Abstract
The significance of patient and donor ethnicity on risk of acute graft-versus-host disease (GVHD) and disease relapse after unrelated donor hematopoietic cell transplantation (HCT) is not known. A total of 4335 patient/donor pairs from the International Histocompatibility Working Group in HCT met the following three criteria: (1) HLA-A, B, C, DRB1 and DQB1 allele matched donor; (2) diagnosis of leukemia, and (3) non-T cell depleted GVHD prophylaxis. Post-transplant risks of acute GVHD and leukemia relapse were defined in Asian/Pacific Islander, Caucasian, African American, Hispanic, and Native American patients transplanted from donors with the same self-described background. Asian patients had a significantly lower incidence of acute GVHD (Japanese patients: 40.0% grades II-IV and 15.3% grades III-IV; non-Japanese Asian patients: 42.1% grades II-IV and 15.7% grades III-IV) compared to Caucasian patients (56.5% grades II-IV and 22.6% grades III-IV) (p< 0.001). The hazard ratio (HR) of acute GVHD for Caucasian patients was significantly higher than for Japanese patients. Unexpectedly, the HR of leukemia relapse in Caucasian patients with early disease status was also significantly higher than that in Japanese patients. These results provide a platform for future investigation into the genetic factors for unrelated donor HCT and clinical implications of diverse ethnic background.
INTRODUCTION
Patients who lack a matched sibling to serve as the transplant donor benefit from allogeneic hematopoietic cell transplantation (HCT) from an HLA matched unrelated donor [1-6]. In both settings, donor recognition of recipient polymorphisms may lead to acute graft-versus-host disease (GVHD), a potentially life-threatening complication that necessitates long-term immunosuppressive therapy. HLA identical siblings are identical by descent and hence, acute GVHD arises from donor recognition of non-HLA polymorphisms located outside of the HLA region [7]. Criteria for unrelated donor selection is based on compatibility for alleles of HLA-A, C, B, DRB1, and DQB1 genes because matching is associated with lower risks of acute GVHD than HLA mismatching. Acute GVHD after HLA matched unrelated donor HCT may result from donor recognition of genome-wide polymorphisms, including undetected variation within the HLA region [8,9].
Clinical outcome after HCT depends on many factors contributed by the patient, the donor, and the specific transplantation procedures employed. In HLA matched sibling transplantation, patient ethnicity has been reported to influence the incidence of GVHD where Caucasian Americans, African Americans, and Irish cohorts were at significantly higher risk for acute GVHD than were Japanese or Scandinavian cohorts [10]. The genetic basis that might explain these different outcomes has not been defined. The distribution of HLA alleles is reflected in the ethnicity of the population [11], and the probability of finding an HLA matched unrelated donor is highest when the donor and patient share the same ethnicity. The results of transplantation from selected populations have been extensively reported. In the US population of Caucasian patients receiving a transplant from a Caucasian unrelated donor [4], the incidence of grades III-IV acute GVHD is 28% compared to 11·8% observed in the Japanese experience [5].These data individually suggest that the risk of acute GVHD after unrelated donor HCT depends on the patient and donor's ethnic backgrounds; however, a formal comparison of outcomes among patients with different ethnic backgrounds has never been undertaken. If the complications after transplantation depend on donor-recipient ethnicity, then this information will facilitate future mapping of polymorphisms responsible for complications such as acute GVHD, and provide insight into the genetic basis of acute GVHD. Furthermore, the information may have practical value in the search for suitable unrelated donors.
We undertook a large-scale international study to define risks after HLA matched unrelated donor HCT performed for patients with different ethnic backgrounds within the International Histocompatibility Working Group (IHWG) in HCT [12]. These data provide a unique opportunity to elucidate the clinical effects of ethnicity on risk of acute GVHD and leukemia relapse in HLA matched unrelated HCT.
PATIENTS AND METHODS
Study Population
A total of 4335 patients from the IHWG database met the following criteria and were included in the current analysis: 1) transplantation from an HLA-A, B, C, DRB1, and DQB1 allele compatible unrelated donor; 2) patient diagnosis of acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML) or chronic myeloid leukemia (CML) and 3) non-T cell depleted stem cell source without use of anti-thymocyte globulin (ATG) for GVHD prophylaxis.
Patient characteristics by ethnicity are described in Table 1. Among these subjects, 1232 patients carried the diagnosis of ALL, 1758 AML and 1345 CML. Low-risk disease was defined as a CML chronic phase (CP) at the time of transplantation. The definition of disease risk was comparable among clinical centers within IHWG. Intermediate-risk was defined as transplantation in the first or second complete remission (CR) of ALL, AML, or the second CP or accelerated phase (AP) of CML. High-risk was defined as transplantation in a more advanced stage than intermediate risk. Early status of disease included patients in first and second complete remission (CR) of acute lymphoblastic leukemia or acute myeloid leukemia at transplantation, first chronic phase of chronic myeloid leukemia at transplantation. For GVHD prophylaxis, a tacrolimus-based regimen was employed in 1536 patients, a cyclosporine-based regimen in 2593 and other regimens in 78. Patients were conditioned for transplantation using either a myeloablative (N=3687) or a non-myeloablative/reduced-intensity regimen (n=381). A total of 3481 patients were transplanted with bone marrow and 854 with peripheral blood stem cells (PBSCs). Informed consent was obtained from patients and donors in accordance with the Declaration of Helsinki in each registry or institution, and consent of participation with the IHWG was obtained from each participating registry or institution before registration of the data or materials. Participating registries and institutions are listed in the supplemental table.
Table 1.
Characteristics of hematopoietic stem cell transplantation from unrelated donor by ethnicity
| Ethnicity of patient-donor pair | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total number of pairs | Japanese* | Asian excluding Japanese | Caucasian | African American | Hispanic | Native American | Mismatched Ethnicity | Unknown Ethnicity | |
| Number of pairs | 4335 | 1734 | 19 | 1794 | 34 | 27 | 1 | 171 | 555 |
| HLA-DPB1 matching status (GVH direction) | |||||||||
| Match | 1053 | 622 | 8 | 320 | 2 | 9 | 0 | 27 | 65 |
| One allele mismatch | 1530 | 798 | 3 | 553 | 12 | 3 | 0 | 45 | 116 |
| Two allele mismatch | 818 | 307 | 1 | 383 | 3 | 5 | 1 | 38 | 80 |
| Unknown | 934 | 7 | 7 | 538 | 17 | 10 | 0 | 61 | 294 |
| Patient age, mean | 33.8 | 30.4 | 33.9 | 36.5 | 34.1 | 30.4 | - | 35.3 | 35.4 |
| Donor age, mean | 34.7 | 34.3 | 35.4 | 35.1 | 38.9 | 34.0 | - | 35.1 | 34.6 |
| Disease | |||||||||
| Acute lymphoblastic leukemia | 1232 | 620 | 11 | 424 | 8 | 9 | 0 | 41 | 119 |
| Acute myeloid leukemia | 1758 | 709 | 4 | 703 | 11 | 8 | 0 | 83 | 240 |
| Chronic myeloid leukemia | 1345 | 405 | 4 | 667 | 15 | 10 | 1 | 47 | 196 |
| Disease risk† | |||||||||
| Low | 938 | 273 | 3 | 480 | 5 | 7 | 1 | 29 | 140 |
| Intermediate | 2441 | 1093 | 10 | 910 | 19 | 16 | 0 | 94 | 299 |
| High | 931 | 352 | 6 | 399 | 10 | 4 | 0 | 46 | 114 |
| Unknown | 25 | 16 | 0 | 5 | 0 | 0 | 0 | 2 | 2 |
| Patient-donor sex | |||||||||
| Female-male | 756 | 302 | 3 | 308 | 9 | 10 | 1 | 33 | 90 |
| Male-female | 944 | 341 | 6 | 435 | 5 | 4 | 0 | 45 | 108 |
| Female-female | 811 | 335 | 3 | 324 | 9 | 3 | 0 | 39 | 98 |
| Male-male | 1789 | 752 | 7 | 725 | 11 | 10 | 0 | 54 | 230 |
| Unknown | 35 | 4 | 0 | 2 | 0 | 0 | 0 | 0 | 29 |
| GVHD prophylaxis | |||||||||
| Cyclosporine based | 2593 | 964 | 9 | 1167 | 19 | 12 | 1 | 97 | 324 |
| Taclorimus based | 1536 | 757 | 10 | 592 | 15 | 14 | 0 | 70 | 78 |
| Other | 78 | 8 | 0 | 23 | 0 | 1 | 0 | 4 | 42 |
| Unknown | 128 | 5 | 0 | 12 | 0 | 0 | 0 | 0 | 111 |
| Conditioning regimen | |||||||||
| Myeloablative | 3687 | 1631 | 18 | 1545 | 27 | 25 | 1 | 149 | 291 |
| Non-myeloablative/reduced intensity | 381 | 103 | 1 | 219 | 7 | 2 | 0 | 21 | 28 |
| Unknown | 267 | 0 | 0 | 30 | 0 | 0 | 0 | 1 | 236 |
| Total body irradiation | |||||||||
| No | 949 | 315 | 1 | 423 | 10 | 5 | 0 | 44 | 151 |
| Yes | 3371 | 1419 | 18 | 1360 | 23 | 22 | 1 | 125 | 403 |
| Unknown | 15 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 |
| Stem cell source | |||||||||
| Bone marrow | 3481 | 1734 | 9 | 1272 | 24 | 19 | 1 | 105 | 317 |
| Peripheral blood stem cells | 854 | 0 | 10 | 522 | 10 | 8 | 0 | 66 | 238 |
| Transplanted year (median) | - | 1993-2005 (2000) | 1991-2005 (2000) | 1984-2007 (1999) | 1991-2005 (2000) | 1996-2006 (2001) | - | - | - |
Japanese include only individuals from the Japan Marrow Donor Program.
Disease status prior to transplantation is categorized as low (chronic phase of chronic myeloid leukemia); intermediate (the first or second remission of acute lymphoblastic leukemia, acute myeloid leukemia, or the second chronic phase [CP] or accelerated phase of chronic myeloid leukemia); high risk (more advanced stage than intermediate risk).
Donor-Recipient Ethnicity
Patients were grouped as having the same, different, or unknown ethnicity as to their HLA matched unrelated donor according to self-designated descriptions as defined elsewhere [13]. A total of 3609 patients had the same ethnicity as their donors (“matched ethnicity”); of these pairs, 1753 were of Asian/Pacific Islander background (of which 1734 were transplants for Japanese pairs performed by the Japan Marrow Donor Program [JMDP] in Japan), 1794 Caucasian, 34 African American, 27 Hispanic, and 1 Native American (Table 1). A total of 171 patients had a different background than their unrelated donor (“mismatched ethnicity”). Most pairs with mismatched ethnicity (162 of 171) were of non-Asian backgrounds. Ethnicity information of either patient or donor was not available for 555 pairs (“unknown ethnicity”).
HLA Typing and HLA Matching
High resolution HLA typing of patient and donor pairs was performed in HLA typing laboratories or registries participating within the IHWG, or in the Division of Clinical Research of Fred Hutchinson Cancer Research Center as described elsewhere [6, 12]. HLA-DPB1 allele mismatching among the donor-recipient pairs was scored when the recipient's alleles were not shared by the donor in GVH direction for all analyses.
Biostatistical Methods
Cumulative incidences of acute GVHD and relapse were assessed by a method described elsewhere to eliminate the effect of competing risk [14]. Overall survival was calculated using the Kaplan-Meier method. A competing event regarding acute GVHD was defined as death without acute GVHD, and a competing event regarding relapse was defined as death without relapse. A log-rank test was applied to assess the impact by the factor of interest. Multivariable Cox regression analyses were conducted to evaluate the impact of acute GVHD, leukemia relapse and mortality after transplantation. Confounders considered were combinations of ethnicity between patient and donor, sex (donor-recipient pair), patient age (linear), donor age (linear), risk of leukemia relapse (low, intermediate and high), GVHD prophylaxis (cyclosporine-based regimen, tacrolimus-based regimen and the other regimen without cyclosporine and tacrolimus), source of stem cell, and preconditioning (myeloablative and non-myeloablative/reduced intensity). Missing events were treated as an unknown group.
RESULTS
Risk of Acute GVHD among Ethnicity Matched Donor-Recipient Pairs
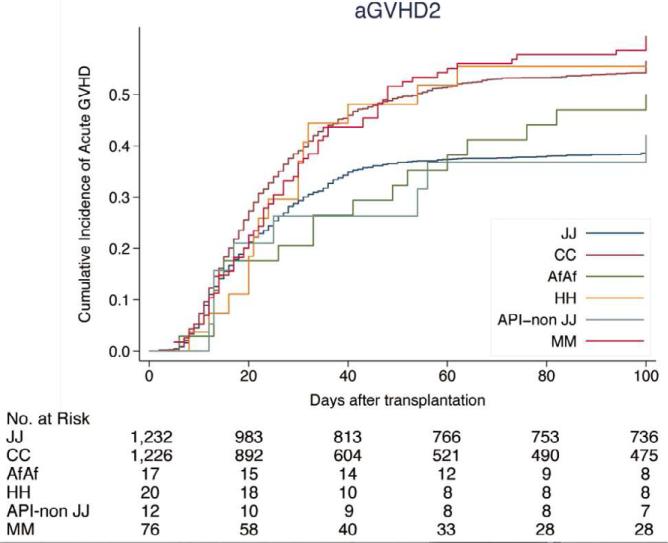
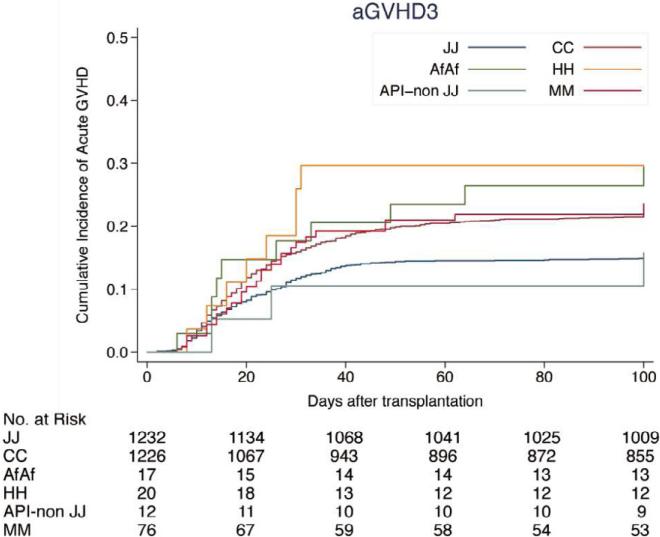
The cumulative incidence of acute GVHD in HLA-A, B, C, DRB1 and DQB1-matched pairs was lower in patients of Asian/Pacific Islander background (Japanese pairs: 40.0% grades II-IV and 15.3% grades III-IV; non-Japanese pairs: 42.1% grades II - IV and 15.7% grades III - IV) compared to Caucasian background (56.5% grades II-IV and 22.6% grades III-IV). The incidence of grades II-IV and III-IV acute GVHD was 50.0% and 29.6% in African American pairs, 55.5% and 29.6% in Hispanic pairs, and 60.0% and 23.5% in patients of mismatched ethnicity, respectively (Figure 1, Supplemental Table S1).
Figure 1.
Cumulative incidence of acute GVHD by ethnicity. (A) Grades II-IV acute GVHD; (B) grades III-IV acute GVHD. JJ, Japanese donor and patient pair; CC, Caucasian donor and patient pair; AfAf, African American donor and patient pair; HH, Hispanic donor and patient pair; API non-JJ, Asian/Pacific Islander donor and patients excluding Japanese pair; MM, Mismatch race/ethnicity pair between donor and patients. Incidence of acute GVHD is shown in Supplemental Table S1.
Of the 4335 pairs, 1053 were matched for HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 alleles (12/12 match). Japanese pairs (n=622) showed a significantly lower incidence of acute GVHD than Caucasian pairs (n=320): 30.7% and 57.0% of grades II-IV (p<0·001); 12.1% and 19.4% grades III-IV (p=0.001), respectively.
Multivariate models adjusted for HLA-DPB1 match status and clinical factors (see Methods) revealed that the HR of acute GVHD in Caucasian pairs was significantly higher compared to Japanese pairs (reference group): HR 1.59 for grades II-IV (p<0.001) and HR 1.48 for grades III-IV (p<0.001). The HR for acute GVHD in non-Japanese Asian pairs was not significantly higher compared to Japanese pairs (reference group): HR 1.07 for grades II-IV (p=0.84) and HR 1.03 for grades III-IV (p=0.96) (Table 2).
Table 2.
Hazard risk (HR) of acute GVHD, leukemia relapse and mortality by ethnicity
| N | Grades II-IV acute GVHD HR (95% CI) | p value | Grades III-IV acute GVHD HR (95% CI) | p value | Relapse HR (95% CI) | p value | Mortality HR (95% CI) | p value | |
|---|---|---|---|---|---|---|---|---|---|
| Ethnicity | |||||||||
| Japanese pair | 1734 | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| Asian pair excluding Japanese | 19 | 1.07 (0.55-2.17) | 0.84 | 1·03 (0.32-3.23) | 0.96 | 2.92 (1.59-5.36) | 0.001 | 1.25 (0.68-2.28) | 0.46 |
| Caucasian pair | 1794 | 1.59 (1.42-1.77) | <0.001 | 1.48 (1.25-1 76) | <0.001 | 1.63 (1.40-1.90) | <0.001 | 1.46 (1.32-1.62) | <0.001 |
| African pair | 34 | 1.21 (0.74-1.98) | 0.43 | 1.81 (0.94-3.45) | 0.07 | 2.35 (1·24-4·47) | 0.04 | 2.51 (1.67-3.76) | <0.001 |
| Hispanic pair | 27 | 1.61 (0.96-2.64) | 0.07 | 2.23 (1.09-4.53) | 0.03 | 3.77 (2.00-7.13) | <0.001 | 2.87 (1.85-4.45) | <0.001 |
| Mismatched race/ethnicity | 171 | 1.71 (1.38-2.11) | <0.001 | 1.57 (1.12-2.22) | 0.009 | 1.80 (1.34-2.42) | <0.001 | 1.62 (1.32-1.98) | <0.001 |
| Unknown ethnicity | 555 | 2.38 (2.01-2.83) | <0.001 | 1.48 (1.10-1.98) | 0.008 | 1.56 (1.21-2.00) | 0.005 | 1.33 (1.11-1.58) | 0.001 |
| HLA-DPB1 matching (GVH direction) | |||||||||
| Match | 1053 | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| 1-allele mismatch | 1530 | 1.34 (1.19-1.51) | <0·001 | 1.40 (1.15-1.69) | 0.001 | 0.66 (0.57-0.77) | <0.001 | 1.02 (0.91-1.13) | 0.69 |
| 2-allele mismatch | 818 | 1.48 (1.29-1.69) | <0·001 | 1.45 (1.16-1.80) | <0.001 | 0.54 (0.44-0.66) | <0.001 | 1.01 (0.89-1.15) | 0.78 |
| Unknown | 934 | 1.20 (1.03-1.40) | 0.01 | 1.12 (0.87-1.43) | 0.30 | 0.62 (0.51-0.75) | <0.001 | 0.74 (0.64-0.85) | <0.001 |
Multivariate analysis adjusted for clinical factors listed in Table 1. Data from one North American pair is not shown in this table.
Risk of Leukemia Relapse among Ethnicity Matched Donor-Recipient Pairs
Patients who experience acute GVHD may have a lower risk of disease recurrence compared to patients without GVHD, a phenomenon termed “graft-versus-leukemia (GVL) effect”.15 To better define GVHD-associated risks after transplantation, we investigated the probability of disease recurrence among the ethnicity-matched group. Using the Japanese pairs as the reference, we unexpectedly found that the HR for relapse in Caucasian patients was significantly higher compared to Japanese patients (HR 1.63, p<0·001) (Table 3). The rates of acute GVHD (grades II-IV and grades III-IV) were not associated with differences in relapse rates either in Japanese pairs or in Caucasian pairs (Supplemental Table S2).
Table 3.
Leukemia relapse rate by ethnicity, leukemia cell type and the status at transplantation
| N | Acute lymphoblastic leukemia | N | Acute myeloid leukemia | N | Chronic myeloid leukemia | |
|---|---|---|---|---|---|---|
| Early status* | ||||||
| Japanese pair | 469 | 23.9† | 465 | 20.9 | 269 | 7.3 |
| p=0.004 | p<0.001 | p=0.02 | ||||
| Caucasian pair | 298 | 30.5 | 436 | 30.7 | 478 | 10.7 |
| Advanced status‡ | ||||||
| Japanese pair | 124 | 54.5 | 205 | 43.7 | 123 | 24.3 |
| p=0.54 | p=0.12 | p=0.30 | ||||
| Caucasian pair | 118 | 51.0 | 249 | 48.4 | 187 | 25.8 |
First and second complete remission (CR) of acute lymphoblastic leukemia or acute myeloid leukemia at transplantation, first chronic phase of chronic myeloid leukemia at transplantation.
Cumulative incidence (%) of leukemia relapse at 5 years after transplantation.
More advanced status than early status.
The cumulative incidence of relapse at five years after transplantation was assessed according to myeloid or lymphoid lineage of the leukemia and disease risk at the time of transplantation (Table 3). In patients transplanted in early status of their disease (AML and ALL in the first CR or 2nd CR, CML in the first chronic phase), Caucasian patients had a significantly higher relapse rate than Japanese patients. In patients transplanted for more advanced status of disease, relapse rates for Caucasian patients and Japanese patients were not significantly different.
Risk of Survival among Ethnicity Matched Donor-Recipient Pairs
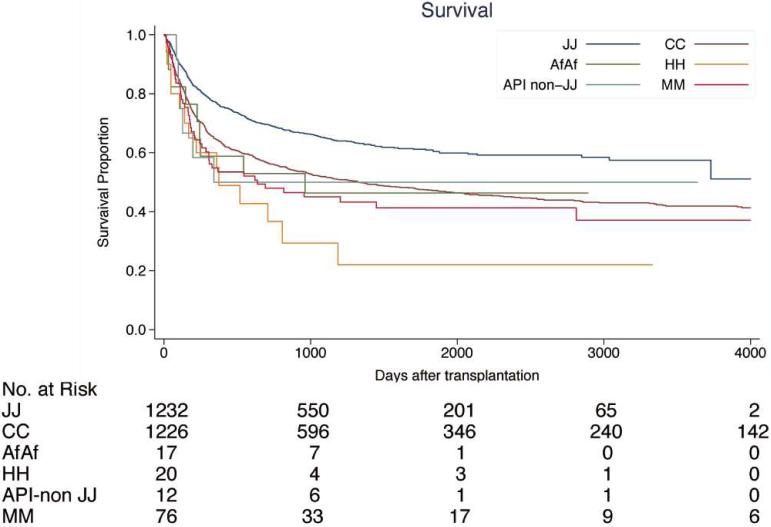
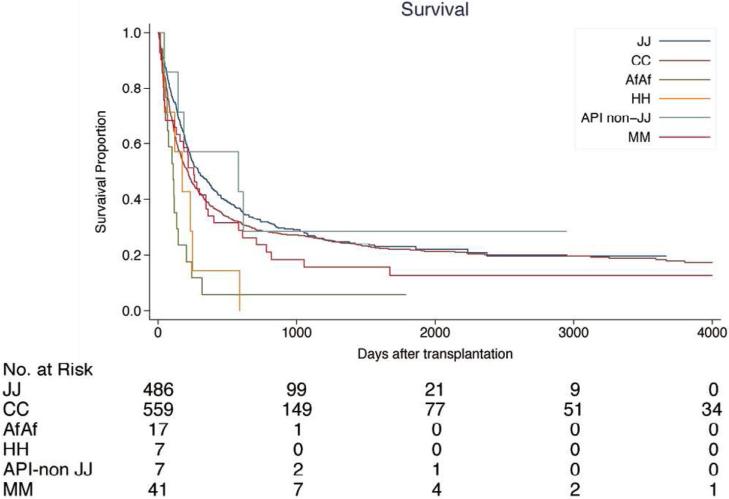
Compared to Japanese patients, patients of Caucasian, African American, Hispanic backgrounds, and patients who received a transplant from donors with a different background, had higher mortality, with the exception of non-Japanese Asian patients (Table 2, Figure 2).
Figure 2.
Survival after transplantation by ethnicity. (A) Patients with early status of leukemia at transplantation; (B) patients with advanced status of leukemia at transplantation. Early status: 1st and 2nd complete remission of ALL or AML at transplantation, 1st chronic phase of CML. Advanced status: more advanced status than early status. JJ, Japanese donor and patient pair; CC, Caucasian donor and patient pair; AfAf, African American donor and patient pair; HH, Hispanic donor and patient pair; API non-JJ, Asian/Pacific Islander donor and patients excluding Japanese pair; MM, Mismatch ethnicity pair between donor and patients. 5-year survival rate after transplantation by ethnicity is provided in Supplemental Table S1.
Factors other than Ethnicity among Ethnicity Matched Donor-Recipient Pairs
The increased hazard of acute GVHD and the decreased hazard of relapse in one or two HLA-DPB1-mismatched transplants did not translate to any difference in survival by ethnicity (Table 2). The HRs of the other factors for acute GVHD, leukemia relapse and mortality by multivariate analysis are shown in Supplemental Table S3.
As all Japanese patients received bone marrow for the grafting source, the association of ethnicity with the hazards of acute GVHD, leukemia relapse and mortality for patients receiving bone marrow transplantation are shown in Supplemental Table S4 and S5. The hazards of acute GVHD, leukemia relapse, mortality and relapse at 5-years after transplantation between Japanese pairs and Caucasian pairs were similar for recipients of bone marrow and peripheral blood stem cell transplantation.
DISCUSSION
Donor HLA matching lowers risks of graft failure, acute GVHD, morbidity and mortality [2-6] and remains the standard for the selection of unrelated donors for HCT. Clinical experience demonstrates that the incidence of GVHD after HLA matched unrelated HCT is not the same across different populations [4,5]. The frequency of HLA phenotypes and haplotypes differs worldwide [11,12] and information regarding the content of HLA region variation is available for selected populations [15,16]. We hypothesize that the risks of GVHD depend on population-specific genetic variation. We undertook the current analysis to compare the risks of GVHD among patients of different ethnic backgrounds. This information provides the necessary basis for future investigation of the impact of specific genetic variation on clinical outcome.
HLA antigens and haplotypes are population specific. The IHWG database is a unique resource for the comparative analysis of transplant outcomes and HLA genetics. In order to analyze acute GVHD and GVL effects clinically, it is essential that non-genetic factors that also influence these transplant-related immunological events are taken into consideration. As HLA compatibility between the patient and donor is a well-recognized risk factor for acute GVHD and leukemia relapse in unrelated donor HCT [1-6,17,18], we restricted the study population to HLA-A, B, C, DRB1, and DQB1 allele-matched pairs. GVHD prophylaxis is also another important factor for acute GVHD. Therefore, only leukemia patients transplanted from non-T cell depleted stem cell sources without ATG were included in the analysis. Caucasian and Asian patients whose donors were also of the same background, were particularly well-represented, and permitted us to assess risks between these two populations. A larger clinical experience will be needed to fully evaluate clinical outcome in patients and donors of African American, Hispanic, Native American heritage.
This study shows that the incidence of acute GVHD in Japanese patients is lower than that in Caucasian patients. Although HLA-DPB1 mismatching is associated with risk of acute GVHD [2,19], ethnicity was an independent risk factor in HLA-A, B, C, DRB1, and DQB1 matched transplants; furthermore, among pairs matched at all six classical loci including HLA-DPB1, we observed a significantly higher incidence of acute GVHD in Caucasian compared to Japanese patients. Multi-SNP analysis of the HLA region has recently been performed in Japanese [9,16] and in Caucasian [8] populations. We surmise that the lowered risk of acute GVHD observed in the Japanese patients might be explained in part by the high degree of conservation of the HLA region for common HLA haplotypes in the Japanese population, and/or the absence of GVHD-risk determinants on haplotypes in the Japanese population, and/or presence of GVHD-risk determinants on haplotypes in Caucasian populations. Although our study population was matched for HLA, unrelated donors are only matched for HLA alleles, and hence, HLA matched pairs may have the same or different HLA haplotypes which could be a source of disparity for undetected haplotype-linked variation [17]. In contrast to the homogeneity of the Japanese transplant population in which three extended HLA haplotypes are observed at very high frequency [16], only 37 Caucasian donor-recipient pairs in the current study were homozygous for the four most commonly observed Caucasian haplotypes . Formal analysis of HLA haplotype-associated variation has uncovered novel untyped variation responsible for GVHD in Caucasian patients [18,19] and comparable analyses are underway for Japanese pairs. Examination of MHC resident variation should shed light on haplotype-linked polymorphisms unique to each ethnic group that could be investigated in the future.
In the HLA region, there exist several candidate genes in which SNPs may be related to immune responses. Genetic variants of TNF-α gene located in the HLA region might influence the risk of developing GVHD [20-22]. In addition, TAP1/TAP2 and LMP2/LMP7 genes encode subunit components of the proteasome implicated in the processing of class I HLA-bound peptides and polymorphisms of these genes may affect antigen presentation on recipient tissues, thus leading to different susceptibility to GVHD.
In HCT from HLA-identical siblings, Caucasian Americans, African Americans, and Irish cohorts were reported to be at significantly higher risk for acute GVHD than Japanese or Scandinavian cohorts [10]. In unrelated donor HCT, there are other polymorphisms other than HLA . These data suggest that other genetic differences might exist outside of the HLA region. Microbe-associated molecules [23], innate immune receptors associated with GVHD, IL-10 [24], and heparanase [25] are all candidates. The HLA alleles of each haplotype might present different immunodominant peptides to T cells and evoke different allo-reactivity in HLA-matched unrelated donor HCT [9]. The critical, but as yet unidentified, minor histocompatibility antigens restricted by major histocompatibility antigens such as common gene deletion polymorphisms, and the differences in ethnically diverse populations should shed light [26]. Other intriguing candidate factors for acute GVHD might include environmental factors, foods, and drug sensitivity of chemotherapeutic regimens used specifically in transplantation and of other pharmacologic agents for health maintenance [27]. These factors could be explored in unrelated as well as HCT from HLA identical related donors.
In the current study, significantly lower incidence of relapse in Japanese pairs than that in Caucasian pairs was observed in patients with early disease status . Japanese patients had a lower relapse rate than Caucasian pairs regardless of leukemia type and the occurrence of acute GVHD. HLA-DPB1 mismatching was also an independent hazard risk for leukemia relapse, and lowered leukemia relapse in both Japanese and Caucasian populations [28,29]. Although these results might be induced in part by GVL effects, GVL effects cannot entirely explain the difference of the risk of leukemia relapse between the two ethnic populations. Although information on pre-transplant induction/consolidation regimens was not available, disease risk stage at the time of transplantation is a surrogate for the risk of relapse. That Japanese patients had both lower relapse and lower GVHD risk compared to Caucasian patients, suggests more complex factors beyond the classic GVL effect, including GVHD-dependent and GVHD-independent pathways. Yang et al reported that leukemia relapse risk after chemotherapy is differ by ethnicity in children with ALL in North American population, showing that Native Americans and Hispanic ethnicity showed the higher incidence of relapse rate [30]. The polymorphisms of chemotherapy resistance for relapse specific to ethnicity might exist among ethnic groups. Identification of non-HLA genes and alleles within the MHC and polymorphisms outside of the HLA region responsible for leukemia relapse remains an important research goal.
Genome-wide analysis of world populations shows intriguing genetic differences that are population-specific [31], however, genetics may be only one of several factors that lead to differences in GVHD and mortality between Japanese and Caucasian patients. The overwhelming majority of Japanese patients included in this analysis were treated in Japan and were therefore exposed to a relatively homogeneous “environment”. The impact of sociocultural and/or dietary factors that could also influence post-transplant risks merits evaluation in future studies of ethnically diverse populations, particularly transplant outcomes for patients of Japanese heritage who reside outside of Japan.
Our study highlights the need for comprehensive analysis of HLA region genetics as a starting point for identification of GVHD-associated polymorphisms. We used broad terms to classify Caucasian, Hispanic and African Americans; however, each of these populations may include individuals of very different ethnic backgrounds, and is one limitation of our study at present. Critical to achieving this goal is the need to define the genetic diversity of all human populations given the highly polymorphic nature of the HLA system. In the US, a larger clinical experience will aid our understanding of the immunogenicity of specific HLA phenotypes representing African American, Hispanic and Native Americans [13,32].
As most Asian/Pacific Islander pairs in our current study (1734 out of 1753) were Japanese pairs registered from the JMDP, the data are representative of the Japanese population rather than Asian/Pacific ethnicities as a whole.
Contribution of data from other populations in the future will aid in the effort to mapping transplantation determinants, information that will benefit all patients. The results of our analysis provide a platform for future international analyses of unrelated HCT outcomes and for international exchange of unrelated donors.
Supplementary Material
Acknowledgments
We thank all the members of the IHWG hematopoietic-cell transplantation component who contributed data to this publication (Supplemental Table S6). We are grateful for support from the following agencies: Grant (H23-Immunology-010) from the Ministry of Health, Labor and Welfare, Japan, Grant (3224-22133011) from the Ministry of Education, Culture, Sports, Science and Technology, Japan; Grants AI069197, CA100019, and CA18029 (EWP, MM), and CA76518 (SS) from the National Institutes of Health, USA. HHSH234200637015C (SS) from the Health Resources and Services Administration, USA; N00014-10-1-0204 and N00014-1-1-0339 (SS) from the Office of Naval Research, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship
Contribution: YM, TK, MM, SM and EWP participated in the design and set up of this study. SS, YM, KK, SK, MM, EWP, AC, JT, DS, AV, and IHWG in HCT participants (a complete list appears in Supplemental Table S6) contributed to data collection and HLA typing; TK performed the statistical analysis; YM, TK, SM, and EWP wrote the paper.
Financial disclosure statement: The authors declare no competing financial interests.
References
- 1.Petersdorf EW. Immunogenomics of unrelated hematopoietic cell transplantation. Curr Opin Immunol. 2006;18(5):559–564. doi: 10.1016/j.coi.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Sasazuki T, Juji T, Morishima Y, et al. Effect of matching of class I HLA alleles on clinical outcome after transplantation of hematopoietic stem cells from an unrelated donor. Japan Marrow Donor Program. N Engl J Med. 1998;339(17):1177–1185. doi: 10.1056/NEJM199810223391701. [DOI] [PubMed] [Google Scholar]
- 3.Petersdorf EW, Gooley TA, Anasetti C, et al. Optimizing outcome after unrelated marrow transplantation by comprehensive matching of HLA class I and II alleles in the donor and recipient. Blood. 1998;92(10):3515–3520. [PubMed] [Google Scholar]
- 4.Lee SJ, Klein J, Haagenson M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110(13):4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 5.Morishima Y, Sasazuki T, Inoko H, et al. The clinical significance of human leukocyte antigen (HLA) allele compatibility in patients receiving a marrow transplant from serologically HLA-A, HLA-B, and HLA-DR matched unrelated donors. Blood. 2002;99(11):4200–2406. doi: 10.1182/blood.v99.11.4200. [DOI] [PubMed] [Google Scholar]
- 6.Spellman SR, Eapen M, Logan BR, et al. A perspective on the selection of unrelated donors and cord blood units for transplantation. Blood. 2012;120(2):259–265. doi: 10.1182/blood-2012-03-379032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goulmy E. Human minor histocompatibility antigens: new concepts for marrow transplantation and adoptive immunotherapy. Immunol Rev. 1997;157:125–140. doi: 10.1111/j.1600-065x.1997.tb00978.x. [DOI] [PubMed] [Google Scholar]
- 8.Baschal EE, Aly TA, Jasinski JM, et al. Defining multiple common “completely” conserved major histocompatibility complex SNP haplotypes. Clin Immunol. 2009;132(2):203–214. doi: 10.1016/j.clim.2009.03.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogawa S, Matsubara A, Onizuka M, et al. Exploration of the genetic basis of GVHD by genetic association studies. Biol Blood Marrow Transplant. 2009;15(1 Suppl):39–41. doi: 10.1016/j.bbmt.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 10.Oh H, Loberiza FR, Jr., Zhang MJ, et al. Comparison of graft-versus-host-disease and survival after HLA-identical sibling bone marrow transplantation in ethnic populations. Blood. 2005;105(4):1408–1416. doi: 10.1182/blood-2004-06-2385. [DOI] [PubMed] [Google Scholar]
- 11.Morishima Y, Kawase T, Malkki M, Petersdorf EW. Effect of HLA-A2 allele disparity on clinical outcome in hematopoietic cell transplantation from unrelated donors. Tissue Antigens. 2007;69(Suppl 1):31–35. doi: 10.1111/j.1399-0039.2006.759_3.x. [DOI] [PubMed] [Google Scholar]
- 12.Petersdorf E, Bardy P, Cambon-Thomsen A, et al. 14th International HLA and Immunogenetics Workshop: report on hematopoietic cell transplantation. Tissue Antigens. 2007;69(Suppl 1):17–24. doi: 10.1111/j.1399-0039.2006.759_1.x. [DOI] [PubMed] [Google Scholar]
- 13.Baker KS, Davies SM, Majhail NS, et al. Race and socioeconomic status influence outcomes of unrelated donor hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2009;15(12):1543–1554. doi: 10.1016/j.bbmt.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94(446):496–509. [Google Scholar]
- 15.Maiers M, Gragert L, Klitz W. High-resolution HLA alleles and haplotypes in the United States population. Hum Immunol. 2007;68(9):779–788. doi: 10.1016/j.humimm.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Morishima S, Ogawa S, Matsubara A, et al. Impact of highly conserved HLA haplotype on acute graft-versus-host disease. Blood. 2010;115(23):4664–4670. doi: 10.1182/blood-2009-10-251157. [DOI] [PubMed] [Google Scholar]
- 17.Petersdorf EW, Malkki M, Gooley TA, Martin PJ, Guo Z. MHC haplotype matching for unrelated hematopoietic cell transplantation. PLoS Med. 2007;4(1):e8. doi: 10.1371/journal.pmed.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersdorf EW, Malkki M, Gooley TA, et al. MHC-resident variation affects risks after unrelated donor hematopoietic cell transplantation. Sci Transl Med. 2012;4(144):144ra101. doi: 10.1126/scitranslmed.3003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersdorf EW, Malkki M, Horowitz MM, et al. Mapping MHC haplotype effects in unrelated donor hematopoietic cell transplantation. Blood. 2013;121:1896–1905. doi: 10.1182/blood-2012-11-465161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen JA, Petersdorf EW, Lin MT, et al. Genetics of allogeneic hematopoietic cell transplantation. Role of HLA matching, functional variation in immune response genes. Immunol Res. 2008;41(1):56–78. doi: 10.1007/s12026-007-0043-x. [DOI] [PubMed] [Google Scholar]
- 21.Bettens F, Passweg J, Schanz U, et al. Impact of HLA-DPB1 haplotypes on outcome of 10/10 matched unrelated hematopoietic stem cell donor transplants depends on MHC-linked microsatellite polymorphisms. Biol Blood Marrow Transplant. 2012;18(4):608–616. doi: 10.1016/j.bbmt.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 22.Harkensee C, Oka A, Onizuka M, et al. Single nucleotide polymorphisms and outcome risk in unrelated mismatched hematopoietic stem cell transplantation: an exploration study. Blood. 2012;119:6365–6372. doi: 10.1182/blood-2012-01-406785. [DOI] [PubMed] [Google Scholar]
- 23.Penack O, Holler E, van den Brink MR. Graft-versus-host disease: regulation by microbe-associated molecules and innate immune receptors. Blood. 2010;115(10):1865–72. doi: 10.1182/blood-2009-09-242784. [DOI] [PubMed] [Google Scholar]
- 24.Lin MT, Storer B, Martin PJ, et al. Relation of an interleukin-10 promoter polymorphism to graft-versus-host disease and survival after hematopoietic-cell transplantation. N Engl J Med. 2003;349(23):2201–2210. doi: 10.1056/NEJMoa022060. [DOI] [PubMed] [Google Scholar]
- 25.Ostrovsky O, Shimoni A, Rand A, Vlodavsky I, Nagler A. Genetic variations in the heparanase gene (HPSE) associate with increased risk of GVHD following allogeneic stem cell transplantation: effect of discrepancy between recipients and donors. Blood. 2010;115(11):2319–2328. doi: 10.1182/blood-2009-08-236455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spierings E, Hendriks M, Absi L, et al. Phenotype frequencies of autosomal minor histocompatibility antigens display significant differences among populations. PLoS Genet. 2007;3(6):e103. doi: 10.1371/journal.pgen.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rotta M, Storer BE, Storb RF, et al. Donor statin treatment protects against severe acute graft-versus-host disease after related allogeneic hematopoietic cell transplantation. Blood. 2010;115(6):1288–1295. doi: 10.1182/blood-2009-08-240358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawase T, Matsuo K, Kashiwase K, et al. HLA mismatch combinations associated with decreased risk of relapse: implications for the molecular mechanism. Blood. 2009;113(12):2851–2858. doi: 10.1182/blood-2008-08-171934. [DOI] [PubMed] [Google Scholar]
- 29.Fleischhauer K, Shaw BE, Gooley T, et al. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haematopoietic-cell transplantation: a retrospective study. Lancet Oncology. 2012;13:366–374. doi: 10.1016/S1470-2045(12)70004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang JJ, Chang C, Devidas M, et al. Ancestry and pharmacogenetics of relapse in acute lymphoblastic leukemia. Nat Genet. 2011;43(3):237–241. doi: 10.1038/ng.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.1000 Genomes Project Consortium. Abecasis GR, Altshuler D, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [Erratum appears in Nature. 2011 May 26;473(7348):544].
- 32.Mielcarek M, Gooley T, Martin PJ, et al. Effects of race on survival after stem cell transplantation. Biol Blood Marrow Transplant. 2005;11(3):231–239. doi: 10.1016/j.bbmt.2004.12.327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.