Abstract
Background
Chronic physical aggression (CPA) is characterized by frequent use of physical aggression from early childhood to adolescence. Observed in approximately 5% of males, CPA is associated with early childhood adverse environments and long-term negative consequences. Alterations in DNA methylation, a covalent modification of DNA that regulates genome function, have been associated with early childhood adversity.
Aims
To test the hypothesis that a trajectory of chronic physical aggression during childhood is associated with a distinct DNA methylation profile during adulthood.
Methods
We analyzed genome-wide promoter DNA methylation profiles of T cells from two groups of adult males assessed annually for frequency of physical aggression between 6 and 15 years of age: a group with CPA and a control group. Methylation profiles covering the promoter regions of 20 000 genes and 400 microRNAs were generated using MeDIP followed by hybridization to microarrays.
Results
In total, 448 distinct gene promoters were differentially methylated in CPA. Functionally, many of these genes have previously been shown to play a role in aggression and were enriched in biological pathways affected by behavior. Their locations in the genome tended to form clusters spanning millions of bases in the genome.
Conclusions
This study provides evidence of clustered and genome-wide variation in promoter DNA methylation in young adults that associates with a history of chronic physical aggression from 6 to 15 years of age. However, longitudinal studies of methylation during early childhood will be necessary to determine if and how this methylation variation in T cells DNA plays a role in early development of chronic physical aggression.
Introduction
Longitudinal studies with birth cohorts have shown that children start to use physical aggression by the end of the first year after birth and frequency peaks between 2–4 years of age [1]–[4]. Longitudinal studies with school age children have found that the frequency of physical aggression decreases for the majority of children between 5 and 15 years of age [5]. However, a minority of children (4–7%) maintain a high frequency of physical aggression from childhood to adolescence [4]–[6]. Males on this chronic physical aggression (CPA) trajectory tend to grow-up in adverse family environments [4], [7]–[9], have lower cognitive abilities [10], tend to be rejected by their peers from early childhood onwards [11] and have numerous physical, mental and social problems such as accidents, hyperactivity, school failure, substance abuse and unemployment [4], [5], [10], [12]–[14].
Twin studies suggest that frequency of childhood physical aggression has a substantial inherited component [15]–[19]. At the molecular level, several polymorphisms were found to be associated with aggressive behavior in humans and animals [20]. Moreover, genetic and environmental factors have been shown to interact in the expression of impulsive aggression in monkeys [15] and violence in humans [21], [22]. However, very little work has been done to identify the mechanisms that might be responsible for these links. We hypothesize that DNA methylation is one such mechanism [4], [23].
It is now well-established that DNA sequence is complemented by epigenetic information including DNA methylation and histone modifications to program gene expression [24]. There is a growing body of evidence suggesting that in addition to the innate endogenous processes sculpting the DNA methylation pattern during gestation, the DNA methylation pattern is responsive to external environmental exposures including the social environment during both intra-uterine development and after birth [25] in animals [26]–[34] and in humans [35]–[37]. For example, early nurturing experiences influence epigenetic programming of the glucocorticoid receptor gene promoter in the hippocampus of rats [31] and humans [37].
DNA methylation patterns are tissue specific and it is therefore anticipated that changes relevant to behavior will only be detected in the brain [38]. Importantly however, DNA methylation alterations associated with social exposures are not restricted to the brain. Previous studies have shown associations between white blood cell (WBC) DNA methylation and various environmental exposures [39]–[42]. Borghol et al., have recently described association between early life socioeconomic position and DNA methylation signatures in adult WBC DNA [43]. Methylation of the ADCYAP1R1 gene in peripheral blood DNA was found to be associated with post-traumatic stress disorder (PTSD) [44] and methylation of FKBP5 in lymphocytes was associated with both genetic risk for PTSD and early life adversity [45]. Importantly, we have recently shown that differential DNA methylation of the serotonin transporter gene promoter (SLC6A4) in T cells and monocytes is associated with in vivo measures of human brain serotonin synthesis and childhood limited physical aggression in men [46]. Moreover, we have shown that young adult males on a chronic physical aggressive (CPA) trajectory between age 6 and 15 years had differential DNA methylated regions located in the genomic loci of cytokines and related transcription factors in T cells and monocytes, compared to males with the same background who did not follow such a high aggression trajectory (control group) [47], [48].
In the study presented here, we tested the hypothesis that a trajectory of chronic physical aggression during childhood would be associated with a distinct DNA methylation profile during adulthood. We compared blood CD3+ T cells genome-wide promoter methylation profiles of two groups: adult males who had been shown to be on a CPA trajectory between 6 and 15 years of age and males with the same background who followed a normal physical aggression trajectory (control group) [9], [12].
Materials and Methods
Participants
The subjects were recruited from participants in two longitudinal studies of child development [9], [49]. We recruited two groups of Caucasian men who were born in families with a low socioeconomic status and were living at the time of the present study within 200 km from our laboratory. The first group had a history of high physical aggression from age 6 to 15 years (chronic physical aggression group, CPA). The second group was recruited from the same longitudinal studies but included only those who did not have a history of high physical aggression from age 6 to 15 (Control group). A total of 65 eligible subjects agreed to participate (8 CPA and 57 controls). All of the 8 CPA subjects were included in the study and for budgetary reasons we reduced the control group to 12. In addition to physical aggression, other behavioral problems, such as hyperactivity, were also rated from age 6 to 15 and violence at 21 years of age. Characteristics of the 2 groups are presented in Table 1 (Additional information on the characteristics of the group can be found in File S1).
Table 1. Characteristics of the chronic physical aggression (CPA) group and control group.
| Variables | Control | CPA | Group | ||
| Mean ± SD or % (n) | |||||
| Age at blood drawn | 25.4±2.71 | (12) | 25.8±2.87 | (8) | t(18) = −0.26, P = 0.80 |
Familial adversity score
|
0.34±0.29 | (12) | 0.51±0.41 | (7) | t(17) = −1.06, P = 0.31 |
| Psychiatric record (21 years old) | 60% | (6/10) | 43% | (3/7) | F exact, 2 tailled: 0.64 |
| Criminal record (21 years old) | 17% | (2/12) | 75% | (6/8) | F exact, 2 tailled: 0.019 |
| Self-reported violence (21 years) | 10% | (1/10) | 57% | (4/7) | F exact, 2 tailled: 0.10 |
| Attention deficit score (6 to 15 years) | 3.23±2.18 | (12) | 4.00±1.89 | (8) | t(18) = −0.81, P = 0.43 |
| Hyperactivity trajectories (6 to 15 years) | 17% | (3/12) | 50% | (4/8) | F exact, 2 tailled: 0.14 |
| Opposition trajectories (6 to 15 years) | 0% | (0/12) | 75% | (6/8) | F exact, 2 tailled: 0.001 |
| Anxiety trajectories (6 to 15 years) | 8% | (1/12) | 13% | (1/8) | F exact, 2 tailled: 0.65 |

include mother and father occupational score, familial status (monoparental vs biparental), mother and father at birth of first child and the years of schooling of the mother and father.
Ethics Statement
After a complete description of the study to the subjects, all participants provided written informed consent. The study was carried out in accordance with the Declaration of Helsinki, and was approved by the research ethics committee of the University of Montreal pediatric hospital (St-Justine Hospital).
DNA methylation analysis
DNA was extracted with Wizard Genomic DNA Purification kit (Promega) from CD3+ T cells isolated from PBMC (whole mononuclear cells from peripheral blood) using CD3 dynabeads (Dynal) following the protocol from Current Protocols in Immunology (1997, sections 7.1 and 7.5.1–7.5.11). A detailed description of the methods and analyses of methylated DNA immunoprecipitation (MeDIP) and microarrays hybridization used in this study were previously described [43] and can be found in File S1. We mapped the methylation state of the promoters of nearly 20 000 genes and 400 microRNAs in triplicate using custom designed 244 K promoter tiling arrays (Agilent technologies) containing probes selected to tile ∼1000 bp upstream to ∼250 bp downstream of the transcription start site.
Microarray analysis
Differential methylation between groups of samples was determined at the probe and promoter levels to ensure both statistical significance and biological relevance as previously described [43] and can be found in File S1. At the probe level, a modified t-statistic was computed for each probe corresponding to probe log-ratio differences between CPA and control groups using the ‘limma’ package [50] of Bioconductor [51]. Then, promoter-level methylation differences were identified as those promoters significantly enriched with probes having positive or negative t-statistics using the Wilcoxon rank-sum test. A probe and the containing promoter were called differentially methylated if the p-value of the probe t-statistic was at most 0.05 (uncorrected for multiple testing), log2-fold change between the groups was at least 0.25, and the false discovery rates (FDR) of the promoter-level statistic was at most 0.2.
The ‘false discovery rate’ (FDR) is used throughout the text to judge the statistical strength of our results. FDR is computed using the Benjamini-Hochberg algorithm, which is designed to control for errors due to multiple statistical tests. Our use of a false discovery rate of 0.2 as a threshold for calling differential methylation implies that we expect at most 20% of our calls to be erroneous. This approach and the threshold of 0.2 allows us to test our hypothesis that T cell DNA methylation is associated with aggression trajectory and to generate further hypotheses about the functions of genes affected by the associated methylation changes. All functional analysis was done using Ingenuity Pathway Analysis with the default parameters as it was shown to be a reliable approach to identify biological pathways that influence disease outcomes [52].
The microarray data are available at http://www.ncbi.nlm.nih.gov/geo under the accession number GSE50674.
Microarray validation
Differentially methylated regions from the microarray analysis with varying p values (between 0.0001 and 0.03 with FDR<0.2) and fold differences (Log2 CPA/Controls between 1.1 and 0.5) at the probe level were selected for validation. Two different techniques were applied on the same subjects used for microarray experiments to validate the regions called differentially methylated from the microarray analysis (n = 8 CPA and n = 12 Controls); quantitative real-time PCR on the immunoprecipitated DNA samples (Q-MeDIP) and pyrosequencing of bisulfite treated DNA (see File S1 for a more detailed description and Table S4 for the list of primers used). Moreover, using a Bayesian deconvolution method to estimate methylation levels from the data [53], we found promoter methylation levels to be significantly inversely correlated (p value = 2.3e-25) with previously published gene expression levels of T cells from human samples (Figure S1). More than 60% of the genes analyzed by the microarray have inversely correlated promoter DNA methylation and expression levels (defined as gene promoters with methylation above 50% and expression below the 50-percentile of expression levels, and vice versa).
Rationale for the MeDIP microarray hybridization approach
MeDIP was selected among many other methods for methylation profiling because it is one of the few methods that is feasible for studying genome-wide methylation differences between groups of subjects. It has been successfully applied in many published studies [53]–[73], and it has been found competitive with the other high-throughput profiling methods that are in use [74]–[80]. Further information about MeDIP and supporting statistics from this study can be found in File S1.
Results
T cell promoter methylation associate with chronic physical aggression
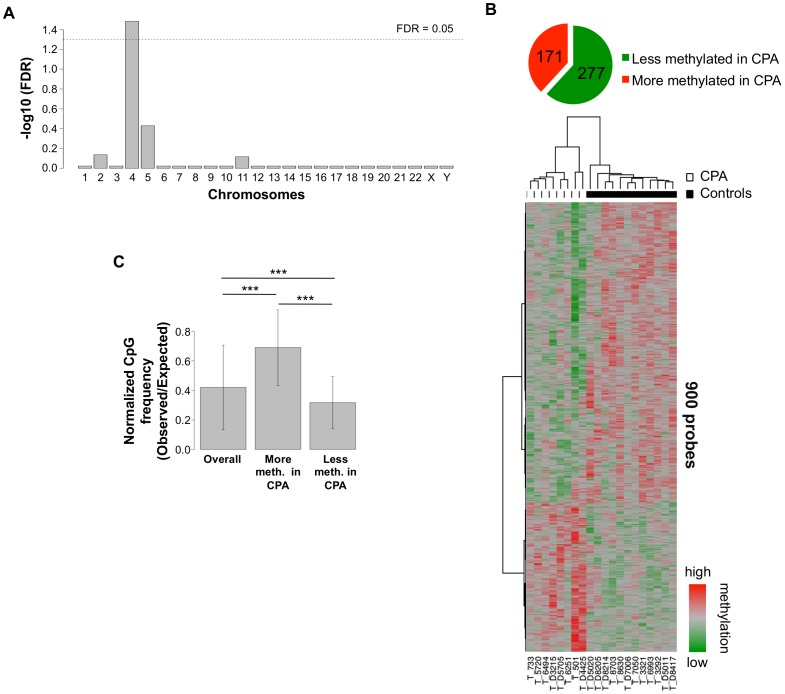
The methylation levels of hundreds of promoters scattered across the genome were found to be associated with CPA with statistically significant overrepresentations of associations in chromosomes 4 and 5 (p ≤ 0.01; hypergeometric, Figure 1A). In all, we found 900 probes from 448 distinct gene promoters whose normalized intensities were significantly associated with chronic aggressive behavior (FDR<0.2, Spreadsheet 1). Of these promoters, 277 were more methylated in the control group, and 171 were more methylated in the CPA group. This differentially methylated list of promoters includes 2 microRNA promoters that are more methylated in the CPA group and 10 microRNA promoters that are less methylated in the CPA group (out of 400 included on the microarray). A heatmap of the probes in each of these gene promoters that best differentiate between the groups is shown in Figure 1B. When applying a more stringent criteria to our list of differentially methylated promoters, such as FDR<0.05, 184 probes located within 60 gene promoters remained significantly associated with CPA (Spreadsheet S1). We have further confirmed methylation differences at several sites using two different techniques, Q-MeDIP (Figures S2A and S2B) and pyrosequencing (Figure S2C) as described in the Methods section.
Figure 1. Gene promoters differentially methylated between CPA (n = 8) and controls (n = 12) in T cells.
A. Bar heights indicate the degree to which each chromosome contains an unexpectedly high number of differentially methylated promoters. This was calculated by Fisher's exact test followed by adjustment for multiple testing by converting p-values to false discovery rates. Bar heights are -log10 values of the false discovery rates, so bars higher than the dashed line have false discovery rates below 0.05. B. Heatmap depicts normalized intensities of microarray probes contained in promoters (at most one per promoter) that best differentiate between CPA and control groups. Rows correspond to promoters and columns to subjects. Red indicates higher methylation in a row and green indicates lower methylation. The 900 differentially methylated probes represent 448 gene promoters where 171 are more methylated in CPA and 277 less methylated. C. CpG density of the differentially methylated promoters. Gene promoters more methylated in CPA have higher CpG density than the average CpG content of all promoters analyzed (p<6.3E-27). In contrast, promoters less methylated in CPA have lower CpG density than the overall average (p<0.00014). Gene promoters more methylated in CPA have also higher CpG density than the promoters less methylated in CPA (p<1.3E-33; Wilcoxon rank-sum test). Normalized CpG density is the density of CpG sites divided by the expected density calculated by multiplying the density of C sites by the density of G sites.
Functional relevance of chronic physical aggression associated with T cells methylation
If changes in methylation play a regulatory role in T cells, they should appear in the regulatory regions of genes that are actively expressed in T cells. Using a publicly available gene expression dataset of 79 different cell types, we found that most of the differentially methylated genes are indeed expressed in T cells (see File S1). Interestingly, the list of genes that are differentially methylated in CPA includes five genes that were previously linked with aggression (Table 2). AVPR1A, HTR1D and GRM5 are less methylated in the CPA group and DRD1 and SLC6A3 genes are more methylated in the CPA group.
Table 2. Differentially methylated genes previously shown to be associated with aggressive behavior.
| Gene ID | P value | FDR | Methylation | Association | Species | Type of aggression | Effects | References |
| GRM5 | 0.00001 | 0.01 | less in CPA | Receptor activity (antagonist) | Mouse | Offensive aggression | Suppressed | Navarro et al., 2006 |
| DRD1 | 0.005 | 0.08 | more in CPA | Genetic (SNP) | Dog | Human-directed canine aggression | Increased | Vage et al., 2010 |
| Genetic (SNP) | Human | Physical aggression | Increased | Sweet et al., 1998 | ||||
| AVPR1A | 0.03 | 0.05 | less in CPA | Receptor activity (antagonist) | Hamster | Offensive aggression | Suppressed | Ferris et al., 2006; Ferris and Potegal, 1988 |
| Genetic (Microsatellite) | Prairie vole | Offensive aggression | Increased | Hammock et al., 2005 | ||||
| HTR1D | 0.04 | 0.05 | less in CPA | Genetic (SNP) | Dog | Human-directed canine aggression | Increased | Vage et al., 2010 |
| SLC6A3 | 0.04 | 0.06 | more in CPA | Genetic (VNTR) | Human (adolescents and young adults) | Violent delinquency | Increased | Guo et al., 2007 |
| Genetic (VNTR) | Human (adolescents) | Chronic criminal and dangerous behavior | Increased | Vaughn et al., 2009 |
We also used the Ingenuity Pathway Analysis software to determine which, if any, gene functions were significantly enriched with genes whose differential promoter methylation levels were associated with chronic aggression. The most enriched functional categories with genes whose promoters were less methylated in the CPA group included behaviour (hyperactivity), metabolic diseases (adiposity) and inflammatory response (chemotaxis of phagocytes). In contrast, the most enriched functional categories with genes whose promoters were more methylated in the CPA group included neurological diseases (encephalopathy), cellular growth and proliferation (cancer and blood cells) and gene expression (Table S1). Specific canonical pathways enriched with such genes included PPAR signalling, cytokine signaling between immune cells and G-protein coupled receptor signalling (Table S2). Furthermore, IPA allows testing for significant enrichment of targets of specific transcription factors. Thirty-four transcription factors were identified to have a significant overlap with our list of affected genes with STAT6, SWI-SNF and FOXH1 being at the top of the list (Table S3). STAT6 is part of the STAT family of transcription factors, its phosphorylation in response to cytokines and growth factors activates the transcription of many genes involved in the immune system such as interleukin 4 (IL-4). Previous studies have shown that STAT6-deficient mice are more hyperactive and have lower levels of midbrain dopamine transporter [77] suggesting that STAT6 is involved in behavior [81].
Differentially methylated promoters in CPA have distinct CpG densities
CpG islands are small regions with unusually high CpG densities that are typically unmethylated [82], particularly when found near the transcription start sites of genes [82]. We observed extremely high CpG densities in promoters with higher methylation in the CPA group in contrast to unusually low CpG densities in promoters with lower methylation in the CPA group (Figure 1C). We note that it was previously shown that genes with high CpG densities in their promoters tend to be involved in housekeeping activities in the cell [83].
Methylation associated with chronic physical aggression clusters by genomic location
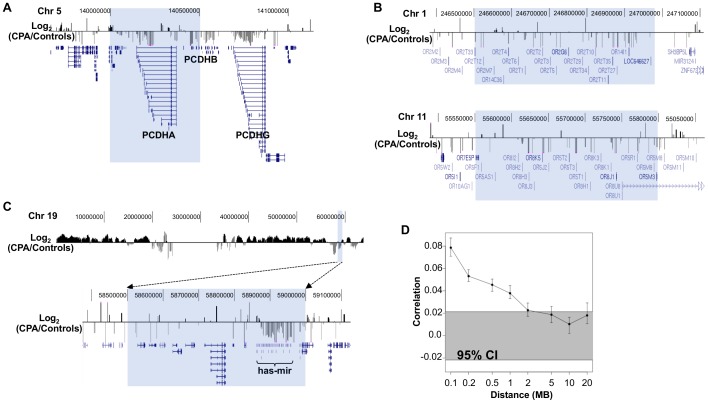
In spite of the fact that differences in DNA methylation associated with aggression groups were distributed uniformly across the chromosomes, with higher density in two chromosomes (4 and 5), probes with similar differences tended to appear in clusters within chromosomes. To measure the strength of this clustering, we partitioned the genome into 500 Kb regions and asked whether each region contained a surprisingly high number of differentially methylated probes. Of the ∼6 K regions, we found that 6 were significantly enriched for decreased methylation in the CPA group (Table 3) even after adjustment for multiple testing. Four of these six regions are located in known gene clusters: the protocadherin alpha cluster, two olfactory receptor clusters and a recently identified human microRNA cluster (Figure 2). From the 718 microRNAs that composed the cluster on chromosome 19, at least 148 (21%) are expressed in T cells according to smirnaDB, a database of microRNA expression profiles [84]. Protocadherins are a superfamily of genes that encode proteins principally involved in cell adhesion. Interestingly, differential methylation of the protocadherin cluster was also found to be associated with childhood socioeconomic position in whole blood [85] and with differential maternal care early in life in hippocampi of rats [33]. In general, methylation differences also tended to cluster across the entire genome. Beyond specific 500 Kb partitions containing clusters of differential methylation, we found that methylation differences as far apart as 2 Mb displayed a small but significant level of interdependence. The statistics are illustrated in Figure 2D and further details can be found in File S1.
Table 3. List of chromosomal regions differentially methylated between CPA and control groups.
| Regions | Locations | FDR | Gene cluster | Gene promoters more or less methylated in CPA |
| 1 | Chr1: 246500001–247000001 | 4.75E-5 | Olfactory receptors | OR2T10 |
| 2 | Chr19: 58500001–59000001 | 2.24E-4 | Has-mir | hsa-mir-517B, hsa-mir-520D, hsa-mir-520G |
| 3 | Chr11: 55500001–56000001 | 1.97E-2 | Olfactory receptors | OR8J1 |
| 4 | Chr12: 10000001–10500001 | 0.10 | KLRC3 | |
| 5 | Chr5: 140000001–140500001 | 0.11 | Protocadherins | PCDHA5, PCDHA10, PCDHA12, PCDHB2 |
| 6 | Chr3: 50000001–50500001 | 0.17 |
Figure 2. Megabase co-clustering of differential methylation between CPA (n = 8) and controls (n = 12).
A. Co-clustering of differential methylation among the protocadherins genes. Positive values (black bars) indicate increased methylation in CPA compared to controls and negative values (grey bars) indicate the opposite. Shaded in blue is a 500 Kb region containing protocadherins family A and B whose promoters are consistently less methylated in CPA than in controls (scale log2 fold differences: −0.2 to 0.2). B. Co-clustering of differentially methylated promoters with common function across megabases of DNA. The olfactory receptor clusters located on chromosome 1 and chromosome 11 are less methylated in CPA compared to controls (scale log2 fold differences: −0.2 to 0.2). C. On chromosome 19, one of the few megabase regions showing decreased methylation in CPA compared to controls contains one of the two human micro-RNA clusters (scale log2 fold differences top and bottom panels: −0.2 to 0.2). D. Methylation dependences across megabases are shown. Pearson correlations of DNA methylation differences between controls and CPA groups at various genomic distances are shown. Error bars show 95% confidence intervals for the correlation values. The grey highlight shows the expected 95% confidence interval if there is no correlation between methylation differences at different genomic sites. This confidence interval does not overlap with the error bars associated with distances less than 2 Mb suggesting the existence of systematic dependencies between methylation differences at distances up to 2 Mb.
Promoter methylation associated with men CPA is also observed in women CPA
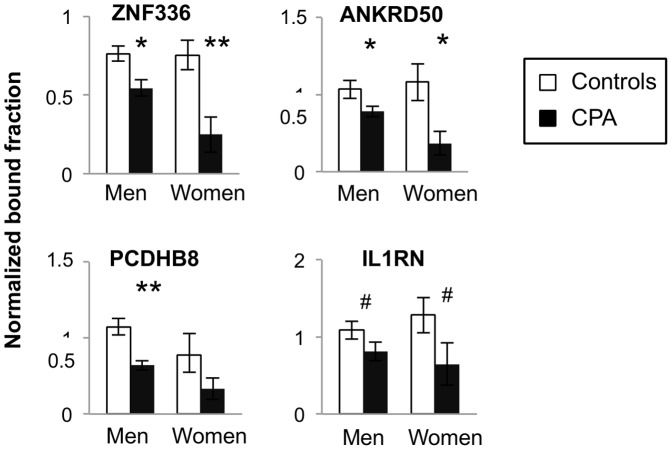
In order to determine whether the methylation differences associated with CPA in men are seen in a different human sample, we used preliminary data from an ongoing study in the laboratory in women selected from the same longitudinal study as the one used here for the aggressive men study. We confirmed by qPCR of the MeDIP DNA that four promoters that are differentially methylated between CPA and control groups in T cells DNA in men are also differentially methylated in women. The Q-MeDIP analysis showed that these promoters were less methylated in the CPA group in men and in women (Figure 3). The validation of these differentially methylated promoters in a distinct human sample, and of a different sex, is consistent with the hypothesis that chronic physical aggression is associated with differential methylation in these promoters.
Figure 3. Q-MeDIP analysis in women T cells of four promoters that are differentially methylated between CPA and control groups in men.
Q-MeDIP analysis of DNA methylation differences between CPA (black) and control (white) groups in men and women T cells samples for four gene promoters predicted to be more methylated in the men CPA group by microarray analysis. Relative bound fraction concentrations obtained in triplicate by Q-MeDIP are shown (see methods). All error bars represent standard error of the mean (SEM). The P value obtained from Mann-Whitney U test is represent by #≤0.1, *≤0.05 and **≤0.01.
Discussion
This study provides evidence of clustered and genome-wide variation in promoter DNA methylation in young adults that associates with a history of chronic physical aggression from 6 to 15 years of age. Probes with similar differences in DNA methylation between the compared groups appear in clusters within chromosomes. Indeed, we found six 500 Kb regions significantly enriched for decreased methylation in the CPA group compared to the control group (Table 3). Four of these regions are well defined gene clusters containing the olfactory receptor and the protocadherins as well as a newly-defined human microRNAs cluster previously shown to be regulated by DNA methylation in cancer cells [86].
Consistent with the fact that these differences are associated with aggression, we validated four of these differentially methylated promoters in a distinct human sample and of a different sex. Moreover, we found 5 genes amongst the 448 differentially methylated promoters that were previously shown to be involved in physical aggression in animals and in humans (Table 2). First, we found that AVPR1A promoter is less methylated in the CPA group, which is consistent with higher activity. Indeed, blocking AVPRIA with a specific antagonist decreases aggressive behavior in hamsters [87] while AVP the agonist of AVPRIA, enhances aggression in animals and humans [88]. Second, DRD1 and SLC6A3 genes have higher promoter methylation in the CPA group and increased brain dopamine levels are thought to be positively associated with aggression [20], [89]. A genetic association was found in the gene coding for DRD1 with psychosis and aggression in Alzheimer patients [90]. Moreover, a variable number of tandem repeat in the 3′UTR of the SLC6A3 gene is associated with various antisocial behaviors such as violent delinquency [91] and propensity to a criminal career [92]. Third, GRM5 promoter is less methylated in CPA and Navarro et al. showed that 2-methyl-6-(phenylethylnyl)pyridine (MPEP), a selective antagonist of GRM5, has anti-aggressive effects in mice [93]. Elevated glutamatergic activity in the brain has been associated with aggression in animals and human [94]. Finally, the inhibitory autoreceptor HTR1D promoter is less methylated in CPA and lower serotonin levels in the brain and the periphery associate with high aggression in animals and humans [95]. Allelic association with aggression in dogs was observed in the gene coding for the receptor HTR1D [96]. Since genetic associations with aggression were observed for AVPR1A, HTR1D, DRD1 and SLC6A3 (Table 2), it is possible that there is an interaction between genetic variations and differential DNA methylation in aggression. Recent studies have found such interactions in asthma [97], chronic fatigue syndrome [98], trauma [99], and gastric cancer [100]. In our samples, the small number of subjects did not allow for testing of genetic association and their interactions with DNA methylation.
It is noteworthy that although these genes are clearly involved in brain function, we observed changes in DNA methylation associated with aggressive behavior in T cells. We obviously don't know whether similar changes occur in the brain of the same subjects. Nevertheless, these observations are consistent with the notion that T cells will be informative not only on immune specific genes that are associated with the HPA axis but also on some genes that are also involved in brain function. A parallel comparison of DNA methylation changes in prefrontal cortex and T cells in response to differential rearing conditions in rhesus monkeys revealed both tissue specific alterations as well as common differentially methylated regions in T cells and prefrontal cortex [101]. We also reported recently that the DNA methylation state of the SLC6A4 promoter in T cells and monocytes inversely associates with positron emission tomography (PET) measures of brain serotonin synthesis and that both measures associate with aggression in humans [46]. However, only a parallel investigation of T cells and brain like the one that we performed in non-human primates could confirm whether specific DNA methylation changes occur in the brain as well.
As anticipated, since we are working with blood samples, the inflammatory and immune response categories with specific signaling pathway such as cytokines signaling between immune cells, IL-6 and IL-10 signaling were identified in our analysis. Indeed, several lines of evidence suggest that cytokines are associated with animal and human aggression [102]–[104] and IL-6 was causally linked to aggression in mice by gene knockout evidence [105]. Specific cytokines and receptors involved in these pathways were previously shown to be involved in aggression and human mood disorders. IL1R1 and IL1RN have been shown to be involved in defensive aggression through their activation by IL-1β [106], [107]. Moreover, our previous research done on the same men as the one studied here, revealed an association between cytokine expression and methylation with chronic physical aggression during childhood [47], [48]. Further work is needed to investigate the exact role of these molecules in the development of chronic aggression, but taken together these data are consistent with the hypothesis that cytokine regulation could be involved in human behavior and behavioral disorders.
The changes in DNA methylation that we observe between CPA and controls are numerous and significant but each individual effect is small (i.e. per-probe fold change is small). Although it is possible to brush off these subtle changes as biologically irrelevant, their consistency and statistical significance point to the possibility of an important biological role whereby the epigenome is modulated by a combination of small changes across functional pathways and chromosomal regions. It is important to note in this respect that DNA methylation is a binary signal, that is, a site is either methylated or unmethylated in a given cell. Therefore a partial methylation such as is observed in our study indicates that a small but statistically significant subpopulation of cells is differentially methylated. A challenge for future experiments is to identify the cellular populations that exhibit these changes in DNA methylation and to understand their biological role.
Although the present study is the first to show an association between chronic physical aggression, and differential DNA methylation, there are several methodological limitations. First, because DNA was available only in adulthood, we cannot establish when chronic physical aggression became associated with the observed methylation patterns and whether they precede or follow the appearance of the aggressive phenotype. To sort out the sequence of events with human samples we will need longitudinal data on DNA methylation from birth and physical aggression from infancy onwards. Second, we do not know the extent to which our observed association between methylation and aggression carries over into the brain. Third, the study was limited by the lack of good quality RNA since we were unable to have the chronically aggressive subjects come to the lab for their blood draw. Fourth, there was no psychometric-physical evaluation at the time of blood draw. The acute psychological and/or physical status might confound our findings. In this respect, longitudinal data on the T-cell methylomes would have been highly valuable but the original study design didn't include brood draws at multiple time points. Future longitudinal studies that include concurrent blood draws and psychometric-physical evaluations are required to address this question.
Taken together, the findings of differentially methylated genes relevant to CPA and clustering of CPA associated methylation across the genome suggests a well-defined, genome-wide epigenetic pattern associated with chronic physical aggression in humans.
Supporting Information
Distributions of promoter methylation levels by published expression level in T cells. Genes are divided into 20 levels by T cells expression percentiles (0–5, 5–10,…, 95–100) based on publicly available expression data(10). Shown are the distributions of methylation levels for each expression percentile. The distributions show that genes with low or no expression (represented in green) tend to have highly methylated promoters, whereas genes with high expression (represented in red) tend to have lower promoter methylation.
(TIF)
Microarray validation by Q-MeDIP and pyrosequencing of differentially methylated sequences between CPA (n = 8) and control groups (n = 12). A. Fold differences between CPA and control groups obtained by either Q-MeDIP or microarray analysis are shown for 20 genes predicted to be either more methylated (n = 1) or less methylated (n = 19) in the CPA individuals by the microarray analysis. B. Correlation of the fold differences between CPA and control obtained by Q-MeDIP and by microarray for the 20 amplicons analyzed in A. C. DNA methylation differences (%) between the groups in the GRM5, ITGB6, OR13C8 and IL-31 genes validated by pyrosequencing. CpG sites near the significant probe were analyzed. For each gene, the mean methylation per aggressive group per CpG is shown in the bar graph. The rightmost bar indicates the mean methylation levels of all CpG sites analyzed in the region. A map of the sites relative to the transcription start site is shown above the bar graph. Each line represents a CpG site. The location of probes whose fold difference is significantly different between CPA and control groups is identified by a grey square. The region analyzed by pyrosequencing is delimited by the red arrows. All error bars represent standard error of the mean (SEM). The P value obtain from Mann-Whitney U test is represent by *≤0.05, **≤0.01 and ***≤0.001.
(TIF)
Supplementary information file containing additional information of the methods use in the study.
(DOCX)
List of the probes differentially methylated between the chronic physical aggression and the control groups.
(XLSX)
Top list of affected biological functions enriched with genes whose methylation is associated with aggression from Ingenuity Pathway (12) analysis (n = 448 genes). All of the p values were calculated using a right tailed Fisher's exact test and corrected for multiple comparison with the Benjamini-Hochberg method. Significance threshold were p = 0.05.
(DOCX)
Top list of canonical pathways enriched with genes whose methylation is associated with aggression from IPA analysis (n = 448 genes). All of the p values were calculated using a right tailed Fisher's exact test and corrected for multiple comparison with the Benjamini-Hochberg method. Significance threshold were p = 0.05.
(DOCX)
List of transcription regulators showing a significant overlap with genes whose methylation is associated with aggression from IPA analysis (n = 448 genes). Transcription regulators differentially methylated between chronic and normal aggression are shown in bold. Significance threshold were p = 0.05.
(DOCX)
Primers and melting temperature (Tm) used for Q-MeDIP and pyrosequencing.
(DOCX)
Acknowledgments
The authors wish to thank all the participants and the staff of the University of Montreal Research Unit on Children's Psychosocial Maladjustment (GRIP) for their valuable assistance, Dongsha Wang and Marilyne Blain for technical support, and Charles-Édouard Giguère for helping with the data banks.
Funding Statement
This work was supported by a fellowship from the Genes, Environment, and Health Training Program from CIHR to NP; a fellowship from the Quebec Training Network in Perinatal Research (QTNPR) Program from CIHR to CG; grants from the Canadian Institutes of Health Research; the Social Sciences Humanities Research Council of Canada; the Quebec Health Research Fund (FRSQ) and the Quebec Social Science and Culture Fund (FQRSC) to RET and SMC; and grants from the Canadian Institutes of Health Research MOP-42411, the Sackler Program in Psychobiology and Epigenetics at McGill University, and the Canadian Institute for Advanced Research to MS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. NICHD Early Child Care Research Network (2004) Trajectories of physical aggression from toddlerhood to middle childhood: predictors, correlates, and outcomes. Monogr Soc Res Child Dev 69: vii, 1–129. [DOI] [PubMed] [Google Scholar]
- 2. Alink LR, Mesman J, van Zeijl J, Stolk MN, Juffer F, et al. (2006) The early childhood aggression curve: development of physical aggression in 10- to 50-month-old children. Child Dev 77: 954–966. [DOI] [PubMed] [Google Scholar]
- 3. Cote SM, Vaillancourt T, LeBlanc JC, Nagin DS, Tremblay RE (2006) The development of physical aggression from toddlerhood to pre-adolescence: a nation wide longitudinal study of Canadian children. J Abnorm Child Psychol 34: 71–85. [DOI] [PubMed] [Google Scholar]
- 4. Tremblay RE (2010) Developmental origins of disruptive behaviour problems: the ‘original sin’ hypothesis, epigenetics and their consequences for prevention. J Child Psychol Psychiatry 51: 341–367. [DOI] [PubMed] [Google Scholar]
- 5. Broidy LM, Nagin DS, Tremblay RE, Bates JE, Brame B, et al. (2003) Developmental trajectories of childhood disruptive behaviors and adolescent delinquency: a six-site, cross-national study. Dev Psychol 39: 222–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Campbell SB, Spieker S, Burchinal M, Poe MD (2006) Trajectories of aggression from toddlerhood to age 9 predict academic and social functioning through age 12. J Child Psychol Psychiatry 47: 791–800. [DOI] [PubMed] [Google Scholar]
- 7. Campbell SB, Spieker S, Vandergrift N, Belsky J, Burchinal M (2010) Predictors and sequelae of trajectories of physical aggression in school-age boys and girls. Dev Psychopathol 22: 133–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cote SM, Boivin M, Nagin DS, Japel C, Xu Q, et al. (2007) The role of maternal education and nonmaternal care services in the prevention of children's physical aggression problems. Arch Gen Psychiatry 64: 1305–1312. [DOI] [PubMed] [Google Scholar]
- 9. Nagin DS, Tremblay RE (2001) Parental and early childhood predictors of persistent physical aggression in boys from kindergarten to high school. Arch Gen Psychiatry 58: 389–394. [DOI] [PubMed] [Google Scholar]
- 10. Barker ED, Seguin JR, White HR, Bates ME, Lacourse E, et al. (2007) Developmental trajectories of male physical violence and theft: relations to neurocognitive performance. Arch Gen Psychiatry 64: 592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barker ED, Boivin M, Brendgen M, Fontaine N, Arseneault L, et al. (2008) Predictive validity and early predictors of peer-victimization trajectories in preschool. Arch Gen Psychiatry 65: 1185–1192. [DOI] [PubMed] [Google Scholar]
- 12. Nagin D, Tremblay RE (1999) Trajectories of boys' physical aggression, opposition, and hyperactivity on the path to physically violent and nonviolent juvenile delinquency. Child Dev 70: 1181–1196. [DOI] [PubMed] [Google Scholar]
- 13. Seguin JR, Boulerice B, Harden PW, Tremblay RE, Pihl RO (1999) Executive functions and physical aggression after controlling for attention deficit hyperactivity disorder, general memory, and IQ. J Child Psychol Psychiatry 40: 1197–1208. [PubMed] [Google Scholar]
- 14. Kokko K, Pulkkinen L, Huesmann LR, Dubow EF, Boxer P (2009) Intensity of Aggression in Childhood as a Predictor of Different Forms of Adult Aggression: A Two-Country (Finland and United States) Analysis. J Res Adolesc 19: 9–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bennett AJ, Lesch KP, Heils A, Long JC, Lorenz JG, et al. (2002) Early experience and serotonin transporter gene variation interact to influence primate CNS function. Mol Psychiatry 7: 118–122. [DOI] [PubMed] [Google Scholar]
- 16. Brendgen M, Boivin M, Vitaro F, Bukowski WM, Dionne G, et al. (2008) Linkages between children's and their friends' social and physical aggression: evidence for a gene-environment interaction? Child Dev 79: 13–29. [DOI] [PubMed] [Google Scholar]
- 17. Dionne G, Tremblay R, Boivin M, Laplante D, Perusse D (2003) Physical aggression and expressive vocabulary in 19-month-old twins. Dev Psychol 39: 261–273. [DOI] [PubMed] [Google Scholar]
- 18. Hicks BM, Krueger RF, Iacono WG, McGue M, Patrick CJ (2004) Family transmission and heritability of externalizing disorders: a twin-family study. Arch Gen Psychiatry 61: 922–928. [DOI] [PubMed] [Google Scholar]
- 19. van Lier PA, van der Ende J, Koot HM, Verhulst FC (2007) Which better predicts conduct problems? The relationship of trajectories of conduct problems with ODD and ADHD symptoms from childhood into adolescence. J Child Psychol Psychiatry 48: 601–608. [DOI] [PubMed] [Google Scholar]
- 20. Pavlov KA, Chistiakov DA, Chekhonin VP (2011) Genetic determinants of aggression and impulsivity in humans. J Appl Genet [DOI] [PubMed] [Google Scholar]
- 21. Caspi A, McClay J, Moffitt TE, Mill J, Martin J, et al. (2002) Role of genotype in the cycle of violence in maltreated children. Science 297: 851–854. [DOI] [PubMed] [Google Scholar]
- 22. Kim-Cohen J, Caspi A, Taylor A, Williams B, Newcombe R, et al. (2006) MAOA, maltreatment, and gene-environment interaction predicting children's mental health: new evidence and a meta-analysis. Mol Psychiatry 11: 903–913. [DOI] [PubMed] [Google Scholar]
- 23. Tremblay RE, Szyf M (2010) Developmental origins of chronic physical aggression and epigenetics. Epigenomics 2: 495–499. [DOI] [PubMed] [Google Scholar]
- 24. Razin A (1998) CpG methylation, chromatin structure and gene silencing-a three-way connection. EMBO J 17: 4905–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Szyf M (2011) The early life social environment and DNA methylation: DNA methylation mediating the long-term impact of social environments early in life. Epigenetics 6: 971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Waterland RA, Jirtle RL (2003) Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23: 5293–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL (2006) Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect 114: 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jirtle RL, Skinner MK (2007) Environmental epigenomics and disease susceptibility. Nat Rev Genet 8: 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. MacLennan NK, James SJ, Melnyk S, Piroozi A, Jernigan S, et al. (2004) Uteroplacental insufficiency alters DNA methylation, one-carbon metabolism, and histone acetylation in IUGR rats. Physiol Genomics 18: 43–50. [DOI] [PubMed] [Google Scholar]
- 30. Ke X, Lei Q, James SJ, Kelleher SL, Melnyk S, et al. (2006) Uteroplacental insufficiency affects epigenetic determinants of chromatin structure in brains of neonatal and juvenile IUGR rats. Physiol Genomics 25: 16–28. [DOI] [PubMed] [Google Scholar]
- 31. Weaver IC, Diorio J, Seckl JR, Szyf M, Meaney MJ (2004) Early environmental regulation of hippocampal glucocorticoid receptor gene expression: characterization of intracellular mediators and potential genomic target sites. Ann N Y Acad Sci 1024: 182–212. [DOI] [PubMed] [Google Scholar]
- 32. Sinclair KD, Allegrucci C, Singh R, Gardner DS, Sebastian S, et al. (2007) DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc Natl Acad Sci U S A 104: 19351–19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McGowan PO, Suderman M, Sasaki A, Huang TC, Hallett M, et al. (2011) Broad epigenetic signature of maternal care in the brain of adult rats. PLoS One 6: e14739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, et al. (2009) Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci 12: 1559–1566. [DOI] [PubMed] [Google Scholar]
- 35. Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, et al. (2008) Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A 105: 17046–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, et al. (2008) Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics 3: 97–106. [DOI] [PubMed] [Google Scholar]
- 37. McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonte B, et al. (2009) Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci 12: 342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Razin A, Szyf M (1984) DNA methylation patterns. Formation and function. Biochim Biophys Acta 782: 331–342. [DOI] [PubMed] [Google Scholar]
- 39. Kinnally EL, Feinberg C, Kim D, Ferguson K, Leibel R, et al. (2011) DNA methylation as a risk factor in the effects of early life stress. Brain Behav Immun [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, et al. (2011) Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci U S A 107: 9470–9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Uddin M, Koenen KC, Aiello AE, Wildman DE, de los Santos R, et al. (2011) Epigenetic and inflammatory marker profiles associated with depression in a community-based epidemiologic sample. Psychol Med 41: 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smith AK, Conneely KN, Kilaru V, Mercer KB, Weiss TE, et al. (2011) Differential immune system DNA methylation and cytokine regulation in post-traumatic stress disorder. Am J Med Genet B Neuropsychiatr Genet 156B: 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Borghol N, Suderman M, McArdle W, Racine A, Hallett M, et al. (2012) Associations with early-life socio-economic position in adult DNA methylation. Int J Epidemiol 41: 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ressler KJ, Mercer KB, Bradley B, Jovanovic T, Mahan A, et al. (2011) Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature 470: 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, et al. (2013) Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci 16: 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang D, Szyf M, Benkelfat C, Provencal N, Turecki G, et al. (2012) Peripheral SLC6A4 DNA Methylation Is Associated with In Vivo Measures of Human Brain Serotonin Synthesis and Childhood Physical Aggression. PLoS One 7: e39501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Provencal N, Suderman MJ, Caramaschi D, Wang D, Hallett M, et al. (2013) Differential DNA methylation regions in cytokine and transcription factor genomic Loci associate with childhood physical aggression. PLoS One 8: e71691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Provencal N, Suderman MJ, Vitaro F, Szyf M, Tremblay RE (2013) Childhood chronic physical aggression associates with adult cytokine levels in plasma. PLoS One 8: e69481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pingault JB, Tremblay RE, Vitaro F, Carbonneau R, Genolini C, et al. (2011) Childhood Trajectories of Inattention and Hyperactivity and Prediction of Educational Attainment in Early Adulthood: A 16-Year Longitudinal Population-Based Study. Am J Psychiatry [DOI] [PubMed] [Google Scholar]
- 50.Smyth GK (2005) Limma: linear models for microarray data. In: R. Gentleman VC, S Dudoit, R Irizarry, W Huber editor. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer,. pp. 397–420. [Google Scholar]
- 51. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ngwa JS, Manning AK, Grimsby JL, Lu C, Zhuang WV, et al. (2011) Pathway analysis following association study. BMC Proc 5 Suppl 9: S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, et al. (2008) A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol 26: 779–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bell CG, Finer S, Lindgren CM, Wilson GA, Rakyan VK, et al. (2010) Integrated genetic and epigenetic analysis identifies haplotype-specific methylation in the FTO type 2 diabetes and obesity susceptibility locus. PLoS One 5: e14040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cheung HH, Lee TL, Davis AJ, Taft DH, Rennert OM, et al. (2010) Genome-wide DNA methylation profiling reveals novel epigenetically regulated genes and non-coding RNAs in human testicular cancer. Br J Cancer 102: 419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Flanagan JM, Cocciardi S, Waddell N, Johnstone CN, Marsh A, et al. (2010) DNA methylome of familial breast cancer identifies distinct profiles defined by mutation status. Am J Hum Genet 86: 420–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guerrero-Bosagna C, Settles M, Lucker B, Skinner MK (2010) Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gunther T, Grundhoff A (2010) The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog 6: e1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hiura H, Sugawara A, Ogawa H, John RM, Miyauchi N, et al. (2010) A tripartite paternally methylated region within the Gpr1-Zdbf2 imprinted domain on mouse chromosome 1 identified by meDIP-on-chip. Nucleic Acids Res 38: 4929–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lempiainen H, Muller A, Brasa S, Teo SS, Roloff TC, et al. (2011) Phenobarbital mediates an epigenetic switch at the constitutive androstane receptor (CAR) target gene Cyp2b10 in the liver of B6C3F1 mice. PLoS One 6: e18216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Morris MR, Ricketts CJ, Gentle D, McRonald F, Carli N, et al. (2011) Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene 30: 1390–1401. [DOI] [PubMed] [Google Scholar]
- 62. Movassagh M, Choy MK, Goddard M, Bennett MR, Down TA, et al. (2010) Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PLoS One 5: e8564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsui DW, Lam YM, Lee WS, Leung TY, Lau TK, et al. (2010) Systematic identification of placental epigenetic signatures for the noninvasive prenatal detection of Edwards syndrome. PLoS One 5: e15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, et al. (2005) Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37: 853–862. [DOI] [PubMed] [Google Scholar]
- 65. Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, et al. (2006) Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet 38: 149–153. [DOI] [PubMed] [Google Scholar]
- 66. Novak JP, Kim SY, Xu J, Modlich O, Volsky DJ, et al. (2006) Generalization of DNA microarray dispersion properties: microarray equivalent of t-distribution. Biol Direct 1: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang S (2006) An improved nonparametric approach for detecting differentially expressed genes with replicated microarray data. Stat Appl Genet Mol Biol 5: Article30. [DOI] [PubMed] [Google Scholar]
- 68. Cheng AS, Culhane AC, Chan MW, Venkataramu CR, Ehrich M, et al. (2008) Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res 68: 1786–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tomazou EM, Rakyan VK, Lefebvre G, Andrews R, Ellis P, et al. (2008) Generation of a genomic tiling array of the human major histocompatibility complex (MHC) and its application for DNA methylation analysis. BMC Med Genomics 1: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liu Y, Balaraman Y, Wang G, Nephew KP, Zhou FC (2009) Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics 4: 500–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Murphy DM, Buckley PG, Bryan K, Das S, Alcock L, et al. (2009) Global MYCN transcription factor binding analysis in neuroblastoma reveals association with distinct E-box motifs and regions of DNA hypermethylation. PLoS One 4: e8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Takeshima H, Yamashita S, Shimazu T, Niwa T, Ushijima T (2009) The presence of RNA polymerase II, active or stalled, predicts epigenetic fate of promoter CpG islands. Genome Res 19: 1974–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Feber A, Wilson GA, Zhang L, Presneau N, Idowu B, et al. (2011) Comparative methylome analysis of benign and malignant peripheral nerve sheath tumors. Genome Res 21: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jia J, Pekowska A, Jaeger S, Benoukraf T, Ferrier P, et al. (2010) Assessing the efficiency and significance of Methylated DNA Immunoprecipitation (MeDIP) assays in using in vitro methylated genomic DNA. BMC Res Notes 3: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jin SG, Kadam S, Pfeifer GP (2010) Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res 38: e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nair SS, Coolen MW, Stirzaker C, Song JZ, Statham AL, et al. (2011) Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics 6: 34–44. [DOI] [PubMed] [Google Scholar]
- 77. Rajendram R, Ferreira JC, Grafodatskaya D, Choufani S, Chiang T, et al. (2011) Assessment of methylation level prediction accuracy in methyl-DNA immunoprecipitation and sodium bisulfite based microarray platforms. Epigenetics 6: 410–415. [DOI] [PubMed] [Google Scholar]
- 78. Robinson MD, Stirzaker C, Statham AL, Coolen MW, Song JZ, et al. (2010) Evaluation of affinity-based genome-wide DNA methylation data: effects of CpG density, amplification bias, and copy number variation. Genome Res 20: 1719–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yang L, Zhang K, Dai W, He X, Zhao Q, et al. (2011) Systematic evaluation of genome-wide methylated DNA enrichment using a CpG island array. BMC Genomics 12: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, et al. (2008) Comprehensive high-throughput arrays for relative methylation (CHARM). Genome Res 18: 780–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yukawa K, Iso H, Tanaka T, Tsubota Y, Owada-Makabe K, et al. (2005) Down-regulation of dopamine transporter and abnormal behavior in STAT6-deficient mice. Int J Mol Med 15: 819–825. [PubMed] [Google Scholar]
- 82. Bird AP (1986) CpG-rich islands and the function of DNA methylation. Nature 321: 209–213. [DOI] [PubMed] [Google Scholar]
- 83. Gardiner-Garden M, Frommer M (1987) CpG islands in vertebrate genomes. J Mol Biol 196: 261–282. [DOI] [PubMed] [Google Scholar]
- 84. Rossi RL, Rossetti G, Wenandy L, Curti S, Ripamonti A, et al. (2011) Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat Immunol 12: 796–803. [DOI] [PubMed] [Google Scholar]
- 85. Borghol N, Lornage J, Blachere T, Sophie Garret A, Lefevre A (2006) Epigenetic status of the H19 locus in human oocytes following in vitro maturation. Genomics 87: 417–426. [DOI] [PubMed] [Google Scholar]
- 86. Tsai KW, Kao HW, Chen HC, Chen SJ, Lin WC (2009) Epigenetic control of the expression of a primate-specific microRNA cluster in human cancer cells. Epigenetics 4: 587–592. [DOI] [PubMed] [Google Scholar]
- 87. Ferris CF, Lu SF, Messenger T, Guillon CD, Heindel N, et al. (2006) Orally active vasopressin V1a receptor antagonist, SRX251, selectively blocks aggressive behavior. Pharmacol Biochem Behav 83: 169–174. [DOI] [PubMed] [Google Scholar]
- 88. Ferris CF (2005) Vasopressin/oxytocin and aggression. Novartis Found Symp 268: 190–198 discussion 198–200, 242–153. [PubMed] [Google Scholar]
- 89. de Almeida RM, Ferrari PF, Parmigiani S, Miczek KA (2005) Escalated aggressive behavior: dopamine, serotonin and GABA. Eur J Pharmacol 526: 51–64. [DOI] [PubMed] [Google Scholar]
- 90. Sweet RA, Nimgaonkar VL, Kamboh MI, Lopez OL, Zhang F, et al. (1998) Dopamine receptor genetic variation, psychosis, and aggression in Alzheimer disease. Arch Neurol 55: 1335–1340. [DOI] [PubMed] [Google Scholar]
- 91. Guo G, Roettger ME, Shih JC (2007) Contributions of the DAT1 and DRD2 genes to serious and violent delinquency among adolescents and young adults. Hum Genet 121: 125–136. [DOI] [PubMed] [Google Scholar]
- 92. Vaughn M, DeLisi M, Beaver K, Wright J (2009) DAT1 and 5HTT are associated with pathological criminal behavior in a nationally representative sample of youth. Crim Justice Behav 36: 1113–1124. [Google Scholar]
- 94.Miczek KA, Fish EW (2005) Monoamines, GABA, Glutamate, and Aggression. In: Nelson RJ, editor. Biology of Aggression: Oxford University Press. [Google Scholar]
- 95. Takahashi A, Quadros IM, de Almeida RM, Miczek KA (2012) Behavioral and Pharmacogenetics of Aggressive Behavior. Curr Top Behav Neurosci [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Vage J, Wade C, Biagi T, Fatjo J, Amat M, et al. (2010) Association of dopamine- and serotonin-related genes with canine aggression. Genes Brain Behav 9: 372–378. [DOI] [PubMed] [Google Scholar]
- 97. Berlivet S, Moussette S, Ouimet M, Verlaan DJ, Koka V, et al. (2012) Interaction between genetic and epigenetic variation defines gene expression patterns at the asthma-associated locus 17q12-q21 in lymphoblastoid cell lines. Hum Genet [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Falkenberg VR, Gurbaxani BM, Unger ER, Rajeevan MS (2011) Functional genomics of serotonin receptor 2A (HTR2A): interaction of polymorphism, methylation, expression and disease association. Neuromolecular Med 13: 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. van IMH, Caspers K, Bakermans-Kranenburg MJ, Beach SR, Philibert R (2010) Methylation matters: interaction between methylation density and serotonin transporter genotype predicts unresolved loss or trauma. Biol Psychiatry 68: 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. David S, Meltzer SJ (2011) Stomach - Genetic and epigenetic alterations of preneoplastic and neoplastic lesions. Cancer Biomark 9: 493–507. [DOI] [PubMed] [Google Scholar]
- 101. Provençal N, Suderman JM, Guillemin C, Massart R, Ruggiero A, et al. (2012) The signature of maternal rearing in the methylome in rhesus macaque prefrontal cortex and T cells. J Neurosci [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Nelson RJ, Chiavegatto S (2001) Molecular basis of aggression. Trends Neurosci 24: 713–719. [DOI] [PubMed] [Google Scholar]
- 103. Siegel A, Bhatt S, Bhatt R, Zalcman SS (2007) The neurobiological bases for development of pharmacological treatments of aggressive disorders. Curr Neuropharmacol 5: 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zalcman SS, Siegel A (2006) The neurobiology of aggression and rage: role of cytokines. Brain Behav Immun 20: 507–514. [DOI] [PubMed] [Google Scholar]
- 105. Alleva E, Cirulli F, Bianchi M, Bondiolotti GP, Chiarotti F, et al. (1998) Behavioural characterization of interleukin-6 overexpressing or deficient mice during agonistic encounters. Eur J Neurosci 10: 3664–3672. [DOI] [PubMed] [Google Scholar]
- 106. Pesce M, Speranza L, Franceschelli S, Ialenti V, Patruno A, et al. (2011) Biological role of interleukin-1beta in defensive-aggressive behaviour. J Biol Regul Homeost Agents 25: 323–329. [PubMed] [Google Scholar]
- 107. Hassanain M, Bhatt S, Zalcman S, Siegel A (2005) Potentiating role of interleukin-1beta (IL-1beta) and IL-1beta type 1 receptors in the medial hypothalamus in defensive rage behavior in the cat. Brain Res 1048: 1–11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distributions of promoter methylation levels by published expression level in T cells. Genes are divided into 20 levels by T cells expression percentiles (0–5, 5–10,…, 95–100) based on publicly available expression data(10). Shown are the distributions of methylation levels for each expression percentile. The distributions show that genes with low or no expression (represented in green) tend to have highly methylated promoters, whereas genes with high expression (represented in red) tend to have lower promoter methylation.
(TIF)
Microarray validation by Q-MeDIP and pyrosequencing of differentially methylated sequences between CPA (n = 8) and control groups (n = 12). A. Fold differences between CPA and control groups obtained by either Q-MeDIP or microarray analysis are shown for 20 genes predicted to be either more methylated (n = 1) or less methylated (n = 19) in the CPA individuals by the microarray analysis. B. Correlation of the fold differences between CPA and control obtained by Q-MeDIP and by microarray for the 20 amplicons analyzed in A. C. DNA methylation differences (%) between the groups in the GRM5, ITGB6, OR13C8 and IL-31 genes validated by pyrosequencing. CpG sites near the significant probe were analyzed. For each gene, the mean methylation per aggressive group per CpG is shown in the bar graph. The rightmost bar indicates the mean methylation levels of all CpG sites analyzed in the region. A map of the sites relative to the transcription start site is shown above the bar graph. Each line represents a CpG site. The location of probes whose fold difference is significantly different between CPA and control groups is identified by a grey square. The region analyzed by pyrosequencing is delimited by the red arrows. All error bars represent standard error of the mean (SEM). The P value obtain from Mann-Whitney U test is represent by *≤0.05, **≤0.01 and ***≤0.001.
(TIF)
Supplementary information file containing additional information of the methods use in the study.
(DOCX)
List of the probes differentially methylated between the chronic physical aggression and the control groups.
(XLSX)
Top list of affected biological functions enriched with genes whose methylation is associated with aggression from Ingenuity Pathway (12) analysis (n = 448 genes). All of the p values were calculated using a right tailed Fisher's exact test and corrected for multiple comparison with the Benjamini-Hochberg method. Significance threshold were p = 0.05.
(DOCX)
Top list of canonical pathways enriched with genes whose methylation is associated with aggression from IPA analysis (n = 448 genes). All of the p values were calculated using a right tailed Fisher's exact test and corrected for multiple comparison with the Benjamini-Hochberg method. Significance threshold were p = 0.05.
(DOCX)
List of transcription regulators showing a significant overlap with genes whose methylation is associated with aggression from IPA analysis (n = 448 genes). Transcription regulators differentially methylated between chronic and normal aggression are shown in bold. Significance threshold were p = 0.05.
(DOCX)
Primers and melting temperature (Tm) used for Q-MeDIP and pyrosequencing.
(DOCX)