Abstract
Previous studies have shown that the DNA repair component Metnase (SETMAR) mediates resistance to DNA damaging cancer chemotherapy. Metnase has a nuclease domain that shares homology with the Transposase family. We therefore virtually screened the tertiary Metnase structure against the 550,000 compound ChemDiv library to identify small molecules that might dock in the active site of the transposase nuclease domain of Metnase. We identified eight compounds as possible Metnase inhibitors. Interestingly, among these candidate inhibitors were quinolone antibiotics and HIV integrase inhibitors, which share common structural features. Previous reports have described possible activity of quinolones as antineoplastic agents. Therefore, we chose the quinolone ciprofloxacin for further study, based on its wide clinical availability and low toxicity. We found that ciprofloxacin inhibits the ability of Metnase to cleave DNA and inhibits Metnase-dependent DNA repair. Ciprofloxacin on its own did not induce DNA damage, but it did reduce repair of chemotherapy-induced DNA damage. Ciprofloxacin increased the sensitivity of cancer cell lines and a xenograft tumor model to clinically relevant chemotherapy. These studies provide a mechanism for the previously postulated antineoplastic activity of quinolones, and suggest that ciprofloxacin might be a simple yet effective adjunct to cancer chemotherapy.
Introduction
Metnase is a recently characterized fusion protein comprising a SET histone methylase domain and a Transposase nuclease domain. Metnase is a DNA repair component present only in anthropoid primates (1–4). The Metnase SET domain di-methylates histone H3 lysine 36 (H3K36), whereas the Transposase nuclease domain has most but not all of the known transposase activities, including 5′-terminal inverted repeats (TIR)-specific DNA binding, DNA looping, assembly of paired end complex, and DNA single-strand cleavage (5, 6).
Metnase enhances nonhomologous end-joining and promotes genomic integration of foreign DNA (3). Both the SET histone methylase and transposase nuclease domains are essential for the enhancement of double-strand break (DSB) repair. The transposase nuclease domain trims free DNA ends to improve end-joining (6, 7), and the SET domain di-methylates H3K36 adjacent to induced DNA DSBs. This di-methylation stabilizes the Ku and MRN complex at the DSB, which enhances DSB repair by nonhomologous end-joining (NHEJ; ref. 8). Interestingly, despite Metnase being present only in primates, it functions seamlessly within the mouse NHEJ repair apparatus to enhance DNA repair when expressed in those cells (9). Metnase also helps suppress chromosomal translocations when 2 simultaneous DSBs are present, probably by speeding proper local intrachromosomal NHEJ (9). Metnase also regulates restart of collapsed replication forks, and promotes Topoisomerase IIα (Topo IIα) mediated chromosome decatenation (7, 10–12).
Metnase is overexpressed in acute leukemia cells relative to normal hematopoietic progenitors (12). Metnase overexpression in acute leukemia cells mediates resistance to etoposide, and repressing Metnase restores sensitivity to this important chemotherapeutic drug. Similarly, repressing Metnase in breast cancer cells increased their sensitivity to the anthracycline Adriamycin (13). Thus, given that Metnase enhances NHEJ DNA repair, repair of collapsed replication forks, and resistance to certain DNA damaging chemotherapies, Metnase represents an attractive clinical target for small molecule inhibition that needs to be validated to get first-in-class anticancer molecules. In theory, small molecule inhibition of Metnase should show an excellent therapeutic index, given that it is overexpressed in malignant cells, and there are few other human Transposase domain proteins with which to cross-react (14).
Therefore, we virtually screened a large chemical library of small compounds for docking into the Metnase nuclease active site. We identified 8 compounds that fit within our docking parameters, including the quinolone gyrase inhibitor antibiotic, ciprofloxacin, and the HIV integrase inhibitors raltegravir and elvitegravir. Quinolones have been reported to have some antineoplastic activity, thought to be due to their ability to inhibit Topo IIα, albeit at high concentrations not achievable clinically (15). In this study, we found that high but clinically achievable concentrations of ciprofloxacin blocked the ability of Metnase to cleave DNA, which is essential for its DSB repair activity (16). Ciprofloxacin inhibited DNA repair of a linearized plasmid only in the presence of Metnase. We also found that ciprofloxacin reduced DNA DSB repair in cells damaged by chemotherapy, and enhanced the sensitivity of cancer cell lines and a xenograft mouse tumor model to clinically relevant chemotherapy. Thus, ciprofloxacin is a Metnase inhibitor that enhances cancer cell sensitivity to chemotherapy by reducing DNA repair.
Materials and Methods
Virtual screening
An approach composed by target-based virtual screening (TBVS) and ligand-based virtual screening (LBVS) was established to select new chemicals (Supplementary Fig. S1). We used LBVS to query the ChemDiv catalog of available chemical structures (more than 550,000) for compounds of potential interest, as described (17). The HIV integrase inhibitors raltegravir and elvitegravir were chosen as ligands for this screen as they virtually docked into the Metnase transposase domain. Two-dimensional models were built using SMDL fingerprints encoding the chemical structures of those compounds. A Tanimoto similarity index then compared the ChemDiv molecules with raltegravir and elvitegravir using a 75% cutoff value. Three-dimensional filters with ROCS v. 2.4.1 (Openeye Inc) were used to choose molecules with the highest similarities to the queries using the Combo similarity index. ChemDiv molecules sharing less than 50% overall similarity with the integrase inhibitors were discarded. Pharmacokinetics/toxicity properties of the compounds were predicted using Volsurf v. 3 (Molecular Discovery Inc). All 3D conformations for the new molecules were generated by the OMEGA software (v. 2.3.2, Openeye Inc.) with the following modifications to the default: wider energy window (eWindow 15) and a lower root mean square cutoff to compare each conformation (RMS of 0.3).
The Metnase transposase domain was modeled by using the bioinformatics tools Fugue and Orchestrar in the Sybyl8.0 software suite (Tripos) using the 3D structure of MOS-1 transposase from Drosophila mauritiana as a template, Supplementary Fig. S2. The high accuracy of the modeled transposase domain structure was verified after a comparison with the published Metnase X-ray crystallographic structures after they became available (PDB codes: 3F2K, 3K9J, 3K9K; ref. 18). All structures were superimposed using DeepView v. 4.0.1 and the root mean square deviation was calculated using all atoms (0.94 Å) and the alpha carbons (0.90 Å), which yielded excellent superimposition, Supplementary Fig. S3.
TBVS was then conducted using the Fast Rigid Exhaustive Docking program (FRED v. 2.2.5, Openeye Inc.) to dock all conformations of compounds into the Metnase Transposase domain. Chemgauss3 was the scoring function, as it was the only one that had a parameterization for the metal atom (Mg2+) present at the active site. A threshold score of −20 (with a tolerance of 2) was kept for the chelation property.
DNA binding and nuclease inhibition assays
Flag-tagged recombinant Metnase was purified as previously described (7), and used to biochemically test the inhibitory activity of the candidate docking compounds. Inhibition of Metnase’s TIR-specific DNA-binding activity was examined using an electrophoretic mobility shift assay as we previously described (7). Inhibition of Metnase’s 5′-overhang cleavage activity was tested using a 5′-32P-labeled oligomer DNA with a pseudo Y structure as we previously described (16).
Colony formation and cell proliferation assays
Colony formation was conducted as we described (13). We monitored proliferation in the presence of chemotherapy and candidate Metnase inhibitors to assess antiproliferative effects on leukemia cells by using cell counting assays conducted in triplicate as described (12).
Generation of stable Metnase over- and underexpressing cells
Stable HEK293T vector control and Metnase overexpressing cells and HEK293 vector control and Metnase knockdown cells were generated as we described previously (10, 13).
Direct NHEJ repair assay
NHEJ DSB repair was directly assayed using intracellular plasmid end-joining as we described (3, 19). All statistical analysis was conducted by using unpaired t tests unless otherwise specified.
Rate of disappearance of γ-H2Ax foci
γ-H2Ax foci were analyzed in A549 cells as we described (10, 19). The data are the composite of 3 experiments ± SEM.
BrdUrd immunofluorescence
A549 cells were seeded and treated above except that at 24 hours intervals, 0.03 mg/mL BrdUrd was added to the cells for 1 hour before fixing and processing for immunostaining using a BrdUrd-specific antibody (Cell Signaling) according to the manufacturer’s instructions. Images were collected using an EVOS fluorescence microscope (Advanced Microscopy Group). Percentage BrdUrd-positive values are averages (+SEM) of 4 to 9 fields with 102 to 642 cells (average = 243) scored per condition.
Xenograft models
5 × 106 A549 human lung cancer cells were injected into each flank of a severe combined immunodeficient (SCID) mouse and allowed to grow for 14 days, then tumor volumes were measured and treatment started. The mice were treated with a 1-week run-in of ciprofloxacin (2 mg/kg daily). Then mice were exposed to concurrent ciprofloxacin (2 mg/kg daily) and cisplatin (2 mg/kg 3 times/week). Tumor volumes were measured 3 times per week. Results were plotted as relative tumor growth for the first day of each week of treatment up to the final day.
Drugs
All drugs were purchased from either ChemDiv or Selleck Chemical with purity higher than 95%.
Results
Virtual screening for Metnase inhibitors
We previously found that the nuclease activity of Metnase resided within its transposase domain, and this domain was required for its DNA repair activity (3, 7). The catalytic core of the Metnase transposase nuclease site comprises a single magnesium ion coordinated by oxygens from D483, E575, and 4 water molecules, based on the 3F2K X-ray structure (18). The remainder of the nuclease pocket comprises neutral or positively charged amino acids that anchor the DNA by means of ionic interactions with phosphates along the DNA backbone (Supplementary Fig. S4).
LBVS was used to screen the ChemDiv library of greater than 550,000 small molecule compounds for potential Metnase nuclease domain inhibitors. The ligands chosen as models for this virtual screening were the HIV integrase inhibitors raltegravir and elvitegravir (20, 21). Three-dimensional structures of the compounds identified in LBVS were then subjected to TBVS for further selection, using FRED as the docking tool (Fig. 1). Eight compounds were selected for biochemical analysis based on docking scores (especially considering their Mg2+ chelation property, depicted in Fig. 1), lack of known toxic moieties, and favorable pharmacokinetics. The compounds in the ChemDiv library chosen for biochemical testing were 5483-0023, 3731-0098 (ciprofloxacin), R092-0025, 8017-3379, D058-0127, D045-0007, raltegravir, and elvitegravir. A carboxylic acid moiety and an aromatic or heteroaromatic ring compose the chemical scaffold of all the compounds except raltegravir, which has a 5-hydroxypyrimidinone moiety that can be partially protonated in pH 7. All molecules share good Mg2+ chelation chemotypes because they were considered as pharmaco-phores throughout the virtual screening procedure. The chelation properties of raltegravir and elvitegravir were already described in X-ray structures of the integrase domain of prototype foamy virus (PFV; ref. 22).
Figure 1.
Target-based virtual screening approach using the modeled Metnase nuclease domain. A, docking pose of ciprofloxacin. Molecular surface representation of the active site of the Metnase nuclease domain showing high-electron density (red) and low-electron density (blue). Valences are displayed to aromatic rings and double bonds. The Mg2+ is embedded into the surface and is not shown here. B, ribbon representation of the active site of the Metnase nuclease domain (dark blue), with ciprofloxacin docked within it, is shown. The pink sphere represents the magnesium ion. C, chemical representation of the final set of molecules chosen from the virtual screen for biochemical screening.
Biochemical analysis of the candidate inhibitors
Inhibition of Metnase’s DNA binding activity by the above compounds was examined using purified recombinant human Metnase protein as we described (6, 7). Metnase binds duplex TIR DNA, a characteristic of transposase domain proteins (1, 5). The ability of these compounds to inhibit TIR DNA binding was assessed using mobility shift assays (Fig. 2A). None of the selected compounds were able to inhibit TIR-specific DNA binding by Metnase. Metnase recognizes and cleaves pseudo Y structures near the 5′ end (6). We therefore determined whether the 8 potential Metnase inhibitors could block this activity. Significantly, raltegravir, elvitegravir, and ciprofloxacin (3731-0098) were highly effective in blocking Metnase cleavage of the 5′ end of the pseudo Y substrate. The remaining compounds either had no effect or seemed to stimulate Metnase nuclease activity. We selected ciprofloxacin for further testing based on (i) its FDA-approval, (ii) its excellent safety profile at high concentrations, (iii) its high bioavailability, and (iv) previous reports that quinolones display antineoplastic activity.
Figure 2.
Effects on Metnase candidate inhibitors on TIR-specific DNA binding and pseudo Y cleavage. The numbers identify ChemDiv compounds with good docking scores. Compound 3731-0098 is the quinolone ciprofloxacin (Cipro). Ralt, HIV integrase inhibitor raltegravir; Elvit, HIV integrase inhibitor elvitegravir. A, mobility shift assay with purified recombinant Metnase and TIR DNA incubated with candidate inhibitors. B, radiolabeled duplex oligomer nuclease assay to examine the inhibition of Metnase’s Transposase nuclease domain by candidate inhibitors. Metnase cleavage of the pseudo Y structure (shown) produces a 20 nt major product and several slightly larger and smaller minor products. Candidate inhibitor concentrations were 2 and 5 μmol/L. DMSO, dimethyl sulfoxide; Ralt, raltegravir; Elvit, elvitegravir.
Metnase inhibition increases sensitivity to chemotherapy
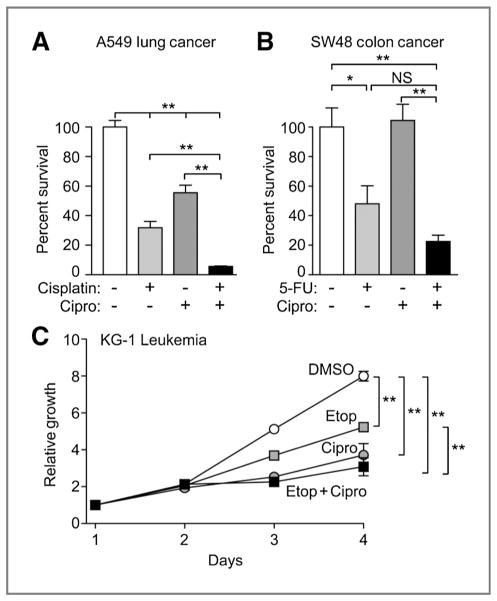
Common solid tumors such as those from lung or colon are difficult to treat once metastatic because of their low sensitivity to chemotherapeutic agents. The solid tumor A549 lung cancer and SW48 colon cancer cell lines were tested for potentiation of cell killing by clinically relevant chemotherapeutic agents with ciprofloxacin by using colony formation assays. Ciprofloxacin concentrations used were clinically achievable, based on early phase clinical trials (23). Ciprofloxacin has shown some activity alone against A549 cells in vitro (P < 0.01), and it significantly enhanced cisplatin-induced cell killing (P < 0.01, Fig. 3A). Ciprofloxacin alone had no effect on SW48 colony formation, and the approximately 2-fold reduction in colony formation with ciprofloxacin plus 5-fluorouracil (5-FU) compared with 5-FU alone did not reach statistical significance (P > 0.1; Fig. 3B).
Figure 3.
Ciprofloxacin (Cipro) reduces cancer cell survival in combination with chemotherapy. A and B, colony formation was conducted with A549 and SW48 cells treated with the clinically relevant chemotherapeutics cisplatin or 5-FU in the presence or absence of 2 μmol/L ciprofloxacin. C, ciprofloxacin at 2 μmol/L decreased proliferation of KG-1 cells and slightly enhanced the effect of etoposide (Etop). *, P < 0.05; **, P < 0.01 (t tests).
We next measured proliferation of the acute myeloid leukemia cell line KG-1 over 4 days in the presence of ciprofloxacin with and without etoposide (Fig. 3C). Ciprofloxacin alone resulted in a significant inhibition of proliferation of this leukemia cell line (P < 0.01), similar to our observations with the THP-1 acute monocytic leukemia cell line when Metnase was repressed by siRNA (12). Acute leukemia cells seem especially reliant on Metnase for proliferation (12). Ciprofloxacin also enhanced the antiproliferative effects of etoposide (P < 0.01), but under these conditions the effect of ciprofloxacin plus etoposide was not significantly different than ciprofloxacin alone (P > 0.1; Fig. 3C).
Therefore we have shown that ciprofloxacin can potentiate the effects of 3 different chemotherapeutic drugs on 3 different types of cognate cancer cells. We did test ciprofloxacin with appropriate chemotherapy on other cancer cell lines, but it was not as effective (data not shown). Therefore, ciprofloxacin has specificity for certain cancers or chemotherapies.
Metnase inhibition decreases NHEJ repair
The in vitro results above confirmed the biologic activity of ciprofloxacin as a Metnase nuclease inhibitor, supporting the virtual screening outcome. The in vivo assays also suggest that ciprofloxacin affects Metnase function to enhance the efficacy of chemotherapy. However, the in vivo assays above do not differentiate between ciprofloxacin acting only on Metnase or whether it is having other effects, such as generating DNA damage by itself.
Previously we had shown that nuclease-deficient Metnase protein cannot enhance DNA repair (3, 16). To examine whether ciprofloxacin specifically inhibited the Metnase nuclease domain’s ability to enhance NHEJ DSB repair, we conducted an NHEJ plasmid repair assay in cells that express varying levels of Metnase protein, in the presence and absence of ciprofloxacin. HEK293T cells were used as they do not express any endogenous Metnase (11). HEK293T parental cells were able to repair the linearized plasmid efficiently but this repair was not significantly affected by ciprofloxacin (Fig. 4B). Metnase transduced HEK293T were also able to efficiently repair the plasmid, but the addition of ciprofloxacin significantly decreased (P < 0.01) the repair of this plasmid (Fig. 4B). Thus, ciprofloxacin inhibits repair only when Metnase is present in the HEK293T cells. Next, we examined whether ciprofloxacin altered the nuclease resection of the free DNA ends of the transfected plasmid subjected to intracellular NHEJ in this assay. Ciprofloxacin decreased the resection of end deletion of the transfected linearized plasmid DNA (Fig. 4C). Then we tested NHEJ repair using the same assay in HEK293 cells (normally expressing Metnase) with Metnase repressed using shRNA (Fig. 4D). As we have seen previously, a reduction in Metnase protein expression results in a significant decrease in DNA repair compared with the parental control cells (Fig. 4D). The presence of ciprofloxacin significantly reduced this repair even further (Fig. 4D), probably as the HEK293-shMetnase cells still express some Metnase protein (Fig. 4A). These results show that ciprofloxacin is likely acting specifically on Metnase as in the complete absence of Metnase the endogenous DNA repair pathway of HEK293T cells is not inhibited by ciprofloxacin.
Figure 4.
Ciprofloxacin (Cipro) suppresses DNA repair in Metnase-expressing cells. A, over- and underexpression of Metnase in HEK293T (which do not normally express Metnase secondary to T-antigen transformation) or HEK293 cells (normally expressing Metnase), respectively, monitored by Western blot analysis with β-actin as loading control. Lanes marked as C are empty vector controls. B, NHEJ repair of linearized, transfected plasmid DNA in HEK293T cells, which do not express Metnase, was not significantly affected by ciprofloxacin (gray bars; P = 0.43, t test). Ciprofloxacin reduced NHEJ by 4- to 5-fold in cells overexpressing Metnase (in A–C and F; *, P < 0.05 and **, P < 0.01). C, in HEK293T cells overexpressing Metnase, a smaller fraction of NHEJ deletions were greater than 100 bp in length (Fisher exact test). D, ciprofloxacin reduces plasmid NHEJ in HEK293 cells (gray bars), and NHEJ is also reduced in untreated cells when Metnase expression is reduced with shRNA. NHEJ levels were not significantly (NS) different in ciprofloxacin-treated HEK293 cells expressing normal versus low levels of Metnase. Ciprofloxacin still reduced NHEJ in HEK293 cells underexpressing Metnase (black bars), probably reflecting inhibition of residual Metnase (A, lower blot). E and F, ciprofloxacin reduces resolution of γ-H2Ax foci after chemotherapy. A549 cells were seeded onto coverslips and allowed to adhere. Ciprofloxacin (2 μmol/L) was added for 18 hours before chemotherapy. Cisplatin was added for 6 hours, and then washed off and new culture medium was added containing ciprofloxacin where appropriate. Cells were fixed and processed for immunofluorescence with an antibody specific for γ-H2Ax and 4′,6-diamidino-2-phenylindole (DAPI) counterstained. Representative images after 48 hours recovery are shown in E, and quantification of all time points is shown in F. Cells with greater than 5 foci per cell were considered positive. Ciprofloxacin alone did not generate DNA damage as shown by unchanged γ-H2Ax foci. The combination of cisplatin and ciprofloxacin resulted in a slower resolution of γ-H2Ax foci than cisplatin alone. G, ciprofloxacin does not affect DNA replication restart after cisplatin. BrdUrd incorporation was scored in A549 cells, treated with cisplatin and ciprofloxacin as in F, at indicated times after release from cisplatin. UT, untreated.
Metnase inhibition blocks repair of chemotherapy-induced DNA damage
We therefore asked whether (i) ciprofloxacin generates DNA damage on its own, (ii) whether it inhibits repair of DNA damage induced by chemotherapeutic agents, and (iii) whether it prevents stalled replication forks from restarting. The presence of γ-H2Ax immunofluorescent foci after DNA damage is a measure of DNA DSBs, and the rate of their disappearance is an assay of DNA DSB repair (24, 25). With cisplatin, these likely represent stalled replication forks that have collapsed to yield free double-strand ends. We previously showed that siRNA knockdown of Metnase slows the rate of disappearance of γ-H2Ax foci after replication fork stalling, indicating that Metnase enhances repair of collapsed forks (10, 19). We scored γ-H2Ax foci in A549 lung cancer cells at various times after treatment with cisplatin, ciprofloxacin, or both (Fig. 4E and F). The percentage of cells with γ-H2Ax foci was the same in the ciprofloxacin-treated compared with untreated A549 cells. Also, ciprofloxacin did not increase the percentage of cells with γ-H2Ax foci immediately after cisplatin relative to cisplatin treatment alone. Thus, ciprofloxacin does not induce DSBs on its own, nor does it enhance the DNA damaging effects of cisplatin. However, γ-H2Ax foci disappeared at a faster rate in cells treated with cisplatin alone compared with the cells treated with both ciprofloxacin and cisplatin (Fig. 4D and E). The differences were statistically significant at 24, 48, and 72 hours after cisplatin exposure. These results have shown that cisplatin DNA DSB damage, likely from collapsed replication forks, is repaired more slowly in cells co-treated with ciprofloxacin, similar to what was seen in cells with repressed Metnase expression (10, 19). This is further evidence that ciprofloxacin inhibits Metnase function.
If free double-strand ends from collapsed replication forks are repaired more slowly when cisplatin is in the presence of ciprofloxacin, it was possible that ciprofloxacin also decreased replication fork progression after cisplatin treatment. To address this A549 cells were treated as above and harvested at the same time points as the γ-H2Ax assay, but were pulsed with BrdUrd 30 minutes before harvesting. Incorporated BrdUrd indicates a cell with replication fork progression after cisplatin exposure. We found that at all time points and for all treatments there was no difference in the fraction of BrdUrd-positive cells (Fig. 4G). Therefore, inhibition of Metnase by ciprofloxacin does not affect the overall fraction of cells with replication fork progression, but does decrease repair of DSBs caused by those forks that have collapsed, which would be in a subset of the BrdUrd-positive cells.
Metnase inhibition increases response of a lung cancer xenograft to chemotherapy
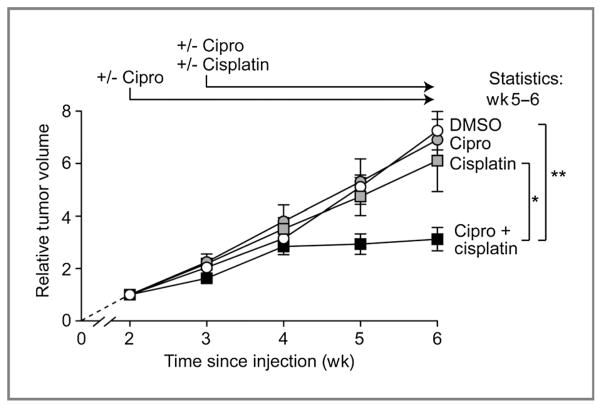
We then addressed whether ciprofloxacin inhibition of Metnase could improve therapeutic efficacy in an in vivo model of malignancy. We used a SCID murine xenograft tumor model with A549 lung cancer cells (26–29). To model human malignancy, tumors were allowed to form for 14 days after injection of the cells into the flanks of mice before chemotherapy was started. Xenograft tumor experiments were conducted 3 independent times with at least 5 mice per group using clinical achievable concentrations of ciprofloxacin and the results are shown in Fig. 5. The vehicle-treated tumors had a volume doubling time of approximately 8 days, which is in accord with previous studies (29, 30). In this system, ciprofloxacin alone had no effect on the rate of growth of the A549 xenografts. We used a clinically relevant cisplatin dose, but it had little effect on the growth of the A549 xenografts. Interestingly, the ciprofloxacin/cisplatin combination significantly reduced growth of the A549 tumors compared with untreated controls (P < 0.01 at weeks 5 and 6). Tumors in mice treated with both drugs showed little or no increase in volume after 1 week of combination therapy, until the experiments were stopped 6 weeks after initial injection because of the large expansions in tumor volumes in control and single drug-treated mice. Moreover, tumors in mice treated with both ciprofloxacin and cisplatin showed significantly reduced growth compared with mice treated with cisplatin alone (P < 0.05 at weeks 5 and 6). The A549 xenograft results parallel our findings with A549 cancer cells in tissue culture (Fig. 3A), and they confirmed that ciprofloxacin enhances sensitivity of A549 lung cancer cells to cisplatin in vivo.
Figure 5.
Ciprofloxacin increases human lung cancer xenograft sensitivity to cisplatin. A549 lung cancer cells, normally resistant to cisplatin, were injected into the flanks of mice, allowed to establish for 2 weeks, then treated daily with ciprofloxacin and 3 times weekly with cisplatin. Tumor volumes were measured 3 times per week and relative tumor volumes at each time point were calculated as tumor volume at that time divided by the tumor volume at the beginning of treatment (2 weeks after injection). Values are average relative tumor volumes (±SEM) for 15 to 26 tumors per condition (average of 23 tumors). Ciprofloxacin alone had no effect on xenograft growth, and cisplatin had a minimal effect on xenograft growth alone. However, the combination showed a statistically significant inhibition of xenograft growth after 4 weeks of treatment. *, P < 0.05;**, P < 0.01 (t tests).
Discussion
We have shown that reducing the expression of Metnase in several cancer cell lines increased their sensitivity to their clinically relevant chemotherapeutic agents (12, 13). Therefore, we hypothesized that targeting Metnase with small-molecule inhibitors would be a novel and potentially important means to enhance cancer chemotherapy. We used both ligand-based and target-based virtual screening tools to identify compounds in the ChemDiv library that could both dock into the Metnase Transposase nuclease active site and that had structural properties common to useful drugs, including lack of toxicity moieties, and that were predicted to have good pharmacokinetic properties. We screened 8 of the top scoring compounds for biochemical inhibition of the nuclease activity of Metnase, and found 3 compounds indeed had inhibitory capability: the HIV integrase inhibitors raltegravir and elvitegravir, and the quinolone antibiotic ciprofloxacin. Elvitegravir and ciprofloxacin are quite similar, sharing a quinolone moiety at one end predicted to interact with the chelated Mg2+ in the HIV integrase and Metnase Transposase domains.
We chose not to pursue elvitegravir because it is not FDA-approved, and thus clinical trials would be more difficult to conduct. Raltegravir and ciprofloxacin are both FDA-approved for clinical use, and they are both very safe, even in high concentrations (31, 32). They are also both highly bioavailable in oral dosing. Ciprofloxacin was chosen for our initial studies because previous reports indicated that quinolones have anti-proliferative activity in several cancer cell lines (15, 33–39). It had been hypothesized that the antiproliferative activity of quinolones were due to inhibition of Topo IIα (15, 39). Because Metnase is in a complex with Topo IIα, and Metnase strongly enhances Topo IIα activity (11), it is possible that the previously reported antineoplastic activity of quinolones attributed to Topo IIα inhibition may be at least in part indirect, from inhibition of Metnase, which in turn resulted in Topo IIα inhibition. We have ongoing studies designed to test whether ciprofloxacin, raltegravir, and/or elvitegravir inhibit Topo IIα-mediated chromosome decatenation, as this is another potential mechanism by which these drugs could enhance tumor killing. Ciprofloxacin could also have off-target activity due to its Mg2+ chelation property. Topo IIα is an example of an essential cellular enzyme that has a magnesium atom at the catalytic site (40). This is consistent with the mechanism for which quinolones were designed, against bacterial topoisomerases (41). However, the studies here showing that ciprofloxacin only decreased DNA DSB repair when Metnase was present imply that Metnase is the main target of ciprofloxacin for its enhancement of cisplatin cytotoxicity seen here. This may be due to the relatively lower concentrations used here compared with other studies. At higher concentrations, it is possible that ciprofloxacin would have additional activities, as has been reported in refs. 33–38. It should be noted that T-antigen–transformed cell lines, such as the HEK293T cells used here, are the only types of cells that do not require Metnase; thus, ciprofloxacin has less effect on repair in those cells. Other cells with Metnase expression seem to be addicted to its presence, requiring it for repair of DSBs and collapsed replication forks (10–13).
We showed that ciprofloxacin alone could slow the growth of the KG-1 leukemia cell line, similar to our previous observation of reduced leukemia cell growth when Metnase protein expression was repressed (12). We also found that ciprofloxacin could increase chemosensitivity to several therapeutic agents in clinically relevant solid tumor cancer cell lines. Clinically relevant concentrations of ciprofloxacin alone did not decrease solid tumor cell line colony formation or induce DNA damage, nor did it increase the amount of DNA damage induced by the chemotherapeutic cisplatin, as revealed by γ-H2Ax foci. However, ciprofloxacin significantly decreased the rate of disappearance of γ-H2Ax foci induced by cisplatin. This finding indicates that ciprofloxacin reduces the rate of repair of damaged DNA, with the likely mechanism being inhibition of Metnase nuclease activity, as this inhibition of repair is present only when Metnase is present. The slow repair rate seen with ciprofloxacin is similar to that seen previously when Metnase was repressed in cells treated with DNA damaging and replication stress agents (10, 12). Therefore, we suggest that ciprofloxacin enhances chemotherapy by inhibiting the ability of Metnase to repair collapsed replication forks. Cancer therapies typically involve direct or indirect induction of DNA damage, and ciprofloxacin may be a useful adjunct to treatments of many types of cancers. Some types of cancers overexpress Metnase (12), and in these cases ciprofloxacin may provide even greater therapeutic benefits. Other quinolones may also be beneficial, although careful preclinical testing is important as several of the candidate quinolones identified in our virtual screen seemed to stimulate rather than inhibit Metnase nuclease activity (Fig. 2B).
Previous studies have shown that ciprofloxacin alone can inhibit proliferation and induce apoptosis in bladder cancer, prostate cancer, colon cancer, and leukemia cell lines. For example, ciprofloxacin decreases proliferation of a number of cancer cell lines, including A549, albeit at concentrations higher than used here (38). Proliferation of human prostate cancer cells and Jurkat acute leukemia cells was repressed by ciprofloxacin (42, 43). Ciprofloxacin also induces apoptosis in colon cancer cells, perhaps by inhibiting repair of endogenous DNA damage (36). All of these studies showed that ciprofloxacin has independent antiproliferative activity; however, higher ciprofloxacin concentrations were used in those studies than in the current study. We propose that ciprofloxacin not be discarded as an antineoplastic agent based on the concentrations used in the prior studies, but rather that it be delivered as an adjunct to DNA damaging chemotherapy or radiotherapy.
Quinolone derivatives are being generated and explored as more potent anticancer drugs. The quinolone derivative voreloxin is being tested in clinical trials in several types of malignancies. Voreloxin alone increases DNA damage by intercalating into DNA. It also acts as a Topo IIα poison, and can be synergistic with known chemotherapy (44). The drug has some activity in acute myeloid leukemia (39). A recent study found that voreloxin had little independent activity in relapsed non–small cell lung cancer (45). However, given the results of the present study, voreloxin might have more efficacy as an inhibitor of cisplatin adducts repair and/or recovery of stalled replication forks. Another quinolone, CHM-1, is being tested for clinical activity in treating human malignancy (46, 47). Interestingly, CHM-1 induces DNA damage and decreases expression of DNA repair genes (48).
A meta-analysis of randomized trials of quinolones for infections during cancer therapy found that mortality in cancer patients was reduced for those on quinolones (49). The authors recognized that it was possible that this reduction in mortality was secondary to reducing infections. However, a follow-up analysis extracted infection-related mortality data from the primary trials used in the previous analysis, and assessed the effect of quinolones on cancer-related mortality. Among trials comparing quinolones with placebo or no treatment, a significant reduction in noninfection-related mortality was observed in the quinolone arm (50). This finding is compatible with an anticancer effect of quinolone antibiotics when combined with other chemotherapy, consistent with the data presented here. It is possible that this effect is due to potentiation of DNA-damaging chemotherapy by inhibition of Metnase. If so, adding adjunct ciprofloxacin represents a novel method of enhancing chemotherapy that is amenable to immediate clinical trial assessment.
Supplementary Material
Acknowledgments
The authors thank Catherine Buchanan-McGrath for excellent technical support.
Grant Support
This research was supported by NIH grants R01 CA092111 (to S.H. Lee), R01 CA151367 (S.H. Lee), R01 GM084020 (J.A. Nickoloff), R01 CA102283 (R. Hromas), R01 HL075783 (R. Hromas), and R01 CA139429 (R. Hromas), and the Leukemia and Lymphoma Society (R. Hromas). L. Sklar was supported by NIH 5U54MH084690-02, Center Driven Initiative 1.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
T. Oprea has a commercial research grant from and ownership interest (including patents) in Sunset Molecular Discovery LLC, has other commercial research support from Givaudan Flavors Corporation, and is a consultant/advisory board member of ChemDiv Inc. No potential Conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: A. Leitao, T. Oprea, J. Bauman, S.H. Lee, R. Hromas
Development of methodology: E.A. Williamson, A. Leitao, H.J. Hathaway, W. Wang, S.H. Lee, R. Hromas
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): E.A. Williamson, H.J. Hathaway, S.H. Lee
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): E.A. Williamson, L. Damiani, A. Leitao, H.J. Hathaway, T. Oprea, W. Wang, J.A. Nickoloff, S.H. Lee, R. Hromas
Writing, review, and/or revision of the manuscript: E.A. Williamson, A. Leitao, L. Sklar, M. Shaheen, J. Bauman, W. Wang, J.A. Nickoloff, S.H. Lee, R. Hromas
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): E.A. Williamson, L. Damiani, C. Hu, S.H. Lee, R. Hromas
Study supervision: T. Oprea, R. Hromas
Identification of small molecules that are biologically active, using computational means: T. Oprea
References
- 1.Cordaux R, Udit S, Batzer MA, Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc Natl Acad Sci U S A. 2006;103:8101–6. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jordan IK. Evolutionary tinkering with transposable elements. Proc Natl Acad Sci U S A. 2006;103:7941–2. doi: 10.1073/pnas.0602656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee SH, Oshige M, Durant ST, Rasila KK, Williamson EA, Ramsey H, et al. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc Natl Acad Sci U S A. 2005;102:18075–80. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robertson HM, Zumpano KL. Molecular evolution of an ancient mariner transposon, Hsmar1, in the human genome. Gene. 1997;205:203–17. doi: 10.1016/s0378-1119(97)00472-1. [DOI] [PubMed] [Google Scholar]
- 5.Liu D, Bischerour J, Siddique A, Buisine N, Bigot Y, Chalmers R. The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol Cell Biol. 2007;27:1125–32. doi: 10.1128/MCB.01899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck BD, Lee SS, Hromas R, Lee SH. Regulation of Metnase’s TIR binding activity by its binding partner, Pso4. Arch Biochem Biophys. 2010;498:89–94. doi: 10.1016/j.abb.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roman Y, Oshige M, Lee Y-J, Goodwin K, Georgiadis MM, Hromas RA, et al. Biochemical characterization of a SET and transposase fusion protein, Metnase (SETMAR) for its DNA binding and DNA cleavage activity. Biochemistry. 2007;46:11369–76. doi: 10.1021/bi7005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, et al. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci U S A. 2011;108:540–5. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wray J, Williamson EA, Farrington J, Chester S, Kwan L, Weinstock D, et al. The transposase domain protein Metnase/SETMAR suppresses chromosomal translocations. Cancer Genet Cytogenet. 2010;200:184–90. doi: 10.1016/j.cancergencyto.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Haro LP, Wray J, Williamson EA, Durant ST, Corwin L, Gentry AC, et al. Metnase promotes restart and repair of stalled and collapsed replication forks. Nucleic Acids Res. 2010;38:5681–91. doi: 10.1093/nar/gkq339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williamson EA, Rasila KK, Corwin LK, Wray J, Beck BD, Severns V, et al. The SET and transposase domain protein Metnase enhances chromosome decatenation: regulation by automethylation. Nucleic Acids Res. 2008;36:5822–31. doi: 10.1093/nar/gkn560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wray J, Williamson EA, Fnu S, Lee S-H, Libby E, Willman CL, et al. Metnase mediates chromosome decatenation in acute leukemia cells. Blood. 2009;114:1852–8. doi: 10.1182/blood-2008-08-175760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wray J, Williamson EA, Royce M, Shaheen M, Beck BD, Lee SH, et al. Metnase mediates resistance to topoisomerase II inhibitors in breast cancer cells. PLoS ONE. 2009;4:e5323. doi: 10.1371/journal.pone.0005323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 15.Hawtin RE, Stockett DE, Byl JA, McDowell RS, Nguyen T, Arkin MR, et al. Voreloxin is an anticancer quinolone derivative that intercalates DNA and poisons topoisomerase II. PLoS ONE. 2010;5:e10186. doi: 10.1371/journal.pone.0010186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck BD, Lee SS, Williamson E, Hromas RA, Lee SH. Biochemical characterization of Metnase’s endonuclease activity and its role in NHEJ repair. Biochemistry. 2011;50:4360–70. doi: 10.1021/bi200333k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leitao A, Andricopulo AD, Montanari CA. In silico screening of HIV-1 non-nucleoside reverse transcriptase and protease inhibitors. Eur J Med Chem. 2008;43:1412–22. doi: 10.1016/j.ejmech.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 18.Goodwin KD, He H, Imasaki T, Lee SH, Georgiadis MM. Crystal structure of the human Hsmar1-derived transposase domain in the DNA repair enzyme Metnase. Biochemistry. 2010;49:5705–13. doi: 10.1021/bi100171x. [DOI] [PubMed] [Google Scholar]
- 19.Hromas R, Wray J, Lee SH, Martinez L, Farrington J, Corwin LK, et al. The human set and transposase domain protein Metnase interacts with DNA ligase IV and enhances the efficiency and accuracy of non-homologous end-joining. DNA Repair. 2008;7:1927–37. doi: 10.1016/j.dnarep.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiu TK, Davies DR. Structure and function of HIV-1 integrase. Curr Top Med Chem. 2004;4:965–77. doi: 10.2174/1568026043388547. [DOI] [PubMed] [Google Scholar]
- 21.Savarino A. In-Silico docking of HIV-1 integrase inhibitors reveals a novel drug type acting on an enzyme/DNA reaction intermediate. Retrovirology. 2007;4:21. doi: 10.1186/1742-4690-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–7. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drusano GL, Standiford HC, Plaisance K, Forrest A, Leslie J, Caldwell J. Absolute oral bioavailability of ciprofloxacin. Antimicrob Agents Chemother. 1986;30:444–6. doi: 10.1128/aac.30.3.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Downey M, Durocher D. γH2AX as a checkpoint maintenance signal. Cell Cycle. 2006;5:1376–81. doi: 10.4161/cc.5.13.2899. [DOI] [PubMed] [Google Scholar]
- 25.Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, et al. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 26.Hardman WE, Moyer MP, Cameron IL. Efficacy of treatment of colon, lung and breast human carcinoma xenografts with: doxorubicin, cisplatin, irinotecan or topotecan. Anticancer Res. 1999;19:2269–74. [PubMed] [Google Scholar]
- 27.Shoemaker AR, Oleksijew A, Bauch J, Belli BA, Borre T, Bruncko M, et al. A small-molecule inhibitor of Bcl-XL potentiates the activity of cytotoxic drugs in vitro and in vivo. Cancer Res. 2006;66:8731–9. doi: 10.1158/0008-5472.CAN-06-0367. [DOI] [PubMed] [Google Scholar]
- 28.Fan T, Li R, Todd NW, Qiu Q, Fang HB, Wang H, et al. Up-regulation of 14-3-3zeta in lung cancer and its implication as prognostic and therapeutic target. Cancer Res. 2007;67:7901–6. doi: 10.1158/0008-5472.CAN-07-0090. [DOI] [PubMed] [Google Scholar]
- 29.Shen H, Hu D, Du J, Wang X, Liu Y, Wang Y, et al. Paclitaxel–octreotide conjugates in tumor growth inhibition of A549 human non–small cell lung cancer xenografted into nude mice. Eur J Pharmacol. 2008;601:23–9. doi: 10.1016/j.ejphar.2008.10.035. [DOI] [PubMed] [Google Scholar]
- 30.Cai KX, Tse LY, Leung C, Tam PK, Xu R, Sham MH. Suppression of lung tumor growth and metastasis in mice by adeno-associated virus-mediated expression of vasostatin. Clin Cancer Res. 2008;14:939–49. doi: 10.1158/1078-0432.CCR-07-1930. [DOI] [PubMed] [Google Scholar]
- 31.Sorgel F, Naber KG, Kinzig M, Mahr G, Muth P. Comparative pharmacokinetics of ciprofloxacin and temafloxacin in humans: a review. Am J Med. 1991;91:51S–66S. doi: 10.1016/0002-9343(91)90312-l. [DOI] [PubMed] [Google Scholar]
- 32.Cocohoba J, Dong BJ. Raltegravir: the first HIV integrase inhibitor. Clin Ther. 2008;30:1747–65. doi: 10.1016/j.clinthera.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 33.Somekh E, Douer D, Shaked N, Rubinstein E. In vitro effects of ciprofloxacin and pefloxacin on growth of normal human hematopoietic progenitor cells and on leukemic cell lines. J Pharmacol Exp Ther. 1989;248:415–8. [PubMed] [Google Scholar]
- 34.Seay TM, Peretsman SJ, Dixon PS. Inhibition of human transitional cell carcinoma in vitro proliferation by fluoroquinolone antibiotics. J Urol. 1996;155:757–62. [PubMed] [Google Scholar]
- 35.Miclau T, Edin ML, Lester GE, Lindsey RW, Dahners LE. Effect of ciprofloxacin on the proliferation of osteoblast-like MG-63 human osteosarcoma cells in vitro. J Orthop Res. 1998;16:509–12. doi: 10.1002/jor.1100160417. [DOI] [PubMed] [Google Scholar]
- 36.Herold C, Ocker M, Ganslmayer M, Gerauer H, Hahn EG, Schuppan D. Ciprofloxacin induces apoptosis and inhibits proliferation of human colorectal carcinoma cells. Br J Cancer. 2002;86:443–8. doi: 10.1038/sj.bjc.6600079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scatena CD, Kumer JL, Arbitrario JP, Howlett AR, Hawtin RE, Fox JA, et al. Voreloxin, a first-in-class anticancer quinolone derivative, acts synergistically with cytarabine in vitro and induces bone marrow aplasia in vivo. Cancer Chemother Pharmacol. 2010;66:881–8. doi: 10.1007/s00280-009-1234-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kloskowski T, Gurtowska N, Nowak M, Joachimiak R, Bajek A, Olkowska J, et al. The influence of ciprofloxacin on viability of A549, HepG2, A375. S2, B16 and C6 cell lines in vitro. Acta Pol Pharm. 2011;68:859–65. [PubMed] [Google Scholar]
- 39.Walsby EJ, Coles SJ, Knapper S, Burnett AK. The topoisomerase II inhibitor voreloxin causes cell cycle arrest and apoptosis in myeloid leukemia cells and acts in synergy with cytarabine. Haematologica. 2011;96:393–9. doi: 10.3324/haematol.2010.032680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei H, Ruthenburg AJ, Bechis SK, Verdine GL. Nucleotide-dependent domain movement in the ATPase domain of a human type IIA DNA topoisomerase. J Biol Chem. 2005;280:37041–7. doi: 10.1074/jbc.M506520200. [DOI] [PubMed] [Google Scholar]
- 41.Drlica K, Malik M, Kerns RJ, Zhao X. Quinolone-mediated bacterial death. Antimicrob Agents Chemother. 2008;52:385–92. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aranha O, Grignon R, Fernandes N, McDonnell TJ, Wood DP, Jr, Sarkar FH. Suppression of human prostate cancer cell growth by ciprofloxacin is associated with cell cycle arrest and apoptosis. Int J Oncol. 2003;22:787–94. [PubMed] [Google Scholar]
- 43.Koziel R, Szczepanowska J, Magalska A, Piwocka K, Duszynski J, Zablocki K. Ciprofloxacin inhibits proliferation and promotes generation of aneuploidy in Jurkat cells. J Physiol Pharmacol. 2010;61:233–9. [PubMed] [Google Scholar]
- 44.Mills DA, Fekrazad HM, Verschraegen CF. SNS-595, a naphthyridine cell cycle inhibitor and stimulator of apoptosis for the treatment of cancers. Curr Opin Investig Drugs. 2008;9:647–57. [PubMed] [Google Scholar]
- 45.Krug LM, Crawford J, Ettinger DS, Shapiro GI, Spigel D, Reiman T, et al. Phase II multicenter trial of voreloxin as second-line therapy in chemotherapy-sensitive or refractory small cell lung cancer. J Thorac Oncol. 2011;6:384–6. doi: 10.1097/JTO.0b013e318200e509. [DOI] [PubMed] [Google Scholar]
- 46.Wang SW, Pan SL, Huang YC, Guh JH, Chiang PC, Huang DY, et al. CHM-1, a novel synthetic quinolone with potent and selective antimitotic antitumor activity against human hepatocellular carcinoma in vitro and in vivo. Mol Cancer Ther. 2008;7:350–60. doi: 10.1158/1535-7163.MCT-07-2000. [DOI] [PubMed] [Google Scholar]
- 47.Hsu SC, Yang JS, Kuo CL, Lo C, Lin JP, Hsia TC, et al. Novel quinolone CHM-1 induces apoptosis and inhibits metastasis in a human osterogenic sarcoma cell line. J Orthop Res. 2009;27:1637–44. doi: 10.1002/jor.20937. [DOI] [PubMed] [Google Scholar]
- 48.Chen HY, Lu HF, Yang JS, Kuo SC, Lo C, Yang MD, et al. The novel quinolone CHM-1 induces DNA damage and inhibits DNA repair gene expressions in a human osterogenic sarcoma cell line. Anticancer Res. 2010;30:4187–92. [PubMed] [Google Scholar]
- 49.Engels EA, Lau J, Barza M. Efficacy of quinolone prophylaxis in neutropenic cancer patients: a meta-analysis. J Clin Oncol. 1998;16:1179–87. doi: 10.1200/JCO.1998.16.3.1179. [DOI] [PubMed] [Google Scholar]
- 50.Paul M, Gafter-Gvili A, Fraser A, Leibovici L. The anti-cancer effects of quinolone antibiotics? Eur J Clin Microbiol Infect Dis. 2007;26:825–31. doi: 10.1007/s10096-007-0375-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.