Abstract
Background and Aims
IL-28B gene polymorphisms predict better therapeutic response and spontaneous clearance of HCV. Moreover, higher expression of IFN-lambda has been reported in patients with the rs12979860 CC favourable genotype. The study aim was to establish possible relationships between IL-28B rs12979860 genotypes and expression of IFN-alpha receptor-1 (IFNAR-1) in naïve HCV patients, and to explore the possible role of IFN-lambda.
Methods
IFNAR-1 mRNA levels were measured in PBMC from naïve patients with chronic hepatitis C with different IL-28 genotypes. The ability of IFN-lambda to up-regulate the expression of IFNAR-1 was established in PBMC from healthy donors carrying different IL-28B genotypes.
Results
Lower IFNAR-1 mRNA levels were observed in PBMC from HCV-infected naïve patients as compared to healthy donors. In healthy donors, IFNAR-1 mRNA levels were independent from IL-28B genotype, while in HCV patients, an increasing gradient was observed in TT vs CT vs CC carriers. In the latter group, a direct correlation between IFNAR-1 and endogenous IL-28B expression was observed. Moreover, IFN-lambda up-regulated IFNAR-1 expression in normal PBMC in a time-and dose-dependent manner, with a more effective response in CC vs TT carriers.
Conclusion
Endogenous levels of IFN-lambda may be responsible for partial restoration of IFNAR-1 expression in HCV patients with favourable IL-28 genotype. This, in turn, may confer to CC carriers a response advantage to either endogenous or exogenous IFN-alpha, representing the biological basis for the observed association between CC genotype and favourable outcome of either natural infection (clearance vs chronicization) or IFN therapy.
Introduction
Hepatitis C virus (HCV) is the leading cause of chronic liver disease, cirrhosis, and hepatocellular carcinoma in developed countries [1]. The ability of HCV to inhibit the activation of the endogenous type I interferon (IFN) system could underlie its success in establishing a chronic infection [2]. Type I IFNs are not only crucial factors in the innate immune system but are also the most important components of current therapy against chronic hepatitis C (CHC). In fact, the standard of care (SOC) for chronic HCV infection consists of pegylated IFN (peg-IFN)-2a or -2b and ribavirin (RBV). This treatment produces sustained virologic response (SVR) in only 40–50% of patients with HCV genotype 1 and approximately 60% in those infected with genotype 4, whereas over 80% of patients with genotype 2 or 3 achieve SVR [3], [4]. Moreover, several studies showed a correlation between pre-treatment expression levels of IFN stimulated genes (ISGs) and response to treatment, using either liver tissues or serum or peripheral blood mononuclear cells (PBMC) [5], [6]. Notably, patients with pre-elevated expression of ISGs (including MxA, PKR and ISG15) in the liver and/or PBMC showed a poor response to SOC as compared to patients with low basal levels [7], [8], [9].
A group of recently discovered cytokines (IFN-lambda1/interleukin-29 [IL-29], IFN-lambda2/IL-28A, and IFN-lambda3/IL-28B), assigned to a new type of IFN (type III IFN) gained increased attention in the HCV field [10].
Genome-wide association studies (GWAS) identified several single-nucleotide polymorphisms (SNPs) in IL-28B gene region, that were strongly related to therapy-induced HCV clearance rate in CHC patients [11], [12], [13]. Numerous investigators confirmed the importance of IL-28B SNPs in the treatment response [14], [15], [16]. Among the identified SNPs, rs12979860, located 3 kb upstream of the IL-28B gene, appeared as the most relevant, being the rs12979860-favorable CC genotype associated with a more than two-fold increased rate of SVR with respect to hapless (CT or TT) genotypes [17]. However, despite the existing clear evidence of association of IL-28B polymorphisms on spontaneous or therapy-induced resolution of the infection, the mechanisms triggered by these polymorphisms and their real biological consequences remain unexplained, and attract intensive investigation. A role for increased endogenous levels of IFN-lambda as the basis for these mechanisms is suggested by a number of studies. For instance, the expression of IL-28B was reported to be lower in whole blood or PBMC from individuals carrying the hapless genotypes as compared to those carrying the CC alleles [12], [13]. In another study by Langhans and others (2011), HCV chronically infected patients showed lower levels of circulating IFN-lambda as compared to spontaneously resolved infections and increased IFN-lambda levels were observed in carriers of the favorable CC rs12979860 [18]. Moreover, the association between increased IL-29 levels and CC allele has been reported in a recent study focused on therapy response [19].
TypeI and III IFNs exhibit a distinct receptor complex but common functions, both in term of broad range of antiviral mechanisms, induction of the same pattern of ISGs, and stimulation of immune effector functions. Despite the functional similarity with type I IFNs, clear evidence exists that type III IFNs have unique mechanisms of action, concerning, in particular, the kinetics of signal transduction after receptor engagement. Conflicting data exist on the relationship between genetic variation of IL-28 and the basal ISGs levels. Recent studies have suggested that baseline hepatic expression of ISGs was associated with genetic variation of IL-28B [20], [21], [22], while two studies reported that ISGs expression was associated with SVR, independently from IL-28B genotype [23], [24].
In a previous study we have shown that reduced expression of the IFN-alpha receptor -1 (IFNAR-1) may represent the biological basis for reduced response to SOC in poorly performing patients [25], such as HIV-coinfected individuals, hampering their ability to mount an appropriate response to exogenously administered IFN-alpha. However, so far no data are available on the correlation between IL-28 genotype and IFNAR-1 expression.
The aim of the present study was to establish whether the IL-28B rs12979860 genotype could influence the endogenous expression IFNAR-1, and to explore the possible role of IFN-lambda.
Methods
Ethics statement
The project was approved by Comitato Etico “Istituto Nazionale per le Malattie Infettive L. Spallanzani, I.R.C.C.S.” and patients agreed to participate to the study by signing informed consent.
Patients
PBMC and plasma samples from 40 treatment-naive patients, chronically infected with HCV (genotype 1 or 4), collected at the National Institute for Infectious Diseases and stored in the Institutional Biorepository, for whom IL-28B genotype at locus rs12979860 was known, were retrospectively selected, in order to include a sufficient number of patients harboring the CC genotype (that is relatively rare in the Italian population) to perform a comparative analysis. The distribution of IL-28B genotypes was: 9 CC (22.5%), 18 CT (45%), 13 TT (32.5%). Clinical characteristics of patients are shown in Table 1. All patients were further characterized for IFN system activation in their PBMC. As control group, 6 healthy donors, matching the anagraphic characteristics of the HCV-positive group, were included (3 donors with IL-28B rs12979860 CC genotype, and 3 donors with TT genotype)
Table 1. Characteristics of 40 treatment naive HCV-infected patients included in the study.
| Age [median(range)] years | 53 (30–81) |
| Sex (Male/Female) | 30/10 |
| AST [median(range)] IU/L | 45 (17–162) |
| ALT [median(range)] IU/L | 62 (13–310) |
| γ-GT [median(range)] IU/L | 40 (9–599) |
| HCV load [median(range)] Log10 IU/ml | 6.12 (4.35–6.84) |
| HCV genotypes (gt1/gt4) | 27/13 |
| rs 12979860 IL28B genotypes (CC/CT/TT) | 9/18/13 |
IL-28B polymorphisms genotyping
IL-28B rs12979860 CC/CT/TT genotype was established on genomic DNA, using a custom made TaqMan assay with the following amplification primers: 5′ GCC TGT CGT GTA CTG AAC CA 3′ and 5′ GCG CGG AGT GCA ATT CAA C 3′, and TaqMan probes: VIC- TGG TTC GCG CCT TC-MGB and FAM-CTG GTT CAC GCC TTC–MGB. Polymerase chain reactions (PCR) were performed with an SDS 7900 ht qPCR thermocycler (Applied Biosystems, Forster City, CA) with the following amplification protocols: denaturation at 95°C for 10 minutes, followed by 40 cycles of denaturation at 92°C for 15 sec, and finished with annealing and extension at 60°C for a 1 minute. Genotype was attributed by SDS 1.3 software for allelic discrimination.
mRNA levels of IFNAR-1 in PBMC of naive HCV-infected patients carrying different IL-28B rs12979860 genotypes before and after exposure to IFN-alpha
PBMC obtained by Ficoll/hyPaque (Pharmacia, Sweden) separation, frozen under liquid nitrogen, were thawed, suspended at 2×106/mL in RPMI medium supplemented with 10% foetal bovine serum and then cultured for 3 h in the absence or presence of 103 IU/ml human recombinant IFN-alpha2b (Intron; Schering Corp., Kenilworth, NJ, USA; specific activity: 400 MIU/mg,1 IU corresponding to 2.5 pg). Total cellular RNA was extracted from PBMC using Trizol (Gibco BRL, Grand Island, NY, USA) and reverse-transcribed by TaqMan Reverse Transcription Reagent kit (Applied Biosystems, Foster City, CA, USA) before and after treatment with IFN-alpha. The quantification of IFNAR-1 mRNA was performed by real-time RT-PCR; the results were normalized using beta-actin as housekeeping gene, and the results were expressed as ratio of IFNAR-1/beta-actin mRNA copy number, as previously described [26]. Basal levels of IFNAR-1 mRNA were measured in freshly thawed PBMC, without further incubation.
IFN-lambda expression in HCV-infected patients
Basal mRNA levels for IFN-lambda were measured in freshly thawed PBMC by quantitative Real-time RT-PCR, according to a previously described method [27]. Standard curves were prepared with serial dilution of recombinant plasmid containing the target region, and the results were expressed as ratio to beta-actin.
Plasma levels of IFN-lambda protein were measured by enzyme-linked immunosorbent assay (ELISA) namely DuoSet human IFN-lambda 1/3, measuring all isotypes of Hu-IFN-lambda (IL-29, IL-28A, IL-28B), purchased from R&D System, Inc, Minneapolis, MN, USA. Result were expressed as pg/ml.
mRNA levels of IFNAR-1 in PBMC of healthy donors before and after exposure to IFN-lambda
PBMC from healthy donors carrying different IL-28B genotypes were exposed either to 10 ng/ml or 100 ng/ml human recombinant IL-28/IFN-lambda2 (PBL, Interferon source, Piscatway, NJ, USA; ED50: 10–50 ng/mL) for different time points. Total cellular RNA was extracted by Trizol and IFNAR-1 mRNA levels were measured by real-time PCR as described above.
Statistical analysis
Statistical analyses were performed by Prism 4 software (GraphPad, San Diego, CA). Differences were evaluated by the non parametric Mann-Whitney U test or by Student's t test, as appropriate. Correlations were analyzed by Pearson r test. Differences with p<0.05 were considered statistically significant.
Results
Levels of IFNAR-1 in PBMC from naive HCV-infected patients carrying different IL-28B rs12979860 genotypes, and in vitro response to IFN-alpha
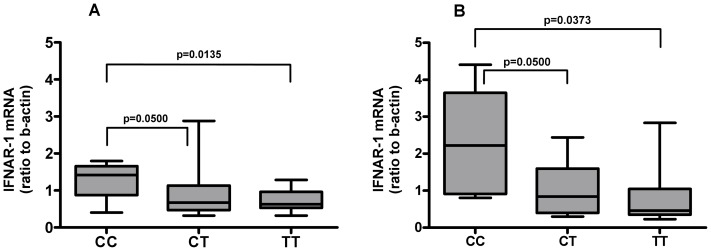
To assess the relationships between the expression of IFNAR-1 mRNA and the IL-28B rs12979860 genotypes, mRNA levels of IFNAR-1 were tested at baseline and after 3 h of exposure to either control medium or IFN-alpha (103 IU/ml) in PBMC obtained from naive HCV-infected patients carrying different IL-28B rs12979860 genotypes. Patients carrying CC genotype showed IFNAR-1 mRNA median basal levels significantly higher than patients with CT/TT genotype: 1.420 (IQR: 0.875–1.655) vs 0.629 (IQR: 0.504–1.005); p = 0.0142; in addition, significantly higher levels in CC vs CT/TT genotypes was observed after exposure to IFN-alpha [median: 2.220 (IQR: 0.908–3.647) vs 0.6280 (IQR: 0.395–1.522); p = 0.0149]. More in detail, the most prominent difference was observed between CC and TT groups, both at basal level (Fig. 1, Panel A) [median: 1.420 (IQR: 0.875–1.655) vs 0.629 (IQR: 0.532–0.925); p = 0.0135] and after exposure to IFN-alpha (Fig. 1, Panel B) [median: 2.220 (IQR: 0.908–3.647) vs 0.461 (IQR: 0.353–1.048); p = 0.0373], while between CC and CT groups a borderline significant difference was observed both at basal level (Fig. 1, Panel A) [median: 1.420 (IQR: 0.875–1.655) vs 0.676 (IQR: 0.468–1.137); p = 0.0500] and after exposure to IFN-alpha (Fig. 1, Panel B) [median values: 2.220 (IQR: 0.908–3.647) vs 0.840 (IQR: 0.397–1.599); p = 0.0500]. IFN-alpha treatment did not significantly affect the levels of IFNAR-1 mRNA in all genotypes (CC, untreated vs IFN-treated, median: 1.420 (IQR: 0.875–1.655) vs 2.034 (IQR: 0.808–2.888); p = 0.3510; CT, untreated vs IFN-treated CT, median: 0.6760 (IQR: 0.468–1.137) vs 1.046 (IQR: 0.411–1.639); p = 0.5510; TT, untreated vs IFN-treated, median: 0.6290 (IQR: 0.532–0.965) vs 0.461 (IQR: 0.353–1.048); p = 0.5240).
Figure 1. IFNAR-1 mRNA levels before and after treatment with IFN-alpha in PBMC from naive HCV-infected patients.
Total cellular RNA was extracted and reverse-transcribed from PBMC of naive HCV-infected patients carrying different IL-28B rs12979869 genotypes CC, TT and CT before (Panel A) and after 3 h of exposure to 103 IU/ml IFN-alpha (Panel B), then mRNA levels for IFNAR-1 were measured. Results are expressed as ratio to beta-actin (median, IQR).
Basal levels of IFN-lambda was also investigated, and no differences were appreciated between the different IL-28B genotypes, at both PBMC mRNA and plasma protein level (data not shown). However, a significant correlation between mRNA levels of IFN-lambda and IFNAR-1 was observed in CC carriers (r = 0.6881, p = 0.0433).
Role of IFN-lambda on IFNAR-1 mRNA expression in PBMC from healthy donors carrying different IL-28B rs12979860 genotypes
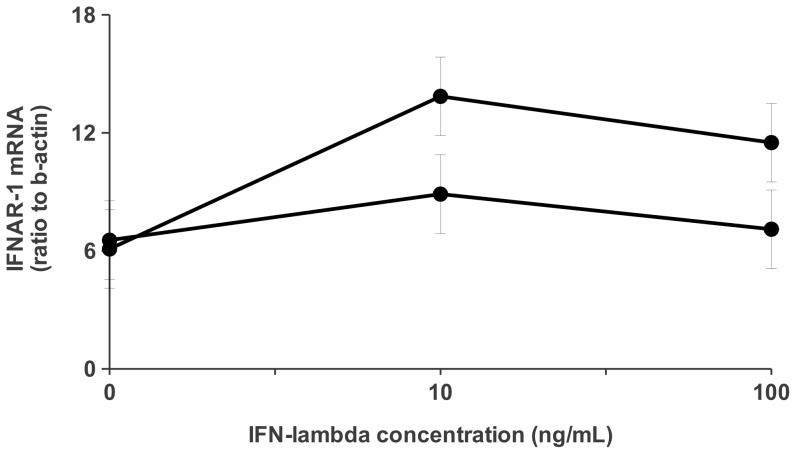
To evaluate the possible role of IFN-lambda on IFNAR-1 mRNA expression, PBMC from 6 healthy donors carrying different IL-28B genotypes (3 rs12979860 CC, and 3 rs12979860 TT) were exposed either to 10 or 100 ng/ml of human recombinant IL-28/IFN-lambda2, and levels of IFNAR-1 mRNA were measured at different time points. The basal levels of IFNAR-1 mRNA in healthy donors were significantly higher than those observed in HCV patients [median: 6.516 (IQR: 4.888–7.556) vs 0.732 (IQR: 0.393–1.829); p = 0.0004], and seemed to be independent from IL-28B genotype. The dose dependent response after 3 h exposure to IFN-lambda is shown in Fig. 2, were up-regulation of IFNAR-1 mRNA was observed in both genotypes, being 10 ng/ml the most effective dose. More interestingly, the IFN-lambda-driven stimulation was more pronounced in subjects carrying the CC genotype at both doses.
Figure 2. Dose dependent induction of IFNAR-1 mRNA levels following treatment with IFN-lambda in PBMC from healthy donors.
PBMC from 6 healthy donors with different IL-28B rs12979860 genotypes (3 CC •; 3 TT ○) were exposed for 3 h to either control medium or IFN-lambda (10 ng/ml, 100 ng/ml), then mRNA levels for IFNAR-1 were measured. Results are expressed as ratio to beta-actin (mean ± SE).
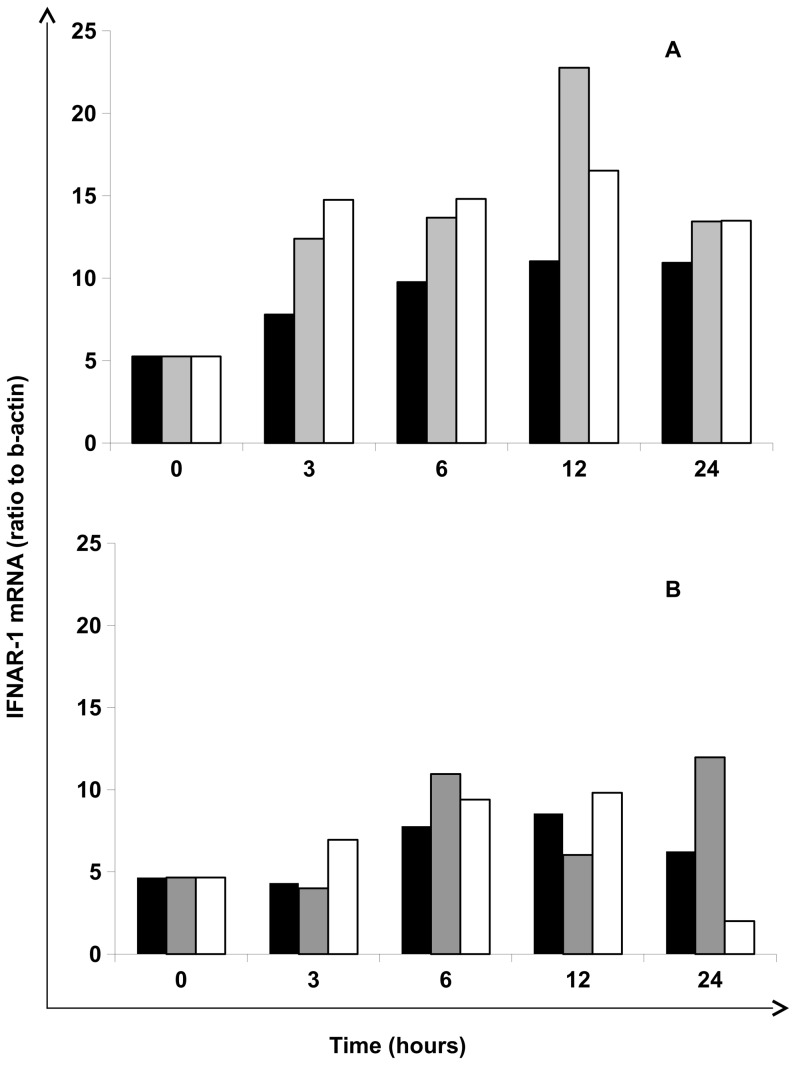
In Fig. 3 time-and dose-dependent response from two representative subjects (CC and TT) is shown, confirming 10 ng/ml of IFN-lambda as optimal dose, and showing, again, a better stimulation in CC genotype. In particular, at 10 ng/ml peak stimulation in CC genotype occurred earlier (12 h) and was more extensive than in TT genotype (24 h).
Figure 3. Time dependent induction of IFNAR-1 mRNA levels following treatment with IFN-lambda in PBMC from healthy donors.
PBMC were exposed to control medium (▪) 10 ng/mL (▪) and 100 ng/mL (□) of IFN-lambda, then mRNA levels for IFNAR-1 were measured at different time points (0, 3, 6, 12 and 24 h). Results are expressed as ratio to beta-actin. Results from one representative experiment performed on PBMC from two healthy donors with IL-28B rs12979860 CC (Panel A) and TT (Panel B) genotype are shown.
Discussion
Since 2009, several studies have shown that there is an important association between IL-28B polymorphisms and the outcome of standard anti-HCV therapy [9], [11]. Among the identified SNPs, rs12979860 appeared the most relevant, being associated with both therapy-induced and spontaneous clearance of HCV. Data so far available indicate that the rs12979860-favorable (CC) genotype is associated with higher expression of IFN-lambda [12], [13] that, in turn, may contribute to viral clearance. This hypothesis is further supported by the evidence that high IFN-lambda peak serum levels during the acute phase are associated with a self-limited course of HCV infection [18]. However, the association between elevated IFN-lambda expression and HCV clearance is not unequivocally reported, since other studies have shown higher levels of IFN-lambda in the liver of patients with the hapless genotype [28]. Inappropriate IFN-lambda activation (altered level, potency or timing) upon HCV infection may explain the apparent contradiction between the two opposite lines of evidence [29]. Based on these evidences and considering a previous study in which our group observed a reduced expression of IFNAR-1 mRNA in poorly performing patients [25] , we explored the possible relationships between IL-28 rs12979860 genotype and the expression of IFNAR-1 in naive HCV patients.
Our results highlighted a significant difference of IFNAR-1 mRNA expression in PBMC from patients carrying rs12979860 CC and CT/TT genotypes, with the most prominent difference in the absence of -C allele. It is to be underlined that the IFN-alpha treatment did not significantly affect the levels of IFNAR-1 mRNA, suggesting a marginal role of this cytokine in the observed differences. A significant correlation between endogenous levels of circulating IFN-lambda and spontaneous levels of IFNAR-1 mRNA in PBMC was observed in patients carrying CC genotype, although no differences of IFN-lambda levels could be appreciated between the different genotypes. The lack of detectable difference is probably due to the chronic infection state of our patients, where the reduced levels of circulating IFN-lambda might have hampered the comparison. This is in line with other groups that described lower levels of circulating IFN-lambda in HCV chronically infected patients as compared to spontaneously resolved infections [18], [27]. We then explored whether IFN-lambda could be able to up-regulate IFNAR-1 expression in normal PBMC. As compared to HCV patients, the basal levels of IFNAR-1 mRNA in healthy donors appeared significantly higher, and seemed independent from IL-28 genotype. On the contrary, the IL-28 genotype was important in the IFN-lambda response, since a stimulation was observed in a time-and dose-dependent manner, and was much more pronounced in CC vs TT carriers.
Our findings suggest that IFN-lambda could play a crucial role in the modulation of IFNAR-1 expression, and that endogenous levels of IL-28B may be responsible for partial restoration of IFNAR-1 expression in HCV patients with favourable IL-28B genotypes. A number of published studies, show that the mRNA levels of IFNAR-1 are correlated with the extent of IFN response both in vivo and in vitro [30], [31], [32]. Therefore, although the present study does not provide direct demonstration that the different IFNAR-1 mRNA levels translate into different expression of IFNAR at cell surface, it is tempting to speculate that this partial restoration may confer to CC carriers a response advantage to either endogenous or exogenous IFN-alpha, representing the biological basis for the observed association between CC genotype and favourable outcome of either natural infection (clearance vs chronicization) or IFN therapy.
In summary, although the findings from the present study are preliminary, since they derive from a limited number of patients and might benefit from larger studies, they provide novel information, contributing to elucidate the mechanisms underlying the strong predictive value of IL-28B polymorphisms on the natural history and on the response to IFN therapy in HCV infection.
Acknowledgments
The contribution of Laboratory of Microbiology and Infectious Disease Biorepository from National Institute for Infectious Diseases “L. Spallanzani”, Rome, Italy, is warmly acknowledged.
Funding Statement
This study has been partly supported by grants from Italian Ministry of Health, Ricerca Corrente and Ricerca Finalizzata. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization (2010) Sixty-Third World Health Assembly. Viral Hepatitis, Report by the Secretariat A63/15
- 2. Strader DB, Wright T, Thomas DL, Seeff LB (2004) American Association for the Study of Liver Diseases. Diagnosis, management, and treatment of hepatitis C. Hepatology 39: 1147–71. [DOI] [PubMed] [Google Scholar]
- 3. European Association for the Study of the Liver (2011) EASL Clinical Practice Guidelines: management of hepatitis C virus infection. J Hepatol 55: 245–64. [DOI] [PubMed] [Google Scholar]
- 4. Hadigan C, Kottilil S (2011) Hepatitis C virus infection and coinfection with human immunodeficiency virus: challenges and advancements in management. JAMA 306: 294–301. [DOI] [PubMed] [Google Scholar]
- 5. Butera D, Marukian S, Iwamaye AE, Hembrador E, Chambers TJ, et al. (2005) Plasma chemokine levels correlate with the outcome of antiviral therapy in patients with hepatitis C. Blood 106: 1175–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lalle E, Calcaterra S, Horejsh D, Abbate I, D'Offizi G, et al. (2008) Ability of peripheral blood mononuclear cells to activate interferon response in vitro is predictive of virological response in HCV patients. J Biol Regul Homeost Agents 22: 153–60. [PubMed] [Google Scholar]
- 7. Giannelli G, Guadagnino G, Dentico P, Antonelli G, Antonaci S (2004) MxA and PKR expression in chronic hepatitis C. J Interferon Cytokine Res 24: 659–63. [DOI] [PubMed] [Google Scholar]
- 8. Gerotto M, Dal Pero F, Bortoletto G, Realdon S, Ferrari A, et al. (2004) PKR gene expression and response to pegylated interferon plus ribavirin therapy in chronic hepatitis C. Antivir Ther 9: 763–70. [PubMed] [Google Scholar]
- 9. Chen L, Borozan I, Feld J, Sun J, Tannis LL, et al. (2005) Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology 128: 1437–44. [DOI] [PubMed] [Google Scholar]
- 10. Donnelly RP, Kotenko SV (2010) Interferon-lambda: a new addition to an old family. J Interferon Cytokine Res 30: 555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, et al. (2009) Genetic variation in IL-28B predicts hepatitis C treatment-induced viral clearance. Nature 461: 399–401. [DOI] [PubMed] [Google Scholar]
- 12. Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, et al. (2009) Genome-wide association of IL-28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet 41: 1105–9. [DOI] [PubMed] [Google Scholar]
- 13. Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, et al. (2009) IL-28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet 41: 1100–4. [DOI] [PubMed] [Google Scholar]
- 14. Mangia A, Thompson AJ, Santoro R, Piazzolla V, Tillmann HL, et al. (2010) An IL-28B polymorphism determines treatment response of hepatitis C virus genotype 2 or 3 patients who do not achieve a rapid virologic response. Gastroenterology 139: 821–7, 827. [DOI] [PubMed] [Google Scholar]
- 15. Riva E, Scagnolari C, Monteleone K, Selvaggi C, Picardi A, et al. (2012) Interleukin-28B (IL-28B) single-nucleotide polymorphisms and interferon plus ribavirin treatment outcome in Italian chronically HCV-infected patients. J Viral Hepat 19: 650–3. [DOI] [PubMed] [Google Scholar]
- 16. McCarthy JJ, Li JH, Thompson A, Suchindran S, Lao XQ, et al. (2010) Replicated association between an IL-28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology 138: 2307–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lange CM, Zeuzem S (2011) IL-28B single nucleotide polymorphisms in the treatment of hepatitis C. J Hepatol 55: 692–701. [DOI] [PubMed] [Google Scholar]
- 18. Langhans B, Kupfer B, Braunschweiger I, Arndt S, Schulte W, et al. (2011) Interferon-lambda serum levels in hepatitis C. J Hepatol 54: 859–65. [DOI] [PubMed] [Google Scholar]
- 19. Torres C, Brahm J, Venegas M (2013) J Lambda Interferon Serum Levels in Patients with Chronic Hepatitis C Virus Infection According to Their Response to Therapy with Pegylated Interferon and Ribavirin. Interferon Cytokine Res Sep 26. [DOI] [PubMed] [Google Scholar]
- 20. Naggie S, Osinusi A, Katsounas A, Lempicki R, Herrmann E, et al. (2012) Dysregulation of innate immunity in hepatitis C virus genotype 1 IL-28B-unfavorable genotype patients: impaired viral kinetics and therapeutic response. Hepatology 56: 444–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, et al. (2010) IL-28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology 52: 1888–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Honda M, Sakai A, Yamashita T, Nakamoto Y, Mizukoshi E, et al. (2010) Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology 139: 499–509. [DOI] [PubMed] [Google Scholar]
- 23. Dill MT, Duong FH, Vogt JE, Bibert S, Bochud PY, et al. (2011) Interferon-induced gene expression is a stronger predictor of treatment response than IL-28B genotype in patients with hepatitis C. Gastroenterology 140: 1021–31. [DOI] [PubMed] [Google Scholar]
- 24. Asahina Y, Tsuchiya K, Muraoka M, Tanaka K, Suzuki Y, et al. (2012) Association of gene expression involving innate immunity and genetic variation in interleukin 28B with antiviral response. Hepatology 55: 20–9. [DOI] [PubMed] [Google Scholar]
- 25. Abbate I, Romano M, Longo R, Cappiello G, Lo Iacono O, et al. (2003) Endogenous levels of mRNA for IFNs and IFN-related genes in hepatic biopsies of chronic HCV-infected and non-alcoholic steatohepatitis patients. J Med Virol 70: 581–7. [DOI] [PubMed] [Google Scholar]
- 26. Capobianchi MR, Lalle E, Martini F, Poccia F, D'Offizi G (2006) Influence of GBV-C infection on the endogenous activation of the IFN system in HIV-coinfected patients. Cell Mol Biol 52: 3–8. [PubMed] [Google Scholar]
- 27. Mihm S, Frese M, Meier V, Wietzke-Braun P, Scharf JG, et al. (2004) Interferon type I gene expression in chronic hepatitis C. Lab Invest 84: 1148–59. [DOI] [PubMed] [Google Scholar]
- 28. Abe H, Hayes CN, Ochi H, Maekawa T, Tsuge M, et al. (2011) IL-28 variation affects expression of interferon stimulated genes and peg-interferon and ribavirin therapy. J Hepatol 54: 1094–101. [DOI] [PubMed] [Google Scholar]
- 29. François-Newton V, Magno de Freitas Almeida G, Payelle-Brogard B, Monneron D, Pichard-Garcia L, et al. (2011) USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon α response. PLoS One 6: e22200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fukuda R, Ishimura N, Kushiyama Y, Moriyama N, Ishihara S, et al. (1997) Effectiveness of interferon-alpha therapy in chronic hepatitis C is associated with the amount of interferon-alpha receptor mRNA in the liver. J Hepatol 26: 455–461. [DOI] [PubMed] [Google Scholar]
- 31. Mathai J, Shimoda K, Banner BF, Mori M, Bonkovsky HL, et al. (1999) IFN-alpha receptor mRNA expression in a United States sample with predominantly genotype 1a/I chronic hepatitis C liver biopsies correlates with response to IFN therapy. J Interferon Cytokine Res 19: 1011–8. [DOI] [PubMed] [Google Scholar]
- 32. Booy S, van Eijck CH, Dogan F, van Koetsveld PM, Hofland LJ (2014) Influence of type-I Interferon receptor expression level on the response to type-I Interferons in human pancreatic cancer cells. J Cell Mol Med 20: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]