-targeting sgRNAs were designated as ‘weak’ or ‘strong’ based their log2 fold change and used to train a support-vector-machine (SVM) classifier. As an independent test, the SVM was used to predict the efficacy of sgRNAs targeting 400 essential non-ribosomal genes. (*p<0.05)
We then infected Cas9-HL60, Cas9-KBM7, and WT KBM7 cells with the entire 73,000-member sgRNA library and used deep sequencing of the sgRNA barcodes to monitor the change in abundance of each sgRNA between the initial seeding and a final population obtained after twelve cell doublings (Fig. S2, A and B).
We began by analyzing ribosomal proteins genes, for which the library contained all possible sgRNAs. We observed strong Cas9-dependent depletion of sgRNAs targeting genes encoding ribosomal proteins, with good concordance between the sets of ribosomal protein genes essential for cell proliferation in the HL60 and KBM7 screens (the median sgRNA fold-change in abundance was used as a measure of gene essentiality) (Fig. 3, B and C). Interestingly, a few ribosomal protein genes were not found to be essential. These were two genes encoded on chromosome Y (RPS4Y2, which is testes-specific (27), and RPS4Y1, which is expressed at low levels compared to its homolog RPS4X on chromosome X (28)), and ‘ribosome-like’ proteins, which may be required only in select tissues (27) and generally are lowly expressed in KBM7 cells (Fig. S3A).
We then turned our attention to other genes within our dataset, for which ten sgRNAs were designed. As for the ribosomal genes, the essentiality scores of these genes were also strongly correlated between the two cells lines (Fig. S3B, Table S4). For the twenty highest scoring genes, we found independent evidence for essentiality, based primarily on data from large-scale functional studies in model organisms (Table S5).
To evaluate the results at a global level, we tested 4722 gene sets to see if they showed strong signatures of essentiality, using Gene Set Enrichment Analysis (29). Gene sets related to fundamental biological processes – including DNA replication, gene transcription, and protein degradation – showed strong depletion, consistent with their essentiality (Fig. 3D, Table S6).
Finally, we sought to understand the features underlying sgRNA efficacy. Although the vast majority of sgRNAs against ribosomal protein genes showed depletion, detailed comparison of sgRNAs targeting the same gene revealed substantial variation in the precise amounts of depletion. These differences are unlikely to be caused by local accessibility to the Cas9/sgRNA complex inasmuch as comparable variability was observed even among sgRNAs targeting neighboring target sites of a given gene (Fig. S4A). Given that our library includes all possible sgRNAs against each of the 84 ribosomal genes, the data allowed us to search for factors that might explain the differential efficacy of sgRNAs. Because the majority of ribosomal proteins genes are essential, we reasoned that the level of depletion of a given ribosomal protein-targeting sgRNA could serve as a proxy for its cleavage efficiency. Applying this approach, we found several trends related to sgRNA efficacy: (1) Single guide sequences with very high or low GC content were less effective against their targets. (2) sgRNAs targeting the last coding exon were less effective than those targeting earlier exons, consistent with the notion that disruption of the terminal exon would be expected to have less impact on gene function. (3) sgRNAs targeting the transcribed strand were less effective than those targeting the non-transcribed strand (Fig. 3E). Although these trends were statistically significant, they explained only a small proportion of differences in sgRNA efficacy (Table S7).
We hypothesized that differences in sgRNA efficacy might also result from sequence features governing interactions with Cas9. To test this, we developed a method to profile the sgRNAs directly bound to Cas9 in a highly parallel manner (Methods). By comparing the abundance of sgRNAs bound to Cas9 relative to the abundance of their corresponding genomic integrants, we found that the nucleotide composition near the 3′-end of the spacer sequence was the most important determinant of Cas9 loading (Fig. 3F). Specifically, Cas9 preferentially bound sgRNAs containing purines in the last 4 nucleotides of the spacer sequence whereas pyrimidines were disfavored. A similar pattern emerged when we examined depletion of ribosomal protein-targeting sgRNAs (r=0.81), suggesting that, in significant part, the cleavage efficiency of a sgRNA was determined by its affinity for Cas9 (Table S7).
We then sought to build an algorithm to discriminate between strong and weak sgRNAs (Fig. 3G). We trained a support-vector-machine classifier based on the target sequences and depletion scores of ribosomal protein-targeting single guide RNAs. As an independent test, we used the classifier to predict the efficacy of sgRNAs targeting the 400 top scoring (i.e. essential) non-ribosomal genes. The top two-thirds of our predictions exhibited 3-fold higher efficacy than the remaining fraction, confirming the accuracy of the algorithm.
Using this algorithm, we designed a whole-genome sgRNA library consisting of sequences predicted to have higher efficacy (Table S8). As with the sgRNA pool used in our screens, this new collection was also filtered for potential off-target matches. This reference set of sgRNAs may be useful both for targeting single genes as well as large-scale sgRNA screening.
Taken together, these results demonstrate the utility of CRISPR/Cas9 for conducting large-scale genetic screens in mammalian cells. Based on our initial experiments, this system appears to offer several powerful features that together provide significant advantages over current functional screening methods.
First, CRISPR/Cas9 inactivates genes at the DNA level, making it possible to study phenotypes that require a complete loss of gene function to be elicited. In addition, the system should also enable functional interrogation of non-transcribed elements, which are inaccessible by RNAi.
Second, a large proportion of sgRNAs successfully generate mutations at their target sites. While this parameter is difficult to directly assess in pooled screens, we can obtain an estimate by examining the ‘hit rate’ at known genes. Applying a z-score analysis of our positive selection screens, we find that over 75% (46/60) of sgRNAs score at a significance threshold that perfectly separates true and false positives on a gene level (Fig. S5, A to D). Together these results show that the effective coverage of our library is very high and that the rate of false negatives should be low even in a large-scale screen.
Third, off-target effects do not appear to seriously hamper our screens, based on several lines of evidence. Direct sequencing of potential off-target loci detected minimal cleavage at secondary sites, which typically reside in non-coding regions and do not impact gene function. Moreover, in the 6-TG screens, the twenty most abundant sgRNAs all targeted one of the four members of the MMR pathway. In total, they represented over 30% of the final pool, a fraction greater the next 400 sgRNAs combined. In the etoposide screen, the two top genes scored far above background levels (p-values 100-fold smaller than the next best gene), enabling clear discrimination between true and false positive hits. Lastly, new versions of the CRISPR/Cas9 system have recently been developed that substantially decrease off-target activity (30, 31).
Although we limited our investigation to proliferation-based phenotypes, our approach can be applied to a much wider range of biological phenomena. With appropriate sgRNA libraries, the method should enable genetic analyses of mammalian cells to be conducted with a degree of rigor and completeness currently possible only in the study of microorganisms.
Supplementary Material
Acknowledgments
We thank all members of the Sabatini and Lander labs especially J. Engreitz, S. Schwartz, A. Shishkin, and Z. Tsun for protocols, reagents and advice; T. Mikkelsen for assistance with oligonucleotide synthesis; L. Gaffney for assistance with figures. This work was supported by the US National Institutes of Health (CA103866) (D.M.S.), National Human Genome Research Institute (2U54HG003067-10) (E.S.L.), the Broad Institute of MIT and Harvard (E.S.L.) and an award from the US National Science Foundation (T.W.). The composition of the sgRNA pools and screening data can be found in the Supporting Online Materials. T.W., D.M.S., and E.S.L. are inventors on a patent application from the Broad Institute for functional genomics using CRISPR-Cas systems. Inducible Cas9 and sgRNA backbone lentiviral vectors and the genome-scale sgRNA plasmid pool are deposited in Addgene.
Footnotes
Materials and Methods
References (33–43)
References and Notes
- 1.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 2.Costanzo M, et al. The Genetic Landscape of a Cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carette JE, et al. Haploid Genetic Screens in Human Cells Identify Host Factors Used by Pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 4.Guo G, Wang W, Bradley A. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature. 2004;429:891–895. doi: 10.1038/nature02653. [DOI] [PubMed] [Google Scholar]
- 5.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Brummelkamp TR, Bernards R, Agami R. A System for Stable Expression of Short Interfering RNAs in Mammalian Cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 7.Moffat J, et al. A Lentiviral RNAi Library for Human and Mouse Genes Applied to an Arrayed Viral High-Content Screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 8.Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 9.Cheung HW, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proceedings of the National Academy of Sciences. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Echeverri CJ, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Meth. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 11.Booker M, et al. False negative rates in Drosophila cell-based RNAi screens: a case study. BMC Genomics. 2011;12:50. doi: 10.1186/1471-2164-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaelin WG. Use and Abuse of RNAi to Study Mammalian Gene Function. Science. 2012;337:421–422. doi: 10.1126/science.1225787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrangou R, et al. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 14.Cong L, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mali P, et al. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotech. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jinek M, et al. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horii T, Tamura D, Morita S, Kimura M, Hatada I. Generation of an ICF Syndrome Model by Efficient Genome Editing of Human Induced Pluripotent Stem Cells Using the CRISPR System. International Journal of Molecular Sciences. 2013;14:19774–19781. doi: 10.3390/ijms141019774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, et al. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotech. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotech. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotech. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan T, Berry SE, Desai AB, Kinsella TJ. DNA Mismatch Repair (MMR) Mediates 6-Thioguanine Genotoxicity by Introducing Single-strand Breaks to Signal a G2-M Arrest in MMR-proficient RKO Cells. Clinical Cancer Research. 2003;9:2327–2334. [PubMed] [Google Scholar]
- 24.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Current Opinion in Genetics & Development. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 25.Burgess DJ, et al. Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proceedings of the National Academy of Sciences. 2008;105:9053–9058. doi: 10.1073/pnas.0803513105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scappini B, et al. Changes associated with the development of resistance to imatinib (STI571) in two leukemia cell lines expressing p210 Bcr/Abl protein. Cancer. 2004;100:1459–1471. doi: 10.1002/cncr.20131. [DOI] [PubMed] [Google Scholar]
- 27.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston CM, et al. Large-Scale Population Study of Human Cell Lines Indicates that Dosage Compensation Is Virtually Complete. PLoS Genet. 2008;4:e9. doi: 10.1371/journal.pgen.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ran FA, et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotech. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshimoto K, et al. Complex DNA repair pathways as possible therapeutic targets to overcome temozolomide resistance in glioblastoma. Frontiers in Oncology. 2012;2 doi: 10.3389/fonc.2012.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.