Abstract
A novel dual cell linear ion trap Fourier transform ion cyclotron resonance mass spectrometer (FT-ICR MS) and its performance characteristics are reported. A linear ion trap-Fourier transform ion cyclotron resonance mass spectrometer has been modified to incorporate a LTQ-Velos mass spectrometer. This modified instrument features efficient ion accumulation and fast MS/MS acquisition capabilities of dual cell linear RF ion trap instruments coupled to the high mass accuracy, resolution, and dynamic range of a FT-ICR for improved proteomic coverage. The ion accumulation efficiency is demonstrated to be an order of magnitude greater than that observed with LTQ-FT Ultra instrumentation. The proteome coverage with yeast was shown to increase over the previous instrument generation by 50% (100% increase on the peptide level). In addition, many lower abundance level yeast proteins were only detected with this modified instrument. This novel configuration also enables beam type CID fragmentation using a dual cell RF ion trap mass spectrometer. This technique involves accelerating ions between traps while applying an elevated DC offset to one of the traps to accelerate ions and induce fragmentation. This instrument design may serve as a useful option for labs currently considering purchasing new instrumentation or upgrading existing instruments.
Keywords: Velos-FT, Dual Cell Fragmentation, Instrumentation, Dynamic Range, Yeast, Duty Cycle
Introduction
The era of modern mass spectrometry has largely been dominated by advancements in instrumentation [1–6], sample handling [7–10], and database searching technologies [11–13]. Innovations made in these areas have made possible incredible improvement in the analysis of complex samples. The drive to decipher information within the proteome [8, 10] has been a major force influencing technological development of mass spectrometry methods for biological applications. The current paradigm in proteome research involves shotgun [9] or bottom-up experiments in which protein samples (pure protein, cell lysates, tissue lysates, etc.) are enzymatically digested into peptide mixtures. The resulting peptide mixtures can have wide dynamic range in peptide concentration [14] and heterogeneity [15]. In fact, the overall complexity of these peptide mixtures exceeds the sensitivity and efficiency in detection of all available modern instruments [4]. The primary impediments to routine whole organism proteome measurements by mass spectrometry are acquisition speed, sensitivity, and dynamic range. Further development of instrumentation technology is required for routine high coverage, in-depth proteome analyses.
Mass spectrometry instrumentation [16] has evolved dramatically over the last twenty years. A current configuration for proteomics research today is a hybrid type and consists of two coupled mass analyzers, each capable of independent data acquisition. A conventional example combines a linear ion trap (LTQ) with a Fourier transform mass spectrometer (FTMS). In this case, the FTMS is either an ion cyclotron resonance mass spectrometer (LTQ-FT) or Orbitrap mass spectrometer. In general, hybrid mass analyzers benefit proteomics research by incorporating desired characteristics from each analyzer to yield unique capabilities.
Here we present results of our efforts to modify the LTQ-FT hybrid by combining a dual cell linear RF ion trap and FT-ICR mass spectrometer, referred to as a Velos-FT mass spectrometer. This instrument has many unique attributes such as the ability to rapidly accumulate ions, MSn analysis at a higher repetition rate, and the ability to perform beam-type CID fragmentation between the dual cell linear RF ion traps. The Velos-FT has improved ability to accumulate ions, with observed ion accumulation time reduction of an order of magnitude or more (~3 ms vs. 40 ms) compared with conventional LTQ-FT instrumentation. When performing MSn, selected ion monitoring (SIM), or data independent acquisition (DIA), this speed improvement can be very significant. Related to this, we show that with the Velos-FT, the number of peptide identifications per run is increased by 100% when compared with the LTQ-FT Ultra (50% on the protein group level). Top down proteomics [17] requires isolation and fragmentation prior to analysis. For top-down experiments, in many cases it is difficult to accumulate enough ions to achieve coherent cyclotron motion for the duration required to resolve high mass ions [18, 19]. Here we show that the ability to accumulate large biomolecules with the custom Velos-FT is greatly improved over those with the LTQ-FT Ultra. The dual cell linear RF ion trap instrument configuration enables operation of a unique fragmentation method, which we call Dual Cell Fragmentation (DCF), performed by transferring ions between the two cells through the background gas using elevated DC potentials to accelerate and induce dissociation. This technique generates fragmentation patterns which share some similarities with spectra acquired on QTOF and triple quadrupole instruments. The so called “1/3 rule” which limits the lower mass limit for product ions generated by resonance excitation in RF based ion traps was shown to be reduced with DCF. The overall peptide identification rate was comparable to that obtained with resonance excitation CID, and many peptides were identified in both methods demonstrating the utility of DCF for peptide identification with spectra that also contain lower m/z ions. Furthermore, a distinct subset of peptides was identified with each method that may be a result of repeated analyses or subtle differences between the fragments observed in DCF and CID. Finally, with the cost of high performance instrumentation currently inflating much faster than available funds from most granting agencies that support mass spectrometry, the upgrade design of the Velos-FT presented here may present a useful option for other labs to consider.
Experimental
Modification of the LTQ-FT Ultra
An LTQ-FT Ultra mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) was modified to include an LTQ Velos mass spectrometer. The linear ion trap (LTQ) was removed and replaced with a dual cell linear radio frequency ion trap mass spectrometer[5] (Velos, Thermo Fisher Scientific, San Jose, CA). The two linear radio frequency ion traps will be referred to throughout the manuscript as the high pressure cell (HPC) and low pressure cell (LPC). No ion optic modifications were necessary for the coupling process as the flange designed to mate the LTQ and Velos to the Orbitrap or the FT mass spectrometer are identical. However, ion trap control software modifications were required for operation of the custom FT-ICR. Software in control of the LPC was modified so that it acts as a simple RF multipole during the transmission of ions from the HPC to the ICR cell for FT-MS acquisition mode. The standard FT ion transmission calibration script was modified to include the center lens, front, center and back sections of the LPC, along with the back lens of the dual cell trap assembly. Optimized DC voltages for each of these elements are applied during the transmission event of an FTMS acquisition. The ion transfer efficiency between the linear ion trap and ICR cell was determined to be similar for both the LTQ-FT Ultra and custom FT-ICR MS based upon unscaled total ion current measurements conducted using the same ion target value for both instruments (see Supplemental data for details).
Ion-Trap Control Language (ITCL) DCF Program
Direct modification of factory installed ITCL code was performed in-house for the implementation of DCF. These modifications operate within the framework of the factory installed code for the LTQ-FT Ultra mass spectrometer. In short, CID collision energy setting of 1.0 in the user interface was used to enable DCF. This allows for the execution of DCF both directly from Tune as well as during method based operation. When DCF is enabled, ion accumulation and isolation proceed in the HPC without alteration of factory installed ITCL code. However, no resonance excitation is applied before initial transfer to the LPC. Ions are then transferred immediately back to the HPC. The HPC center section potential is adjusted to induce fragmentation through application of a large negative DC bias. This DC potential bias is determined as a function of precursor ion m/z, in a similar fashion as with beam-type CID collision cell experiments[20]. This equation relating m/z to DC potential bias of the center section of the HPC was determined empirically to have a slope of −0.141 (V/Da) and an intercept of −20.00 (V) based upon collision energy optimization using a bovine serum albumin (BSA) digest. A q-value of 0.205 was used for fragmentation of Angiotensin I, which permitted detection of low mass fragment ions down to 25% of the precursor m/z.
Yeast Sample
Saccharomyces cerevisiae strain S288C (Baker’s yeast) was grown in glucose rich media to mid-log phase. Cells were collected by centrifugation and resuspended in 100 mM ammonium bicarbonate buffer at an optical density (OD) of 0.90. The cells were lysed using a bead-beater[21]. The lysate was centrifuged at 1000g for five minutes to remove debris (cell wall particles and unbroken cells). The lysate was centrifuged again for 30 minutes at 15,000g to separate the soluble and membrane protein fractions. The soluble fraction was assayed for protein concentration using Coomassie Plus Protein assay (Pierce, Rockford, IL) and found to be ~5.0 mg/mL. The insoluble fraction was not utilized in this experiment. The sample was reduced with 15 mM dithiothreitol (Sigma-Aldrich, St. Louis, MO) at room temperature for the duration of 30 minutes. The cysteine residues were blocked using iodoacetamide (Sigma-Aldrich, St. Louis, MO) 15 mM at room temperature for the duration of 30 minutes. The soluble fraction of the yeast lysate was digested using 1:250 ratio of sequencing grade trypsin (Promega, Madison, WI). The reaction was allowed to proceed for 2 hours at 37°C while under constant agitation via orbital shaking. The digest was quenched and frozen at −20°C. This sample was desalted using C18 Sepak (Waters Corporation, Milford, MA). The desalted peptides were lyophilized and redissolved in mobile phase A (99.9% ultrapure water, 0.1% formic acid). Injections of 1 µg of total protein were loaded for LC-MS/MS analysis.
Liquid Chromatography
Liquid chromatography was performed using a Waters NanoAcquity UPLC (Waters Corporation, Milford, MA). Pulled tip columns were constructed in-house using a laser-pulling device (Sutter Instrument Company, Novato, CA). A column of 30 cm in length was constructed with 75 um ID×360 um OD fused silica capillary. The packing material used for peptide separation was 100 Å C18 magic beads (Microm Bioresources Inc., Auburn, CA). A fused silica trap column was constructed from 100 um ID×360 um OD fused silica capillary. The frit was made on one end of the trap with Kasil (PQ Corporation, Valley Forge, PA) to contain C18 packing material. The packing material used in the trap was 200 Å C18 magic beads (Microm Bioresources Inc., Auburn, CA). A binary solvent gradient was used for peptide separation. Mobile phase A consisted of 99.9% water with 0.1% formic acid. Mobile phase B consisted of 95% acetonitrile with 0.1% formic acid. The gradient was setup as follows 5%–35% B in 30 minutes. Column washing was done with 80% B for 20 minutes, followed by re-equilibration for 30 minutes using 5% B.
Mass Spectrometry
All data were acquired on the LTQ-FT Ultra or the custom FT-ICR mass spectrometers (Thermo Fisher Scientific, San Jose, CA.). Data dependent acquisition (DDA) experiments for identification comparison were conducted using a “top 5” approach in which a high resolution FTMS acquisition (50,000 resolution at 400 m/z) was followed by 5 ion trap MS/MS acquisitions for the LTQ-FT Ultra or a “top 10” approach was used in the case of the custom FT-ICR MS with the same resolution settings. The AGC target value for the precursor scan was set to 1x106 counts and for ion trap MS/MS scans to 1x104 counts. All MS/MS targets were chosen from the high resolution FTMS scan. Charge state screening was applied with consideration of only 2+ and 3+ isotope distributions. Dynamic exclusion was used with the following parameters: one repeat count, 15 second repeat duration, 500 exclusion list size, and 90 second exclusion duration. Preview mode was enabled for all LC-MS/MS experiments which allowed both the ICR cell and the LTQ to acquire concomitantly.
Top down experiments
Myoglobin was used as received from Sigma-Aldrich (St. Louis, MO) and was dissolved in an electrospray ionization solution consists of HPLC grade methanol (Sigma-Aldrich, St. Louis, MO), 18 M Ohm water, and glacial acetic acid (Sigma-Aldrich, St. Louis, MO) in the ratio of 49:49:2. This solution was directly infused into the mass spectrometer at a flow rate of 0.500 uL/min. The myoglobin concentration of the infusion solution was 500 nM.
Data Handling and Searching
Data was extracted from the raw files and converted to mzXML format using ReAdW(ver 4.2.1). The mzXML2search (ver 4.1) was used to convert the data to Mascot generic format. All database searches were performed using Mascot Server (ver 2.2). The FASTA sequence database was downloaded from NCBI (Saccharomyces cerevisiae). Precursor mass tolerance of 25 ppm and fragment mass tolerance of 0.6 Da were used for data-dependent experiments. Protein groups were assigned within the latest version of the Mascot[11] or Sequest[12, 13] software and represent the simplest explanation of the proteins given the identified the peptide sequences. Reverse sequence searching was used as a strategy to estimate false discovery rate (FDR). Ion times and raw precursor ion intensities were extracted directly from the raw file headers utilizing the Thermo SDK. Maximum ion intensities for identified peptides were calculated from monoisotope peaks determined using Hardklör[22].
Results and Discussion
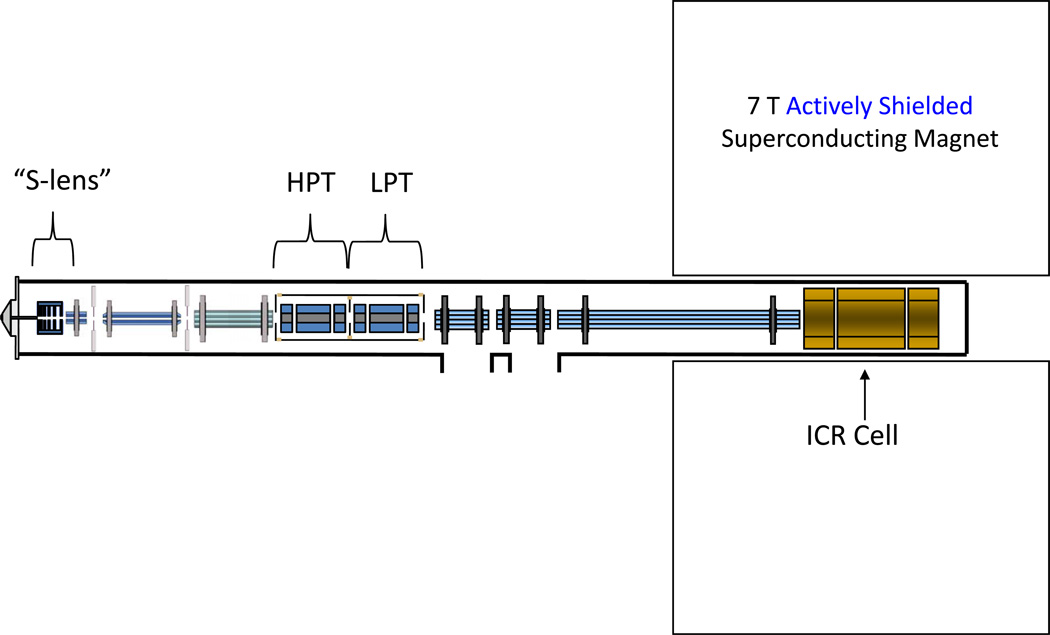
The use of dual cell linear ion trap instrument configuration has gained popularity recently. The first of these instruments released was simply a stand-alone, bench-top dual cell linear ion trap[5] (Velos) and is currently available coupled to the Orbitrap mass spectrometer [4]. This manuscript is the first published report of a dual cell linear ion trap-FT-ICR hybrid mass spectrometer and represents a useful reconfiguration option for existing LTQ-FT instruments. The monetary expense of this upgrade is equivalent to upgrading from an LTQ to an LTQ-Velos. No additional hardware is required for the upgrade. Other laboratories have subsequently adopted this technology[23]. A conceptual diagram of the dual cell linear ion trap FT-ICR mass spectrometer reported in this manuscript is shown in Figure 1. The unique features of this instrument which provide significant performance enhancement are contained within the dual cell linear ion trap mass spectrometer. This instrument has been described in detail by Second[5] et al. In brief, the S-lens or stacked ring ion guide and the high pressure cell contribute to increased ionization source transmission efficiency and increased trapping efficiency respectively. Operation of the LPC for mass scanning allows for faster MS/MS acquisition while the HPC allows for efficient accumulation and fragmentation of ions. The increased accumulation efficiency and faster scan speeds are manifest in faster MS/MS repetition rates. For example the predecessor to this instrument, the LTQ, could perform MS/MS at 4 Hz. With the dual cell linear RF ion trap front end, an MS/MS acquisition rate of ~10 Hz is achievable.
Figure 1.
A conceptual diagram of the Velos-FT. This hybrid instrument consists of a dual linear ion trap “front end” coupled to a 7 Tesla FT-ICR high resolution mass spectrometer. Notable aspects include the stacked ring ion guide in the source, as well as the tandem linear ion trap arrangement.
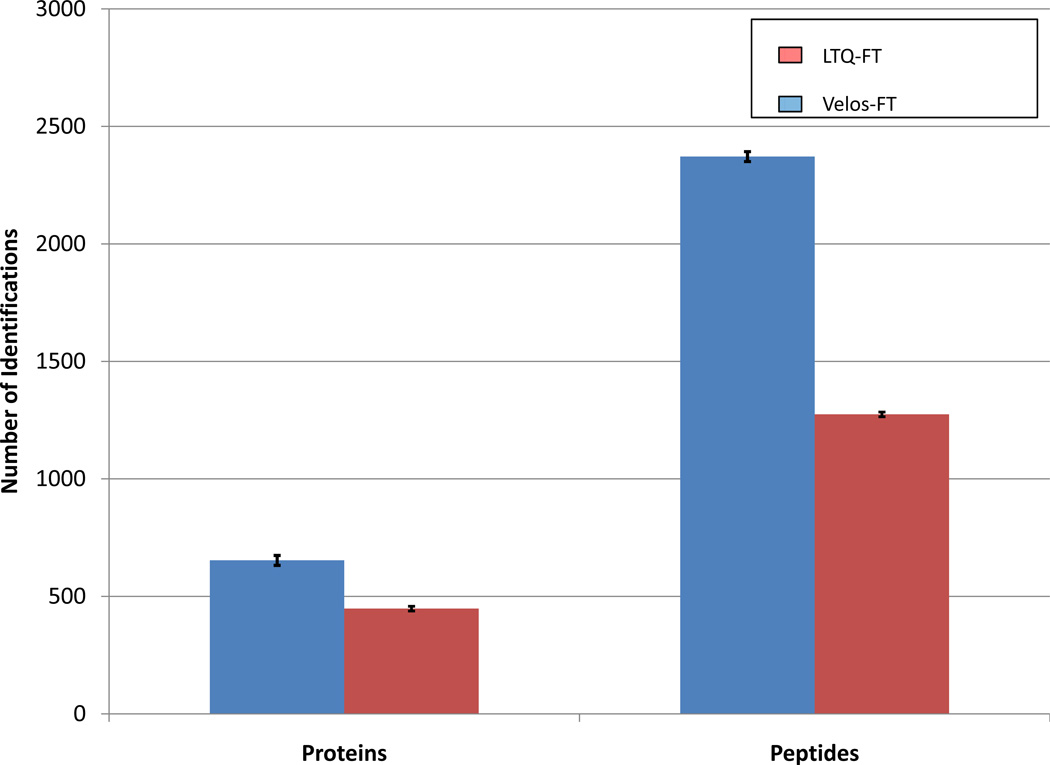
Faster MS/MS acquisition rate should lead to more peptides being identified from complex samples. Yeast lysate digest[14, 24] provides a useful benchmark sample for mass spectrometry based proteomics because of its complexity and wide dynamic range of protein concentration. In this comparison, the custom FT-ICR MS and the LTQ-FT Ultra were set to operate in data-dependent acquisition mode. The top ten most abundant precursors were selected for MS/MS analysis in the Velos-FT, whereas, the top five most abundant precursors were selected in the LTQ-FT Ultra. The number of MS/MS events in each DDA experiment was selected based on the number of events which could be executed during the simultaneous high resolution precursor acquisition. This number is approximately two-fold larger for the Velos-FT due to the combination of shorter injection times and the higher linear trap scan rates. The results of three technical replicates, all searched with Mascot, are presented in Figure 2A. The Velos-FT identified an average of 653 +- 21 protein groups, which is a 46% increase over the 448 +- 10 protein groups identified with the LTQ FT Ultra. For unique peptides, 100% more identifications are observed for the Velos-FT. This can be rationalized by considering that the additional unique peptides are attributed to proteins which have already been identified, thus providing on average more complete sequence coverage. This instrument performs very similarly to the Velos-Orbitrap. The number of ion trap MS/MS events achievable for the Velos-FT in an equivalent analysis time is roughly double that of the immediate predecessor the LTQ-FT, as was observed in the initial evaluation of the Velos-Orbitrap (compared with the LTQ-Orbitrap). In fact, gains were observed by Second et al.[5] (95% more unique peptides, C. elegans digest) and Olsen et al.[4] (28% more unique proteins, HeLa cell digest) in initial experiments describing the Velos and the Velos-Orbitrap instruments.
Figure 2.
Comparison of the improvement in number of identifications achieved with the Velos-FT over the LTQ-FT Ultra. Both protein group and peptide identifications were made from three replicate analyses of yeast lysate digest. Mascot was used for protein group and peptide identification. The error bars represent one standard deviation.
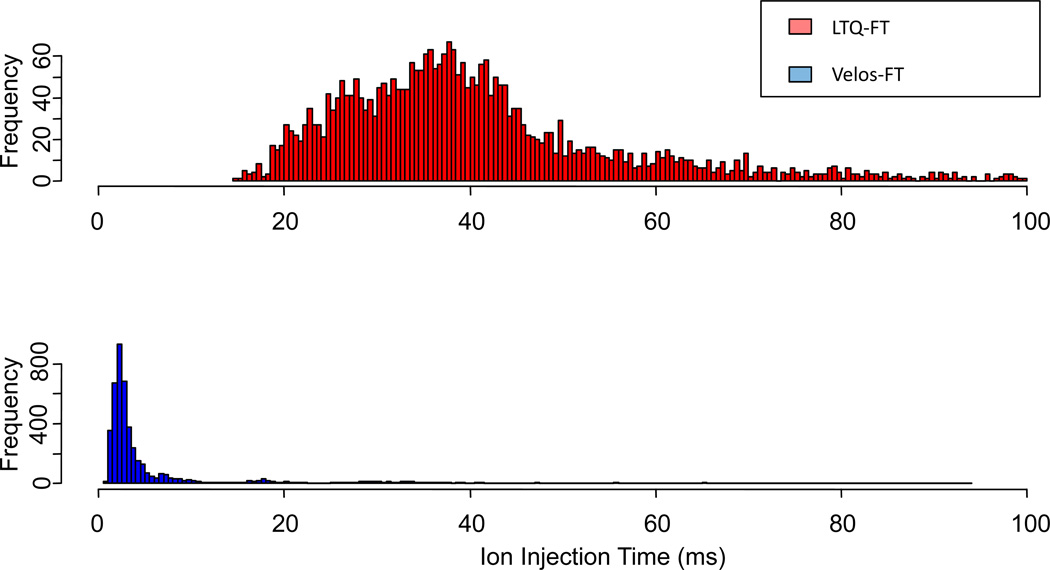
The increase in number of identifications has been attributed partially to the increased ion transmission efficiency of the source of this instrument. The electrospray source of this instrument has been shown by others to be 5–10 times more efficient for ion transmission and accumulation[4, 5]. This is attributed to the radio frequency stacked ring ion guide and the high pressure cell. For the yeast lysate, we observe average ion injection times of ~3 ms for the high resolution precursor mass measurement Figure 3. This is an order of magnitude lower than that of the LTQ-FT Ultra (~40 ms) for the same sample. Others have observed similar improvements for injection times when replacing the standard LTQ source optics with an ion funnel[25]. Shorter ion accumulation times and faster analytical scan rates enable sampling depths not obtainable with the previous generation of instruments.
Figure 3.
Histogram of ion injection times associated with the FT-MS precursor scans of a yeast lysate tryptic digest analysis. Average ion injection time for the LTQ-FT Ultra is ~40 ms whereas with the Velos-FT the average resides at <3 ms.
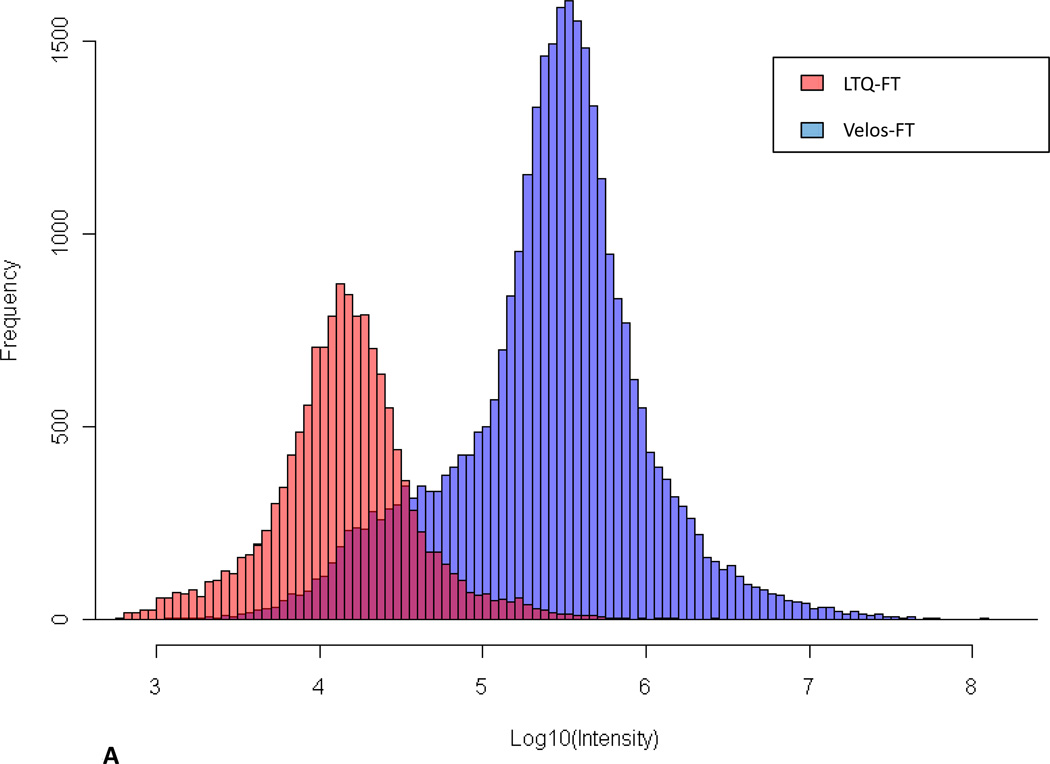
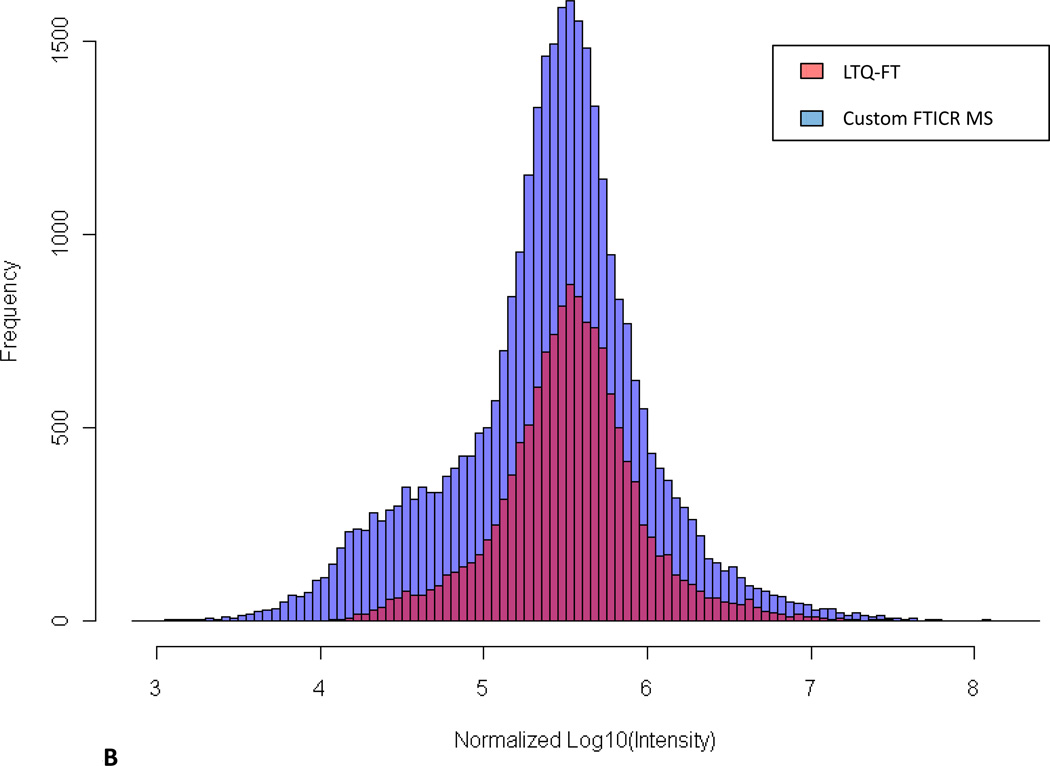
To allow demonstration of improved ion injection times possible with the custom FT-ICR MS, the maximum intensities for every precursor chosen for data-dependent MS/MS from the two instruments were extracted from data files and compared (Figure 4). Differences in the intensity distributions are due primarily to a 10-fold decrease in ion accumulation time for the Velos-FT, but also are influenced by other factors, such as ion transfer efficiency from the linear trap to ICR cell along with excitation and detection efficiencies. With automated gain control enabled, signal intensity is calculated by scaling inversely with the accumulation time in seconds (Figure 4A). To account for this difference, we normalized the data to the average peptide intensity (Figure 4B). Figure 4B illustrate the broader MS/MS sampling range, particularly the low intensity precursor tail, and increased overall sampling frequency achieved with the Velos-FT. These data indicate that when using the Velos-FT, an order of magnitude greater depth in sampling can be achieved over its predecessor.
Figure 4.
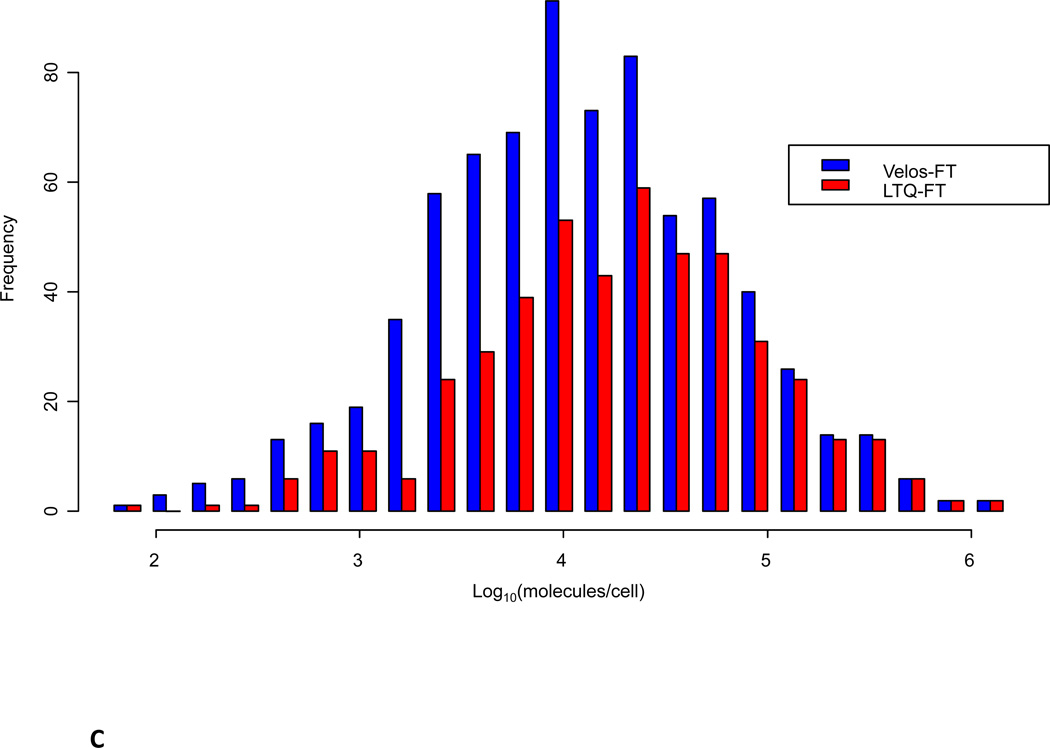
A) Comparison of distribution of the ion intensity of precursors selected for data-dependent MS/MS sequencing. B) Normalized comparison of ion intensity of precursors selected for data-dependent MS/MS sequencing. C) Comparison of identified peptide maximum ion intensities between the LTQ-FT Ultra and the Velos-FT sampled across the observed intensity range. D) Maximum intensity range over which peptides were identified.
Although the Velos-FT is able to interrogate peptide species over a broader intensity range, this may not correspond with actual cellular protein and peptide abundance. In an effort to understand if peptide and protein identifications using the Velos-FT were extended to lower abundance species, proteins were mapped back to the yeast quantitative western blotting results of Ghaemmaghami et al .(Figure 4C). This analysis revealed an enhancement in the identification of lower abundance species using the Velos-FT relative to the LTQ-FT Ultra.
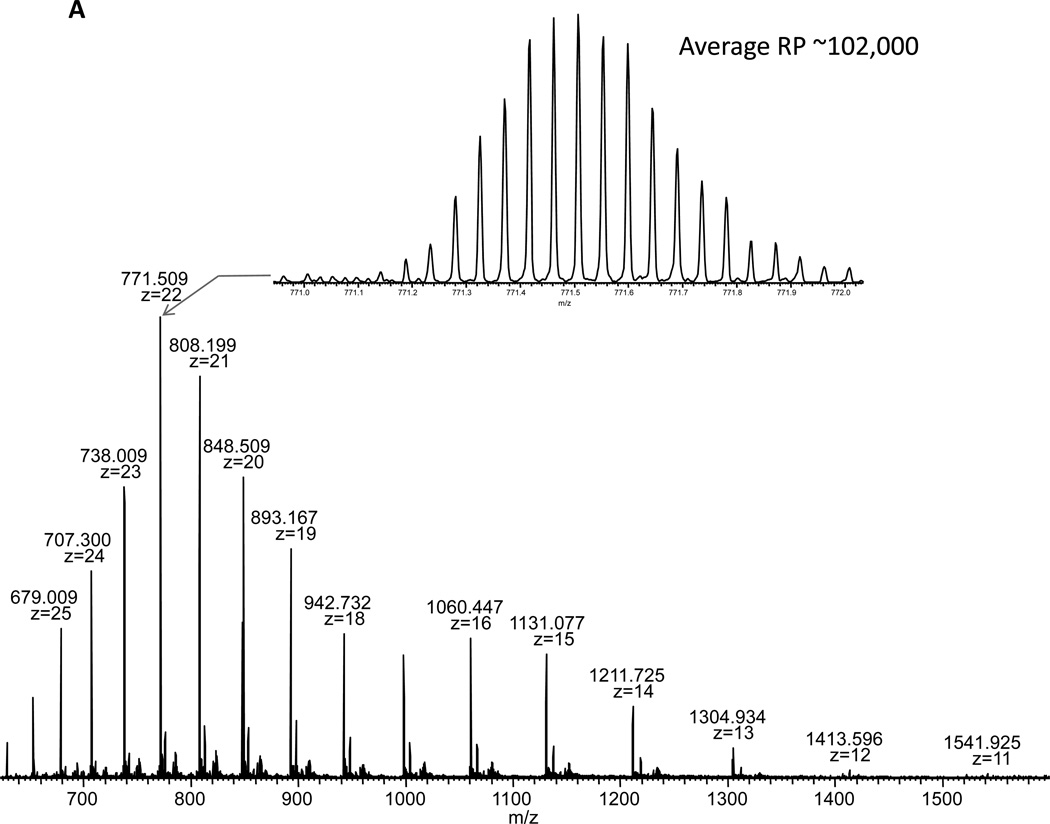
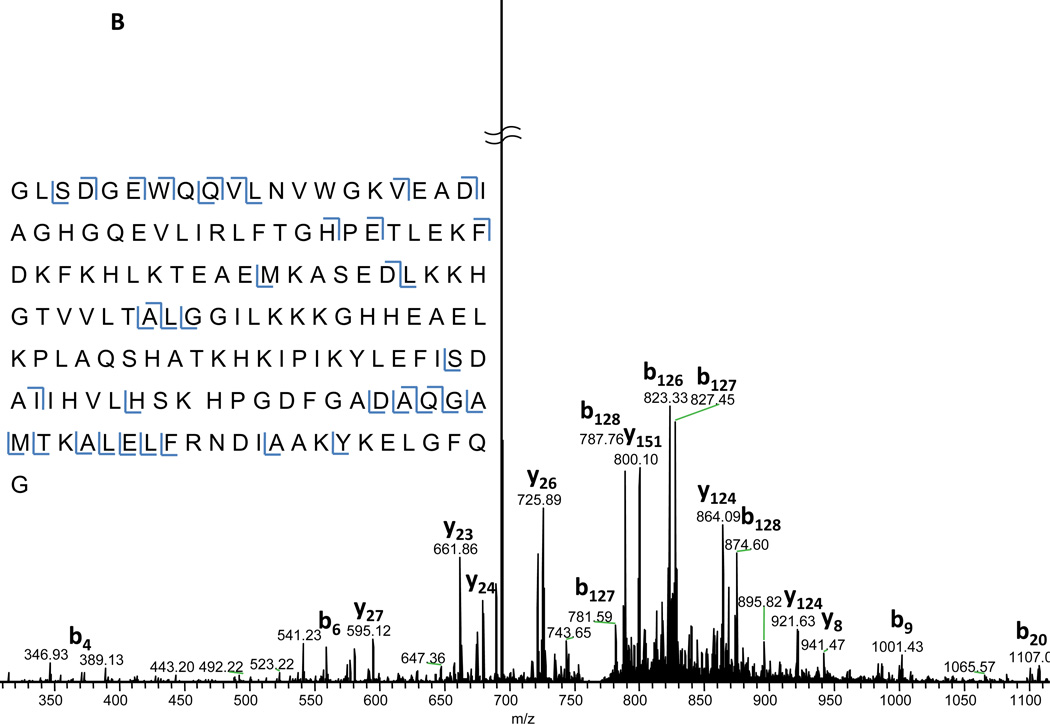
Top-down proteomics, the study of intact proteins, has been pursued by many as a method for a more complete protein characterization[26–28]. This technique potentially allows one to not only identify proteins, but to also interrogate which isoforms, post-translational modifications, and other variants are present in the sample. Although top-down proteomics has great potential, intact protein analysis by mass spectrometry presents a unique set of challenges. One such challenge [29, 30] is increased susceptibility to systematic signal distribution. This is in part due to properties of large biomolecules, such as proteins, occupying a large number of charge states and isotopic forms. This charge state distribution effectively divides the analyte amongst the observed mass spectral peaks. Therefore, it becomes more likely for any single peak to appear at or below the detection limit of the analyzer. Decreased ion injection times as observed using the Velos-FT are beneficial to all proteomics analyses, particularly for top-down MS/MS. In Fig. 5A, the phenomenon described above can be observed even for the relatively small, 17 kDa protein, myoglobin. Here charge states 11–27 are all simultaneously observed in the full spectrum along with greater than 15 isotope peaks for each charge state (isotope distribution shown in the inset of Figure 5A). Isolation and fragmentation of the most abundant charge state requires only 7.9 ms to accumulate 1,000,000 ion charges in the Velos-FT. Large targets such as this are typical for top-down experiments due to the numerous possible fragments along with the isotopic complexity for each of these fragments. Given the flow rate and concentration, this is corresponds to approximately 33 attomoles for this acquisition. In Figure 5B, the fragmentation pattern for myoglobin [M+22H]22+ is shown. Upon deconvolution and searching using Mascot Top-down against the entire SwissProt database, myoglobin is the most probable identification with an E-value of 5.0×10−5 (73 of 897possible b and y ion matches) demonstrating that useful and effective fragmentation of large biomolecules can be achieved in the HPC on the Velos-FT.
Figure 5.
A) Full scan spectrum of myoglobin collected using the Velos-FT. The isotopic envelope is for myoglobin [M+H]22+ is shown in the inset. B) Annotated FT-MS/MS spectrum of myoglobin [M+H]22+. The ion injection time required for this acquisition was 7.9 ms.
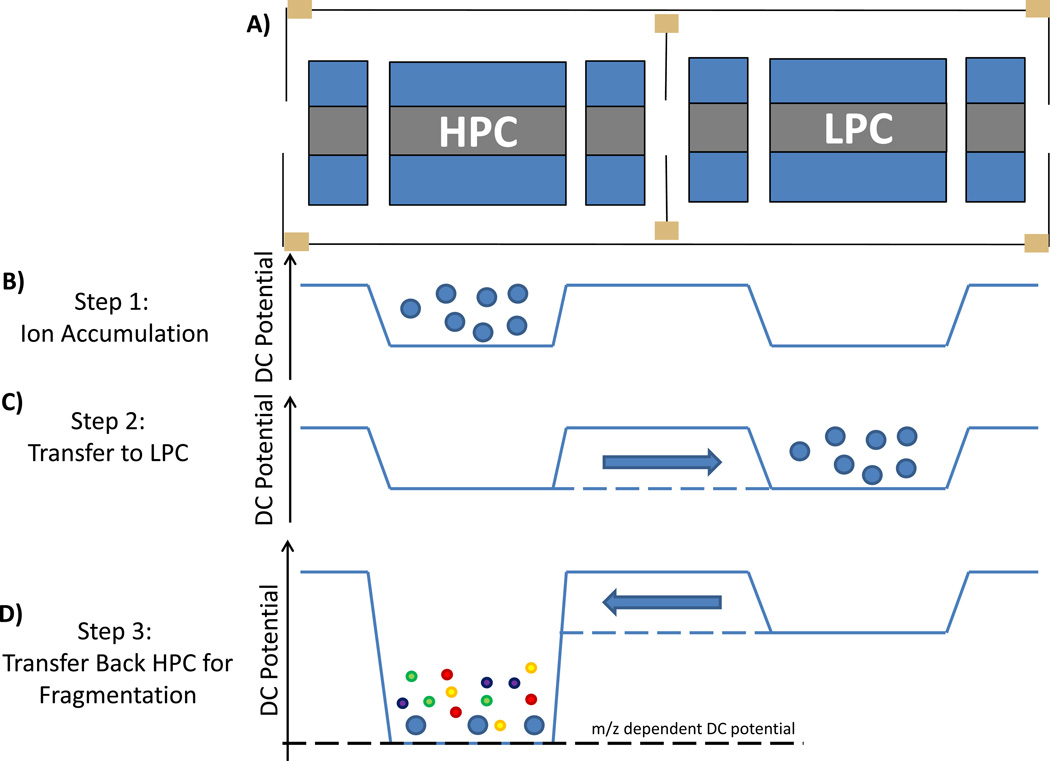
The configuration of two linear RF ion traps in series enables unique mass spectrometry experiments to be conducted with the Velos-FT. One such experiment, DCF, involves beam-type fragmentation of isolated precursor ions. A conceptual representation of DCF operation on the custom FT-ICR MS mass spectrometer is shown in Figure 6. During ion accumulation in the HPC, a DC potential barrier is applied to prevent ions from “leaking” from the HPC to the LPC. At this stage of a standard experiment, mass isolation and resonance excitation would be performed on the ion population in the HPC, followed by transfer to the LPC for mass analysis. In the DCF experiment, after accumulation in the HPC, isolation is performed but the resonance excitation is skipped, leaving the selected precursors. These precursors are transferred to the LPC, but then almost immediately transferred back to the HPC. Between transfers, the DC voltages applied to the HPC have been adjusted to create a large potential offset between the two cells. As the ion population is accelerated back to the HPC it encounters a much higher density of helium, with the higher energy collisions inducing collisional dissociation. Although collisional dissociation in an ion trap is not novel, DCF allows unique experimental capabilities previously unrealized with Velos, Velos Orbitrap, and Velos-FT instruments.
Figure 6.
A) A conceptual representation of the dual cell linear ion traps used to enable fragmentation during transfer between the LPC and the HPC. B) DC potentials applied during ion accumulation in the HPC. C) DC potentials applied during transfer and trapping of the ion population in the LPC (solid lines = trapping potential, dashed lines = transfer potential). D) DC potentials applied during higher energy transfer back to HPC.
During the preparation of this manuscript iHCD was described by McAlister et al.[31] to enable similar fragmentation utilizing the inlet of an LTQ. DCF was developed independently without the endorsement or support of Thermo Fisher Scientific. iHCD is different in that it is performed with air as the collision medium whereas DCF is performed primarily with helium. In iHCD ions are accumulated in the LTQ and transferred back to a multipole near the inlet to induce fragmentation subsequent to ion manipulation. Although collisions with nitrogen are more effective, DCF may benefit from reduced ion losses due to the short, simple ion path traversed during the activation process.
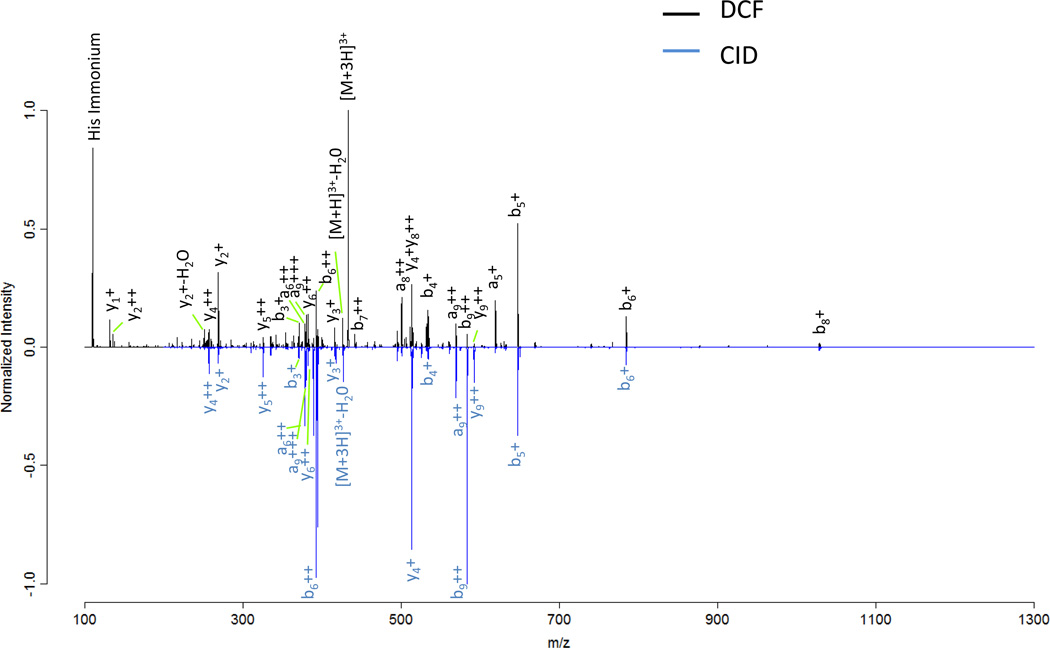
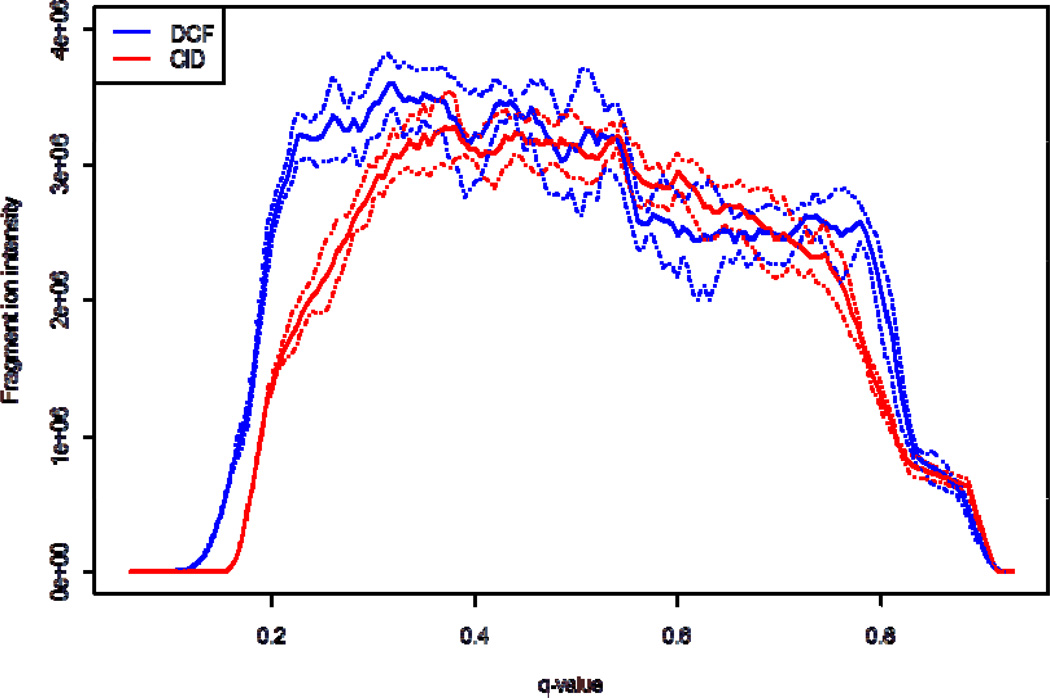
Product ion scanning using CID in RF ion traps is a sensitive method for obtaining sequence information from peptides and proteins. CID fragmentation in RF ion traps is performed through application of a m/z dependent secular or “tickle” frequency which selectively excites the oscillatory motion of a particular m/z species. This excitation induces multiple collisions with the background gas in the trap until the peptide fragments. Although this technique has been shown to be highly efficient, usually a single bond is dissociated per peptide ion since as dissociation occurs, the applied frequency is non-resonant with most product ions. When the RF ion trap is operated with the default q of 0.25 product ions with m/z values less than 28% of the precursor m/z will not be stable. The default q represents a compromise between containing low m/z product ions and confining/inducing fragmentation of the parent. This cut-off is commonly rounded up and referred to as the “1/3 rule” and is the reason that small y1 fragment ions, immonium ions, iTRAQ[32] , and other low mass species are typically not observed in peptide MS/MS spectra with RF ion traps. DCF experiments are less dependent on the q-value applied during activation, and therefore are less susceptible to the low m/z limitation then resonant excitation CID. In Figure 7, two MS/MS fragmentation patterns of angiotensin I were acquired; one using resonant excitation CID (blue) with a q-value of 0.250 and the other using DCF(black) with a q-value of 0.205. The major differences between the two methods include the observance of more a ions (DCF), His immonium ion (DCF), amount of precursor remaining, and different relative b and y fragment ion intensity ratios (DCF vs. CID). The overall cycle times for each fragmentation method are comparable, however, DCF has a slightly shorter activation time (1 ms for DCF vs. 10 ms for CID). The dependence of the fragment ion yield from angiotensin I [M+3H]3+ (y4, b5, and b8 ions) on the q-value applied during fragmentation is shown in Figure 8. CID results in a q-value of 0.380 for optimum fragment ion yield. The optimum q-value for DCF fragmentation with the applied acceleration energy was 0.320. In general, DCF can be operated at lower q-values, inaccessible to CID (see Fig. 8), without sacrificing fragment ion yield. The use of lower q-values permits observance of low m/z fragment ions. In theory, the use of low m/z ions characteristic of peptides with certain amino acids should increase the probability of peptide identifications in complex samples. These ions should be useful with database searching algorithms to optimize peptide identification rates, as suggested by McAllister et al[31].
Figure 7.
A comparison of MS/MS spectra acquired from fragmentation of angiotensin I [M+3H]3+ using DCF (black) and CID (blue). Notably, the DCF spectrum contains more a ion series, y1, and histidine immonium ions.
Figure 8.
A plot of the fragment ion yield of angiotensin I [M+3H]3+ as function of applied q-value during DCF and CID fragmentation. The solid traces represent an average of five technical replicates and the dashed traces above and below each solid trace represent ±σ.
Differences between relative fragment ion ratios or fragment ion yields between CID and beam-type CID has long been known. This has presented significant difficulty in the transition between discovery based proteomic measurements on a trapping instrument to quantitative measurements using selected reaction monitoring (SRM) on quadrupole based instruments. We feel that b and y fragment ion intensity obtained with DCF may provide a more reliable selection of transitions when shifting to triple quadrupole SRM based quantitative proteomics(see Supplemental Figures).
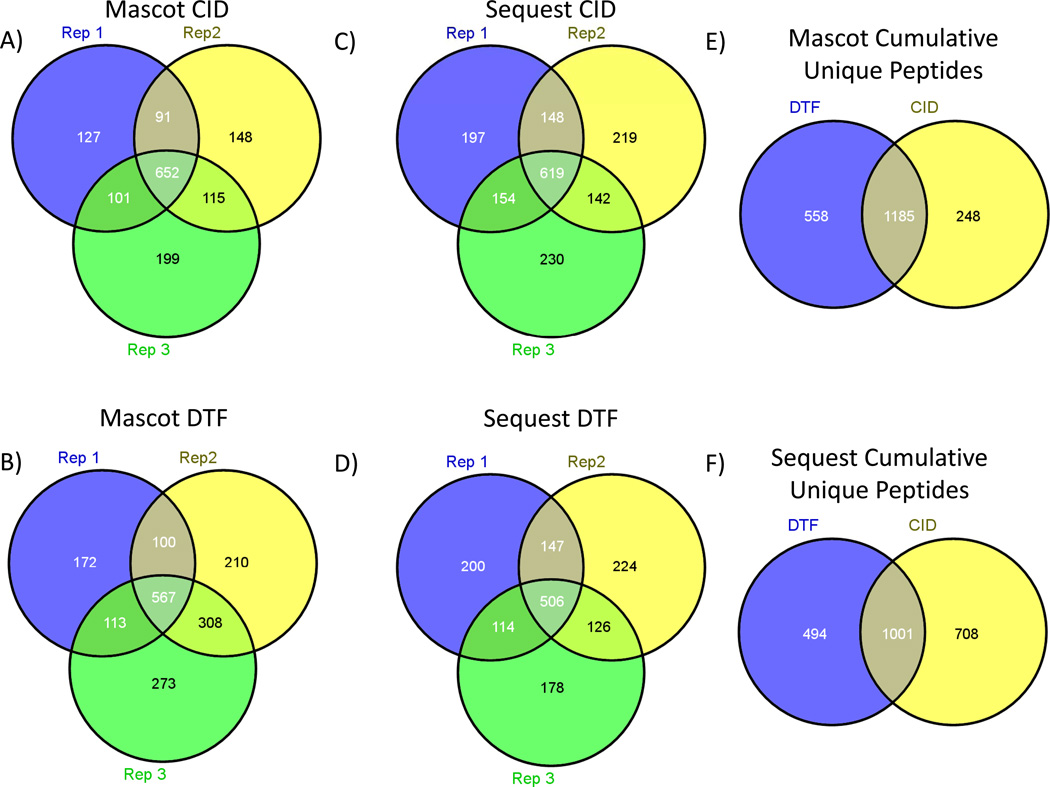
The peptide identification rate for CID vs. DCF was evaluated using a tryptic digest of the soluble fraction of S. Cerviseae. The peptide identifications resulting from technical triplicate analyses using both Mascot[11] and Sequest[12, 13] database search algorithms are reported to show the capability of DCF compared to CID on a full-scale biological sample(Figure 9). No modification or tailoring of the search algorithms was performed in the case of DCF, although exploitation of unique fragmentation characteristics of DCF may improve scores and identification rates[31]. All numbers reported reflect identifications made at <5% estimated FDR using a reverse database search strategy. Mascot search results produced 1743 unique peptides for DCF, a 22% increase relative to the 1433 unique peptides found for CID. Sequest search results produce 1495 unique peptides for DCF and 1709 unique peptides for CID. Optimization of each search algorithm parameters was not performed in either case, which may be the reason that CID data outperforms DCF data in the case of Sequest searches. The number of uniquely identified peptides between technical triplicates using the Sequest pipeline for each fragmentation type was found to be 215 +/− 14 peptides using CID and 201 +/− 19 peptides using DCF. In addition, these two means were not found to be significantly different by Student’s t-test. These results indicate that the reproducibility of peptide identifications using both CID and DCF are not statistically different, even when processed through two separate database searching algorithms. However, after a significant number of technical replicates unique peptides are found with both DCF and CID, indicating that this technique may provide complementary peptide identifications not accessible to CID methods alone.
Figure 9.
A+B) Venn diagram of technical triplicate LC-MS/MS Mascot uniquely identified peptides for CID and DTF respectively (>=5% FDR). C+D) Venn diagram of technical triplicate LC-MS/MS Sequest uniquely identified peptides for CID and DTF respectively (>=5 % FDR). E) Venn diagram of total unique peptides identified in CID and DTF using Mascot. F) Venn diagram of total unique peptides identified in CID and DTF using Sequest.
Conclusions
Here we described a dual cell linear ion trap Fourier transform ion cyclotron resonance mass spectrometer, which we call the Velos-FT. This instrument is a viable upgrade option for those labs with LTQ-FT instruments. Performance improvement from the LTQ-FT Ultra to the Velos-FT are comparable to the improvements observed between the LTQ-Orbitrap and the Velos-Orbitrap. The key technological advancements which increase performance are the radio frequency stacked ring ion guide and the high pressure dual cell linear ion trap. The stacked ring ion guide provides a 5–10 fold increase in ion transmission efficiency in the source, the HPC provides more efficient ion accumulation and fragmentation, and the LPC allows for faster scan rates. The overall data dependent repetition rate increases from about 4 Hz for the LTQ-FT Ultra to 10 Hz for the Velos-FT. The increased scan speed and accumulation efficiency directly result in an increased number of identifications. The observed increase in identifications achieved using the Velos-FT over the LTQ-FT Ultra is ~100% when considering unique peptide sequences. When protein group identifications are considered ~50% more increase in identifications was observed. This instrument is well suited to bottom-up proteomics; however, we show that accumulation and fragmentation of intact proteins with the Velos-FT is highly efficient and should enable acquisition of higher quality on-line top-down proteomic measurements. This instrument configuration allows the execution of DCF experiments, providing a significantly reduced low m/z cut-off. Also, a unique subset of identified peptides are obtained using DCF when compared directly to CID.
Supplementary Material
Acknowledgments
This work is supported in part by National Institutes of Health grants 7S10RR025107, 5R01GM086688, 5R01RR023334 and the University of Washington's Proteomics Resource (UWPR95794). The authors also would like to thank Dr. Priska D. von Haller and Jimmy K. Eng for helpful discussion.
Bibliography
- 1.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science (New York, N.Y.) 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 2.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu Q, Noll RJ, Li H, Makarov A, Hardman M, Graham Cooks R. The Orbitrap: a new mass spectrometer. Journal of mass spectrometry : JMS. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 4.Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, Wouters ER, Senko M, Makarov A, Mann M, Horning S. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Molecular & cellular proteomics : MCP. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Second TP, Blethrow JD, Schwartz JC, Merrihew GE, MacCoss MJ, Swaney DL, Russell JD, Coon JJ, Zabrouskov V. Dual-pressure linear ion trap mass spectrometer improving the analysis of complex protein mixtures. Analytical chemistry. 2009;81:7757–7765. doi: 10.1021/ac901278y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comisarow MB, Marshall AG. Fourier transform ion cyclotron resonance spectroscopy. Chemical Physics Letters. 1974;25:282–283. [Google Scholar]
- 7.Wilm M, Neubauer G, Mann M. Parent ion scans of unseparated peptide mixtures. Analytical chemistry. 1996;68:527–533. doi: 10.1021/ac950875+. [DOI] [PubMed] [Google Scholar]
- 8.Yates JR., 3rd Mass spectrometry. From genomics to proteomics. Trends in genetics. 2000;16:5–8. doi: 10.1016/s0168-9525(99)01879-x. [DOI] [PubMed] [Google Scholar]
- 9.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nature biotechnology. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 10.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 11.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 12.Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 13.Eng JK, Fischer B, Grossmann J, Maccoss MJ. A fast SEQUEST cross correlation algorithm. Journal of proteome research. 2008;7:4598–4602. doi: 10.1021/pr800420s. [DOI] [PubMed] [Google Scholar]
- 14.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nature biotechnology. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 16.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science (New York, N.Y.) 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 17.Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. Top Down versus Bottom Up Protein Characterization by Tandem High-Resolution Mass Spectrometry. Journal of the American Society for Mass Spectrometry. 1999;121:806–812. [Google Scholar]
- 18.Bruce JE, Anderson GA, Hofstadler SA, Van Orden SL, Sherman MS, Rockwood AL, Smith RD. Selected-ion accumulation from an external electrospray ionization source with a Fourier-transform ion cyclotron resonance mass spectrometer. Rapid communications in mass spectrometry. 1993;7:914–919. [Google Scholar]
- 19.Mitchell DW, Smith RD. Prediction of a space charge induced upper molecular mass limit towards achieving unit mass resolution in Fourier transform ion cyclotron resonance mass spectrometry. Journal of Mass Spectrometry. 1996;31:771–790. [Google Scholar]
- 20.Haller I, Mirza UA, Chait BT. Collision induced decomposition of peptides. Choice of collision parameters. Journal of the American Society for Mass Spectrometry. 1996;7:677–681. doi: 10.1016/1044-0305(96)85613-3. [DOI] [PubMed] [Google Scholar]
- 21.Hurley SS, Splitter GA, Welch RA. Rapid lysis technique for mycobacterial species. Journal of clinical microbiology. 1987;25:2227–2229. doi: 10.1128/jcm.25.11.2227-2229.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoopmann MR, Finney GL, MacCoss MJ. High-speed data reduction, feature detection, and MS/MS spectrum quality assessment of shotgun proteomics data sets using high-resolution mass spectrometry. Analytical chemistry. 2007;79:5620–5632. doi: 10.1021/ac0700833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Catherman A, Ahlf D, Li M, Tran JC, Berg A, Valaskovic G, Kelleher NL. Top Down Characterization of Integral Membrane Proteins. 59th ASMS Conference on Mass Spectrometry and Allied Topics; JASMS, Denver, CO. 2011. [Google Scholar]
- 24.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 25.Kelly RT, Tolmachev AV, Page JS, Tang K, Smith RD. The ion funnel: theory, implementations, and applications. Mass spectrometry reviews. 29:294. doi: 10.1002/mas.20232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han X, Jin M, Breuker K, McLafferty FW. Extending top-down mass spectrometry to proteins with masses greater than 200 kilodaltons. Science (New York, N.Y.) 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 27.McLafferty FW, Breuker K, Jin M, Han X, Infusini G, Jiang H, Kong X, Begley TP. Top-down MS, a powerful complement to the high capabilities of proteolysis proteomics. The FEBS journal. 2007;274:6256–6268. doi: 10.1111/j.1742-4658.2007.06147.x. [DOI] [PubMed] [Google Scholar]
- 28.Zabrouskov V, Giacomelli L, van Wijk KJ, McLafferty FW. A new approach for plant proteomics: characterization of chloroplast proteins of Arabidopsis thaliana by top-down mass spectrometry. Molecular & cellular proteomics : MCP. 2003;2:1253–1260. doi: 10.1074/mcp.M300069-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Kelleher NL. Top-down proteomics. Analytical chemistry. 2004;76:197A–203A. [PubMed] [Google Scholar]
- 30.Patrie SM, Charlebois JP, Whipple D, Kelleher NL, Hendrickson CL, Quinn JP, Marshall AG, Mukhopadhyay B. Construction of a hybrid quadrupole/Fourier transform ion cyclotron resonance mass spectrometer for versatile MS/MS above 10 kDa. Journal of the American Society for Mass Spectrometry. 2004;15:1099–1108. doi: 10.1016/j.jasms.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 31.McAlister GC, Phanstiel DH, Westphall MS, Coon JJ. Higher-energy collision-activated dissociation without a dedicated collision cell. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.O111.009456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.