Abstract
Cetuximab is a monoclonal antibody that targets the human epidermal growth factor receptor (EGFR). Although approved for use in EGFR over-expressing advanced colorectal cancer, recent studies have demonstrated a lack of association between EGFR over-expression and cetuximab response, requiring the identification of novel biomarkers predictive of response to this agent. To do so, 22 colon cancer cell lines were screened for cetuximab response in-vitro and sensitive and resistant lines identified. In sensitive cell lines cetuximab induced a G0/G1 arrest without inducing apoptosis. Notably, cetuximab sensitive but not resistant cell lines were preferentially responsive to EGF-stimulated growth. While neither EGFR protein/mRNA expression nor gene copy number correlated with cetuximab response, examination of the mutation status of signaling components downstream of EGFR demonstrated that cells lines with activating PIK3CA mutations or loss of PTEN expression (PTEN null) were more resistant to cetuximab than PIK3CA wild type/PTEN expressing cell lines (14±5.0% versus 38.5±6.4% growth inhibition, mean ± SEM, p=0.008). Consistently, PIK3CA mutant isogenic HCT116 cells showed increased resistance to cetuximab compared to PIK3CA wild type controls. Furthermore, cell lines that were PIK3CA mutant/PTEN null and Ras/BRAF mutant were highly resistant to cetuximab compared to those without dual mutations / PTEN loss (10.8±4.3% versus 38.8±5.9% growth inhibition, respectively, p=0.002), indicating constitutive and simultaneous activation of the Ras and PIK3CA pathways confers maximal resistance to this agent. A priori screening of colon tumors for PTEN expression status and PIK3CA and Ras/BRAF mutation status could help stratify patients likely to benefit from this therapy.
Keywords: cetuximab, colon cancer cell lines, EGFR expression, EGFR copy number, PIK3CA, PTEN, Ras, BRAF
INTRODUCTION
The EGFR signaling pathway is commonly activated in colorectal cancer, and has been explored for several years as a target for cancer therapy (1). EGFR is expressed in 30–85% of CRC’s, and the intensity of its expression has been linked to reduced survival (2, 3). Dysregulation of EGFR signaling has been shown to stimulate cell proliferation, angiogenesis and metastatic spread and to inhibit apoptosis (1, 4). Activation of this pathway occurs following ligand (EGF, TGF, amphiregulin) binding to EGFR, which leads to EGFR phosphorylation and oligodimerization at the plasma membrane. This in turn triggers a chain of downstream signaling events that include activation of the Ras/Raf/mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K)-Akt, and STAT pathways (4, 5). Cetuximab is a chimeric IgG1 monoclonal antibody that targets the extra-cellular domain of EGFR, blocking ligand binding to the receptor (1). Based on a randomized phase II clinical trial, cetuximab was approved in the US in 2004 for use in combination with irinotecan, or as monotherapy in EGFR-positive irinotecan-refractory colorectal cancer (6).
Cetuximab however has an objective response rate of only 9% when used as a single agent, along with toxicities of diarrhea, skin rash and infusion reactions (7). There is therefore a clear need for biomarkers predictive of response to cetuximab, in order to maximize likelihood of response while minimizing toxicities and cost.
While cetuximab was initially approved for treatment of patients with EGFR overexpression, the utility of EGFR as a predictive biomarker for cetuximab response has become increasingly controversial (6, 8, 9). One limitation of these studies is that EGFR expression status was frequently measured in the primary tumor, while objective response is measured in metastatic lesions. Recent studies have demonstrated variability in EGFR expression between primary and metastatic tumors from the same patient (10, 11), suggesting the need to determine EGFR expression status at the stage and site at which response is determined. The first objective of this study therefore, was to directly compare EGFR expression and cetuximab response in the same colon cancer cell lines.
In addition to EGFR expression, a further determinant of cetuximab sensitivity may be the presence of absence of mutations that result in constitutive activation of EGFR-mediated signaling. For example, in lung cancer, patients with activating mutations in the EGFR tyrosine kinase domain (encoded by exons 18–21) show significantly greater response to the small molecule inhibitors of EGFR tyrosine kinase activity, gefitinib and erlotinib (12–14). While mutations in the EGFR kinase domain are extremely rare in colon cancer (15), mutations which constitutively activate key signaling mediators downstream of EGFR, particularly K-Ras/BRAF and PTEN/PIK3CA, are more common (16–19). We hypothesized therefore that colon tumors with constitutively activated downstream signaling mediators of the EGFR pathway would be refractory to inhibition of the pathway at the receptor level. Indeed, several recent studies have demonstrated a link between K-Ras mutation status and cetuximab response, with tumors WT for K-Ras showing improved response to this agent (20–22). However, other studies failed to do so (23).
In the present study we observed that separation of cell lines according to PIK3CA and PTEN mutation status significantly distinguished cell lines according to cetuximab response. Particularly, cell lines harboring mutations in both PIK3CA/PTEN and Ras/BRAF were highly resistant to cetuximab. Therefore, a priori screening of colon tumors for PIK3CA/PTEN and Ras/BRAF mutation status may help identify patients likely to benefit from this therapy.
MATERIALS AND METHODS
Determination of sensitivity of colon cancer cell lines to cetuximab and EGF
The sources and maintenance of the colon cancer cell lines used in this study have been previously described (24), with the exception of the GEO cell line which was kindly provided by Dr. Z Fan (M.D. Anderson Cancer Center, TX).
For determination of cetuximab sensitivity, 5000–50,000 cells per well (24) were seeded in 96-well plates and treated with 0, 0.01, 0.1, 1, 5, 10, 20, 50, and 100 µg/ml cetuximab for 72h. For each cell line, one plate was harvested for determination of t=0 absorbance values. Viable cells were determined 72h post-treatment using the MTT assay by measurement of absorbance at 570 nm. The relative rate of cell growth for each cell line was factored into the analysis by subtracting the absorbance at time zero from both the control and treatment groups. All the experiments were replicated a minimum of 3 times.
For determination of sensitivity to EGF, cells were serum starved for 4h then treated with 0, 0.5 or 5 ng/ml EGF for 24–72h. For cetuximab/EGF co-treatment experiments, cells were pre-treated with cetuximab for 4h prior to EGF addition.
HCT116 K-Ras and PIK3CA isogenic cell lines
HCT116 colon cancer cells harbor both activating K-Ras and PIK3CA mutations. Isogenic HCT116 K-Ras and PIK3CA WT and mutant cells were generously provided by the Sasuzaki and Vogelstein/Velculescu laboratories, respectively (25, 26).
Cell cycle distribution-Fluorescent Accelerated Cell Sorting (FACS) analysis
For assessment of the effect of cetuximab and EGF on cell cycle distribution, cells were stained with 50 µg/ml propidium iodide overnight, and FACS analyses performed as previously described (27).
Determination of cetuximab response in vivo
For xenograft experiments, 5×106 GEO or LIM2405 cells in 200 µl of PBS/matrigel (1:1) were injected sub-cutaneously into the right flank of SCID mice. Tumors were allowed to form for approximately 1 week. Animals were then injected with either PBS or cetuximab (10 mg/kg or approximately 300 µg per mouse), intraperitoneally, biweekly for 2 weeks as previously described (28). Upon sacrifice, tumor volume was calculated from measurements of the smallest (s) and longest (l) diameter based on the following formula: Volume = [(s2 × l) × π] / 6
Determination of EGFR protein and mRNA expression in colon cancer cell lines
EGFR protein expression was determined by Western blot, using an anti-EGFR antibody (Cell signaling Technology™). EGFR mRNA expression was determined by quantitative real time PCR. EGFR specific primers were as follows: (F: ATGCTCTACAACCCCACCAC, R: GCCCTTCGCACTTCTTACAC). Results were expressed relative to GAPDH (GAPDH primers: F:TCGGAGTCAACGGATTTGG, R: GAATTTGCCATGGGTGGAAT).
Determination of EGFR gene copy number by Fluorescence in situ Hybridization (FISH)
EGFR copy number in was assessed in colcemid treated cells by standard cytogenetic methods using 0.075M KCl and Carnoy's fixative (methanol:acetic acid, 3:1). Metaphase chromosomes were hybridized overnight with the dual color LSI EGFR/CEP 7 (Vysis locus specific identifier DNA) probe and counterstained with DAPI. This probe has a specific EGFR Spectrum Orange probe and a CEP 7 probe, labeled in Spectrum Green, which hybridizes to the alpha satellite DNA located at the centromere of chromosome 7 (7p11.1-q11.1). Images were acquired with an epifluorescence microscope (Olympus BX51) connected to a Sensicam QE CCD cooled camera. Ten metaphases for each cell line were analyzed using the FISH view software (Spectral Imaging). EGFR gene amplification was defined as >1 copy of EGFR locus per chromosome 7, and EGFR polysomy defined as >2 EGFR loci per nucleus.
Identification of K-Ras, BRAF, PIK3CA and PTEN mutations in colon cancer cell lines
The mutation status of K-Ras, BRAF (Exon 15), PIK3CA (Exon 9 and 20) and PTEN for a subset of the cell lines was obtained from the Wellcome Trust Sanger Institute Cancer Genome Project web site, http://www.sanger.ac.uk/genetics/CGP/cosmic/, or from previous publications. For cell lines for which the mutation status of one or more of these genes was unknown, genomic DNA was isolated using the Qiagen DNA extraction kit. Primers used for amplification of exon 2 of K-Ras were: F: AGGCCTGCTGAAAATGACTGAATA, and R: CTGTATCAAAGAATGGTCCTGCAC. Primers used for amplification of exon 15 of BRAF were F: AACACATTTCAAGCCCCAAA, and R: GAAACTGGTTTCAAAATATTCGTT, for Exon 9 of PIK3CA were F: GCTTTTTCTGTAAATCATCTGTG, and R: CTGAGATCAGCCAAATTCAGT, for exon 20 of PIK3CA were, F: CATTTGCTCCAAACTGACCA and R: TACTCCAAAGCCTCTTGCTC (for codon 1023 mutation) and F: ACATTCGAAAGACCCTAGCC and R: CAATTCCTATGCAATCGGTCT (for codon 1047 mutation). PTEN expression status was determined by western blot, using an anti-PTEN antibody (Cell Signaling).
Statistical analyses
Differences between two groups was analyzed using an unpaired student’s t test with p<0.05 considered statistically significant. Differences in cetuximab sensitivity between cell lines wild type or mutant for specific genes or gene combinations was determined using an unpaired student’s t test with Bonferroni adjustment. Since we performed 5 separate analysis, p<0.01 was considered statistically significant. For correlative analyses, a Pearson’s correlation coefficient was computed with P<0.05 considered statistically significant.
RESULTS
Determination of sensitivity of colon cancer cell lines to cetuximab
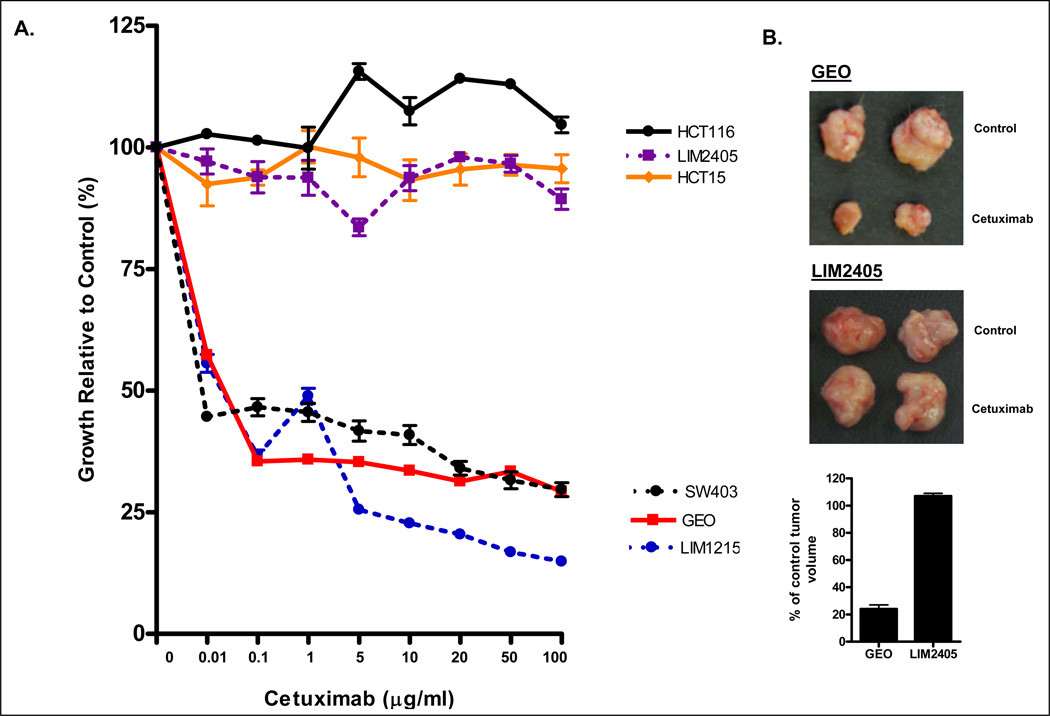
To identify colon cancer cell lines with differential response to cetuximab, a panel of 22 colon cancer cell lines was screened for cetuximab response using the MTT assay. As shown in Figure 1 and Supplementary Figure 1, a spectrum of sensitivity to cetuximab was identified. Maximal response was observed in the LIM1215, GEO and SW403 cell lines, where growth was inhibited >70% at the maximum concentration of cetuximab tested. An intermediate response (approximately 50% growth inhibition), was observed in the Caco-2 and SW948 cell lines while minimal response (<35% growth inhibition) was observed in the remaining cell lines (Figure 1, Supplementary Figure 1).
Figure 1.
(A) Differential sensitivity of colon cancer cell lines to 72 h cetuximab treatment. Shown for simplicity are the 3 most sensitive and resistant cell lines of the 22 cell lines screened for cetuximab response. Values shown are the mean ± SEM of n=3–5 experiments. (B) Differential sensitivity of colon cancer cells to cetuximab in vivo. 5×106 cells of the cetuximab sensitive (GEO) and resistant (LIM2405) colon cancer cell lines were injected in SCID mice. Once palpable tumors had formed animals were injected with cetuximab (10 mg/kg) or PBS (control), biweekly, for 2 weeks following which animals were sacrificed, tumors excised and tumor volume calculated as in Materials and Methods.
Differential sensitivity of colon cancer cell lines to cetuximab in vitro is also observed in vivo
To confirm the differential sensitivity of colon cancer cell lines to cetuximab in vivo, the sensitive GEO, and resistant LIM2405, cell lines were grown as xenografts in SCID mice and treated with 10 mg/kg cetuximab biweekly for 2 weeks. Consistent with the in vitro findings, cetuximab inhibited growth of GEO cells by approximately 75% (205±36 mm3 compared to 854±201 mm3 in cetuximab treated and control mice respectively), whereas no growth inhibitory effect was observed in LIM2405 cells (999±32 mm3 versus 934±66 mm3 in cetuximab treated and control mice respectively) (Figure 1B).
Comparable sensitivity profiles of colon cancer cells to cetuximab and erlotinb
To confirm that the sensitivity spectrum of colon cancer cells to cetuximab reflected inhibition of EGFR and its downstream signaling pathway, we assessed the response of 12 of the cell lines to erlotinib (5µg/ml), a small molecule tyrosine kinase inhibitor that targets the intracellular domain of EGFR. As shown in Supplementary Figure 2, a significant correlation between response of colon cancer cell lines to cetuximab and erlotinib was observed, consistent with both agents mediating growth inhibition through inhibition of EGFR signaling (r2=0.542, p=0.006). In contrast, no significant correlation was observed between cetuximab response and response to the mechanistically distinct chemotherapeutic agent, 5FU (r2=0.019, p=0.678, Supplementary Figure 2).
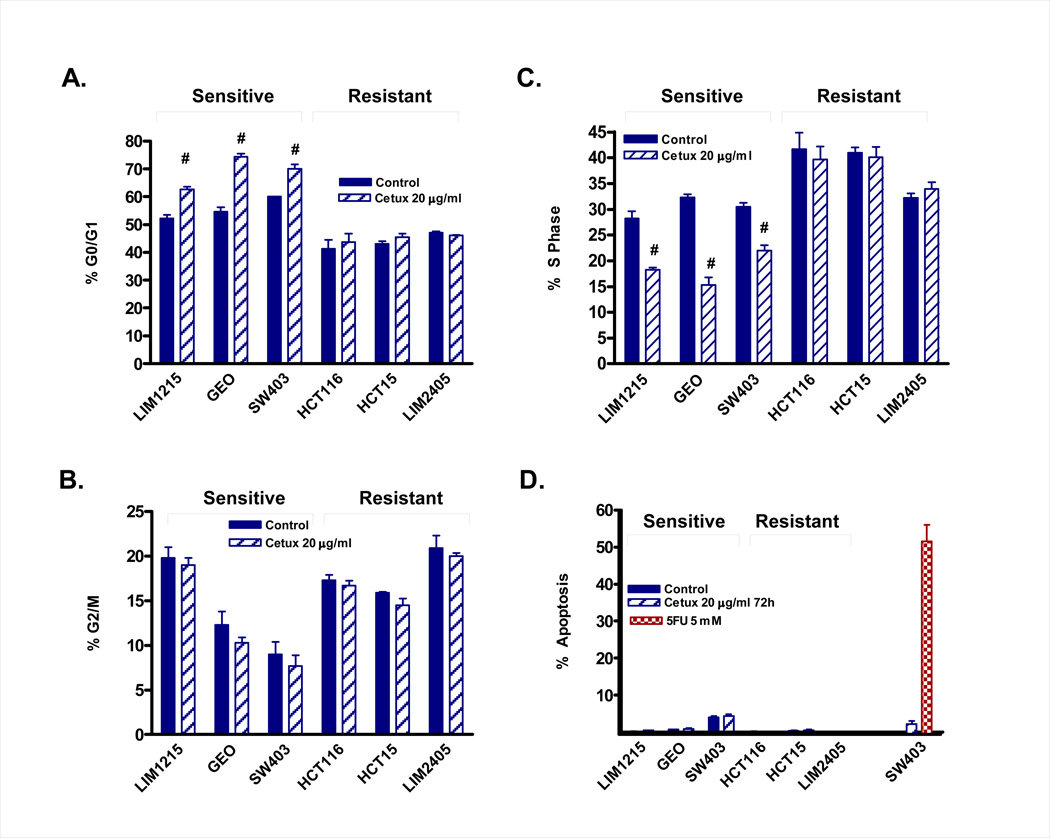
Cetuximab induces a G0/G1 arrest in colon cancer cells
To confirm the findings of the MTT assay, and to further assess the effect of cetuximab on cell cycle distribution, we determined the effect of cetuximab on cell cycle distribution in the 3 most sensitive and 3 most resistant cell lines (Figure 2A–C). Consistent with the MTT data, an increase in the percentage of cells in G0/G1 and a concomitant decrease in the percentage of cells in S phase was observed in the 3 sensitive cell lines (LIM1215, GEO and SW403). No difference in the percentage of cells in G2/M was observed. In comparison, minimal change in cell cycle distribution was observed in the resistant cell lines (LIM2405, HCT116 and HCT15). Importantly, minimal effects on apoptosis were observed in this cell line panel either at 24 or 72 hours following cetuximab treatment (Figure 2D), indicating cetuximab elicits a predominantly cytostatic effect in colon cancer cells. In contrast, treatment of SW403 cells with 5 µM 5-fluoruracil induced approximately 50% apoptosis following 72 hours treatment, demonstrating that these cell lines are not inherently resistant to apoptosis (Figure 2D).
Figure 2.
Summary of cell cycle analysis of cetuximab sensitive (GEO, LIM1215, SW403) and resistant (LIM2405, HCT116, HCT15) colon cancer cell lines. For assessment of cell cycle distribution, cells were treated for 24 hours with 20 µg/ml cetuximab (A–C). For assessment of apoptosis, cells were treated with 20 µg/ml cetuximab or 5 µM 5FU for 72h (D). Cell cycle distribution and apoptosis was assessed by PI staining and FACS analysis. Values shown are mean ± SEM, n=3, #P<0.05.
EGFR mRNA, protein expression or gene copy number do not correlate with cetuximab response in colon cancer cell lines
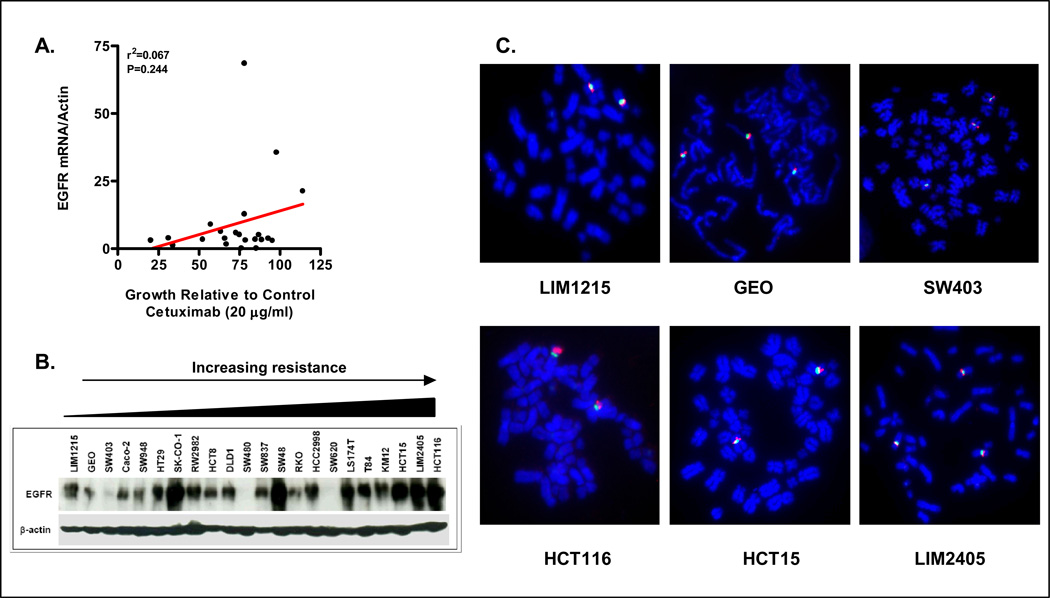
Since cetuximab targets EGFR, we determined whether basal EGFR mRNA and or protein expression in the cell line panel correlated with cetuximab response. EGFR mRNA in the 22 cell line panel was assessed by QRT-PCR. As shown in Figure 3A, no correlation between basal EGFR mRNA expression and cetuximab response was observed (R2=0. 067, p=0.244). To determine whether basal EGFR protein expression correlated with cetuximab response, EGFR protein expression was assessed in the 22 cell lines. While significant correlation between EGFR mRNA and protein expression was observed (R2=0.35, p=0.003), no correlation between basal EGFR protein expression and cetuximab response was observed (R= −0.36, R2=0.13, p=0.099, Figure 3B).
Figure 3.
Basal EGFR mRNA, protein expression, or EGFR copy number does not correlate with cetuximab response. (A) Correlation of basal EGFR mRNA expression as assessed in exponentially growing colon cancer cells by Q-RT-PCR and cetuximab response at the 20 µg/ml dose. (B) EGFR protein expression in the 22 cell lines as assessed by western blot analysis. (C) EGFR (red) copy number as determined by FISH analysis in the 3 sensitive and 3 resistant cell lines. CEP7 (green) was used as marker of chromosome 7.
Finally, studies by Moroni et al suggested that EGFR gene amplification status correlates with response to cetuximab in patients with colon cancer (23). Hence, the EGFR amplification status of the 3-most sensitive and 3-most resistant colon cancer cell lines was determined by FISH analysis. As shown in Figure 3C, no EGFR gene amplification was seen in any of the 6 colon cancer cell lines tested. Two sensitive (GEO and SW403) and one resistant cell line (LIM2405) contained 3 copies of EGFR per nucleus, however, in each case 3 copies of the chromosome 7 marker, CEP7 were also observed indicating this reflected polysomy as opposed to amplification of the EGFR locus. Collectively therefore, no correlation between cetuximab response and EGFR expression of copy number was observed in the colon cancer cell line panel.
Cetuximab sensitivity correlates with growth response to EGF
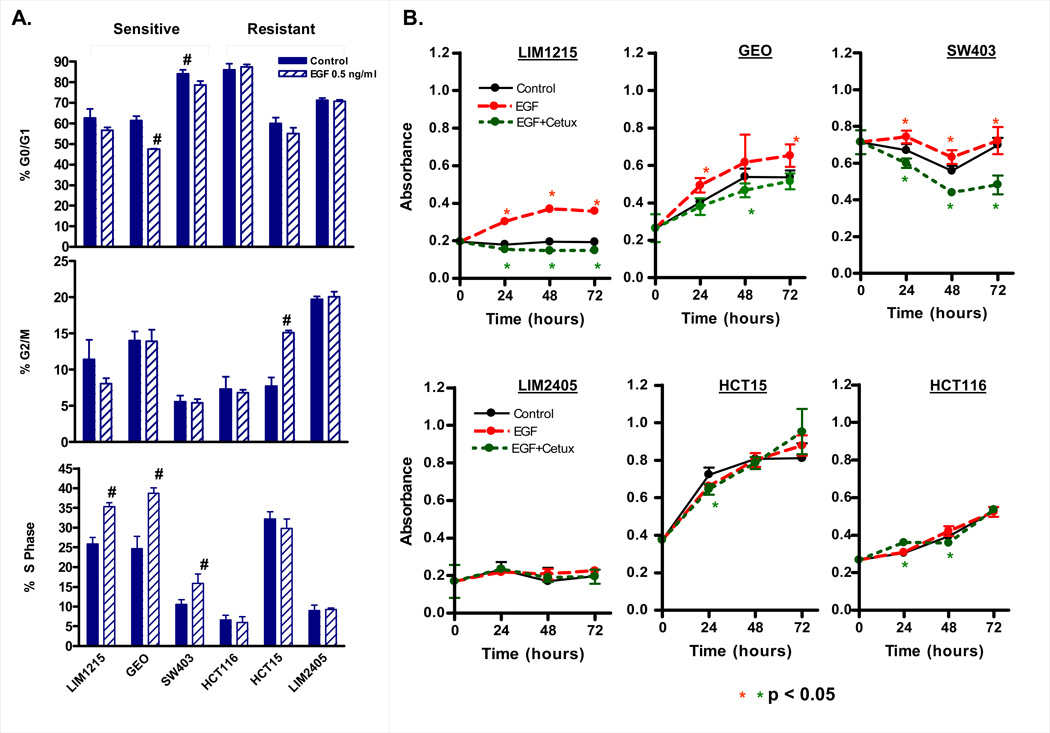
Signaling via the EGFR receptor is initiated upon ligand binding (EGF, TGFalpha, amphiregulin), with signal transduction primarily through the PTEN/PI3K/AKT and / or Ras/Raf/MEK/ERK pathways. We speculated therefore, that cell lines responsive to ligand mediated canonical activation of this pathway for their growth would be most sensitive to cetuximab.
To test this, the proliferative response of the 3 most cetuximab-sensitive and 3 most cetuximab-resistant cell lines to exogenous EGF-treatment was examined under serum-free conditions (Figure 4). A significant increase in S-phase was observed 24h post EGF treatment (0.5 ng/ml) in the 3 cetuximab sensitive cell lines, GEO, LIM1215, and SW403, but not in the 3 most cetuximab resistant cell lines, LIM2405, HCT15, and HCT116 (Figure 4A). To confirm this result we also performed MTT assays. Treatment with 0.5 or 5 ng/ml EGF preferentially increased growth of the cetuximab sensitive cell lines, establishing a clear link between response to the mitogenic effects of EGF and the growth inhibitory effects of cetuximab (Fig 4B). Similar results were observed when response to a 10-fold higher concentration of EGF (5 ng/ml) was tested (Supplementary Figure 3). Importantly, pre-treatment of the EGF responsive cell lines with cetuximab significantly attenuated the mitogenic effect of EGF (Figure 4B).
Figure 4.
EGF selectively stimulates cell growth in cetuximab sensitive cell lines. (A) Effect of EGF treatment on cell cycle progression. Cells were treated for 24 hours with 0.5 ng/ml EGF and cell cycle distribution determined by PI staining and FACS analysis. Values shown are mean ± SEM from a representative experiment, # p<0.05. Experiments were repeated 3 separate times. (B) The 3-most cetuximab sensitive and resistant cell lines were treated for 24–72 hours with 0.5 ng/ml EGF, EGF + cetuximab (20 µg/ml) or left untreated (control). Cell growth was assayed by MTT assay. Values shown are mean ± SEM from a representative experiment, # p<0.05. Experiments were repeated 3 separate times.
Mutation status of PIK3CA and/or PTEN predicts response to cetuximab
Ligand binding to EGFR results in signal transduction via the Ras/Raf/MEK/MAPK and the PI3K/AKT pathway. As mutations that result in constitutive activation of each of these pathways occur at high frequencies in colon cancer, we hypothesized that colon cancer cell lines with constitutively activated signaling downstream of EGFR would not be dependent on ligand binding to EGFR for their growth, and in turn, would be refractory to cetuximab.
Mutation driven constitutive activation of the PI3K signaling pathway has been reported to occur in approximately 30% of colon tumors, primarily due to activating mutations in exons 9 and 20 of the PIK3CA gene (18), and to a lesser extent due to inactivating PTEN mutations or PTEN promoter methylation (29). The presence of activating PIK3CA mutations in the cell line panel was assessed by literature searches, from the COSMIC database and by direct sequencing of exons 9 and 20 of the PIK3CA gene. Mutations in PIK3CA were identified in 8 of the 22 cell lines (Table 1, Supplementary Figure 5). Separation of cell lines according to PIK3CA mutation status alone did not distinguish cetuximab sensitive from resistant cell lines (16.3±6.0% versus 33.7±6.4% growth inhibition for PIK3CA mutant versus WT cell lines respectively, p=0.08).
Table 1.
Correlation of mutation status of cell lines and response to cetuximab.
| Cell line | % Inhibition relative to control (Mean±SEM) |
Mutant K-Ras Ex 2 |
Mutant BRAF Ex 15 |
Mutant PIK3CA Ex 9 |
Mutant PIK3CA Ex 20 |
Mutant Total PIK3CA |
PTEN Null | Mutant RAS/BRAF |
Mutant PIK3CA/PTEN |
RAS/BRAF and PIK3CA/PTEN Mutant |
|---|---|---|---|---|---|---|---|---|---|---|
| LIM12151 | 79.6 ± 3.5 | |||||||||
| GEO3 | 68.7 ± 1.6 | + | + | |||||||
| SW4032 | 66.0 ± 4.9 | + | + | |||||||
| CAC022 | 47.7 ± 4.3 | |||||||||
| SW9482 | 42.7 ± 1.4 | + | + | + | ||||||
| HT292 | 36.5 ± 8.2 | + | + | |||||||
| SKCO12 | 33.9 ± 3.5 | + | + | |||||||
| RW29821 | 33.0 ± 1.3 | |||||||||
| HCT81 | 27.1 ± 4.0 | + | + | + | + | + | + | |||
| DLD3 | 24. 9 ± 7.4 | + | + | + | + | + | + | |||
| SW4803 | 23.7 ± 3.0 | + | + | |||||||
| SW8372 | 21.8 ± 8.5 | + | + | |||||||
| SW482 | 21.8 ± 1.2 | |||||||||
| RKO2 | 21.2 ± 6.9 | + | + | + | + | + | + | |||
| HCC29982,4 | 15.2 ± 3.3 | |||||||||
| SW6202 | 14.5 ± 2.2 | + | + | |||||||
| LS174T2 | 13.0 ± 3.9 | + | + | + | + | + | + | |||
| T842 | 11.2 ± 9.2 | + | + | + | + | + | + | |||
| KM122 | 7.1 ± 9.1 | + | + | + | + | + | ||||
| HCT152 | 4.5 ± 6.5 | + | + | + | + | + | + | |||
| LIM24051 | 2.0 ± 2.2 | + | + | + | + | + | ||||
| HCT1162 | −14.1 ± 1.3 | + | + | + | + | + | + |
Present study
From cosmic database http://www.sanger.ac.uk/genetics/CGP/cosmic/
Ras/BRAF from cosmic; PIK3CA by present study
PTEN is a tumor suppressor that acts as a negative regulator of PI3K signaling by converting PIP3 to PIP2, and truncating mutations which result in loss of PTEN expression have been reported in approximately 20% of MSI colon cancers (30, 31). We noted that the KM12 cell line that was PIK3CA wild type yet highly resistant to cetuximab, harbors a truncating mutation in PTEN (Wellcome Trust Sanger Institute Cancer Genome Project, http://www.sanger.ac.uk/genetics/CGP). The PTEN expression status of the cell line panel was therefore determined by western blot (Supplementary Figure 4). In addition to KM12, loss of PTEN expression was also observed in the cetuximab resistant LIM2405 line (Supplementary Figure 4). Consistent with previous reports, both lines with loss of PTEN expression were derived from MSI colon cancers. Furthermore, the occurrence of PIK3CA mutations and loss of PTEN expression in the cell line was mutually exclusive as previously reported (32).
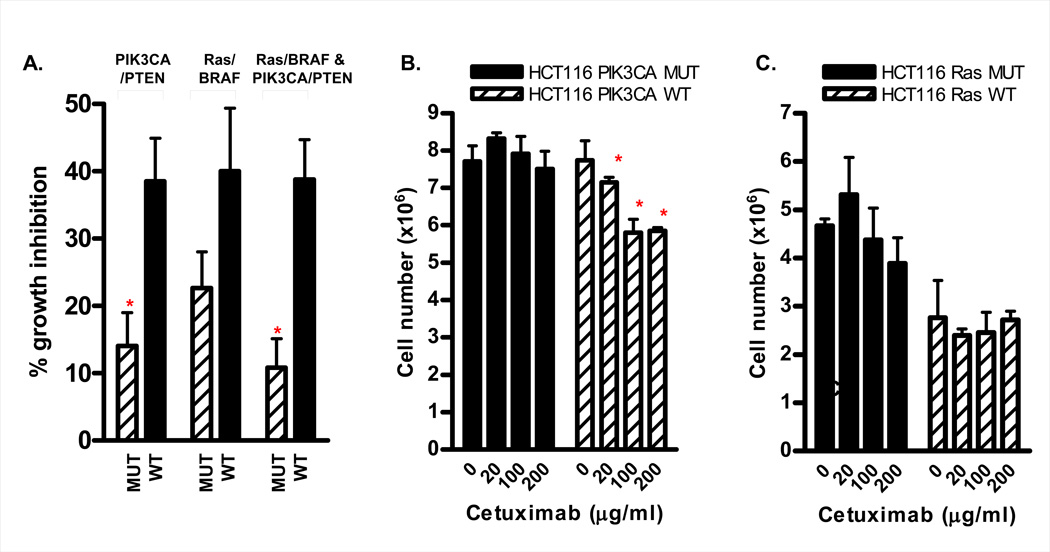
Separation of cell lines according to PIK3CA mutation and/or PTEN expression status identified a significant difference in cetuximab sensitivity, with PIK3CA mutant / PTEN null cell lines being significantly more refractory to cetuximab treatment (14.0±5.0% versus 38.5±6.4% growth inhibition for PIK3CA mutant/PTEN null versus PIK3CA WT/PTEN expressing cell lines respectively, p=0.008, figure 5A).
Figure 5.
Mutation status and response to cetuximab in vitro. (A) Cetuximab response in the 22 colon cancer cell line panel separated according to PIK3CA mutation/PTEN expression status (*p=0.008), Ras/BRAF mutation status (p=0.11), or according to presence or absence of synchronous mutations in the PIK3CA/PTEN and the K-Ras/BRAF pathways (*p=0.002). (B) Cetuximab response in isogenic PIK3CA mutant and WT HCT116 cell lines. Cells were serum starved overnight then treated with 20 or 100 µg/ml cetuximab for 24h in medium containing 0.5% serum. Differential sensitivity was assessed by direct counting of cell number. (C) Cetuximab response in isogenic Ras mutant and WT HCT116 cells. Cetuximab response was determined 24h post treatment by counting cell number.
To further confirm this finding, we examined cetuximab response in a pair of isogenic HCT116 cells provided by the Velculescu / Vogelstein laboratories, in which either the mutant or WT PIK3CA allele has been deleted by homologous recombination (26). While the PIK3CA mutant HCT116 isogenic cell line was highly resistant to cetuximab, a modest though statistically significant response to cetuximab was observed in the PIK3CA WT isogenic line (Figure 5B). Collectively, these findings demonstrate that colon cancer cell lines with constitutively active PI3K signaling are refractory to cetuximab.
Mutation status of K-Ras or BRAF does not predict response to cetuximab
Several clinical studies have examined the relationship between Ras mutation status of colon tumors and response to cetuximab. While an initial study did not observe an association (23), more recent studies have demonstrated that colon tumors mutant for K-Ras are less responsive to cetuximab compared to wild-type tumors (20–22).
To validate and extend these findings, the K-Ras mutation status of the cell line panel was determined by literature searches, searching of the Sanger database, or direct sequencing of codons 12 and 13 in exon 2 of the K-Ras gene (Table 1, Supplementary Figure 5). Of the 22 cell lines in the panel, 13 were mutant and 9 were wild type for K-Ras. Separation of the cell lines according to Ras mutation status did not result in a differential response to cetuximab (22.2±6.6% versus 34.7±6.8% growth inhibition for Ras mutant vs WT cell lines respectively, p=0.21). The lack of an association between Ras mutation status and resistance to cetuximab was underscored by the observation that the established cetuximab sensitive cell line, GEO (28) and SW403, harbored a mutant K-Ras allele (Figure 1). To further confirm this finding we compared cetuximab response in parental HCT116 cells (Ras mutant) and the HCT116 Ras WT isogenic derivative (Hke3), generated by targeted deletion of the mutant K-Ras allele. No difference in cetuximab response, as assessed by cell number 24h post cetuximab treatment was observed between the Ras mutant and WT clones (Figure 5C).
Mutations in BRAF are frequently observed in colon tumors wild type for K-Ras. The BRAF mutation status of the cell line panel was therefore determined by direct sequencing of exon 15. Three cell lines harboring BRAF mutations were identified in the cell line panel (RKO, HT29 and LIM2405). Consistent with the previously observed mutual exclusion of Ras and BRAF mutations in colon tumors, the 3 cell lines with BRAF mutations were wild type for K-Ras. Separation of the cell lines according to Ras/BRAF mutation status resulted in a tendency for Ras/BRAF WT cell lines to be more sensitive to cetuximab compared to Ras/BRAF mutant lines, although this effect was not statistically significant (22.6±5.4% versus 40.0±9.4 % growth inhibition for Ras/BRAF mutant versus WT cell lines respectively, p=0.11; Figure 5A).
Collective consideration of PIK3CA/PTEN and RAS/BRAF mutation status further predicts cetuximab response
Notably our mutation screening analysis demonstrated that while 9 of 16 (56%) cell lines with Ras/BRAF mutations harbored a synchronous PIK3CA/PTEN mutation, 9 of the 10 cell lines (90%) harboring PIK3CA/PTEN mutations contained synchronous Ras/BRAF mutations (Table 1). This finding is consistent with previous reports indicating significant overlap between the presence of PIK3CA and K-Ras/BRAF mutations within the same colorectal tumor (19, 33). Importantly, separation of cell lines that were mutant for both PIK3CA/PTEN and Ras/BRAF versus those that were not, further discriminated between cetuximab sensitive and resistant cell lines, with PIK3CA/PTEN;Ras/BRAF mutant lines (10.8±4.3 % growth inhibition) being significantly more resistant to cetuximab than cell lines that did not harbor mutations in both of these pathways (percent growth 38.8±5.9% growth inhibition, p=0.002; Figure 5A).
DISCUSSION
The development and approval of novel therapies including the monoclonal antibodies bevacizumab (anti-VEGF, Avastin) and cetuximab (anti-EGFR), has increased median survival of patients with metastatic colon cancer to approximately 24 months (34, 35).
Cetuximab was initially approved for use in CRC patients with EGFR overexpression, however, recent retrospective clinical reports found no evidence for limiting cetuximab therapy to patients with EGFR overexpressing tumors (8, 9). Similarly, our present findings failed to demonstrate an association between EGFR expression and cetuximab response in colon cancer cell lines, indicating that factors other than EGFR expression are primarily responsible for determining response to this agent.
In addition to EGFR expression, a link between EGFR amplification and response to anti-EGFR monoclonal antibody-based treatment has been demonstrated (23). For example, the Difi colon cancer cell line which has high level EGFR amplification, is highly sensitive to cetuximab, undergoing both growth arrest and apoptosis at cetuximab concentrations several orders of magnitude lower than those required for response in other cell lines (6, 36). Lievre et al also described a complete response in a patient with high level EGFR amplification (20 copies / nucleus) (22). EGFR amplification however, is a rare event in colon cancer, occurring at a frequency of less than 1% of cases (37–39). Consistent with this finding, examination of EGFR amplification status in cetuximab-sensitive and resistant cell lines in the present study failed to identify any lines with EGFR amplification. These findings demonstrates that while rare tumors and cell lines with EGFR amplification are highly sensitive to cetuximab, more modest, cytostatic responses to this agent are obtained in the absence of EGFR amplification.
Two of the cell lines identified as cetuximab sensitive, LIM1215 and SW403, have previously been shown to be dependent upon activation of EGFR in an autocrine manner for their growth (40, 41). A previous study also demonstrated a link between the proliferative response of colon adenocarcinoma cell lines to EGF and response to EGFR inhibition (42). We confirmed and extended these observations by demonstrating that cetuximab-sensitive cell lines were more sensitive to EGF-induced growth promotion than cetuximab resistant lines. Importantly, the growth stimulatory effect of EGF could be completely inhibited by pre-treatment with cetuximab. These findings indicate that a subset of colon tumors exist that are dependent upon ligand activation of EGFR for their growth, and it is these cell lines that are growth inhibited by blockade of ligand binding to the EGFR.
As recently demonstrated, tumor cells can lose their dependence on growth factors via mutation driven, constitutive activation, of signaling pathways downstream of growth factor receptors, specifically the PI3 kinase and Ras/MAPK pathways (43). We observed that stratification of cell lines according to PIK3CA/PTEN mutation status identified a significant difference in cetuximab response, with PIK3CA/PTEN mutant lines being consistently more resistant to this agent compared to wild type lines. This finding was further confirmed using the HCT116 PIK3CA WT and mutant isogenic cell lines, where increased sensitivity to cetuximab was observed in the PIK3CA WT line. This finding is consistent with the reported observation that HCT116 PIK3CA WT cells are more dependent on serum derived growth factors for their growth, and are more responsive to EGF ligand induced signaling compared to the mutant line (26). Collectively, these findings imply that colon cancer cell lines which acquire mutations that result in constitutive activation of the PI3K pathway have a diminished dependence on canonical EGFR ligand-induced signaling for their growth, and are, therefore, more resistant to EGFR-targeted therapies.
Two prior studies failed to observe a link between PIK3CA mutation status and cetuximab response in patients with colon cancer (22, 23). However, in both of these studies PTEN mutation status of the tumors was not examined, and very few patients with PIK3CA mutations - 3/31 (23) and 2/30 (22) – were identified. Notably, while the low PIK3CA mutation frequency may have precluded the overall ability of these studies to detect differences in cetuximab response between PIK3CA WT and mutant tumors, 4 of the 5 patients with PIK3CA mutations did not respond to cetuximab. Furthermore, the single patient with a PIK3CA mutation that did respond had a tumor with low level EGFR amplification. Collectively considered therefore, the current data available in patient samples are relatively consistent with the present findings, although the need for validation in a larger patient study clearly remains.
Consistent with the present findings, Frattini et al recently reported that colon tumors with loss of PTEN expression have significantly reduced response to cetuximab (44). Likewise, breast cancers with either activating mutations in PIK3CA or with loss of PTEN expression respond poorly to treatment with the Her2/Neu targeting antibody, trastuzumab (45). Collectively, these studies provide additional clinical evidence that the mutation status of the PI3K signaling pathway should be considered prior to treatment with EGF receptor family antagonists.
Several (20–22, 46), though not all studies (23) that have examined the link between Ras mutation status and cetuximab response, have shown that patients with tumors WT for both K-Ras and BRAF have improved response to cetuximab. It is notable however, that in some of these studies, patients with mutant Ras tumors who showed clinical response to cetuximab treatment were identified (20). In the present study, separation of cell lines according to Ras and or BRAF mutation status did not stratify cell lines according to cetuximab response, although a clear tendency for Ras/BRAF mutant lines to be more resistant to cetuximab was observed (p=0.11). We also observed that two Ras mutant cell lines, GEO and SW403, showed significant response to cetuximab.
While it remains to be clarified whether determination of Ras/BRAF mutation status alone is sufficient to stratify patients for cetuximab treatment (20), we observed that the 6 most cetuximab resistant cell lines harbored mutations in both the PIK3CA/PTEN and Ras/BRAF pathways. This finding is consistent with previous reports indicating the co-existence of Ras and PIK3CA mutations within the same tumor (19, 33). Importantly, collective consideration of PIK3CA/PTEN and Ras/BRAF mutation status provided the most robust determinant of cetuximab response, with cell lines harboring mutations in both of these pathways being highly resistant to cetuximab. In terms of therapeutic implications, these findings suggest that patients whose tumors harbor simultaneous mutations in both the PIK3CA/PTEN and Ras/BRAF pathways are unlikely to benefit from cetuximab treatment. Instead, these patients may be suitable candidates for treatment with newer targeted drugs currently in clinical trial, which inhibit signaling mediators further downstream, including PI3 kinase, AKT or mTOR inhibitors and Ras, Raf or MEK inhibitors.
Importantly, while cell lines with mutations in PIK3CA/PTEN and Ras/BRAF were consistently resistant to cetuximab, not all cell lines wild type at the four loci tested, were sensitive to cetuximab. A possible mechanism of resistance to cetuximab of these cell lines may be the existence of alternate mutations in the Ras/BRAF and or PIK3CA/PTEN pathway, other than those screened for in the present analysis. For example, with regards to the PI3K pathway, less frequently occurring mutations have been described in exons 1 and 2 of PIK3CA which encode the p85 interacting domain (18). Mutations in p85α (47), PDK1, AKT2, PAK4, and INSRR (19), as well as amplifications in AKT2 and IRS2 (19) have also been described, as have less frequently occurring mutations in codon 146 (A146T) of the K-Ras gene (48). In this regard, it is notable that mutations in both codon 146 of K-Ras, and in p85alpha have been reported in the relatively resistant HCC2998 cell line (48, 49), which is wild type at the 4 hot spot loci examined in the present analysis. Comprehensive screening of all known components of the PI3K and Ras signaling pathways may therefore be required to further improve prediction of cetuximab response.
In addition to inhibition of EGFR signaling, cetuximab can stimulate antibody-dependent cell mediated cytotoxicity (ADCC) and complement dependent cytotoxicity (CDC) in vivo (50), which may contribute to its anti-tumor activity. As our current in vitro model does not assess the role of ADCC, additional validation of these findings in patient samples is clearly required.
In conclusion, this study demonstrates that cell lines responsive to canonical EGFR signaling-mediated growth are also responsive to cetuximab. We observed that cell lines mutant for PIK3CA/PTEN are significantly more resistant to cetuximab compared to PIK3CA/PTEN wild type lines. Furthermore, cell lines with both constitutively active PIK3CA and Ras/BRAF signaling were highly refractory to cetuximab. Determination of the mutation status of signaling mediators downstream of EGFR may help stratify patients likely to benefit from cetuximab.
Supplementary Material
ACKNOWLEDGEMENTS
Dr. Z. Fan (M.D. Anderson Cancer Center) for kindly providing the GEO colon cancer cell lines. The AECOM Genome Imaging shared resource, DNA sequencing Facility and Animal Facilities were used to generate data for this study.
REFERENCES
- 1.Mendelsohn J, Baselga J. The EGF receptor family as targets for cancer therapy. Oncogene. 2000;19:6550–6565. doi: 10.1038/sj.onc.1204082. [DOI] [PubMed] [Google Scholar]
- 2.Galizia G, Lieto E, Orditura M, et al. Epidermal growth factor receptor (EGFR) expression is associated with a worse prognosis in gastric cancer patients undergoing curative. surgery. World J Surg. 2007;31:1458–1468. doi: 10.1007/s00268-007-9016-4. [DOI] [PubMed] [Google Scholar]
- 3.McKay JA, Murray LJ, Curran S, et al. Evaluation of the epidermal growth factor receptor (EGFR) in colorectal tumours and lymph node metastases. Eur J Cancer. 2002;38:2258–2264. doi: 10.1016/s0959-8049(02)00234-4. [DOI] [PubMed] [Google Scholar]
- 4.Jorissen RN, Walker F, Pouliot N, et al. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- 5.Quesnelle KM, Boehm AL, Grandis JR. STAT-mediated EGFR signaling in cancer. J Cell Biochem. 2007;102:311–319. doi: 10.1002/jcb.21475. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 7.Saltz LB, Meropol NJ, Loehrer PJ, Sr, et al. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. 2004;22:1201–1208. doi: 10.1200/JCO.2004.10.182. [DOI] [PubMed] [Google Scholar]
- 8.Chung KY, Shia J, Kemeny NE, et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol. 2005;23:1803–1810. doi: 10.1200/JCO.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 9.Vallbohmer D, Zhang W, Gordon M, et al. Molecular determinants of cetuximab efficacy. J Clin Oncol. 2005;23:3536–3544. doi: 10.1200/JCO.2005.09.100. [DOI] [PubMed] [Google Scholar]
- 10.Bralet MP, Paule B, Falissard B, Adam R, Guettier C. Immunohistochemical variability of epidermal growth factor receptor (EGFR) in liver metastases from colonic carcinomas. Histopathology. 2007;50:210–216. doi: 10.1111/j.1365-2559.2007.02578.x. [DOI] [PubMed] [Google Scholar]
- 11.Scartozzi M, Bearzi I, Berardi R, et al. Epidermal growth factor receptor (EGFR) status in primary colorectal tumors does not correlate with EGFR expression in related metastatic sites: implications for treatment with EGFR-targeted monoclonal antibodies. J Clin Oncol. 2004;22:4772–4778. doi: 10.1200/JCO.2004.00.117. [DOI] [PubMed] [Google Scholar]
- 12.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 13.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 14.Takano T, Ohe Y, Sakamoto H, et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;23:6829–6837. doi: 10.1200/JCO.2005.01.0793. [DOI] [PubMed] [Google Scholar]
- 15.Barber TD, Vogelstein B, Kinzler KW, Velculescu VE. Somatic mutations of EGFR in colorectal cancers and glioblastomas. N Engl J Med. 2004;351:2883. doi: 10.1056/NEJM200412303512724. [DOI] [PubMed] [Google Scholar]
- 16.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 17.Forrester K, Almoguera C, Han K, Grizzle WE, Perucho M. Detection of high incidence of K-ras oncogenes during human colon tumorigenesis. Nature. 1987;327:298–303. doi: 10.1038/327298a0. [DOI] [PubMed] [Google Scholar]
- 18.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 19.Parsons DW, Wang TL, Samuels Y, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 20.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 21.Di Fiore F, Blanchard F, Charbonnier F, et al. Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by Cetuximab plus chemotherapy. Br J Cancer. 2007;96:1166–1169. doi: 10.1038/sj.bjc.6603685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lievre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 23.Moroni M, Veronese S, Benvenuti S, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 24.Mariadason JM, Arango D, Shi Q, et al. Gene expression profiling-based prediction of response of colon carcinoma cells to 5-fluorouracil and camptothecin. Cancer Res. 2003;63:8791–8812. [PubMed] [Google Scholar]
- 25.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 26.Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 27.Mariadason JM, Rickard KL, Barkla DH, Augenlicht LH, Gibson PR. Divergent phenotypic patterns and commitment to apoptosis of Caco-2 cells during spontaneous and butyrate-induced differentiation. J. Cell. Physiol. 2000;183(3):347–354. doi: 10.1002/(SICI)1097-4652(200006)183:3<347::AID-JCP7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 28.Luo FR, Yang Z, Dong H, et al. Correlation of pharmacokinetics with the antitumor activity of Cetuximab in nude mice bearing the GEO human colon carcinoma xenograft. Cancer Chemother. Pharmacol. 2005;56:455–464. doi: 10.1007/s00280-005-1022-3. [DOI] [PubMed] [Google Scholar]
- 29.Goel A, Arnold CN, Niedzwiecki D, et al. Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability-high sporadic colorectal cancers. Cancer Res. 2004;64:3014–3021. doi: 10.1158/0008-5472.can-2401-2. [DOI] [PubMed] [Google Scholar]
- 30.Guanti G, Resta N, Simone C, et al. Involvement of PTEN mutations in the genetic pathways of colorectal cancerogenesis. Hum Mol Genet. 2000;9:283–287. doi: 10.1093/hmg/9.2.283. [DOI] [PubMed] [Google Scholar]
- 31.Shin KH, Park YJ, Park JG. PTEN gene mutations in colorectal cancers displaying microsatellite instability. Cancer Lett. 2001;174:189–194. doi: 10.1016/s0304-3835(01)00691-7. [DOI] [PubMed] [Google Scholar]
- 32.Frattini M, Signoroni S, Pilotti S, et al. Phosphatase protein homologue to tensin expression and phosphatidylinositol-3 phosphate kinase mutations in colorectal cancer. Cancer Res. 2005;65:11227. doi: 10.1158/0008-5472.CAN-05-2780. [DOI] [PubMed] [Google Scholar]
- 33.Velho S, Oliveira C, Ferreira A, et al. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur J Cancer. 2005;41:1649–1654. doi: 10.1016/j.ejca.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 34.Goyle S, Maraveyas A. Chemotherapy for colorectal cancer. Dig Surg. 2005;22:401–414. doi: 10.1159/000091441. [DOI] [PubMed] [Google Scholar]
- 35.Venook A. Critical evaluation of current treatments in metastatic colorectal cancer. Oncologist. 2005;10:250–261. doi: 10.1634/theoncologist.10-4-250. [DOI] [PubMed] [Google Scholar]
- 36.Liu B, Fang M, Schmidt M, et al. Induction of apoptosis and activation of the caspase cascade by anti-EGF receptor monoclonal antibodies in DiFi human colon cancer cells do not involve the c-jun N-terminal kinase activity. Br J Cancer. 2000;82:1991–1999. doi: 10.1054/bjoc.2000.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Layfield LJ, Bernard PS, Goldstein NS. Color multiplex polymerase chain reaction for quantitative analysis of epidermal growth factor receptor genes in colorectal adenocarcinoma. J Surg Oncol. 2003;83:227–231. doi: 10.1002/jso.10272. [DOI] [PubMed] [Google Scholar]
- 38.Petrova D, Jankova R, Yosifova A, et al. Tissue microarray analysis of EGFR gene amplification and gain in Bulgarian patients with colorectal cancer. Onkologie. 2006;29:198–200. doi: 10.1159/000092646. [DOI] [PubMed] [Google Scholar]
- 39.Shia J, Klimstra DS, Li AR, et al. Epidermal growth factor receptor expression and gene amplification in colorectal carcinoma: an immunohistochemical and chromogenic in situ hybridization study. Mod Pathol. 2005;18:1350–1356. doi: 10.1038/modpathol.3800417. [DOI] [PubMed] [Google Scholar]
- 40.Hirsch T, Eggstein S, Frank S, Farthmann E, von Specht BU. Autocrine growth stimulation of SW403 colon carcinoma cell line is caused by transforming-growth-factor-alpha-mediated epidermal growth factor receptor activation. J. Cancer Res. Clin. Oncol. 1996;122:328–334. doi: 10.1007/BF01220799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pouliot N, Burgess AW. Multiple autocrine factors including an extracellular matrix protein are required for the proliferation and spreading of human colon carcinoma cells in vitro. Growth Factors. 2000;18:31–49. doi: 10.3109/08977190009003232. [DOI] [PubMed] [Google Scholar]
- 42.Karnes WE, Jr, Walsh JH, Wu SV, et al. Autonomous proliferation of colon cancer cells that coexpress transforming growth factor alpha and its receptor. Variable effects of receptor-blocking antibody. Gastroenterology. 1992;102:474–485. doi: 10.1016/0016-5085(92)90093-e. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Kuropatwinski K, Hauser J, et al. Colon carcinoma cells harboring PIK3CA mutations display resistance to growth factor deprivation induced apoptosis. Mol Cancer Ther. 2007;6:1143–1150. doi: 10.1158/1535-7163.MCT-06-0555. [DOI] [PubMed] [Google Scholar]
- 44.Frattini M, Saletti P, Romagnani E, et al. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer. 2007;97:1139–1145. doi: 10.1038/sj.bjc.6604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berns K, Horlings HM, Hennessy BT, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 46.Khambata-Ford S, Garrett CR, Meropol NJ, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 47.Philp AJ, Campbell IG, Leet C, et al. The phosphatidylinositol 3'-kinase p85alpha gene is an oncogene in human ovarian and colon tumors. Cancer Res. 2001;61:7426–7429. [PubMed] [Google Scholar]
- 48.Edkins S, O'Meara S, Parker A, et al. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol. Ther. 2006;5:928–932. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi BH, Nashimoto T, Andoh R, et al. Mutation of the PI3' kinase gene in a human colon carcinoma cell line, HCC2998. DNA Cell Biol. 2006;25:399–405. doi: 10.1089/dna.2006.25.399. [DOI] [PubMed] [Google Scholar]
- 50.Zhang W, Gordon M, Schultheis AM, et al. FCGR2A and FCGR3A polymorphisms associated with clinical outcome of epidermal growth factor receptor expressing metastatic colorectal cancer patients treated with single-agent cetuximab. J Clin Oncol. 2007;25:3712–3718. doi: 10.1200/JCO.2006.08.8021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.