Abstract
Rare single-gene disorders cause chronic disease. However, half of the 6,000 recessive single gene causes of disease are still unknown. Because recessive disease genes can illuminate, at least in part, disease pathomechanism, their identification offers direct opportunities for improved clinical management and potentially treatment. Rare diseases comprise the majority of chronic kidney disease (CKD) in children but are notoriously difficult to diagnose. Whole exome resequencing facilitates identification of recessive disease genes. However, its utility is impeded by the large number of genetic variants detected. We here overcome this limitation by combining homozygosity mapping with whole exome resequencing in 10 sib pairs with a nephronophthisis-related ciliopathy, which represents the most frequent genetic cause of CKD in the first three decades of life. In 7 of 10 sib-ships with a histologic or ultrasonographic diagnosis of nephronophthisis-related ciliopathy we detect the causative gene. In six sib-ships we identify mutations of known nephronophthisis-related ciliopathy genes, while in two additional sib-ships we found mutations in the known CKD-causing genes SLC4A1 and AGXT as phenocopies of nephronophthisis-related ciliopathy. Thus whole exome resequencing establishes an efficient, non-invasive approach towards early detection and causation-based diagnosis of rare kidney diseases. This approach can be extended to other rare recessive disorders, thereby providing accurate diagnosis and facilitating the study of disease mechanisms.
INTRODUCTION
Rare recessive diseases cause chronic diseases that often require hospitalization.1 For example, rare chronic kidney diseases (CKD) comprise the majority of cases treated within chronic dialysis and renal transplantation programs in the first 3 decades of life, but are notoriously difficult to diagnose.2 However, the genetic basis of approximately half of recessive diseases including CKD is still unknown (http://omim.org/statistics/entries). As recessive mutations represent directly the primary disease cause, gene identification offers a powerful approach to revealing disease mechanisms in such disorders. Furthermore, because recessive mutations predominantly convey loss of function, recessive single-gene defects can be transferred directly into animal models, to study the related disease mechanisms and to screen for small molecules as possible treatment modalities.
Nephronophthisis (NPHP) is a recessive cystic kidney disease that represents the most frequent genetic cause of CKD in the first three decades of life. NPHP-related ciliopathies (NPHP-RC) are typically recessive single-gene disorders that affect kidney, retina, brain and liver by prenatal-onset dysplasia or by organ degeneration and fibrosis in early adulthood.3 Ultrasonographically, NPHP are characterized by increased echogenicity and cyst formation at the corticomedullary junction in small or normal-sized kidneys (Figure 1).4 And renal histology exhibits a characteristic triad of renal corticomedullary cysts, tubular basement membrane disruption, and tubulointerstitial inflitrations.5 Regarding renal, retinal and hepatic involvement there is phenotypic overlap of NPHP-RC with Bardet-Biedl syndrome (BBS).6 Identification of recessive mutations in 15 different genes (NPHP1-NPHP15)7–20 revealed that the encoded proteins share localization at the primary cilia-centrosomes complex, characterizing them as retinal-renal “ciliopathies”.3, 21 However, the 15 known NPHP-RC genes explain less than 50% of all cases with NPHP-RC, indicating that many of the genetic causes of NPHP-RC are still elusive.22, 23
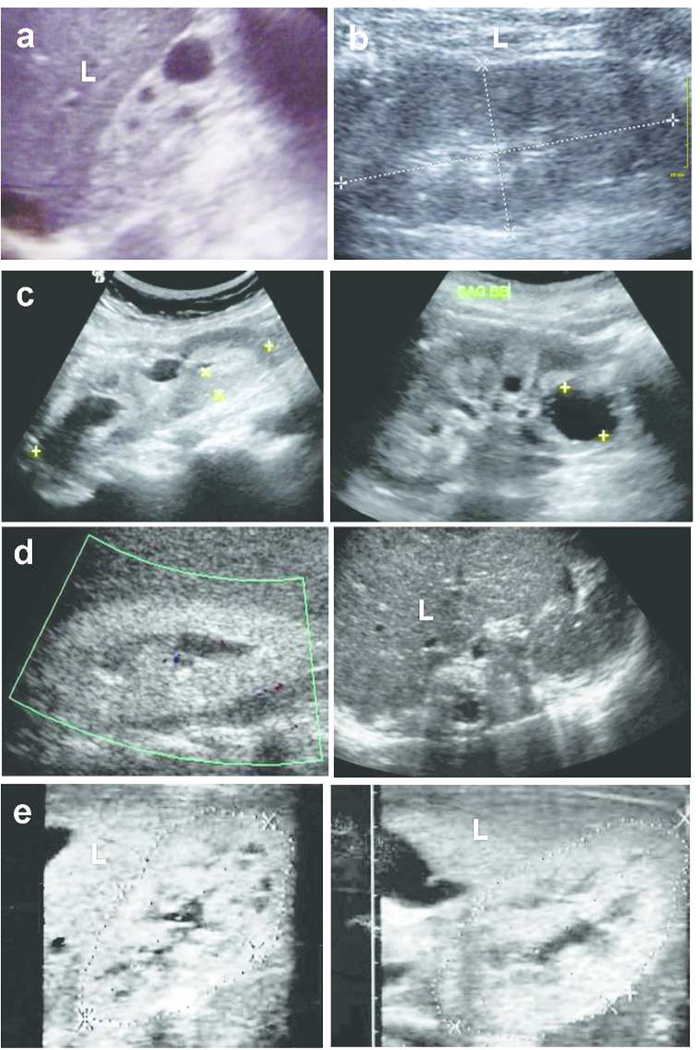
Figure 1. Images of representative renal ultrasound (RUS) and renal biopsy findings in individuals with an initial diagnosis of “NPHP-RC”.
(a) In A2557-21 with a mutation in NPHP4, RUS showed a normal-sized kidney with increased echogenicity when compared to liver (L), corticomedullary cysts (CMC) and loss of corticomedullary differentiation (CMD).
(b) In F93-29 with homozygosity mapping implicating the PKHD1 locus, RUS showed normal sized kidneys with small CMC and diminished CMD.
(c) In both siblings, F650-21 (left panel) and F650-22 (right panel) with dRTA as indicated by a mutation in SLC4A1, RUS exhibits increased echogenicity and CMC in normal sized kidneys with loss of CMD, which prompted the diagnosis of NPHP-RC early in the course of disease.
(d) In A3254 (left panel) and A3255 (right panel) with the molecular diagnosis of hyperoxaluria type 1 as indicated by a mutation in AGXT, RUS of A3255 exhibited CMC. RUS of A3254 showed mild distention of the collecting ducts.
(e) Right kidneys of siblings F838-21 (left panel) and -22 (right panel) harboring a heterozygous mutation in INPP5E exhibited CMC and increased echogenicity comparable to liver (L) signal.
Some of the more recently identified genetic causes of NPHP-RC are exceedingly rare.15 This observation necessitates a strategy to identify additional genetic causes of NPHP-RC in single affected families. In this context whole exome capture with consecutive massively parallel sequencing, (here referred to as whole exome resequencing, WER), theoretically offers a powerful approach towards gene identification in rare recessive diseases.24 However, the utility of WER is hampered by the large number of genetic variants that result from whole exome sequencing in any given individual.18, 25
To overcome the difficulty of variant prioritization in WER, we developed a strategy that combines WER18 with homozygosity mapping.26 We here apply this approach to 10 families with siblings with the diagnosis of “NPHP-RC”, based on clinical, renal sonographic, and/or histologic findings. Using this strategy we identified the primary causative mutations in 7 of the 10 sib pairs (70%). In six families we detect mutations of known NPHP-RC genes. In two additional families we revise the erroneous clinical diagnosis of NPHP-RC through identification of mutations in SLC4A1 and AGXT. This established the correct diagnoses of distal renal tubular acidosis and hyperoxaluria, respectively, which had appeared as clinical phenocopies of NPHP-RC.
We hereby establish a non-invasive molecular genetic approach towards early detection and causation-based diagnosis of rare kidney diseases by applying WER and homozygosity mapping to sibling cases. The approach is efficient and can be extended to all rare recessive diseases, thereby facilitating the study of disease mechanisms.
RESULTS
Clinical features of sibs with an NPHP-RC phenotype
From over 500 families with a diagnosis on NPHP-RC that were referred to us from worldwide sources for molecular genetic diagnosis we selected sibling cases with no known primary mutations from 10 different families (Table 1). Inclusion criteria were a diagnosis of NPHP-RC in both siblings based on renal ultrasonographic4 (Figure 1) and/or histologic5 findings characteristic for NPHP or a related ciliopathy. Many cases had extrarenal symptoms typical for NPHP-RC, including retinitis pigmentosa and neurologic involvement (Table 1).
Table 1. Primay causal mutations and clinical phenotypes of 10 sibships with diagnosis of a “nephronophthisis-related ciliopathy”.
Highlights denote known NPHP-RC genes (blue) and known NPHP-RC phenocopying genes (red).
| Family -Individuala |
Ethnic origin |
Causative Gene |
Nucleotide alterationb,c |
Deduced protein change |
Exon (state) |
Continuous amino acid sequence conservation in evolution |
Parental consanguinity |
Kidney (age at ESKD) |
Eye (age at RD) |
Other |
|---|---|---|---|---|---|---|---|---|---|---|
| Mutation of known NPHP-RC genes | ||||||||||
|
A2204 −21 −23 |
Arab | INVS/NPHP2 | c.2719C>Td | p.R907X | 13 (Hom) | - | Yes |
−21: (4 yr) −23: (4 yr?) |
nl | - |
|
A2557 −21 −31 (cousin) |
Arab African | NPHP4 | c.402delG | p.I135SfsX43 | 4 (Hom) | - | Yes |
Bx:
NPHP −11: died 13 yr −21: (at 9 yr, Creat. 7 mg/dL) (Figure 1a) −31: (at 32 yr Creat. 7 mg/dL) RUS: echogenic kidneys, CMC |
nl | polyuria, failure to thrive, salt craving |
|
A2882
(KK7) −21 (03) −22 (04) |
Saudi Arabian | BBS1 | c.1062 +58C>Td | cryptic splice site activation | Intron 10 (Hom) | - | Yes |
−21: ND −22: nl |
Retinitis pigmentosa | −21, −22: BBS, postaxial
polydactyly, obesity −21: webbed thumbs −22: speech delay |
|
A2888 (R1) −21 (04) −22 (05) |
Latino | BBS9 | c.1536A>G | p.T512T, 60% conserved splice donor site | 14 (Hom) | - | Yes |
−21: ND −22: ND |
Retinitis pigmentosa | BBS |
|
A2841
(AR245) −21 (03) −22 (04) |
Europe | ALMS1 | c.5900C>G c.8383insA |
p.S1967X p.L2797fsX3 |
7 (het) 9 (het) |
- | No | −21: left and right kidney
enlargement −22: kidney enlargement |
Nystagmus Retinitis pigmentosa |
−21, −22: Alström
syndreome, obesity, insulin resistance,
cardiomyopathy −21: recurrent otitis media, developmental delaly −22: microcephaly, asthma |
| Mutation of known NPHP-RC-phenocopying genes | ||||||||||
|
F650 −21 −22 |
Turkey | SLC4A1 | c.1571–1573delTCT | p.delF524 | 13 (Hom) | C. elegans | Yes (1st cousins) | −21, −22: Bx at 19 yr, 18 yr: NPHP (global sclerosis, cystic ectasia) RUS at 35 yr, 34 yr: ↑ EG, CMC, nl size (Figure 1c) |
−21: coloboma of iris, choroid | −21, −22: polyuria, failure to thrive, blood pH <7.35, oral intake of NaHCO3, 3 g /day |
|
A3254 A3255 (cousins) |
Saudi Arabia | AGXT | c.584T>Gd | p.M195R | 5 (Hom) | D. melanogaster | Yes | A3254: (ESKD stage
5) RUS: A3254: ↑ EG, nl size, mild distention of the collecting system A3255 (3 mo): ↑ EG, echogenic kidneys, died at 19 mo (Figure 1d) |
A3254: retinal
pigmentation A3255: retinal pigmentation |
A3254: brain atrophy, developmental delay;
hypotonia A3255: brain atrophy (MRI), short stature, CHD, respiratory failure, bone disease |
| Genetically unsolved cases | ||||||||||
|
F93
(A3223) −21 −24 −25 −29 |
Germany | e | NDe | NDe | ND (Hom)e | - | Yes |
Bx (−21,
−24, −25): NPHP −21: (15 yr) −24: (10 yr) −25: (died 5 yr) −29: (14 yr) RUS (−24, −29): small kidneys, CMC (Figure 1b) |
−24:coloboma |
−24: thorax deformity |
|
F838 −21 −22 |
Poland | INPP5E | c.925C>T | p.Q309X | (het) | - | No | Bx: NPHP −21: (6 yr) −22: (7 yr) RUS: echogenic kidneys, CMC (Figure 1e) |
nl | - |
|
A2059 −21 −22 −23 |
Turkey | - | - | - | - | - | Yes |
−21: (at 25 yr Creat. 1.0 mg/dL) −23: (at 19 yr Creat. 7.8 mg/dL) RUS: small kidneys |
nl |
−23: heart anomaly |
Individual with exome sequencing data is underlined in first column.
For GenBank accession numbers see Online Methods in Supplementary Material)
All mutations were absent from >270 healthy control individuals.
Mutation published in BIOBASE (http://www.biobase-international.com).
Although no mutation was detected, linkage mapping excluded all loci but the PKHD1 locus (see Figure S1h).
BBS, Bardet-Biedl syndrome; Bx, Kidney biopsy demonstrates nephronophthisis; CHD, congenital heart defect; CMC, corticomedullary cysts; Creat., serum creatinine; EG, echogenicity; ERG, electroretinogram; ESKD, end-stage kidney disease; GFR, glomerular filtration rate; Hom, homozygous mutation; het, heterozygous mutation; mo, months; ND no data; nl, normal; RD, retinal degeneration; RUS, renal ultrasound; yr, year(s); -, not applicable.
Homozygosity mapping in 10 sibs with a diagnosis of NPHP-RC
The finding that most of the known NPHP-RC genes (NPHP2-NPHP13) contribute causative mutations in only a small number of cases each (<1–3%)15 necessitates the ability to map and identify disease-causing genes in single families. We therefore employed a previously developed strategy,18, 26 that combines homozygosity mapping in single families with WER. We performed genome wide homozygosity mapping in the 10 sibships with NPHP-RC as described (see Figure S1).26 Eight families were known to be consanguineous and two had no evidence for consanguinity (Table 1). Homozygosity mapping yielded segments of likely homozygosity by descent (“homozygosity peaks”)26 in all eight families with consanguinity, but in none of the two families (A2841 and F838) without consanguinity (see Figure S1). This is consistent with our previous finding that individuals with known consanguinity exhibit segments of homozygosity upon mapping, whereas segments of homozygosity are rare in outbred families.26 In the eight consanguineous families the number of homozygosity peaks ranged from one to fifteen (Table 1 and Figure S1).
Mutations in six known NPHP-RC genes
Following homozygosity mapping and WER (Figure S1 and Table S1–S2), we identified recessive mutations in the known ciliopathy genes INVS/NPHP2, NPHP4, BBS1, BBS9, and ALMS1 in five families with multiple affected sibs with NHPH-RC (families A2204, A2557, A2882, A2888, and A2841) respectively (Table 1, Figure 2 and Table S1). Individual A2557-21 with a homozygous truncating mutation in NPHP4 had characteristic clinical signs (Table 1) and renal ultrasound features (Figure 1a) of NPHP. Interestingly, individual A2557-31, who is a cousin of A2557-21 and has the same mutation, developed end-stage kidney disease (ESKD) at 32 years. This late manifestation with ESKD beyond age 25 years is unusual in NPHP. Individuals A2882-21 and -22, who both carry a mutation in BBS1, presented with postaxial polydactyly and obesity. Mutations in ALMS1 cause Alström syndrome of which clinical features include blindness, obesity, type 2 diabetes, renal dysfunction, and hypertension. Individuals A2841-21 and -22, who have two truncating compound heterozygous mutations in ALMS1, presented with obesity, insulin resistance, retinitis pigmentosa and kidney enlargement which are consistent with the genetic findings.
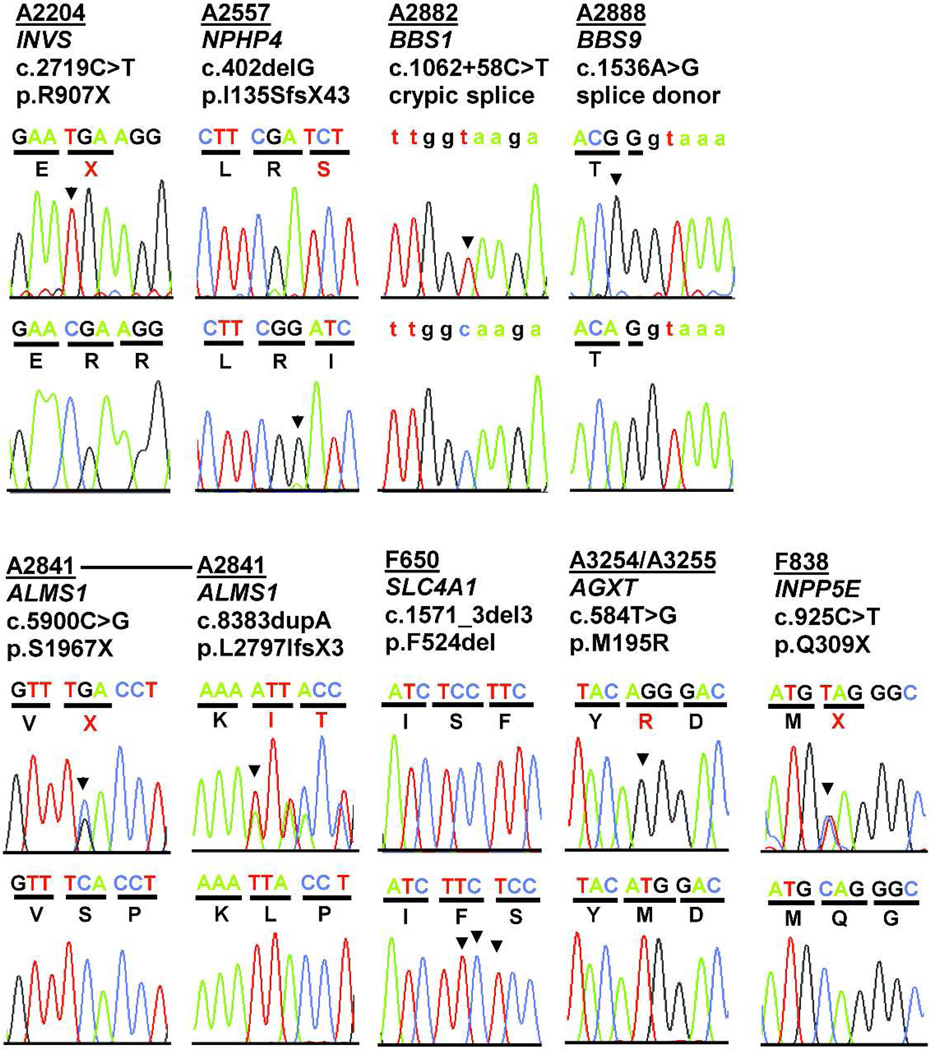
Figure 2. Recessive mutations detected by WER in 10 sibling cases with an NPHP-RC phenotype.
Families are listed in the same order as in Table 1. Family numbers (underlined), mutated gene, altered nucleotides and amino acid changes are given above sequence traces. Wild type control sequences are shown below mutated sequences. Codon triplets are underlined to indicate reading frame. Non-coding sequence is in lower case. Mutated nucleotides are denoted by an arrow head. All mutations were absent from >270 ethnically matched healthy controls. Five families have mutations in the known ciliopathy genes INVS/NPHP2, NPHP4, BBS1, BBS9, and ALMS1. Two families have mutations in known NPHP-RC phenocopying genes (SLC4A1 and AGXT). In F838 a heterozygous mutation was detected in INPP5E.
Mutations in two known CKD genes phenocopy NPHP-RC
Surprisingly, in families F650 and A3254 we identified mutations in the known CKD-causing genes SLC4A1 and AGXT1, respectively, that apparently represent phenocopies of NPHP-RC (Table 1).First, renal biopsy performed in both male siblings of family F650 at 19 and 18 years of age, respectively, revealed the suspected diagnosis of NPHP-RC with cystic tubular ectasia (Table 1). This diagnosis was supported by the findings of polyuria, polydipsia, failure to thrive, coloboma of the eye, and metabolic acidosis, which was thought to be secondary to renal failure from NPHP. Subsequent renal ultrasound performed at 35 and 34 years of age, respectively, also showed features characteristic of NPHP, including increased echogenicity and corticomedullary cysts in kidneys of normal size (Figure 1c). However, over the years both brothers developed requirement of oral bicarbonate supplementation of 3 g/day. They did not develop terminal renal failure by the ages of 35 and 34 years, respectively, and this late age of onset is not typical of NPHP. In addition, renal ultrasound showed increased echogenicity that was pronounced in the rims surrounding the corticomedullary renal cysts and in the pyramids (Figure 1d), a feature unusual for NPHP. Identification of a homozygous mutation that deletes a highly conserved amino acid residue in SLC4A1, which encodes the anion exchange protein 1 (AE1), enabled us to make the unexpected diagnosis of distal renal tubular acidosis (dRTA) (Table 1, Figure 2 and Table S1). Recessive mutations of SLC4A1 have been reported previously to cause dRTA with and without red blood cell dysmorphology.27
In another family with two affected cousins, A3254 and A3255, we suspected infantile-onset NPHP-RC (Table 1). Individual A3254 had end-stage kidney disease (ESKD) at three months with small echogenic kidneys on renal ultrasound (Figure 1d). Individual A3255 developed ESKD at 3 months of age, had brain atrophy and developmental delay, and died age 19 months. Both cousins displayed retinal pigmentation (Table 1). WER revealed a homozygous mutation in AGXT which encodes alanine-glyoxylate transferase 1, thereby establishing the diagnosis of hyperoxaluria type 1 (Table 1, Figure 1 and Table S1).28 Thus, in both families, we established an accurate molecular diagnosis by WER, which was previously incorrectly ascribed to NPHP-RC early in the disease course, even following detailed evaluation by renal biopsy or ultrasound.
In family F93 with four children with NPHP-RC and typical renal ultrasonographic features (Figure 1b), genetic mapping excluded the entire genome from linkage with a disease locus with the exception of the PKHD1 locus (Figure S1h). Although no mutations were detected in PKHD1 by WER, the mapping result implicates PKHD1 as the most likely causative gene, which is known to cause autosomal recessive polycystic kidney disease (ARPKD). The four affected children of family F93 had a phenotype unusual for ARPKD, because the kidneys were not enlarged, and there was extrarenal involvement with retinal coloboma.
Finally, two additional families, F838 and A2059 were non-consanguineous (Table 1) and did not yield homozygosity peaks upon genetic mapping (Figure S1i–j). In family F838 for which both affected individuals had a renal ultrasound consistent with NPHP (Figure 1e) we detected a heterozygous nonsense mutation in the ciliopathy gene INPP5E (Table 1 and Figure 2), but we were unable to detect any additional mutations in trans at the same locus. Finally, we were unable to detect a likely primary causal locus in family A2059 (Table 1 and Table S1). In addition, we examined variants in known ciliopathy genes in WER data of all 10 families. The included genes were NPHP1, INVS, NPHP3, NPHP4, IQCB1, CEP290, GLIS2, RPGRIP1L, NEK8, SDCCAG8, TMEM67, TTC21B, WDR19, ZNF423, CEP164, BBS1, BBS2, ARL6, BBS4, BBS5, MKSS, TTC8, BBS9, BBS10, TRIM32, BBS12, MKS1, WDPCP, TMEM216, AHI1, and CCDC28B. However, we could not detect any additional pathogenic variants in these genes in the seven solved and three unsolved cases. Furthermore, we checked genomic structural variants including large deletions and insertion, inversions, replacements, and translocations for the three unsolved cases based on WER, but there was no significant structural abnormality observed.
Taken together, we identified the disease-causing gene in 7 of 10 (70%) sibships, suggesting that homozygosity mapping with WER provides an efficient approach for molecular genetic diagnostics in diseases such as NPHP-RC and other ciliopathies where there is broad genetic locus heterogeneity.
DISCUSSION
Here, we demonstrate that WER, when combined with homozygosity mapping in sibling cases, represents a high-yield approach towards identification of primary causal mutations in rare recessive diseases. From our findings, we draw several conclusions: First, WER offers a viable, non-invasive approach for molecular diagnosis of rare recessive diseases. Second, however, to reduce the multitude of variants generated by WER, an a priori method to restrict this number is still required. Here, we show that the study of sib cases and the use of homozygosity mapping provides a robust solution to this problem. Third, using this approach, we achieved a high success rate for disease gene identification of 70%. In monogenic diseases about 85% of all recessive mutations are thought to reside within exons and adjacent intronic regions29 which are target regions of WER, so mutations in deep introns and promoter regions are not covered by WER. In addition, WER can miss a causal variant because of inadequate coverage (e.g. poor capture or poor sequencing) or inaccurate variant calling (e.g. a small but complex indel).30 Fourth, our study demonstrates that for individuals with childhood-onset renal failure, clinical diagnosis, renal ultrasound, and even renal histology represent relatively blunt diagnostic tools, which can be incapable of establishing the correct diagnosis. In this setting WER offers a powerful, non-invasive, cost-efficient diagnostic tool for arriving at a correct, unequivocal, etiology-based diagnosis.31 Fifth, rare, genetically heterogeneous chronic kidney diseases comprise the majority of cases of CKD in children but are notoriously difficult to diagnose. The use of WER will be beneficial for these individuals, because it will be possible to accurately assign them to therapeutic studies in larger cohorts. Sixth, our approach of combining homozygosity with WER can be applied to other rare recessive diseases. This may be of great clinical utility, as rare recessive disorders together cause a very high percentage of chronic diseases that require inpatient treatment in pediatrics. Finally, because WER reveals the major etiologic cause of a disease, gene identification will facilitate the elucidation of altered biological pathways, as well as the generation of animal models for testing of new treatment modalities.
WER now costs about $1,000 each per sample from several providers due to the substantial cost reductions associated with next-generation sequencing technologies. It usually takes four to eight weeks to get WER data after samples are submitted. Then, another four to eight weeks are required to analyze the WER data including alignments, variant filtering, confirmation and segregation analysis by Sanger sequencing. Therefore, the overall process usually takes at least two to three months. This is only valid when analysis of WER is combined with HM. When mapping data are not available, more time is necessary for evaluation and there is no standard protocol to filter variants from WER. Many laboratories are using their own way to filter variants and are evaluating WER differently. Therefore, to use WER widely as a diagnostic tool, a standard analytic pipeline should be established.
MATERIALS AND METHODS
Study Participants
From worldwide sources we obtained blood samples, clinical and pedigree data following informed consent from individuals with NPHP-RC and/or their parents. Approval for human subjects’ research was obtained from the University of Michigan Institutional Review Board and relevant local Review Boards. The diagnosis of NPHP-RC was made by (pediatric) nephrologists based on standardized clinical32, 33 and renal ultrasound4 criteria. Renal biopsies were evaluated by renal pathologists.5 Clinical data were obtained using a standardized questionnaire (http://www.renalgenes.org). The presence of retinal degeneration or neurologic involvement was evaluated by ophthalmologists and (pediatric) neurologists, respectively. In about 500 different families with NPHP-RC we excluded homozygous deletions of the NPHP1 gene. In a subset of these families we excluded mutations in selected known NPHP-RC genes using an approach of high-throughput mutation analysis.34, 35 The remaining 10 families with multiple affected siblings without a molecular genetic diagnosis were entered into this study for homozygosity mapping and WER.
Homozygosity mapping
For genome-wide homozygosity mapping26 the ‘Human Mapping 250k StyI’ array or the ‘Genome-wide Human SNP 6.0 Array’ from Affymetrix™ were utilized. Genomic DNA samples were hybridized, and scanned using the manufacturer’s standard protocol at the University of Michigan Core Facility (www.michiganmicroarray.com). Non-parametric LOD scores were calculated using a modified version of the program GENEHUNTER 2.136, 37 through stepwise use of a sliding window with sets of 110 SNPs using the program ALLEGRO.38 Genetic regions of homozygosity by descent (‘homozygosity peaks’) were plotted across the genome as candidate regions for recessive genes (see Figure S1), as described.18, 39 Disease allele frequency was set at 0.0001, and Caucasian marker allele frequencies were used.
Whole exome resequencing (WER)
Exome enrichment was conducted following the manufacturer’s protocol for the ‘NimbleGen™ SeqCap EZ Exome v2’ beads (Roche NimbleGen Inc.). The kit interrogates a total of approximately 30,000 genes (~330,000 CCDS exons). Massively parallel sequencing was performed largely as described in Bentley et al.40 For ten WER samples included in this study, the average of 118 million reads (100 bp) per each WER was obtained and the average coverage on target regions (exons) was 42.3 ± 13.4. For detail see Online Methods in Supplementary Material, available with the full text of this article at http://www.nature.com/ki.
Mutation calling
Sequence reads were mapped to the human reference genome assembly (NCBI build 36/hg18) using CLC Genomics Workbench™ (version 4.7.2) software (CLC bio, Aarhus, Denmark) as described in Online Methods in Supplementary Material. Mutation calling was performed in parallel with a team of geneticists/cell biologists, who had knowledge of the clinical phenotypes and pedigree structure, as well as experience with homozygosity mapping and exome evaluation. Because exon capture with subsequent massively parallel sequencing yields too many variants from normal reference sequence (VRSs) to make a confident decision regarding the disease-causing mutation of a single recessive disease-causing gene18, 25, we devised a strategy of a priori reduction of VRSs (see Online Methods (‘Filtering of variants from normal reference sequence’) and Table S1 in Supplementary Material).18
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the families who contributed to this study and the physicians who contributed clinical data, Davut Pehlivan, MD; Clifford Kashtan, MD; Judy Henry, MD; K.E. Bonzel, MD; Volker Klingmueller, MD; and Richard A. Lewis, MD. We thank Robert H. Lyons for excellent Sanger sequencing.
This research was supported by grants from the National Institutes of Health to F.H. (DK1069274, DK1068306, DK064614) and to N.K. (HD042601, DK075972, DK072301) and by grants from the European Community's Seventh Framework Programme FP7/2009 under grant agreement no: 241955, SYSCILIA to N.K.
H.Y.G. is a Research Fellow of the American Society of Nephrology (ASN). N.K. is a distinguished Jean and George Brumley Professor. F.H. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
DISCLOSURE
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Bell CJ, Dinwiddie DL, Miller NA, et al. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med. 2011;3:65ra64. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith JM, Stablein DM, Munoz R, et al. Contributions of the Transplant Registry: The 2006 Annual Report of the North American Pediatric Renal Trials and Collaborative Studies (NAPRTCS) Pediatr Transplant. 2007;11:366–373. doi: 10.1111/j.1399-3046.2007.00704.x. [DOI] [PubMed] [Google Scholar]
- 3.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blowey DL, Querfeld U, Geary D, et al. Ultrasound findings in juvenile nephronophthisis. Pediatr Nephrol. 1996;10:22–24. doi: 10.1007/BF00863431. [DOI] [PubMed] [Google Scholar]
- 5.Zollinger HU, Mihatsch MJ, Edefonti A, et al. Nephronophthisis (medullary cystic disease of the kidney). A study using electron microscopy, immunofluorescence, and a review of the morphological findings. Helv Paediatr Acta. 1980;35:509–530. [PubMed] [Google Scholar]
- 6.Badano JL, Mitsuma N, Beales PL, et al. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 7.Hildebrandt F, Otto E, Rensing C, et al. A novel gene encoding an SH3 domain protein is mutated in nephronophthisis type 1. Nat Genet. 1997;17:149–153. doi: 10.1038/ng1097-149. [DOI] [PubMed] [Google Scholar]
- 8.Otto EA, Schermer B, Obara T, et al. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat Genet. 2003;34:413–420. doi: 10.1038/ng1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olbrich H, Fliegauf M, Hoefele J, et al. Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto-retinal degeneration and hepatic fibrosis. Nat Genet. 2003;34:455–459. doi: 10.1038/ng1216. [DOI] [PubMed] [Google Scholar]
- 10.Otto E, Hoefele J, Ruf R, et al. A gene mutated in nephronophthisis and retinitis pigmentosa encodes a novel protein, nephroretinin, conserved in evolution. Am J Hum Genet. 2002;71:1167–1171. doi: 10.1086/344395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mollet G, Salomon R, Gribouval O, et al. The gene mutated in juvenile nephronophthisis type 4 encodes a novel protein that interacts with nephrocystin. Nat Genet. 2002;32:300–305. doi: 10.1038/ng996. [DOI] [PubMed] [Google Scholar]
- 12.Otto E, Loeys B, Khanna H, Hellemans J, Sudbrak R, Fan S, Muerb U, O'Toole JF, Helou J, Attanasio M, Utsch B, Sayer JA, Lillo C, Jimeno D, Coucke P, De Paepe A, Reinhardt R, Klages S, Tsuda M, Kawakami I, Kusakabe T, Omran H, Imm A, Tippens M, Raymond PA, Hill J, Beales P, He S, Kispert A, Margolis B, Williams DS, Swaroop A, Hildebrandt F. A novel ciliary IQ domain protein, NPHP5, is mutated in Senior-Loken syndrome (nephronophthisis with retinitis pigmentosa), and interacts with RPGR and calmodulin. Nat Genet. 2005;37:282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 13.Sayer JA, Otto EA, O'Toole JF, et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 14.Valente EM, Silhavy JL, Brancati F, et al. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- 15.Attanasio M, Uhlenhaut NH, Sousa VH, et al. Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nat Genet. 2007;39:1018–1024. doi: 10.1038/ng2072. [DOI] [PubMed] [Google Scholar]
- 16.Delous M, Baala L, Salomon R, et al. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat Genet. 2007;39:875–881. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- 17.Otto EA, Trapp ML, Schultheiss UT, et al. NEK8 mutations affect ciliary and centrosomal localization and may cause nephronophthisis. J Am Soc Nephrol. 2008;19:587–592. doi: 10.1681/ASN.2007040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otto EA, Hurd TW, Airik R, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet. 2010 doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bredrup C, Saunier S, Oud Machteld M, et al. Ciliopathies with Skeletal Anomalies and Renal Insufficiency due to Mutations in the IFT-A Gene WDR19. The American Journal of Human Genetics. 2011;89:634–643. doi: 10.1016/j.ajhg.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaki M, Airik R, Ghosh Amiya K, et al. Exome Capture Reveals ZNF423 and CEP164 Mutations, Linking Renal Ciliopathies to DNA Damage Response Signaling. Cell. 2012;150:533–548. doi: 10.1016/j.cell.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ansley SJ, Badano JL, Blacque OE, et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- 22.Otto EA, Ramaswami G, Janssen S, et al. Mutation analysis of 18 nephronophthisis associated ciliopathy disease genes using a DNA pooling and next generation sequencing strategy. J Med Genet. 2010 doi: 10.1136/jmg.2010.082552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halbritter J, Porath J, Diaz K, et al. Identification of 99 novel mutations in a worldwide cohort of 1,056 patients with a nephronophthisis-related ciliopathy. Hum Genet. 2013:1–20. doi: 10.1007/s00439-013-1297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ku C-S, Cooper DN, Polychronakos C, et al. Exome sequencing: Dual role as a discovery and diagnostic tool. Annals of Neurology. 2012;71:5–14. doi: 10.1002/ana.22647. [DOI] [PubMed] [Google Scholar]
- 25.Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hildebrandt F, Heeringa SF, Rüschendorf F, et al. A Systematic Approach to Mapping Recessive Disease Genes in Individuals from Outbred Populations. PloS Genetics. 2009;5:31000353. doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alper SL. Familial renal tubular acidosis. J Nephrol. 2010;23(Suppl 16):S57–S76. [PubMed] [Google Scholar]
- 28.Frishberg Y, Rinat C, Shalata A, et al. Intra-familial clinical heterogeneity: absence of genotype-phenotype correlation in primary hyperoxaluria type 1 in Israel. Am J Nephrol. 2005;25:269–275. doi: 10.1159/000086357. [DOI] [PubMed] [Google Scholar]
- 29.Lupski JR, Reid JG, Gonzaga-Jauregui C, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 31.Mistry K, Ireland JH, Ng RC, et al. Novel mutations in NPHP4 in a consanguineous family with histological findings of focal segmental glomerulosclerosis. Am J Kidney Dis. 2007;50:855–864. doi: 10.1053/j.ajkd.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Waldherr R, Lennert T, Weber HP, et al. The nephronophthisis complex. A clinicopathologic study in children. Virchows Arch A Pathol Anat Histol. 1982;394:235–254. doi: 10.1007/BF00430668. [DOI] [PubMed] [Google Scholar]
- 33.Hildebrandt F, Jungers P, Robino C, et al. Nephronophthisis, medullary cystic kidney disease and medullary sponge kidney disease. In: Schrier RW, editor. Diseases of the kidney and urinary tract. Philadelphia: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- 34.Otto EA, Ramaswami G, Janssen S, et al. Mutation analysis of 18 nephronophthisis associated ciliopathy disease genes using a DNA pooling and next generation sequencing strategy. J Med Genet. 2011;48:105–116. doi: 10.1136/jmg.2010.082552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harville HM, Held S, Diaz-Font A, et al. Identification of 11 novel mutations in eight BBS genes by high-resolution homozygosity mapping. J Med Genet. 2010;47:262–267. doi: 10.1136/jmg.2009.071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kruglyak L, Daly MJ, Reeve-Daly MP, et al. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 37.Strauch K, Fimmers R, Kurz T, et al. Parametric and nonparametric multipoint linkage analysis with imprinting and two-locus-trait models: application to mite sensitization. Am J Hum Genet. 2000;66:1945–1957. doi: 10.1086/302911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gudbjartsson DF, Jonasson K, Frigge ML, et al. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000;25:12–13. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- 39.Hildebrandt F, Heeringa SF, Ruschendorf F, et al. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 2009;5:e1000353. doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.