Abstract
Rapid three-dimensional imaging of embryos to better understand the complex process of morphogenesis has been challenging. Recently introduced iodine staining protocols (I2KI and alcoholic iodine stains) combined with microscopic X-ray computed tomography allows visualization of soft tissues in diverse small organisms and tissue specimens. I2KI protocols have been developed specifically for small animals, with a limited number of quantitative studies of soft tissue contrasts. To take full advantage of the low X-ray attenuation of ethanol and retain bound iodine while dehydrating the specimen in ethanol, we developed an ethanol I2KI protocol. We present comparative microscopic X-ray computed tomography analyses of ethanol I2KI and I2KI staining protocols to assess the performance of this new protocol to visualize soft tissue anatomy in late stage Japanese quail embryos using quantitative measurements of soft tissue contrasts and sample shrinkage. Both protocols had only 5% shrinkage compared with the original harvested specimen, supporting the use of whole mounts to minimize tissue shrinkage effects. Discrimination within and among the selected organs with each staining protocol and microscopic X-ray computed tomography imaging were comparable to those of a gray scale histological section. Tissue discrimination was assessed using calibrated computed tomography values and a new discrimination index to quantify the degree of computed tomography value overlaps between selected soft tissue regions. Tissue contrasts were dependent on the depth of the tissue within the embryos before the embryos were saturated with each stain solution, and optimal stain saturations for the entire embryo were achieved at 14 and 28 days staining for I2KI and ethanol I2KI, respectively. Ethanol I2KI provided superior soft tissue contrasts by reducing overstaining of fluid-filled spaces and differentially modulating staining of some tissues, such as bronchial and esophageal walls and spinal cord. Delineating the selected soft tissues using optimal threshold ranges derived from the quantitative analyses of the contrast enhancement in optimally stained embryos is possible. The protocols presented here are expected to be applicable to other organisms with modifications to staining time and contribute toward rapid and more efficient segmentation of soft tissues for three-dimensional visualization.
Keywords: avian embryonic development, calibrated computed tomography values, iodine, microscopic X-ray computed tomography, three-dimensional quantitative imaging, X-ray computed tomography
Introduction
Methods of capturing complex developmental processes have transformed from histological preparations and images of whole and dissected embryos to current technologies including microscopic magnetic resonance imaging (microMRI), microscopic X-ray computed tomography (microCT) and optical projection tomography (OPT). Compared with traditional methods using serial sections of specimens and subsequent three-dimensional (3D) reconstructions, microMRI, microCT, and OPT technologies are much faster and provide inherently registered images of biological samples but each approach has tradeoffs, depending on the final imaging goals (e.g. see reviews Paulus et al. 2001; Sharpe, 2004). OPT machines can capture 3D images of gene and protein expressions using in situ hybridization and immunohistochemical methods and fluorescence ex vivo (e.g. Sharpe et al. 2002) and in vivo (e.g. Boot et al. 2008). However, major limitations of OPT machines include small specimen sizes (< 10 × 15 mm: Bioptonics) and the need for specimens to be optically cleared for light to completely penetrate the specimen. MicroMRI is capable of visualizing tissues of optically opaque samples ex vivo (e.g. Dhenain et al. 2001; Johnson et al. 2002; Ruffins et al. 2007; Petiet et al. 2008) and in vivo (e.g. Bain et al. 2007; Li et al. 2007; Hogers et al. 2009; Duce et al. 2011). Currently, microMRI machines are limited to approximately 20–25 μm resolution with relatively longer scanning times (generally, hours per scan), whereas OPT and microCT achieve much higher resolutions with shorter image acquisition times. Many microCT machines have a greater range of scanning dimensions and achieve scanning resolutions of 1–50 μm (Ritman, 2011) with short acquisition times, depending on the size of the specimen and machine type. For example, the SkyScan 1172 machine used here has a scanning envelope of up to approximately 75 mm in height and 68 mm in width. A major constraint of X-ray computed tomography is the low intrinsic contrast of soft tissues measured by the attenuated X-ray intensities (Kalender, 2005). MicroCT has been widely used for bone and tooth research but has only recently been used for soft tissue imaging with the aid of contrast agents.
Contrast agents have been used orally, intravenously or rectally in medicine to highlight particular fluid-filled cavities, such as the gastrointestinal tract and vasculature (Woźniak, 2008). MicroCT with an aid of contrast agents has become an important tool for small animal soft tissue imaging, such as renal organs, abdominal tumors, and vasculature both in vivo and ex vivo (reviewed in Paulus et al. 2001; Schambach et al. 2010; Ritman, 2011; Gregg & Butcher, 2012). There are several types of iodine-based contrast agents, including organ specific iodinate contrast agents, injected intravascularly or intraperitoneally (e.g. for organ specific contrast agents, see Henning et al. 2008; Willekens et al. 2009). Commercially available iodine-based contrast agents have been commonly used intravascularly in vivo with microCT imaging of vascular spaces in model animals (reviewed by Badea et al. 2008a). These media do not permeate quickly across tissue boundaries, remain localized to the injected vasculature and then are cleared by the kidney or liver. The transient nature of these contrast media enables visualization of living vasculature and abdominal organs through time. These contrast media have been widely used in live adult mice (e.g. Badea et al. 2005, 2008b; Ford et al. 2006; Almajdub et al. 2008; Schambach et al. 2010; Prajapati & Keller, 2011) and their use has been recently demonstrated in live chick embryos (Henning et al. 2011). Other contrast media can be diffused into tissues ex vivo by soaking fixed samples (rabbit and mice brains: de Crespigny et al. 2008; mice brains: Saito & Murase, 2012).
Ex vivo microCT soft tissue imaging has used similar techniques as in vivo imaging. As an alternative to commonly used contrast medium, osmium tetroxide, which is toxic and volatile, phosphotungstic acid and iodine-based contrast agents (I2KI, alcoholic iodine) have been introduced to microCT imaging of diverse organisms that allowed high soft tissue contrast (Metscher, 2009a,2009b, 2011; see review of Mizutani & Suzuki, 2012 for other contrast agents). Iodine solutions permeate soft tissues rapidly as a general cellular stain (Kiernan, 2008) and allow for nearly histological quality imaging. Osmium tetroxide binds preferentially to lipids, such as cell membranes and nerves (Kiernan, 2008), and has been successfully demonstrated in mice, chicks, rabbits, and honey bees (Johnson et al. 2006; Litzlbauer et al. 2006; Bentley et al. 2007; Carney et al. 2007; Zhu et al. 2007; Ribi et al. 2008; Martinez et al. 2009; Kim et al. 2011; Pai et al. 2012). Phosphotungstic and phosphomolybdic acids (both known as heteropolyacids) bind preferentially to proteins and collagen (Kiernan, 2008) and the latter has been applied to image molluscan anatomy (Golding & Jones, 2007; Golding et al. 2009). Although phosphotungstic acids successfully generate high soft tissue contrasts, their usage requires caution because of possible decalcification or other chemical effects on stained tissues due to their acidic nature (Metscher, 2009b). The dominance of iodine staining in microCT imaging (e.g. Baverstock et al. 2013; Vickerton et al. 2013) justify limiting our discussion to only iodine staining of soft tissues.
A range of iodine staining protocols has been developed for individual studies: mice hearts in embryos and neonates (Degenhardt et al. 2010), adult mice cranial muscles (Jeffery et al. 2011), squirrel, guinea pig, and rat cranial muscles (Cox & Jeffery, 2011), adult rat and rabbit hearts (Stephenson et al. 2012), and adult dog heart (Aslanidi et al. 2012). Many of these studies reported that muscle tissue is remarkably stained with iodine compared with connective tissue. Thus, staining techniques have been generally employed to study muscles. The three-dimensionally reconstructed morphologies of these structures are useful to further examine biomechanical aspects of these morphologies (see Baverstock et al. 2013; Vickerton et al. 2013 for complete reviews of publications examining iodine enhanced muscles via microCT, and references therein). Only a few have studied the effects of staining concentrations and/or durations on tissue contrast enhancement qualitatively (Jeffery et al. 2011; Aslanidi et al. 2012) or quantitatively (early chick embryo: Metscher, 2009b; Degenhardt et al. 2010). Comparing concentrations of iodine solutions is complicated because some authors describe diluted percentages from various available concentrations of iodine [e.g. I2KI is available in 1 and 5% (w/v)]. To avoid confusion arising from describing concentrations of iodine and quantifying soft tissue contrasts between different iodine solutions over time, we define our iodine solution by molar concentration. Quantitative studies of soft tissue contrasts with I2KI and alcoholic iodine staining via microCT have been limited by: (i) the use of uncalibrated pixel intensity values (gray scale) (Metscher, 2009b); (ii) the absence of the CT values; (iii) using average pixel intensities of two regions to estimate signal-to-noise ratios (Wicklein et al. 2012) and K factors; (iv) K factors that are not reported (but see Degenhardt et al. (2010); for the first report of this value); and (v) not considering the relationship between tissue or organ types and depths within the stained sample. CT values are calibrated to water as Hounsfield units (HU) in most medical CT scanners but not in microCT scanners. MicroCT scanners can be calibrated with phantoms but this is rarely reported in non-clinical research and is not necessary for qualitative results. CT values must be calibrated to identify specific tissues, but universal usage of CT values as a contrast measurement index is complicated by the fact that values vary with sample size (Siegel et al. 2004), voltage (Nakayama et al. 2005; Miles et al. 2007), and between CT systems (Miles et al. 2007). Furthermore, comparisons of CT values between clinical and microCT systems is complicated by differences in X-ray beam geometries (Siewerdsen & Jaffray, 2001; Kalender, 2005; Katsumata et al. 2007; Kachelrieß, 2008; Mah et al. 2010; Chindasombatjaroen et al. 2011). An attempt has been made to provide an alternative standard value, called a K factor, which is the slope of the linear relationship between the CT values and iodine concentration, in order to overcome these factors with iodine-based staining methods (Garrett et al. 1985; Miles et al. 2007).
Two kinds of iodine staining solutions are currently used in ex vivo microCT imaging due to their accessibility and low toxicity. I2KI (Lugol's solution) is made of iodine metal and potassium iodine dissolved in distilled water, and I2E or I2M (alcoholic iodine) is made of iodine dissolved in ethanol or methanol. Generally, I2KI staining begins with soaking an already fixed sample in Lugol's solution, which is subsequently washed in either water or an alcohol series for scanning. Alcoholic iodine staining first dehydrates the sample in an alcohol series, followed by soaking in alcoholic iodine with a final washing in alcohol before scanning. We focus on protocols that make use of the low X-ray attenuation of alcohol because it better discriminates tissues from background noise (Metscher, 2009a,b). In the former protocol, the dehydration step may wash out some iodine from the stained sample, whereas in the latter, the lengthy submersion in alcohol may cause stiffness and shrinkage of the sample (Fig. 1). Shrinkage associated with iodine staining has been quantified with a comparison of vessel diameters between I2KI-stained samples and histological sections (Degenhardt et al. 2010) and isolated muscle and brain tissues stained with I2KI (Vickerton et al. 2013). Washing samples stained with Lugol's solution with ethanol may remove the iodine differentially depending on factors such as sample size and tissue and cell types. To overcome these limitations of the currently available iodine staining protocols but keeping the advantage of ethanol-based staining that enables discrimination between the soft tissues and background, we developed an ethanol I2KI solution using a mixture of I2KI and ethanol (see Methods). Our objective is to present an analysis of this solution and optimal protocols (staining time) to discriminate organs via microCT in bird embryos and present a method to efficiently delineate soft tissues for rapid 3D reconstruction based on calibrated pixel intensities. To optimize this solution and protocol, we compare it with I2KI with and without ethanol dehydration protocols using the same molar concentration of iodine. All experiments used late-stage Japanese quail embryos to determine the best protocol for a relatively well developed but small-sized vertebrate. We examine some staining dynamics in the context of depth within the sample, organ type, and staining durations. Comparisons are made qualitatively, with reference to a histological section of the same region, and quantitatively with a new discrimination index we devised to quantify how CT and gray scale values are discriminated between and within selected soft tissues. We also provide an iodine calibration factor as a standard value to compare iodine contrast enhancements of microCT images across different protocols, imaging systems, and samples. We also test whether subsequent ethanol washing of I2KI stained specimens differentially removes iodine among and within selected soft tissue regions using quantitative measurements of soft tissue contrasts. We also quantify shrinkage effects on embryos in the ethanol I2KI compared to I2KI without ethanol washing.
Figure 1.
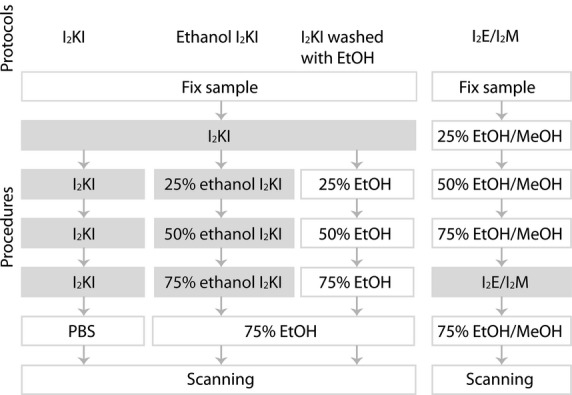
Overview of standard iodine staining procedures. The three protocols (I2KI, ethanol I2KI, and I2KI washed with EtOH) on the left were used in this study. Each step positioned at the same horizontal level was performed for the same duration. Note that the I2E/I2M protocol can use either ethanol (EtOH) or methanol (MeOH). All steps that used an iodine solution are highlighted with a gray background for clarity.
Materials and methods
Japanese quail (Coturnix coturnix) eggs were obtained from a local hatchery (Simetin, Quebec) and incubated for 15–16 days to obtain the largest possible embryo for this species. These embryos are approximately 50 mm long and 13 mm in diameter across their thorax. Extracted embryos were fixed overnight with a mixture of 4% paraformaldehyde in phosphate-buffered saline (1× PBS) and 1% glutaraldehyde because this fixative causes minimal tissue shrinkage (Schmidt et al. 2010). Embryos were washed with 1× PBS and feather germs were removed prior to staining to prevent excessive iodine staining on feathers. Ten embryos were used: one unstained embryo, and four for I2KI staining protocol for four different staining times (2, 7, 14, and 28 days), one I2KI washed with ethanol, and four for ethanol I2KI staining protocol for four different staining times (2, 7, 14, and 28 days).
Staining solutions
Four staining solutions were made from stock Lugol's solution (I2KI). Stock Lugol's solution was made of 5 g I2 and 10 g KI dissolved in 100 mL H2O. Because 3.75% I2KI (w/v) (249.1 mm) causes the least shrinkage of embryos due to its similar osmolarity to biological tissues (Degenhardt et al. 2010), the stock solution was diluted to 3.75% (w/v) in water and 50, 75, and 95% ethanol. Hereafter, we refer the former as our I2KI solution and the last three as our ethanol I2KI solutions (25, 50, and 75% ethanol I2KI).
Staining protocols
Both staining protocols were diffusion-based; vascular injection of staining solutions was not performed (Fig. 1).
Ethanol I2KI staining protocol was performed as follows. Embryos were first soaked in 3.75% I2KI solution and dehydrated to a series of ethanol I2KI solutions (25, 50, 75% ethanol I2KI solutions) for several hours before scanning. Total staining time performed in this protocol was 2, 7, 14, and 28 days.
I2KI staining protocol was performed as follows. Embryos were first soaked in 3.75% I2KI solution as above. To maintain consistent iodine concentrations and to make a comparison between each staining procedure, 3.75% I2KI baths were refreshed in I2KI samples every time embryos were dehydrated to ethanol I2KI solutions. Total staining time performed in this protocol was 2, 7, 14, and 28 days.
Testing differential washing effects of ethanol after I2KI staining
We tested whether ethanol washes of I2KI stained embryos differentially removed bound iodine (Fig. 1). An embryo was first stained with 3.75% I2KI solution for 14 days and subsequently dehydrated to ethanol (25, 50, and 75% ethanol) with the same dehydration duration as the ethanol I2KI samples. This experiment was undertaken in a sufficiently stained embryo (14 days staining).
Scanning parameters
All samples were scanned using a SkyScan 1172 machine (Burker microCT, Belgium). To avoid imaging excessive staining on the external surfaces of embryos, the samples were washed prior to scanning with 1× PBS for I2KI staining or 75% ethanol for ethanol I2KI staining twice for 10 min each time in an orbital nutator. The samples were inserted into a low-density plastic tube (24 mm external diameter) and wrapped with parafilm to prevent specimen drying and rotational motion artifacts during scanning. A field of view (FOV, 25.92 mm diameter and 13.58 mm height) for scanning was set to encompass the diameter of the tube to avoid inaccuracies of tomographic reconstruction and conversion of gray values to CT values. Raw projection data for all samples were acquired with the following parameters: 80 kV, 80 mA, 0.5 mm aluminum filter, 400 image projection data at 0.45º rotation steps for 180º of rotation scanning, 885 ms exposure time, 2 × 2 pixel binning, 13 μm voxel size, and eight-frame averaging. Image quality is highly dependent on the numbers of projection images and average frames per image; however, both parameters increase scanning time. Based on successful imaging of vessels in live mice with constraints of short scanning times (360 projections with eight frames per view and 720 projections with four frames per view: Kindlmann et al. 2005), 400 projections with eight frames per view were adopted in this study. One third of a whole specimen was captured within a single FOV and scanned in approximately 1 h with these parameters.
To convert the gray scale into the HU scale, a calibration of distilled water was required, given that the CT value of water is defined as zero. To replicate the physical scanning parameters of the embryos, various I2KI solutions diluted with water or ethanol were scanned in a low-density plastic tube (19.38 mm external diameter) wrapped with parafilm corresponding to the diameter of the quail embryos. To obtain a calibration of distilled water in HU scale and an iodine calibration factor for I2KI and ethanol I2KI used in our protocols, distilled water, 75% ethanol, and the four contrast media including the same iodine molar concentration used in this study were scanned. The contrast media scanned were: 3.75% I2KI, and 7.5% I2KI, 3.75% ethanol I2KI, and 7.5% ethanol I2KI. Scanning parameters were identical between embryonic samples and solutions.
Tomographic reconstructions
All raw projection data including embryonic samples and solutions were reconstructed by the Feldkamp cone-beam filtered back-projection algorithm using nrecon version 1.6.4.1 supplied by Burker microCT. During the tomographic reconstruction, ring artifacts and beam-hardening were corrected and the data were smoothed using identical numeric parameters in all samples. Smoothing was performed to qualitatively improve the image data for subsequent 3D reconstruction and had negligible changes to the distribution of pixel intensities (Fig. 2). The same dynamic ranges of the pixel intensities were also applied to projection data of all samples. The intensity values of the reconstructed cross-section images (2000 × 2000 pixels) were initially provided only as gray values.
Figure 2.
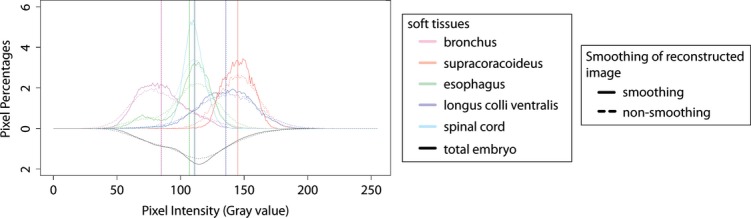
Comparison of histograms of pixel percentages in gray value with and without a smoothing function. The data are derived from an embryo stained with ethanol I2KI for 28 days. The full range of pixel intensities is presented, 0–255. The histogram of the total embryo is flipped along the x-axes. Vertical solid and dashed lines indicate mean pixel intensity of each selected soft tissue and total embryo in the reconstructed images with and without smoothing.
Calculating the calibration factor
Circular regions with a diameter of 13.5 mm were selected across 100 sections in scans of the four contrast media, distilled water, and 75% ethanol to estimate the average intensities of those solutions, using ctan version 1.11.4.2 + (Burker microCT), and to obtain the calibration factors for I2KI and ethanol I2KI. A scan of water was first used to calibrate the CT value of water. The gray value of air was set to –1000 HU and the average gray value of water set to 0 HU. This water calibration allows for a linear conversion of the gray values, which range from 0 to 255, to a range of the CT values from –1000 to 8585 HU. This HU range was applied to all the samples used in this study. To calculate calibration factors for I2KI and ethanol I2KI, all the converted CT values of the four contrast media, distilled water, and 75% ethanol were plotted against their concentration of iodine (mm) and regression values were estimated using r (R Core Team, 2012; Fig. 3).
Figure 3.
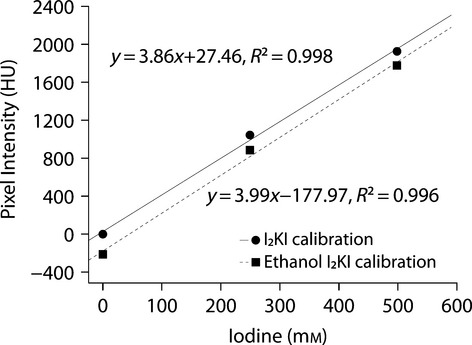
Calibration of I2KI and ethanol I2KI stains to mean CT values of various concentrations of iodine solutions, water, and 75% ethanol. CT values were initially calibrated to water to facilitate stain comparison. 95% confidence intervals of all the solutions, provided by CTAn, are not shown in the graph because their ranges are smaller than the plotted symbols (< 0.1 HU).
Histological preparation
To assess the anatomical discrimination of multiple organs qualitatively in microCT images achieved using our protocol, we compare a histological section with the corresponding microCT image. An embryonic 15-day quail was fixed overnight with 4% paraformaldehyde in 1× PBS and only pectoral regions of the embryo were processed for paraffin embedding after decalcifying the mineralized tissues with DeCal solutions (Fisher Scientific, Pittsburgh, PA; Presnell & Schreibman, 1997). The paraffin embedded sample was sectioned coronally at 10 μm with microtome and stained with Mallory Trichrome (Presnell & Schreibman, 1997). The section image was captured under a Nikon SMZ1000 dissecting microscope with 1616 × 1216 pixel images.
Iodine diffusion rate and anatomical discrimination
We used two methods to quantitatively and qualitatively describe the enhancement of soft tissue discrimination between I2KI and ethanol I2KI staining protocols over staining time. First, we compared the time to iodine saturation, which we determined as the point at which CT values for selected regions remain stable. Secondly, we described the relative disparity of contrasts within and between the selected soft tissues. A common set of image stacks were chosen for each specimen. Image stacks contained 10 consecutive slices and the first image represents the anteriormost separation of the primary bronchus from the trachea. For both methods of quantifying the contrast enhancement of the soft tissue and the relative disparities of contrasts within and between these soft tissues in the embryos, five organs and the whole embryo were selected based on their histological complexity from all embryos throughout these 10 section ranges: bronchus (including its lumen), esophagus (including its lumen), spinal cord, and two muscles (supracoracoideus and longus colli ventralis). The histological complexities include their hollow fluid-filled spaces of the bronchus and esophagus, the two substances (gray and white matter) of the spinal cord, and the transverse (supracoracoideus) and longitudinal (longus colli ventralis) muscle fibers in the transverse planes of the section images. All but the supracoracoideus were selected by tracing their entire outlines throughout all 10 sequential images. Only the supracoracoideus was selected as a 0.65-mm diameter circle because of the difficulty of delineating it from the deep pectoral muscle laterally. Selected organs in sections of poorly discriminated specimens, in particular the unstained and 2-day stained samples, were identified only based on position, using the better discriminated samples as guides. A circular region within the bronchial wall was also used when comparing staining saturation times to function as an indicator of sufficient diffusion time into the center of the embryo.
To estimate relative distance of the selected soft tissue regions from the embryo periphery, each embryo was outlined, excluding the limbs. Centroids for the entire embryo and each measured selected region were estimated, and averaged over the 10 sections used through the analysis. The distance from the embryo surface to its centroid was calculated by assuming a cylindrical shape for the embryo with a cross-sectional area equivalent to the average cross-sectional area of the embryo in these sections. The distance of the selected region from the embryo surface was estimated as a ratio of the distance from the surface of this cylinder to the selected region centroid divided by the total embryo radius.
Secondly, the entire ranges of all pixels of gray and calibrated CT values were obtained for each selected organ and entire embryo across all 10 sections for each sample. The pixel values were standardized as a percentage of each selected volume to make comparisons across selections of different sizes. We devised an index to quantify the degrees of overlap between the selected organs that we call the discrimination index (DI). DI was calculated as the total range of overlap between two volumes (based on the range between the minimum and maximum pixel intensity value of overlap) divided by the total percentage of overlap between the two volumes. The range of overlap was used as the numerator to emphasize the broader overlap ranges but relatively low overlap in total percentages present in longer stain durations.
All selections were processed in ctan version 1.11.4.2 + (Burker microCT) and the gray scale pixel values of the selected regions were linearly converted to HU scales using the calibration function of water in ctan in all the samples. Gray and CT values of all selected regions and whole embryos were analyzed and graphed using r (R Core Team, 2012).
Histogram-based 3D reconstruction of soft tissues
An embryo stained with ethanol I2KI for 28 days was used to derive the optimal threshold of the selected soft tissues and to assess if these thresholds can contribute to rapid 3D reconstruction of the soft tissues. For 3D reconstruction, volume rendering was used because it represents original voxel information by assigning the opacity and color values during rendering. Thus, resulting 3D reconstructed images are highly dependent on those two values and the parameters used here for 3D reconstruction are optimized to this embryonic sample. 3D volume rendering of the embryo was processed with avizo version 7.0.0 (Visualization Science Group). Parameters for volume rendering visualization used were: voxel size = 13 μm, alpha scale = 0.15, opacity = 50%. Color was assigned to each threshold range described below.
With the exception of the cartilage and bone threshold ranges, the results of quantitative analysis of contrast enhancement of the selected organs (bronchus, supracoracoideus, esophagus, longus colli ventralis, spinal cord) were used to derive their optimal threshold ranges. The ranges were constrained to be narrow rather than the entire distributions of the pixel values of the selected soft tissue to emphasize their discrimination. Optimal ranges for white and gray matter are narrow and overlap with the bronchial and esophageal walls. Five optimal soft tissue ranges in gray scales used here are: 65–75 for the cartilage and bone, 96–101 for white matter and bronchial wall, 105–110 for white matter and esophageal and bronchial walls, 112–118 for gray matter and esophageal and bronchial walls, and 131–145 for two muscles (supracoracoideus and longus colli ventralis). The values above 145 are displayed in red to show other soft tissues.
Measuring shrinkage
Ethanol I2KI solutions alter osmolarity, thus differences in the shrinkage effect on embryos stained with ethanol I2KI and I2KI solutions are expected. We measured the shrinkage effects on 20 embryos (incubation 15 days), 10 in each solution (14 days of staining time). Embryos were photographed after initial harvest and after staining with a Nikon SMZ1000 dissecting microscope. Embryos were positioned with their cranial sagittal plane normal to the image plane. Three replicates of positioning and imaging per embryo were taken to derive a mean of each measurement and to reduce any artificial errors introduced by positioning. Skull length was measured from the tip of the beak to the back of the skull through the center of the ear aperture. Mean ratios of skull lengths at harvest and after staining were calculated to present the relative shrinkage on embryos. Although softer tissues (such as the brain or eyeball) would have provided potentially more dramatic shrinkage proxies, they were not accurately measurable in whole mount embryos.
Results and Discussion
Iodine calibration
Various iodine concentrations of I2KI and ethanol I2KI stain (including 0% iodine: distilled water and 75% ethanol) yielded linear equations to derive calibrations for each stain (Fig. 3). These results provide a K factor of 3.9 with an intercept of 27.5 for I2KI and a K factor of 4.0 with an intercept of –178.0 for ethanol I2KI. Although a similar K factor was reported by Degenhardt et al. (2010), the disparity of tissue staining within specimens is not simply a single number and we advocate using the methods described below to assess the discrimination of anatomical staining.
Iodine diffusion rate
Iodine solutions were not found to adequately stain the selected regions until at least 7 days of staining (Fig. 4–6). Only the supracoracoideus stained with I2KI seems to have reached a saturated level at 7 days staining. The similar times to I2KI and ethanol I2KI stain saturation of the bronchus and bronchial wall suggest that both staining solutions did not enter through the trachea but diffused through the embryo to reach this region. A general relationship of higher staining intensity and faster stain saturation with more superficial organs was found (Fig. 4). Although this result was expected, different organ types also appeared to saturate at different times. Both muscles seemed to be saturated after 14 days of staining, whereas the other organs were saturated at 28 days of staining. A notable exception is that between 2 and 7 days staining in ethanol I2KI, the supracoracoideus showed no change in stain intensity. We cannot explain this result.
Figure 4.
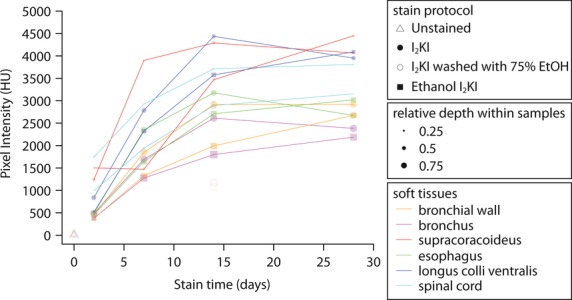
Iodine staining intensities for different staining durations and regions within late stage quail embryos. 95% confidence intervals, provided by CTAn, are not shown in the graph because they are too small (< 39.0 HU). Note that the mean CT value of each region is used to present the overall trends of the diffusion rate of the regions because some regions present bimodal CT value distribution.
Figure 6.
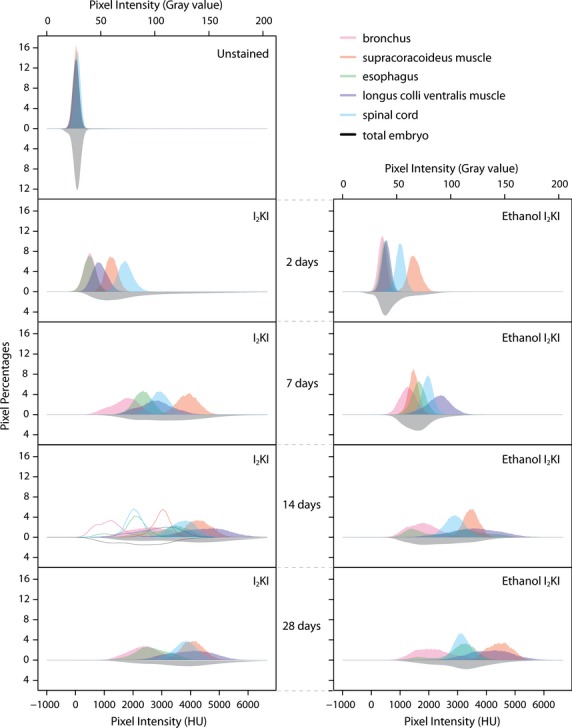
Histograms of pixel percentages in both CT and gray values for selected organs and whole quail embryos for unstained and I2KI, I2KI washed with ethanol, and ethanol I2KI stained specimens. The histogram of the total embryo in all samples is flipped along the x-axes. The range in pixel intensity was cut in the graphs at 6668 HU and 204 gray value for clarity, even though all the ranges for all organs and whole embryos extended to the calculated limit of CT values of 8585 HU and gray values of 255, but at near zero percentages. Histograms of an embryo washed with 75% ethanol after 14 days of I2KI staining, are presented as outlines in the same graph as 14 days of I2KI staining to show the effect of an ethanol wash of the stained embryo.
I2KI stain was found to saturate all the selected soft tissue regions completely at 14 days of staining whereas the ethanol I2KI stain did not reach tissue stain saturation, although their mean CT values between 14 and 28 days are close to each other. The mean CT value of the selected regions at 28 days of ethanol I2KI staining are similar to the saturated values of those regions in I2KI stain when taking into account that the mean CT value of 75% ethanol is approximately 200 HU lower than water. Thus, we considered the embryos to be saturated by 28 days of ethanol I2KI staining. After sufficient staining time, the longus colli ventralis and supracoracoideus have similar mean CT values despite their different distances from the embryo periphery. Ethanol I2KI stain required longer staining times compared with I2KI stain because of the general washing effects of ethanol. However, comparison of the resulting cross-sectional image at 14 days staining revealed a higher contrast between organs and internal spaces using ethanol I2KI stain.
Stain effects on cranial shrinkage
Shrinkage of the measured skull lengths (n = 10 + 10) decreased by only 5% from harvest to each staining solution. Shrinkage was probably mostly due to tissue fixation, given that a 4.4% shrinkage was recorded in harvested mice embryos (10.5 days post-conception) measured by cranial centroid size (Schmidt et al. 2010). Our data suggest that both I2KI and ethanol I2KI staining solutions have the same shrinkage effects. However, soft tissues may experience more shrinkage, given that the skull length measured here is composed primarily of ossified bones. Although a 5% shrinkage was found in tissue processing for microCT imaging, we note that the fixed, paraffin-embedded serial section in Fig. 5 is enlarged to 1.8 times to be comparable to the microCT section images. Dramatic shrinkage of up to 70% volume was reported for isolated cardiac muscle samples stained with 20% I2KI (Vickerton et al. 2013). However, as Vickerton et al. (2013) suggested, the minimal shrinkage recovered here indicates shrinkage effects may be minimized by maintaining tissues in whole mounts. These observations are consistent with the nearly identical muscle volumes of dissected muscles compared with those reconstructed after whole mount staining with I2KI and microCT imaging (Baverstock et al. 2013).
Figure 5.
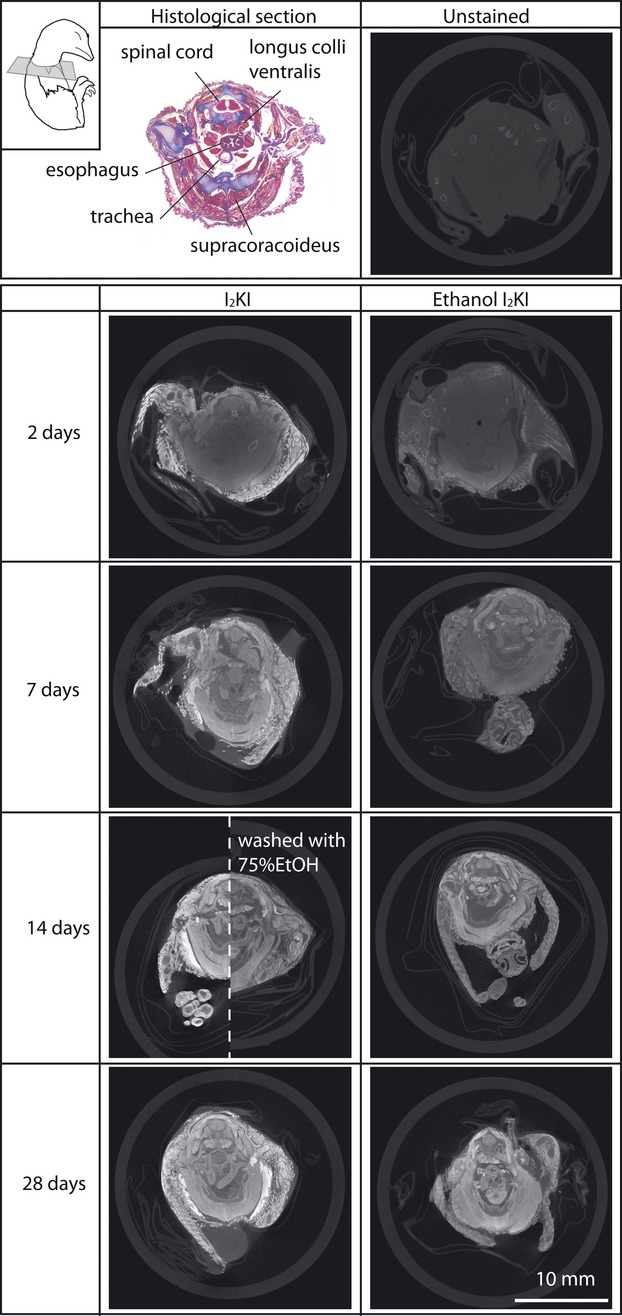
MicroCT images of quail embryos stained with 3.75% I2KI and ethanol I2KI at various stain durations and a Mallory Trichrome histological section for comparison. All CT images are unmodified images obtained from tomographic reconstruction software and at the same scale. An embryo stained with I2KI for 14 days and subsequently washed with 75% ethanol is presented in the right side of an embryo stained with I2KI for 14 days for direct comparison. All the microCT images are from approximately the same cross-sectional level illustrated in the top left. The histological image is magnified 1.8 times to be comparable to a size of all microCT images. The extreme shrinkage of the histological sample is an artifact of dehydration and paraffin processing.
Qualitative comparison of microCT image and histological section
Comparative sections of samples scanned with our protocols are shown in Fig. 5. In general, the greater staining times yielded more histologically interpretable data. As expected, only the ossifying bones are observed in an unstained embryo and their CT values range between 500 and 1700 HU. At 28 days staining in I2KI, the CT values of the bones increased to between 4000 and 6000 HU, probably because of the high degree of vascularization of developing bones. However, iodine seems to have been unable to diffuse into the cartilages and only the perichondrium was stained to CT values between 1500 and 4000 HU. These iodine staining patterns for the bones and cartilages are consistent with figures reported by Metscher (2009a). The probable reason for the inability of iodine to diffuse into the cartilage is the avascular nature of this tissue and relatively low cell density. In the embryonic samples of this study, remaining feathers were intensely stained, much more than any other tissue in I2KI stain, whereas they were stained much less in ethanol I2KI stain. Enhancement of perichondrial tissues with iodine can be beneficial to extract unossified skeletal morphologies automatically using 3D software relative to other enhanced soft tissues, as demonstrated in molluscan cartilages with phosphomolybdic acid stain (Golding et al. 2009).
Although the iodine staining mechanism is not clearly understood, high contrast enhancements of non-keratinized squamous cells of the esophageal epithelial tissue and muscles are observed in this study. The former is derived from iodine binding to glycogen-rich cells and has been used to demarcate mucosal abnormalities from the normal epithelia of the esophagus using Lugol's solution during endoscopy (Wolfsen & Wallace, 2009; Smith & Lightdale, 2010; Squier & Brogden, 2011). Similarly, glycogen-rich cells in muscles may explain the enhancement of muscle via iodine staining (Jeffery et al. 2011; Stephenson et al. 2012) because iodine and glycogen are known to form glycogen iodine complex (Lecker et al. 1997). Additionally, the multinucleated muscle fibers may also contribute to enhancing iodine binding. Both skeletal muscles examined here were uniformly stained well enough to identify some structures. In transverse aspect, the fiber directions are visible in the supracoracoideus muscle and the fascicles within the longitudinal longus colli ventralis muscle after 7 days of both staining protocols. The voxel size of these samples was 13 μm and is within the range of muscle fiber diameters (10–100 μm: Ross et al. 2003) but not small enough to resolve individual fibers. Six-micron voxel images of a pig hind limb muscle stained with the same concentration of I2KI solutions were reported to provide a better resolution of each transverse fiber (Jeffery et al. 2011).
The gray and white matter of the spinal cord consist of regions of densely concentrated large nerve cell bodies and numerous small supporting cells and no nerve cell bodies, respectively (Henrikson et al. 1997). The different densities of these regions are evident in brain CT images in which their CT values range from 20 to 30 HU for white matter and from 37 to 45 HU for gray matter (Toennies, 2012). These differential contrasts are identifiable in the embryos stained for more than 14 days in both staining solutions. The 14-day I2KI stained embryo washed in ethanol lost any discrimination between these two regions. Mallory Trichrome stained all nervous tissue evenly and did not distinguish white from gray matter in the histological section.
In the esophagus, five different tissue layers are visible in the histological section but only three different intensities are visible in the microCT sections. The bright internal layer of the esophagus bounding the lumen is comparable to the squamous cells of the epithelial tissue lining the internal surface of the esophagus in the histological section. The bright intensities of the epithelial tissue are explained by its glycogen-rich cells and the dense packing of cells. The enhanced external layer of the esophagus in the microCT images appears to be the striated muscle tissue visible in the histological section. The lighter stained intermediate layer of the esophagus appears to be comparable to the cartilages, thin smooth muscles, and connective tissues in the histological section. The boundary between these tissues within the intermediate layer in the microCT images is not resolved at 13 μm resolution.
The bronchus in the microCT images has three identifiable layers of intensities that correspond to the internal and external epithelial tissues and the connective tissue composed of cartilage and loose connective tissue. The internal epithelial tissue of the bronchus is well stained through the epithelial wall and does not contain glycogen. Its intense staining may be attributed to its dense columnar arrangements of cells and juxtaposition to the empty lumen. Smaller voxel size images are required to further explore the differential staining of these tissues. The external epithelial layer is only visible after 28 days staining in ethanol I2KI.
Quantifying anatomical discrimination
The unstained embryo has no discrimination among the selected organs, resulting in low DI values (Fig. 6, Table 1). The CT values are near those of water in all the selected organs and total embryo. Over the course of staining, the CT values of the total embryo tended to distribute into higher values. Average DI values for each stain and staining time increased from 0.25 in the unstained embryo up to 3.41 in the ethanol I2KI embryo stained for 28 days. Although the average DI increased, there are many subtle differential staining levels that contribute to a more complicated staining pattern.
Table 1.
Discrimination indices (DI) for the selected organs for unstained embryos and embryos stained with I2KI, I2KI washed with ethanol and ethanol I2KI staining protocols. DI values for I2KI and ethanol I2KI for 2 and 7 days staining are excluded in this table because all the selected organs are not saturated with each staining and highly dependent on their depths within the embryos (Fig. 4)
| Stain | Time (days) | Discrimination Index |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| b – m.s | b – e | b – m.l | b – s | m.s – e | m.s – m.l | m.s – s | e – m.l | e – s | m.l – s | Mean DI | ||
| Unstained | 0 | 0.24 | 0.24 | 0.25 | 0.27 | 0.24 | 0.24 | 0.25 | 0.25 | 0.27 | 0.26 | 0.25 |
| I2KI | 14 | 4.56 | 1.63 | 4.21 | 2.68 | 1.92 | 1.28 | 1.19 | 2.50 | 1.39 | 1.94 | 2.33 |
| I2KI washed with 75%EtOH | 14 | 9.13 | 2.51 | 3.87 | 2.63 | 2.62 | 1.10 | 3.13 | 1.96 | 0.74 | 1.83 | 2.95 |
| Ethanol I2KI | 14 | 12.08 | 1.98 | 4.32 | 3.36 | 1.42 | 1.12 | 1.28 | 1.72 | 1.66 | 1.57 | 3.05 |
| I2KI | 28 | 6.39 | 1.17 | 3.73 | 4.54 | 3.18 | 1.17 | 0.94 | 2.63 | 2.64 | 1.39 | 2.78 |
| Ethanol I2KI | 28 | 9.54 | 2.24 | 4.42 | 2.36 | 3.71 | 1.12 | 5.34 | 2.41 | 0.89 | 2.02 | 3.41 |
b, bronchus; e, esophagus; m.l, musculus longus colli ventralis; m.s, musculus supracoracoideus; s, spinal cord.
I2KI stain
At 2 days of staining with I2KI, the distributions of the CT values of all the selected organs are higher than the unstained embryo but still remain narrow. At 7 days of staining, these distributions occupy a broader range and become complex, with the exception of the spinal cord, indicating discriminated staining within and among the organs. At 2 and 7 days of staining, the CT values of the selected organs are highly dependent on their depth within the embryos as discussed above (Fig. 4).
At 14 days of staining, CT values of all the selected organs are high and form complex and broad distributions. With this stain time, the selected organs of the embryo are saturated (Fig. 4) and thus present organ-specific absorptions of iodine regardless of their depths within the embryo and all their DI values represent accurate discriminations between them (Table 1). Both of the two muscles present high mean CT values. Interestingly, these values for the longus colli ventralis distribute over a broad range of approximately 4000 HU (between 2500 and 6500 HU), whereas those of the supracoracoideus encompass only a half of this range (between 3000 and 5000 HU). Stephenson et al. (2012) discussed whether the different absorption of iodine between the specialized muscles fibers for the cardiac conduction system and the myocardium in rabbits and rats may be derived from the density of the muscles (Stephenson et al. 2012). However, both of the selected muscles in this study are dense skeletal muscles. Thus, differential staining of these muscles is more likely to be associated with the muscle structures. The more spatially divided fascicles within the longus colli ventralis may generate more available surfaces to absorb the iodine than the relatively tighter fascicles of the supracoracoideus.
At 14 days, staining yields the first obvious discriminations within the organs. For example, the bimodal distribution of the CT values in the esophagus is due to a high peak associated with the esophageal tissues and a lower peak associated with its lumen. On the other hand, the bronchus shows CT values ranging over nearly 3000 HU without having a bimodal distribution, although the highlighted epithelial layer lining the internal space provides a good demarcation of the bronchial wall from its lumen (Fig. 5). The white and gray matter of the spinal cord is identified in the microCT image but does not contribute to an obvious bimodal distribution of CT values.
Distributions of the CT values of all the selected organs at 28 days of staining are similar to those at 14 days of staining but are generally shifted to lower values. This reduction is unexplained, but overall, DI values remain slightly higher than those of 14 days staining. The apparent higher DI values are largely attributable to the reduction of the right tail distributions of the bronchus and esophagus. The two muscles, however, do not appear to be more stained, indicating they had reached stain saturation at 14 days.
The bimodal distribution of the CT values of the esophagus at 14 days of staining is not observed at 28 days and reflects further staining of the lumen (Fig. 6). The bronchus achieves higher CT values at 28 days staining, probably due to overstaining of its lumen. Enhancements of the internal spaces is probably derived from free iodine within the space because there were no more iodine binding sites available in the stain-saturated tissues. This result suggests that determining minimum stain times will help to limit the background noise. Although the three pairs of DI values (bronchus and esophagus, supracoracoideus and longus colli ventralis, supracoracoideus and spinal cord) at 28 days staining are relatively low, these morphologies are spatially distributed in the embryos and they can be easily identified.
Ethanol I2KI stain
At 2 and 7 days of ethanol I2KI staining, the distributions of the CT values of the selected organs are dependent on their depths within the embryos, as with the same staining times with I2KI (Fig. 4). Ethanol I2KI stain reached saturation levels about 7 days after I2KI staining, and only CT values of the longus colli ventralis formed a complex pattern at 7 days staining.
At 14 days of staining, all the selected organs present broad and complex distributions of CT values, similar to 14 days of I2KI staining. The distinctive bimodal distribution of the CT values is observed in the esophagus. A dark staining substance is present in the sample presented in Fig. 5 and may be displaced yolk within the esophagus. The bronchus is clear but, as with I2KI staining, still presents a unimodal CT value distribution. Flushing PBS through selected fluid-filled spaces before iodine staining may help to lessen background noise. A mean DI value of ethanol I2KI staining is slightly higher than that of I2KI staining; however, because, with this stain time, none of the selected organs reaches saturation, this value does not truly suggest that this stain time with ethanol I2KI is better at discriminating the organs overall than I2KI staining. The difficulties of diffusing iodine into the gray matter of the spinal cord appear to be solved with this longer staining time.
At 28 days of staining, the distribution of the CT values of all the selected organs show similar patterns to those at 14 days of staining but at higher CT values. Similar to 28 days of I2KI staining, the low CT values of the esophageal and bronchial lumens are shifted to higher values compared with 14 days of ethanol I2KI staining. This is again probably due to higher concentrations of free iodine within the bronchial and esophageal lumens after tissues throughout the sample have become stain-saturated.
Twenty-eight days of staining in ethanol I2KI is most comparable to 14 days staining in I2KI, as these times appear to reach levels of general stain saturation for these solutions in these samples. The larger DI value between the supracoracoideus and spinal cord with 28 days ethanol I2KI staining is the result of this stain preferentially limiting excess iodine in the spinal cord. Although the supracoracoideus reaches CT values of between 3500 and 5000 in both cases, CT values of the spinal cord at 14 days I2KI staining range between 3000 and 4800, whereas 28 days ethanol I2KI staining limits the spinal cord to between 2500 and 3500. A similar restriction of upper CT values in ethanol I2KI staining is observed in the esophageal and bronchial walls. The lumen of these organs still remained relatively understained at 28 days ethanol I2KI, indicating this solution does limit overstaining effects of internal spaces. Addition of ethanol to the stain solution appears to yield important benefits, such as reducing overstaining of internal spaces and limiting overstaining of some tissues, such as bronchial and esophageal walls and the spinal cord.
Differential washing effects of ethanol after I2KI staining
Comparisons of the means and distributions of CT values of the selected soft tissue regions between the I2KI-stained embryo and a similarly stained embryo wash in ethanol reveal differential washing effects of ethanol (Figs 4 and 6). The mean intensities of CT values of all the selected regions in an embryo washed with 75% ethanol after 14 days of I2KI staining were reduced by between 1500 and 2000 HU. Distributions of the CT values of the longus colli ventralis in the ethanol-washed sample remained broad, whereas those of the other organs are narrowed. These CT value changes translate to particularly low DI values in the longus colli ventralis because it broadly overlaps the other organs. Distributions of the CT values of the selected organs indicate differential washing effects of ethanol, such that these effects do not apply from the peripheral to the central organs (Fig. 4). Thus, the amount of iodine loss in terms of morphology is difficult to predict with this simple ethanol washing protocol after staining of the embryos. Discriminations of the internal spaces of organs, such as the bronchus, are similar to those of 14 days of ethanol I2KI staining but unlike those of 14 days of I2KI staining. The mean CT values of the selected regions in this sample washed with ethanol reduced stain levels to less than those of 7 days I2KI staining (Fig. 4).
Means and distributions of the CT and DI values of the selected organs between the ethanol washed embryo and ethanol I2KI stained embryo for same stain time are difficult to compare directly because the washing effects of ethanol after staining are quite different from the effects of ethanol during staining (Figs 4 and 6, Table 1). The higher mean CT values of the selected soft tissue regions stained with ethanol I2KI indicate it has a much greater tissue-staining effect than simply washing a stained sample in ethanol.
Histogram-based 3D reconstruction of soft tissues
Histogram-based optimal threshold ranges for the selected soft tissues successfully demonstrate their basic delineation in the volume-rendered embryo, although pixel intensities in each selected soft tissue may be found to bridge optimal ranges between some soft tissues (Fig. 7). A slight discrimination of white and gray matter is identifiable in the gray scale images, as reported by Degenhardt et al. (2010), but the threshold ranges of each morphology allow better visualization between these two in the 3D reconstruction. Esophageal and bronchial walls are involved in two and three ranges, respectively, and yet are visualized clearly in contrast with the lower pixel values of their lumens. Two selected muscles (supracoracoideus and longus colli ventralis) occupy high pixel value ranges, whereas other muscles have slightly lower gray values. The various pixel values observed in cardiac muscle helped delineate the heart and are consistent with the results of Stephenson et al. (2012). Lungs are easily identified with differentially colored soft tissue and alveolae.
Figure 7.
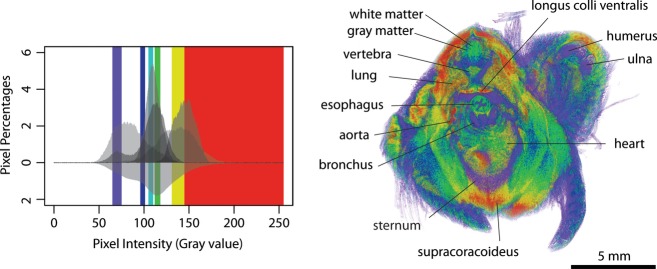
Histogram-based segmentation via volume rendering of a quail embryo stained with ethanol I2KI for 28 days. The histogram is from Fig. 6. The color bars in the histogram represent optimal threshold ranges for the selected soft tissues: cartilage and bone in purple (65–75 gray value), white matter and bronchial wall in blue (96–101 gray value), white matter and esophageal and bronchial walls in light blue (105–110 gray value), gray matter and esophageal and bronchial walls in green (112–118 gray value) and two muscles (supracoracoideus and longus colli ventralis) in yellow (131–145 gray value). The values above 145 were displayed in red to show other soft tissues. The full range of pixel intensities is presented, 0–255.
Stained perichondria and the surface of the bones contrast with their unstained internal tissues. However, the enhanced bone surfaces are often difficult to discriminate from the wide range of gray values of the soft tissues. If only bone is of interest, we do not recommend iodine staining of the sample. The ranges of gray value of the esophageal and bronchial lumens, other internal spaces, and feathers overlap with bone and cartilage and make accurate visualizations of the non-enhanced bone and cartilage difficult. Skinning embryos may help improve clear 3D visualization of the samples stained with iodine.
Conclusions
We presented analyses of two iodine protocols using I2KI and ethanol I2KI solutions that enable discrimination of soft tissues in microCT images specific to Japanese quail embryos using both qualitative and new quantitative methods and calibrated CT values. After the samples were stain-saturated for 14 and 28 days, respectively, both protocols allowed discriminations among and within the selected organs. We demonstrate that a new solution of ethanol I2KI has superior properties to both the normal I2KI solution and normal I2KI staining followed by washing in ethanol, while retaining the same minimal shrinkage effects. In general, ethanol I2KI provides larger DI values and better discriminates CT value ranges among and within soft tissues. Addition of ethanol to I2KI appears to limit overstaining of internal spaces and some tissues, such as bronchial and esophageal walls and the spinal cord. However, the stain reductions in these organs are differential, in that the gray matter in the spinal cord is most discernible in samples stained for 28 days with ethanol I2KI. We used this optimal stain protocol, 28 days of ethanol I2KI, to determine the optimal threshold ranges for the selected soft tissue based on their pixel distribution in the embryo. These optimal threshold ranges of the soft tissue demonstrated preliminary delineations of these morphologies. The ethanol I2KI protocol developed in this study contributes toward quantifying specific gray value ranges for particular soft tissues, and providing faster and more efficient segmentation of soft tissues in microCT images for 3D visualization.
Iodine-binding properties facilitate good staining of soft tissues composed of glycogen-rich cells, such as the stratified muscle and kidney, and packed cell arrangements of epithelial tissue. Cell densities may play a large role in iodine-based CT imaging, but higher resolution images are required to test these associations. Although we only tested these staining protocols in late stage Japanese quail embryos, the protocols are applicable to other organisms, with modifications as to staining time. Iodine staining via diffusion may be more suitable for small samples due to limitations of diffusion rates and damage to the specimen. Larger or less permeable specimens may require injection of iodine solutions to overcome these problems (Cox & Jeffery, 2011).
Acknowledgments
We thank the McGill Institute for Advanced Materials Facilities in the Department of Mining and Materials Engineering for access to their microCT machine. We thank two anonymous reviewers for critical comments that dramatically improved the manuscript. We thank members of the Larsson lab for discussions and critical suggestions on the manuscript. This project was supported by a Japan Student Services Organization, Student Exchange Support Program (Scholarship for Long-term Study Abroad) to R.T. and a Canada Research Chair and Natural Sciences and Engineering Research Council of Canada grant to H.C.E.L.
Conflict of interest
The authors have no conflicts of interest.
Author contributions
R.T. performed the acquisitions of microCT data, a histological section of the sample and measurements of the samples and processed the microCT data for quantitative analysis. R.T. and H.C.E.L. designed this study, analyzed the data and wrote the manuscript.
References
- Almajdub M, Magnier L, Juillard L. Kidney volume quantification using contrast-enhanced in vivo X-ray micro-CT in mice. Contrast Media Mol Imaging. 2008;3:120–126. doi: 10.1002/cmmi.238. [DOI] [PubMed] [Google Scholar]
- Aslanidi OV, Nikolaidou T, Zhao J. Application of micro-computed tomography with iodine staining to cardiac imaging, segmentation, and computational model development. IEEE Trans Med Imaging. 2012;32:8–17. doi: 10.1109/TMI.2012.2209183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea CT, Fubara B, Hedlund LW. 4-D micro-CT of the mouse heart. Mol Imaging. 2005;4:110–116. doi: 10.1162/15353500200504187. [DOI] [PubMed] [Google Scholar]
- Badea CT, Drangova M, Holdsworth DW. In vivo small-animal imaging using micro-CT and digital subtraction angiography. Phys Med Biol. 2008a;53:R319–R350. doi: 10.1088/0031-9155/53/19/R01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea CT, Wetzel AW, Mistry N. Left ventricle volume measurements in cardiac micro-CT: the impact of radiation dose and contrast agent. Comput Med Imaging Graph. 2008b;32:239–250. doi: 10.1016/j.compmedimag.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain MM, Fagan AJ, Mullin JM. Noninvasive monitoring of chick development in ovo using a 7T MRI system from day 12 of incubation through to hatching. J Magn Reson Imaging. 2007;26:198–201. doi: 10.1002/jmri.20963. [DOI] [PubMed] [Google Scholar]
- Baverstock H, Jeffery NS, Cobb SN. The morphology of the mouse masticatory musculature. J Anat. 2013;223:46–60. doi: 10.1111/joa.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley MD, Jorgensen SM, Lerman LO. Visualization of three-dimensional nephron structure with microcomputed tomography. Anat Rec (Hoboken) 2007;290:277–283. doi: 10.1002/ar.20422. [DOI] [PubMed] [Google Scholar]
- Boot MJ, Westerberg CH, Sanz-Ezquerro J. In vitro whole-organ imaging: 4D quantification of growing mouse limb buds. Nat Methods. 2008;5:609–612. doi: 10.1038/nmeth.1219. [DOI] [PubMed] [Google Scholar]
- Carney EW, Tornesi B, Keller C. Refinement of a morphological scoring system for postimplantation rabbit conceptuses. Birth Defects Res B Dev Reprod Toxicol. 2007;80:213–222. doi: 10.1002/bdrb.20118. [DOI] [PubMed] [Google Scholar]
- Chindasombatjaroen J, Kakimoto N, Shimamoto H. Correlation between pixel values in a cone-beam computed tomographic scanner and the computed tomographic values in a multidetector row computed tomographic scanner. J Comput Assist Tomogr. 2011;35:662–665. doi: 10.1097/RCT.0b013e31822d9725. [DOI] [PubMed] [Google Scholar]
- Cox PG, Jeffery N. Reviewing the morphology of the jaw-closing musculature in squirrels, rats, and guinea pigs with contrast-enhanced microCT. Anat Rec (Hoboken) 2011;294:915–928. doi: 10.1002/ar.21381. [DOI] [PubMed] [Google Scholar]
- de Crespigny A, Bou-Reslan H, Nishimura MC. 3D micro-CT imaging of the postmortem brain. J Neurosci Methods. 2008;171:207–213. doi: 10.1016/j.jneumeth.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Wright AC, Horng D. Rapid 3D phenotyping of cardiovascular development in mouse embryos by micro-CT with iodine staining. Circ Cardiovasc Imaging. 2010;3:314–322. doi: 10.1161/CIRCIMAGING.109.918482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhenain M, Ruffins SW, Jacobs RE. Three-dimensional digital mouse atlas using high-resolution MRI. Dev Biol. 2001;232:458–470. doi: 10.1006/dbio.2001.0189. Available at: http://mouseatlas.caltech.edu/index.html. [DOI] [PubMed] [Google Scholar]
- Duce S, Morrison F, Welten M. Micro-magnetic resonance imaging study of live quail embryos during embryonic development. Magn Reson Imaging. 2011;29:132–139. doi: 10.1016/j.mri.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford NL, Graham KC, Groom AC. Time-course leftacterization of the computed tomography contrast enhancement of an iodinated blood-pool contrast agent in mice using a volumetric flat-panel equipped computed tomography scanner. Invest Radiol. 2006;41:384–390. doi: 10.1097/01.rli.0000197981.66537.48. [DOI] [PubMed] [Google Scholar]
- Garrett JS, Lanzer P, Jaschke W. Measurement of cardiac output by cine computed tomography. Am J Cardiol. 1985;56:657–661. doi: 10.1016/0002-9149(85)91030-6. [DOI] [PubMed] [Google Scholar]
- Golding RE, Jones AS. Micro-CT as a novel technique for 3D reconstruction of molluscan anatomy. Molluscan Res. 2007;27:123–128. [Google Scholar]
- Golding RE, Ponder WF, Byrne M. Three-dimensional reconstruction of the odontophoral cartilages of Caenogastropoda (Mollusca: Gastropoda) using micro-CT: morphology and phylogenetic significance. J Morphol. 2009;270:558–587. doi: 10.1002/jmor.10699. [DOI] [PubMed] [Google Scholar]
- Gregg CL, Butcher JT. Quantitative in vivo imaging of embryonic development: opportunities and challenges. Differentiation. 2012;84:149–162. doi: 10.1016/j.diff.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning T, Weber AW, Bauer JS. Imaging leftacteristics of DHOG, a hepatobiliary contrast agent for preclinical microCT in mice. Acad Radiol. 2008;15:342–349. doi: 10.1016/j.acra.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Henning AL, Jiang MX, Yalcin HC. Quantitative three-dimensional imaging of live avian embryonic morphogenesis via micro-computed tomography. Dev Dyn. 2011;240:1949–1957. doi: 10.1002/dvdy.22694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrikson RC, Kaye GI, Mazurkiewicz JE. Histology. Baltimore: Williams and Wilkins; 1997. [Google Scholar]
- Hogers B, van der Weerd L, Olofsen H. Non-invasive tracking of avian development in vivo by MRI. NMR Biomed. 2009;22:365–373. doi: 10.1002/nbm.1346. [DOI] [PubMed] [Google Scholar]
- Jeffery NS, Stephenson RS, Gallagher JA. Micro-computed tomography with iodine staining resolves the arrangement of muscle fibres. J Biomech. 2011;44:189–192. doi: 10.1016/j.jbiomech.2010.08.027. [DOI] [PubMed] [Google Scholar]
- Johnson GA, Cofer GP, Gewalt SL. Morphologic phenotyping with MR microscopy: the visible mouse. Radiology. 2002;222:789–793. doi: 10.1148/radiol.2223010531. [DOI] [PubMed] [Google Scholar]
- Johnson J, Hansen M, Wu I. Virtual histology of transgenic mouse embryos for high-throughput phenotyping. PLoS Genet. 2006;2:e61. doi: 10.1371/journal.pgen.0020061. Available at: http://www.civm.duhs.duke.edu/Radiology/index.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachelrieß M. Micro-CT. In: Semmler W, Schwaiger M, editors. Molecular Imaging I. Berlin: Springer-Verlag; 2008. pp. 23–52. [Google Scholar]
- Kalender WA. Computed Tomography: Fundamentals, System Technology, Image Quality, Applications. Erlangen: Publicis Corporate Publishing; 2005. [Google Scholar]
- Katsumata A, Hirukawa A, Okumura S. Effects of image artifacts on gray-value density in limited-volume cone-beam computerized tomography. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2007;104:829–836. doi: 10.1016/j.tripleo.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Kiernan JA. Histological and Histochemical Methods: Theory and Practice. Bloxham: Scion; 2008. [Google Scholar]
- Kim JS, Min J, Recknagel AK. Quantitative three-dimensional analysis of embryonic chick morphogenesis via microcomputed tomography. Anat Rec (Hoboken) 2011;294:1–10. doi: 10.1002/ar.21336. [DOI] [PubMed] [Google Scholar]
- Kindlmann GL, Weinstein DM, Jones GM. Practical vessel imaging by computed tomography in live transgenic mouse models for human tumors. Mol Imaging. 2005;4:417–424. doi: 10.2310/7290.2005.05166. [DOI] [PubMed] [Google Scholar]
- Lecker DN, Kumari S, Khan A. Iodine binding capacity and iodine binding energy of glycogen. J Polym Sci A Polym Chem. 1997;35:1409–1412. [Google Scholar]
- Li X, Liu J, Davey M. Micro-magnetic resonance imaging of avian embryos. J Anat. 2007;211:798–809. doi: 10.1111/j.1469-7580.2007.00825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litzlbauer HD, Neuhaeuser C, Moell A. Three-dimensional imaging and morphometric analysis of alveolar tissue from microfocal X-ray-computed tomography. Am J Physiol Lung Cell Mol Physiol. 2006;291:L535–L545. doi: 10.1152/ajplung.00088.2005. [DOI] [PubMed] [Google Scholar]
- Mah P, Reeves TE, McDavid WD. Deriving Hounsfield units using grey levels in cone beam computed tomography. Dentomaxillofac Radiol. 2010;39:323–335. doi: 10.1259/dmfr/19603304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez HG, Prajapati SI, Estrada CA. Microscopic computed tomography-based virtual histology for visualization and morphometry of atherosclerosis in diabetic apolipoprotein E mutant mice. Circulation. 2009;120:821–822. doi: 10.1161/CIRCULATIONAHA.108.829531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metscher B. MicroCT for comparative morphology: simple staining methods allow high-contrast 3D imaging of diverse non-mineralized animal tissues. BMC Physiol. 2009a;9:11–24. doi: 10.1186/1472-6793-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metscher B. MicroCT for developmental biology: a versatile tool for high-contrast 3D imaging at histological resolutions. Dev Dyn. 2009b;238:632–640. doi: 10.1002/dvdy.21857. [DOI] [PubMed] [Google Scholar]
- Metscher BD. X-ray microtomographic imaging of vertebrate embryos. In: James S, Wong RO, Rafael Y, editors. Imaging in Developmental Biology: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2011. pp. 1462–1471. [DOI] [PubMed] [Google Scholar]
- Miles K, Young H, Chica S. Quantitative contrast-enhanced computed tomography: is there a need for system calibration? Eur Radiol. 2007;17:919–926. doi: 10.1007/s00330-006-0424-x. [DOI] [PubMed] [Google Scholar]
- Mizutani R, Suzuki Y. X-ray microtomography in biology. Micron. 2012;43:104–115. doi: 10.1016/j.micron.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Nakayama Y, Awai K, Funama Y. Abdominal CT with low tube voltage: preliminary observations about radiation dose, contrast enhancement, image quality, and noise. Radiology. 2005;237:945–951. doi: 10.1148/radiol.2373041655. [DOI] [PubMed] [Google Scholar]
- Pai VM, Kozlowski M, Donahue D. Coronary artery wall imaging in mice using osmium tetroxide and micro-computed tomography (micro-CT) J Anat. 2012;220:514–524. doi: 10.1111/j.1469-7580.2012.01483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus MJ, Gleason SS, Easterly ME. A review of high resolution X-ray computed tomography and other imaging modalities for small animal research. Lab Anim. 2001;30:36–45. [PubMed] [Google Scholar]
- Petiet AE, Kaufman MH, Goddeeris MM. High-resolution magnetic resonance histology of the embryonic and neonatal mouse: a 4D atlas and morphologic database. Proc Natl Acad Sci U S A. 2008;105:12331–12336. doi: 10.1073/pnas.0805747105. Available at: http://www.civm.duhs.duke.edu/devatlas/index.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati SI, Keller C. Contrast enhanced vessel imaging using microCT. J Vis Exp. 2011;47:e2377. doi: 10.3791/2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnell JK, Schreibman MP. Humason's Animal Tissue Techniques. Baltimore: Johns Hopkins University Press; 1997. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2012. Available at: http://www.R-project.org/ [Google Scholar]
- Ribi W, Senden TJ, Sakellariou A. Imaging honey bee brain anatomy with micro-X-ray-computed tomography. J Neurosci Methods. 2008;171:93–97. doi: 10.1016/j.jneumeth.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Ritman EL. Current status of developments and applications of micro-CT. Annu Rev Biomed Eng. 2011;13:531–552. doi: 10.1146/annurev-bioeng-071910-124717. [DOI] [PubMed] [Google Scholar]
- Ross MH, Kaye GI, Pawlina W. Histology: A Text and Atlas. Philadelphia: Lippincott Williams and Wilkins; 2003. [Google Scholar]
- Ruffins SW, Martin M, Keough L. Digital three-dimensional atlas of quail development using high-resolution MRI. Sci World J. 2007;7:592–604. doi: 10.1100/tsw.2007.125. Available at: http://131.215.15.121/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Murase K. Ex vivo imaging of mouse brain using micro-CT with non-ionic iodinated contrast agent: a comparison with myelin staining. Br J Radiol. 2012;85:e973–e978. doi: 10.1259/bjr/13040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schambach SJ, Bag S, Schilling L. Application of micro-CT in small animal imaging. Methods. 2010;50:2–13. doi: 10.1016/j.ymeth.2009.08.007. [DOI] [PubMed] [Google Scholar]
- Schmidt E, Parsons T, Jamniczky H. Micro-computed tomography-based phenotypic approaches in embryology: procedural artifacts on assessments of embryonic craniofacial growth and development. BMC Dev Biol. 2010;10:18. doi: 10.1186/1471-213X-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe J. Optical projection tomography. Annu Rev Biomed Eng. 2004;6:209–228. doi: 10.1146/annurev.bioeng.6.040803.140210. [DOI] [PubMed] [Google Scholar]
- Sharpe J, Ahlgren U, Perry P. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- Siegel MJ, Schmidt B, Bradley D. Radiation dose and image quality in pediatric CT: effect of technical factors and phantom size and shape. Radiology. 2004;233:515–522. doi: 10.1148/radiol.2332032107. [DOI] [PubMed] [Google Scholar]
- Siewerdsen JH, Jaffray DA. Cone-beam computed tomography with a flat-panel imager: magnitude and effects of x-ray scatter. Med Phys. 2001;28:220–231. doi: 10.1118/1.1339879. [DOI] [PubMed] [Google Scholar]
- Smith MS, Lightdale CJ. Esophageal cancer. In: Talley NJ, DeVault KR, Fleischer DE, editors. Practical Gastroenterology and Hepatology: Esophagus and Stomach. Chichester: Wiley-Blackwell; 2010. pp. 315–324. [Google Scholar]
- Squier C, Brogden K. Human Oral Mucosa: Development, Structure and Function. Chichester: Wiley-Blackwell; 2011. [Google Scholar]
- Stephenson RS, Boyett MR, Hart G. Contrast enhanced micro-computed tomography resolves the 3-dimensional morphology of the cardiac conduction system in mammalian hearts. PLoS One. 2012;7:e35299. doi: 10.1371/journal.pone.0035299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toennies KD. Guide to Medical Image Analysis: Methods and Algorithms. London: Springer; 2012. [Google Scholar]
- Vickerton P, Jarvis J, Jeffery N. Concentration-dependent specimen shrinkage in iodine-enhanced microCT. J Anat. 2013;223:185–193. doi: 10.1111/joa.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicklein M, Schwyn DA, Simonsen TJ. Radiocontrast micro-CT imaging optimised for differential tissue segmentation in Calliphora vicina (blowfly) In: Society EM, editor. The 15th European Microscopy Congress. UK: Manchester Central; 2012. Available at http://www.emc2012.org.uk//documents/Abstracts/Abstracts/EMC2012_0874.pdf. [Google Scholar]
- Willekens I, Lahoutte T, Buls N. Time-course of contrast enhancement in spleen and liver with Exia 160, Fenestra LC, and VC. Mol Imaging Biol. 2009;11:128–135. doi: 10.1007/s11307-008-0186-8. [DOI] [PubMed] [Google Scholar]
- Wolfsen H, Wallace M. New technologies for imaging of Barrett's esophagus. In: Sampliner RE, editor. Endoscopic Therapy for Barrett's Esophagus. Dordrecht: Humana Press; 2009. pp. 1–20. [Google Scholar]
- Woźniak M. Contrast media: posology, risks and side effects. In: Tröger J, Seidensticker P, editors. Paediatric Imaging Manual. Berlin: Springer; 2008. pp. 5–12. [Google Scholar]
- Zhu X-Y, Bentley MD, Chade AR. Early changes in coronary artery wall structure detected by microcomputed tomography in experimental hypercholesterolemia. Am J Physiol Heart Circ Physiol. 2007;293:H1997–H2003. doi: 10.1152/ajpheart.00362.2007. [DOI] [PubMed] [Google Scholar]


