Abstract
The neural crest and craniofacial placodes are two distinct progenitor populations that arise at the border of the vertebrate neural plate. This border region develops through a series of inductive interactions that begin before gastrulation and progressively divide embryonic ectoderm into neural and non-neural regions, followed by the emergence of neural crest and placodal progenitors. In this review, we describe how a limited repertoire of inductive signals – principally FGFs, Wnts and BMPs – set up domains of transcription factors in the border region which establish these progenitor territories by both cross-inhibitory and cross-autoregulatory interactions. The gradual assembly of different cohorts of transcription factors that results from these interactions is one mechanism to provide the competence to respond to inductive signals in different ways, ultimately generating the neural crest and cranial placodes.
INTRODUCTION
The entire peripheral nervous system is derived from two multipotent progenitor domains that arise at the border of the future neural plate and epidermis. The neural crest, which forms along almost the entire length of the neuraxis, will generate the neurons and glia of the sensory and autonomic nervous systems, secretory, pigmented and mesenchymal cells as well as the bone and cartilage of much of the face (Betancur et al., 2010; Le Douarin and Kalcheim, 1999; Milet and Monsoro-Burq, 2012; Prasad et al., 2012; Stuhlmiller and Garcia-Castro, 2012a). In the head, the craniofacial placodes are a second population of progenitors that give rise to sensory structures such as the olfactory epithelium, the entire inner ear, neurons in a variety of cranial sensory ganglia, the lateral line system in anamniotes and accessory sensory structures such as the lens of the eye (Baker and Bronner-Fraser, 2001; Graham and Shimeld, 2013; Schlosser, 2005, 2010). They derive from a molecularly distinct domain in the neural plate border termed the pre-placodal region (Bailey and Streit, 2006; Bhattacharyya and Bronner-Fraser, 2004; Grocott et al., 2012; Schlosser, 2006; Streit, 2007). The development of neural crest and placodes is intimately associated with the induction of the neural plate. At early stages in neural induction, the border between the future anterior neural plate and epidermis contains cells capable of forming neural tissue, neural crest, placodal derivatives and epidermis (Baker et al., 1999; Basch et al., 2000; Bhattacharyya and Bronner-Fraser, 2008; Gallagher et al., 1996; Groves and Bronner-Fraser, 2000; Hans et al., 2007; Köster et al., 2000; Kwon et al., 2010; Pieper et al., 2012; Streit and Stern, 1999). A series of inductive interactions between border cells and the neural plate, epidermis and underlying mesoderm gradually partition the border region into two spatially and molecularly distinct domains, with neural crest forming immediately adjacent to the neural plate and the pre-placodal region forming slightly more laterally.
Neural crest and cranial placodes share some superficial similarities: they both originate from the border region, they can generate multiple cell types including sensory neurons and secretory cells, and are capable of producing migratory cells. The weight of recent molecular data and comparative studies of non-vertebrate chordates suggests they may be separate vertebrate innovations with independent evolutionary origins (Bronner and LeDouarin, 2012; Gasparini et al., 2013; Graham and Shimeld, 2013; Schlosser, 2005, 2008). The fact that two distinct progenitor populations differentiate from a common embryonic region at similar times raises the question of how these two cell populations become distinct from one another and from the surrounding neural and epidermal tissue when presented with a similar limited array of inducing signals.
In this review, we first summarize what is known about the cell-intrinsic transcription factors and environmental signals that establish neural and non-neural ectoderm early in development, and then describe some of the similarities and differences between placode and neural crest induction at the border of the neural plate. A number of comprehensive reviews of neural crest and cranial placode formation have appeared in the last few years (Betancur et al., 2010; Grocott et al., 2012; McCabe and Bronner-Fraser, 2009; Milet and Monsoro-Burq, 2012; Prasad et al., 2012; Sauka-Spengler and Bronner-Fraser, 2008; Schlosser, 2006, 2010; Stuhlmiller and Garcia-Castro, 2012a), and we refer the reader to these reviews for a detailed discussion of these inductive events. Here, we focus particularly on the ways in which patterns of transcription factors are gradually established and spatially refined within the developing border region, and how different cohorts of transcription factors confer competence on different cell populations to respond to inducing signals in different ways. Analysis of early events in neural crest and placode development has benefited from studies in zebrafish, Xenopus, chicken and mouse embryos. Although the precise timing of events and the identity of genes involved in these inductive processes can differ somewhat between species, we draw on studies in all these model organisms to give a consensus view of the major findings and unresolved problems in the field.
THE INDUCTION OF NEURAL AND NON-NEURAL ECTODERM
Most vertebrate embryos show some early division of the blastula into presumptive neural (or “pre-neural”) and non-neural tissue before the onset of gastrulation. Pre-neural markers such as ERNI, Geminin, Otx2 and Sox3 are expressed in future neural (dorsal) ectoderm (Bally-Cuif et al., 1995; Kroll et al., 1998; Papanayotou et al., 2008; Rex et al., 1997; Streit et al., 2000) and can be induced by neural inducing signals - FGF signals from underlying hypoblast, anterior visceral endoderm or presumptive endoderm in amniotes, and Wnt and BMP antagonists (Albazerchi and Stern, 2007; Papanayotou et al., 2008; Rogers et al., 2011; Stern and Downs, 2012; Streit et al., 2000; Wilson and Edlund, 2001). At the same time, a number of transcription factors that will become restricted to non-neural ectoderm are expressed more laterally, including members of the Ap2, Dlx, Foxi, Gata2/3, and Msx transcription factor families (Brown et al., 2005; Hans et al., 2007; Hoffman et al., 2007; Knight et al., 2003; Li and Cornell, 2007; Luo et al., 2001a; Matsuo-Takasaki et al., 2005; McLarren et al., 2003; Ohyama and Groves, 2004; Papalopulu and Kintner, 1993; Pera et al., 1999; Phillips et al., 2006; Pieper et al., 2012; Sheng and Stern, 1999; Woda et al., 2003). Consistent with their expression in non-neural tissue, these transcription factors are regulated by BMP and Wnt family members that are expressed in or adjacent to non-neural ectoderm (Beanan and Sargent, 2000; Bhat et al., 2013; Hoffman et al., 2007; Hong and Saint-Jeannet, 2007; Kwon et al., 2010; Matsuo-Takasaki et al., 2005; Pera et al., 1999; Skromne and Stern, 2001; Wilson et al., 2001), although the precise time at which these non-neural factors are expressed differs between species. Moreover, the requirement of these non-neural genes for inductive signals can change rapidly over time – for example, Ap2, Foxi1 and Gata2 require BMP signals for their expression in non-neural ectoderm before, but not after gastrulation in zebrafish (Bhat et al., 2013; Kwon et al., 2010).
The domains of pre-neural and non-neural genes can overlap significantly prior to gastrulation, but these domains become far more distinct as definitive neural tissue, marked by genes such as Sox2, is induced by signals from the organizer (Rex et al., 1997; Streit et al., 1997; Uchikawa et al., 2003). At this time, many non-neural genes become restricted to regions close to the developing neural plate (Feledy et al., 1999b; Khudyakov and Bronner-Fraser, 2009; Kwon et al., 2010; Pieper et al., 2012; Streit, 2002; Woda et al., 2003) under the influence of specific levels of BMP and FGF signaling. A number of genes associated with the early neural crest, such as Foxd3 also begin to be expressed in the border region (Khudyakov and Bronner-Fraser, 2009) together with other markers of the border region such as Zic genes (Hong and Saint-Jeannet, 2007; Monsoro-Burq et al., 2003). BMP, FGF and Wnt signals all participate in the positioning of these genes at the neural plate border. It should be stressed that the expression of genes in the border region at this stage is not uniform, with some sets of genes being expressed more laterally in the border region than others. The significance of these subdivisions at this stage is not clear and can vary between different species. Nevertheless, it is notable that loss of function of many of the genes expressed in the border region at this stage can produce phenotypes in neural, neural crest or placodal derivatives (for example (Esterberg and Fritz, 2009; Hoffman et al., 2007; Li and Cornell, 2007; Lillevali et al., 2006; Robledo and Lufkin, 2006; Woda et al., 2003), although none of these genes appears to affect all border derivatives when disrupted individually. This suggests that at this early stage in border formation, combinations of transcription factor are preparing or priming the border region to respond to lineage-specific inducing signals, but are not yet distinguishing the lineages one from the other. We will discuss this in more detail later in the review.
Despite the heterogeneity that is already apparent in the border region at this stage, a number of consensus conclusions regarding the decision between neural and non-neural ectoderm emerge from studies in different species. First, non-neural genes are typically induced in primitive ectoderm by BMP and Wnt signaling, whereas FGF signals promote early neural markers and the subsequent differentiation of the neural plate in concert with Wnt and BMP antagonists from the organizer (Levine and Brivanlou, 2007; Rogers et al., 2009; Wilson and Edlund, 2001). Second, as definitive neural tissue emerges, mutually repressive interactions between neural plate genes and border genes serve to refine and sharpen the neural plate boundary. For example, over-expression of Dlx, Gata, Msx, Foxi and Ap2 factors can repress neural markers such as Sox2, whereas loss-of-function experiments with the same genes expand the neural plate at the expense of non-neural ectoderm (Feledy et al., 1999a; Kwon et al., 2010; Linker et al., 2009; Luo et al., 2001b; Matsuo-Takasaki et al., 2005; McLarren et al., 2003; Pieper et al., 2012; Tribulo et al., 2003; Woda et al., 2003). Finally, positive autoregulatory interactions between different non-neural genes act to strengthen and maintain the boundary between neural and non-neural domains (Kwon et al., 2010; Pieper et al., 2012). For example, although Ap2, Foxi1 and Gata2 require BMP signals for their induction in non-neural ectoderm, they do not require BMP for their subsequent maintenance, and instead positively regulate one another’s expression (Bhat et al., 2013; Kwon et al., 2010; Pieper et al., 2012). This positive autoregulation can be attenuated and stabilized by auto-inhibitory interactions of Foxi1 and Gata3 with their own enhancers (Bhat et al., 2013).
INDUCTION OF THE PRE-PLACODAL REGION
After the establishment of neural and non-neural territories in embryonic ectoderm, future neural crest and placodal lineages begin to be determined. We deal first with the pre-placodal region, which is operationally defined by members of the Six and Eya families (Grocott et al., 2012). The Six gene family (Six1-6) are homeodomain-containing transcription factors that are homologous to the Drosophila sine oculis gene, and which can form activating complexes with Eya (eyes absent) co-factors, or repressive complexes with Groucho or Dach proteins (Christensen et al., 2008; Donner and Maas, 2004; Hanson, 2001; Kawakami et al., 2000; Kumar, 2009; Wawersik and Maas, 2000). Six and Eya genes are expressed in the anterior neural plate border region, extending from approximately the first pair of somites to the most anterior regions of the neural plate (Ahrens and Schlosser, 2005; Bessarab et al., 2004; Brugmann et al., 2004; Esteve and Bovolenta, 1999; Ishihara et al., 2008; Kobayashi et al., 2000; Litsiou et al., 2005; McLarren et al., 2003; Mishima and Tomarev, 1998; Pandur and Moody, 2000). By the early stages of neurulation this expression domain lies slightly lateral to the neural crest progenitor domain at the edge of the neural plate (Christophorou et al., 2009; Grocott et al., 2012; Litsiou et al., 2005; Streit, 2002). The Six-Eya-expressing region is believed to encompass the progenitors of all craniofacial placodes and Six and Eya gene family members continue to be expressed in many placodal derivatives as they differentiate (Ahmed et al., 2012a; Ahmed et al., 2012b; Bessarab et al., 2004; Purcell et al., 2005; Schlosser, 2007; Xu et al., 1999; Zhang et al., 2004; Zheng et al., 2003; Zhu et al., 2002; Zou et al., 2004; Zou et al., 2006). Just as Drosophila sine oculis and eyes absent form a regulatory network with the Pax gene homologue, eyeless (Chen et al., 1997; Cheyette et al., 1994; Pappu and Mardon, 2004; Pignoni et al., 1997a; Pignoni et al., 1997b; Serikaku and O’Tousa, 1994), vertebrate Six genes can regulate the expression of Pax gene family members in differentiating placodes (Christophorou et al., 2009; Lagutin et al., 2001; Liu et al., 2006; Purcell et al., 2005), although the vertebrate Six genes appear to act upstream of Pax genes, rather than downstream as in Drosophila. Loss of different Six or Eya family members in different species (including humans) can cause specific defects in one or more placodal derivatives (Abdelhak et al., 1997; Bricaud and Collazo, 2006; Chen et al., 2009; Johnson et al., 1999; Konishi et al., 2006; Kozlowski et al., 2005; Laclef et al., 2003; Li et al., 2003; Ozaki et al., 2004; Ruf et al., 2004; Wayne et al., 2001; Winchester et al., 1999; Xu et al., 1999; Zhang et al., 2004; Zheng et al., 2003; Zou et al., 2004; Zou et al., 2006). Mutation, knock-down or dominant-negative mutants of Six and Eya family members can also cause defects in the pre-placodal region, whereas over-expression of Six1 and 4 genes, in some cases without their Eya1 or 2 co-factors, can expand the pre-placodal region at the expense of neural crest and epidermis (Brugmann et al., 2004; Christophorou et al., 2009). Given these results, it is somewhat surprising that to date, no combination of Six and Eya mouse knockouts or knock-down or dominant-negative mutants of these gene family members in other vertebrates has resulted in the complete absence of one or more placodes (see references above). It is not clear whether this is due to redundancy and compensation between different Six or Eya family members, to the use of techniques such as morpholino knockdown that can give an incomplete loss of function for technical reasons, or that these genes are not necessary for the first stages in placode formation, but rather regulate later aspects of the differentiation of specific placodes.
Induction of pre-placodal region genes is regulated by similar sets of signals that act at earlier stages to establish the boundary between neural and non-neural ectoderm. FGF family members, including FGF8 and FGF4, are expressed in cranial mesoderm underlying the neural plate border region. In Xenopus and chick, removal of this mesoderm can prevent induction of Six and Eya genes, while cranial mesoderm grafts can activate Six and Eya gene expression in non-neural ectoderm (Ahrens and Schlosser, 2005; Litsiou et al., 2005). Blockade of FGF signaling in chick and Xenopus can down-regulate Six and Eya gene expression, and FGF8 is sufficient to induce at least some pre-placodal genes such as Eya2 in competent ectoderm (Ahrens and Schlosser, 2005; Brugmann et al., 2004; Litsiou et al., 2005). Several members of the Wnt and BMP families are expressed in non-neural ectoderm both lateral and posterior to the pre-placodal region (Wnt8c and Wnt6; (Garcia-Castro et al., 2002; Jayasena et al., 2008; Kil et al., 2005; Litsiou et al., 2005; Schubert et al., 2002); BMP4 and 7; (Fainsod et al., 1994; Streit et al., 1998)), whereas several antagonists of both BMPs and Wnts are expressed in the mesoderm underlying the pre-placodal region (Cerberus and DAN; (Chapman et al., 2004; Ogita et al., 2001; Rodriguez Esteban et al., 1999)), or in the pre-placodal region itself (Cv2; (Esterberg and Fritz, 2009)). This suggests that inhibition of both signaling pathways may promote pre-placodal gene expression at the neural plate border. Accordingly, activation of the Wnt signaling pathway blocks or attenuates expression of the pre-placodal genes Eya2, Six1 and Six4, whereas inhibition of Wnt signaling by ectopic expression of Wnt antagonists can expand the expression domain of these genes (Brugmann et al., 2004; Litsiou et al., 2005). Similarly, BMPs can block expression of pre-placodal genes, whereas expression of SMAD intracellular effectors that inhibit BMP signaling, or of secreted antagonists of BMPs such as Noggin can expand the expression of several pre-placodal genes (Ahrens and Schlosser, 2005; Kwon et al., 2010; Litsiou et al., 2005). As will be discussed below, Wnts and BMPs have very different effects on the induction of the pre-placodal region and neural crest, and it is likely that the first differentiation steps that distinguish these two progenitor populations establish alternative competent states that underlie their differing responses to these signals.
At present it is unclear whether the effects of FGFs, Wnts and BMPs on the expression of pre-placodal genes are direct or indirect. With the exception of Six1, almost nothing is known about the regulatory enhancers controlling expression of pre-placodal genes. In the case of Six1, the single enhancer identified to date that can direct expression to the pre-placodal region does not contain binding sites for the direct transcriptional effectors of the FGF, BMP or Wnt pathways (Sato et al., 2012; Sato et al., 2010), although it does contain a binding site for Msx1 which is frequently regulated by BMP signaling (Monsoro-Burq et al., 1996). While evidence for a direct regulation of pre-placodal genes by these signaling cascades must await better characterization of their regulatory regions, there is good evidence for their regulation by other transcription factors that are expressed earlier in the neural plate border region. For example, Dlx3 and 5 gene family members are able to up-regulate expression of both Six and Eya genes in zebrafish, Xenopus and chick, and knock-down of these Dlx genes can attenuate pre-placodal gene expression (Esterberg and Fritz, 2009; Kaji and Artinger, 2004; McLarren et al., 2003; Pieper et al., 2012; Solomon and Fritz, 2002). Dlx5 may act directly in this context, as it is able to bind directly to the Six1 enhancer (Sato et al., 2010). Markers of non-neural ectoderm in the early gastrula, such as Ap2, Foxi1 and Gata2/3 genes can be regulated by BMP, and simultaneous but not individual knock-down of these factors causes a loss of both Six and Eya family members from the pre-placodal region (Kwon et al., 2010). Moreover, the expression of these factors in non-neural ectoderm seems to be necessary for this tissue to activate pre-placodal gene expression in response to inducing signals such as FGFs and BMP inhibition (Bhat et al., 2013; Kwon et al., 2010; Pieper et al., 2012). Other transcription factors such as the Iroquois factors Irx1/Xiro1 are expressed in the pre-placodal region immediately before Six and Eya genes and can positively regulate their expression (Glavic et al., 2002; Glavic et al., 2004; Gomez-Skarmeta et al., 1998; Goriely et al., 1999; Khudyakov and Bronner-Fraser, 2009). Irx genes can be regulated by both FGF and BMP signaling in some circumstances (Bellefroid et al., 1998; Glavic et al., 2004), and so it is possible that they act as direct mediators of these signals to activate pre-placodal genes.
As described above, the early boundary between neural and non-neural ectoderm is sharpened by positive autoregulation between non-neural genes and mutual inhibition between neural and non-neural genes. Once pre-placodal region gene expression is established adjacent to the neural plate, similar strategies of mutual positive autoregulation and cross-inhibition are used to sharpen and reinforce this domain. For example, a Six1;Eya1/2 complex can repress expression of Dlx5 and Gata3, whose expression is initially required to induce Six and Eya genes (Bhat et al., 2013; Brugmann et al., 2004; Christophorou et al., 2009; Kwon et al., 2010; Pieper et al., 2012). Over-expression of Six1 and Eya2 can up-regulate endogenous Eya2, and also other Six genes such as Six4 (Christophorou et al., 2009), although it is again unclear whether this positive regulation is direct or mediated through other transcription factors.
NEURAL CREST INDUCTION AND ITS RELATIONSHIP TO THE PRE-PLACODAL REGION
Neural crest progenitors begin to be induced at the neural plate border region somewhat earlier than the pre-placodal region (Milet and Monsoro-Burq, 2012), although induction of both populations overlaps temporally. Pax3 (Xenopus) or Pax7 (chick), are expressed shortly after gastrulation at the future neural plate border (Basch et al., 2006; Milet et al., 2013; Murdoch et al., 2010, 2012) in a pattern overlapping genes such as Dlx5/6, Gata2/3, Foxi1/3, Msx1/2, Zic1, Gbx2 and Ap2 (Grocott et al., 2012; Khudyakov and Bronner-Fraser, 2009). Foxd3 is often thought of as a defining neural crest marker (Labosky and Kaestner, 1998; Sasai et al., 2001; Sauka-Spengler and Bronner-Fraser, 2008; Stewart et al., 2006), although as described above, it is also expressed in early pre-neural ectoderm, in the organizer and in epiblast and trophectoderm lineages in mice (Arduini and Brivanlou, 2012; Steiner et al., 2006; Tompers et al., 2005; Xu et al., 2009) and in the presumptive posterior neurectoderm in Xenopus (Sasai et al., 2001). Some of these genes such as Msx1/2 and Foxd3 will eventually localize with Pax3 or Pax7 to the developing neural folds where nascent neural crest cells will form, marked by the expression of Snail and SoxE family genes (Aoki et al., 2003; Betancur et al., 2010; Grocott et al., 2012; Hong and Saint-Jeannet, 2005; Lee et al., 2004; Milet and Monsoro-Burq, 2012). Others, such as Foxi1/3, Dlx5/6 and Gata2/3 will localize with pre-placodal region genes such as Six1/4 and Eya1/2 (Grocott et al., 2012). At the neural fold stage, neural crest markers and the pre-placodal region are finally distinct, with neural crest markers expressed in the neural folds and pre-placodal genes expressed slightly more laterally in the ectoderm. There is good evidence that mutually repressive interactions occur between transcription factors of the neural crest and placode lineages, just as earlier interactions demarcate the boundary between neural and non-neural ectoderm. For example, the pre-placodal gene Six1 can repress the neural crest factors Msx1 and Foxd3, whereas Pax7 and Msx1 in turn repress Six1 (Sato et al., 2010). We will discuss the implications of this segregation of neural, non-neural, neural crest and pre-placodal genes later in the review.
The tissue interactions that promote neural crest formation have been well characterized (reviewed in (Milet and Monsoro-Burq, 2012; Prasad et al., 2012; Sauka-Spengler and Bronner-Fraser, 2008). Since neural crest cells emigrate from the junction of the neural folds with the adjacent ectoderm, an interaction between these tissues has long been proposed to induce neural crest formation. Juxtaposition of neural plate tissue with epidermis results in the generation of migratory neural crest cells (Dickinson et al., 1995), although there is still debate as to whether neural crest cells are generated exclusively from neural plate tissue or from both neural and epidermal tissue in vivo (Pieper et al., 2012; Schlosser, 2008). The precise contribution of neural crest cells in different experiments may be due to differences in the age of tissue used, or the environment (in vitro versus in ectopic grafts in vivo) in which the apposition of neural and epidermal tissue was performed.
There is also good evidence that mesoderm may induce the formation of neural crest tissue (Bonstein et al., 1998; Marchant et al., 1998; Mayor et al., 1995; Monsoro-Burq et al., 2003; Steventon et al., 2009). Some of the experiments implicating mesoderm in neural crest specification were performed in young embryos prior to the overt formation of the neural plate. For example, expression of Pax7 in chick embryos is observed in early gastrulating embryos adjacent to pre-neural (Sox3+) ectoderm and lying above early mesoderm (Basch et al., 2006). It is therefore likely that induction of neural crest involves both early signals emanating from the mesoderm and subsequent interactions between ectodermally-derived cells that may function to strengthen and maintain a neural crest precursor state. A large body of recent work on the secreted signals promoting neural crest specification support this view (Milet and Monsoro-Burq, 2012). An early requirement for both FGF and Wnt signaling has been demonstrated in zebrafish, Xenopus and chick (LaBonne and Bronner-Fraser, 1998; Mayor et al., 1997; Monsoro-Burq et al., 2003; Monsoro-Burq et al., 2005; Stuhlmiller and Garcia-Castro, 2012b; Villanueva et al., 2002), and this phase of neural crest induction may also involve partial inhibition of BMP signaling from non-neural ectoderm. Such inhibition may be mediated in part by FGF signaling, which is known to trigger the phosphorylation and attenuate the activity of Smad proteins transducing BMP signals (Pera et al., 2003). A second stage of signaling occurs at neurula stages that maintains neural crest fate and promotes the expression of later neural crest markers in response to Wnts from the neural folds, and BMPs in the dorsal neural folds and surrounding epidermis (Garcia-Castro et al., 2002; Steventon et al., 2009).
TROUBLE AT THE BORDER: UNRESOLVED ISSUES WITH THE SEGREGATION OF BORDER PROGENITORS
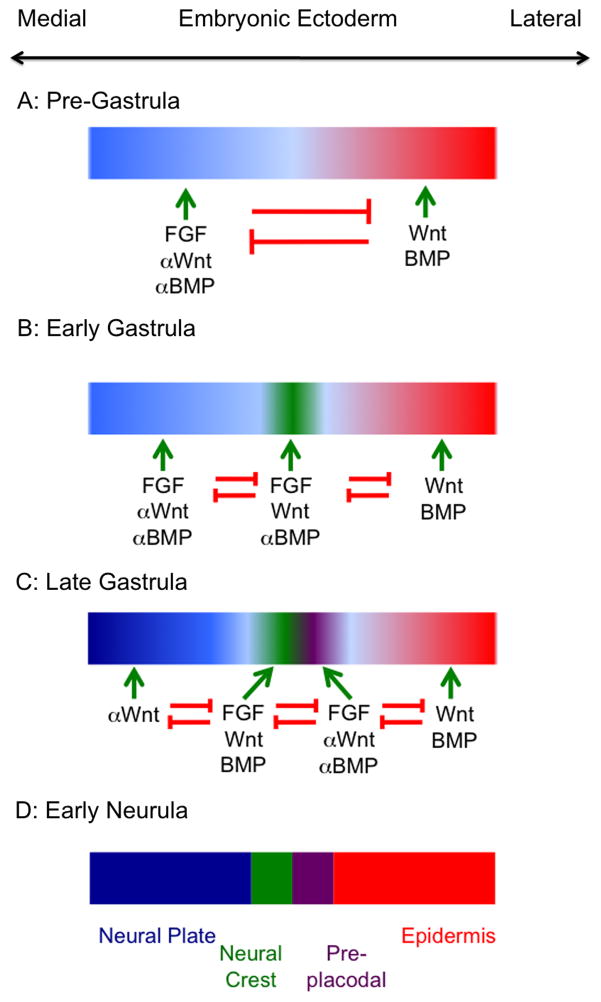
The above sections provide a rudimentary outline of the developmental decisions that mediate the emergence of neural versus non-neural ectoderm and the appearance of neural crest and placodal progenitors at the neural plate border. In brief, pre-neural ectoderm begins to develop under the influence of FGFs and the suppression of BMP and Wnt signaling, while non-neural ectoderm genes are regulated by Wnt and BMP signals. As definitive neural tissue begins to form in response to FGFs and the suppression of Wnt and BMP signaling, neural crest progenitors become specified under the influence of FGF and Wnt signals and the suppression of BMP signals. At the same time, pre-placodal tissue becomes distinguishable from non-neural ectoderm under the influence of FGFs and the suppression of both Wnt and BMP signals. Finally, as neurulation begins, Wnts and BMPs expressed at the edge of the neural plate stabilize and maintain a neural crest cell state, whilst distinct sets of signals along the anterior-posterior axis induce formation of specific placodes. (Figure 1)
Figure 1.
A graphical summary of the signals received by embryonic ectoderm during the establishment of the neural plate, neural crest, pre-placodal region and epidermis. The diagram is a consensus of data taken from zebrafish, Xenopus, chick and mouse studies and is not intended to be an accurate representation of any one species. (A): Wnt and BMP signals in the ectoderm initiate differentiation of non-neural ectoderm, while these signals are counteracted by FGFs and BMP and Wnt inhibitors from the organizer or hypoblast. (B): Wnt and FGF signals start to induce the first neural crest genes; BMP signaling is not required for this step and may be actively inhibited. (C) Pre-placodal genes begin to be induced by FGFs and by an attenuation of Wnt and BMP signals. Wnts and BMPs begin to be expressed at the edge of the neural plate and continue to induce neural crest tissue. (D): The final resolution of the border region into four distinct regions.
Although there is much evidence from zebrafish, Xenopus and chick for this consensus view of neural plate border development, a number of important questions remain. In particular, the mechanism by which distinct levels of FGF, Wnt and BMP signaling are interpreted by closely opposed or intermingled progenitors to yield distinct developmental outcomes remains unclear. Cell labeling studies from chick and zebrafish support the notion that progenitors for the neural plate, epidermis, neural crest and placodes are initially tightly intermingled and gradually segregate from one another as development proceeds (Bhattacharyya et al., 2004; Bhattacharyya and Bronner-Fraser, 2008; Chen and Streit, 2013; Ezin et al., 2009; Fernandez-Garre et al., 2002; Hatada and Stern, 1994; Kozlowski et al., 1997; Streit, 2001; Xu et al., 2008). Since cells in the border region do not express lineage–specific markers at this stage, the state of commitment of these progenitors is not clear. At one extreme, most cells in the early border region might be multipotent and would gradually receive specific signals to determine their fate at later times. Alternatively, border cells at this early stage may already have received signals committing them to a particular fate, and the committed progenitors would simply need to segregate or actively migrate into different territories. Although directed movements resulting from interactions between neural crest and placodal cells have been characterized at later stages of development in Xenopus (Theveneau et al., 2013), there is no good evidence of directed migration of progenitors in the boundary region. Part of the difficulty in understanding how intermingled progenitors might respond differently to inducing or migratory signals are due to technical limitations in determining cell fate and tracking gene expression in this region, and understanding to what extent individual cells are responding to particular signals. We discuss some of these issues below.
Although the changing patterns of gene expression at the neural plate border have been well-characterized (Figure 2), we know almost nothing about how gene expression at the border correlates with progenitor fate, or indeed whether it is predictive of cell fate at all, especially at early stages. Very little is known of the fate of individual progenitor cells at the neural plate border in any species, nor of how the range of potential fates of these progenitors might change over time. As an illustration, consider the transcription factors Foxd3 and Gata3 that are initially considered to be expressed in pre-neural and non-neural ectoderm respectively at the onset of gastrulation, and which end up being expressed by neural crest and pre-placodal ectoderm respectively at neurulation stages (Khudyakov and Bronner-Fraser, 2009; McLarren et al., 2003). If it were possible to identify individual Foxd3- or Gata3-expressing cells at different points in development and follow their fates faithfully, when and how would the gene expression patterns and cell identities of their progeny change? Do the progeny of Foxd3-expressing cells contribute to neural plate, neural crest, placode and epidermis, and do some of these fates become restricted before others? Alternatively, are Foxd3-expressing cells already committed to a neural crest fate at these early stages? At present, we have no information of how the fate of single cells at the border region correlates with their early gene expression, and how the developmental potential of these cells may change over time in response to signals from neighboring cells and tissues.
Figure 2.
A graphical summary of the expression pattern of different transcription factors in the establishment of the neural plate border. Panels A-D show the transition from the establishment of neural and non-neural ectoderm domains prior to gastrulation (A), to the early neurula stage where distinct neural, epidermal, placodal and neural crest territories can be observed. Shading of colored blocks implies that the edges of a particular gene expression domain is not sharply defined.
A second problem with interpreting gene expression in terms of cell fate at the neural plate border– or anywhere else in the early embryo for that matter – are the limitations of techniques to visualize gene expression. RNA in situ hybridization is used almost exclusively to visualize gene expression at the neural plate border, but suffers from the limitation that different mRNA species have different stabilities and that the stability of their protein products can vary from hours to days. This wide variation is true even for transcription factors that are the main class of genes used to identify neural plate border progenitors. In an attempt to pinpoint the time at which cells at the border region receive inductive signals, some groups have made use of specification assays, in which small pieces of embryonic tissue are removed from specific regions of the embryo and cultured alone in “simple” defined media. If the tissue adopts a particular fate in culture, it is said to be specified, i.e. that it has already received sufficient inducing signals to allow a particular fate to be adopted without further inductive events. Such assays must be interpreted with caution, particularly when carried out at early embryonic stages, because many regions of the early embryo, including the neural plate border region, are heterogeneous collections of different progenitors. It is therefore difficult to exclude the possibility that ongoing inductive interactions between different progenitors in the explants contributed to the observed fate in culture, or determine if the over-representation of one derivative over another was a consequence of cell competition, differential proliferation or cell death. The difficulty in interpreting specification assays is most problematic with very young embryonic tissue, extended periods of culture or both.
A final problem in understanding the individuation of neural crest and placodes at the border region in response to FGF, BMP and Wnt signals is that it is currently not possible to interrogate individual cells at the border region in real time to determine the levels to which different signaling pathways have been activated. Nevertheless, it is likely that individual cells within the neural plate border region experience constantly fluctuating levels of FGF, Wnt and BMP signaling as the cell types that will arise from this domain begin to segregate. Consequently, the expression of transcription factors that will ultimately mark neural, epidermal, neural crest or placode lineages is also likely to fluctuate at the single cell level. How, then, does this developmentally malleable set of progenitors ultimately segregate into defined territories?
As described above, there is clear evidence for both positive and negative feedback interactions among transcription factors expressed in the border region: neural factors repress non-neural factors and reinforce their own expression, and vice versa (Bhat et al., 2013; Kwon et al., 2010; Pieper et al., 2012).. This has also been seen for neural crest and pre-placodal transcription factors (Bhat et al., 2013; Esterberg and Fritz, 2009; Kaji and Artinger, 2004; McLarren et al., 2003; Pieper et al., 2012; Solomon and Fritz, 2002). At the same time, there is abundant evidence that the BMP, FGF and Wnt signaling pathways can positively and negatively cross-regulate each other at multiple levels (Bilican et al., 2008; Cho et al., 2013; Ille et al., 2007; Inui et al., 2012; Itasaki and Hoppler, 2010; Liu et al., 2012; Pera et al., 2003; Retting et al., 2009; Squarzoni et al., 2011; Tirosh-Finkel et al., 2010). It is therefore feasible that positive and negative feedback loops between these signaling pathways, and the transcription factors they regulate, may drive the segregation of neural crest and placodal progenitors in the border region. It is also possible that cell-cell interactions may reinforce these positive and negative feedback loops, although there are currently no known examples of this occurring at the neural plate border. Moreover, a number of labeling and time lapse studies suggest there is considerable movement within and between the future neural, neural crest and placode territories at the neural plate border (for example, Streit, 2001; Ezin et al., 2009) and the contributions these cell rearrangements may make to ultimate fate segregations remains unknown.
TRANSCRIPTION FACTORS IN DEVELOPMENTAL COMPETENCE: BUILDING COMBINATORIAL CODES FOR LINEAGE-SPECIFIC DIFFERENTIATION
Despite gains in our understanding of the signals that play instructive roles at the border, and transcriptional responses to these signals, it remains unclear whether individual border cells are competent to give rise to all border-derived cell types, how this competence is conferred at the molecular level, and how long it is maintained. Changes in competence are also linked to changes in the response to the signals that set up the border region, as these signals are all used reiteratively to different effect. For example, once the pre-placodal domain has formed, cells within this region appear to be uniquely competent to respond to FGF signaling by expressing early markers of the otic placode (Martin and Groves, 2006; Ohyama et al., 2007). In this final section, we briefly consider some of the factors that can contribute to developmental competence and propose a model in which the gradual accumulation of transcription factors may prime progenitor cells for lineage-specific differentiation into neural crest or placode derivatives.
Competence can be operationally defined as the capacity of a particular cell to respond to an inducing signal by adopting a defined fate or set of fates (Groves and Bronner-Fraser, 2000). There are also different levels of competence that must be considered. At the most basic level, competence involves being able receive a signal (often by having appropriate cell surface receptors), transduce the signal in the cytoplasm, and control the activity of one or more nuclear and/or cytoskeletal/cytoplasmic effectors. Cells can also be differentially competent to respond to the same signal with different outcomes, and the response of a given cell can change over developmental time. Competence of both types can be regulated in many ways including the numbers and types of cell surface receptors, the presence or activity of specific components of the signaling cascade, and the possible transcriptional responses in the nucleus given the cell’s current transcriptional or epigenetic state. There are many examples of competence being regulated in each of these ways. For example, the response to FGFs during mesoderm or lens induction in Xenopus can be regulated by the presence and numbers of FGF receptors (Arresta et al., 2005; Gillespie et al., 1989). The competence of endoderm to form particular pancreas derivatives can be regulated by Fgf10 in mice and ectopic expression of fgf10 in zebrafish can cause pancreas and gut tissue to become competent to differentiate as liver (Kobberup et al., 2010; Shin et al., 2011). The presence of Sprouty proteins can reduce FGF signal amplitude or perdurance by controlling the activation of the RAS-MAPK pathway (Cabrita and Christofori, 2008). In Drosophila, Wnt and Dpp signaling can regulate signal transduction components downstream of Ras (Halfon et al., 2000) and can promote competence for muscle or heart development by differential regulation of tinman or twist (Furlong, 2004; Lee and Frasch, 2005).
Competence to respond to signals in a cell context specific way is frequently regulated by the transcriptional regulatory factors available in the nucleus. For example, differential expression of Irx1 and Six3 in different territories of the forebrain alters the competence of forebrain progenitors to respond to both Shh and FGF8 (Kobayashi et al., 2002). Differential competence can also be controlled by the levels at which a given transcription factor is expressed, rather than its presence or absence. For example, the levels of Sox2 in retinal progenitors can regulate production of different derivatives in response to Notch signaling (Taranova et al., 2006). Transcription factors can also be sequestered in sub-domains of the nucleus to prevent their activation of lineage-specific fates. For example, localization of Drosophila Hunchback to the perinuclear lamina correlates with the loss of competence for neuroblasts to make early neuronal types (Kohwi et al., 2013; Tran et al., 2010).
In addition to the activity of transcription factors in conferring competence to respond to inductive signals, evidence from a variety of stem and progenitor cells suggests that the chromatin of differentiation genes needs to be organized in a competent state before transcription can be activated. For example, competence to respond to Nodal and Activin signaling during early mesoderm induction in Xenopus can also be regulated by titrating the levels of histone H3 with the linker histone H1 (Lim et al., 2013). One class of transcription factors – called pioneer factors – has been shown to interact with compact chromatin at silent gene loci and organize chromatin in a transcriptionally competent state (Smale, 2010; Zaret and Carroll, 2011). Pioneer factors can penetrate repressive chromatin and bind to enhancers of differentiation genes, marking them for activation at later stages. Binding of pioneer factors can be followed by the post-translational modification of histones at these enhancers that convert them from a silent state to one which is “poised” for transcription (Ram and Meshorer, 2009; Serandour et al., 2011). These enhancers become rapidly activated once a cell receives differentiation signals. For example, a number of studies have identified the histone mark H3K4me1 as an indicator of poised enhancers. When these enhancers become transcriptionally active, they typically acquire H3K27Ac as an additional histone mark (Creyghton et al., 2010; Rada-Iglesias et al., 2011; Rada-Iglesias and Wysocka, 2011).
Members of the Forkhead transcription factor family can act as pioneer factors (Lalmansingh et al., 2012; Zaret et al., 2008). The winged helix DNA binding domains of many Fox genes have structural similarities to linker histones (Clark et al., 1993; Ramakrishnan et al., 1993), allowing them to bind directly to nucleosomes, open chromatin and promote accessibility to the DNA, characterized by DNA hypersensitivity. Importantly, binding of Fox pioneer factors has been associated with the establishment of epigenetic histone marks associated with poised or active enhancers such as H3K4me1 (Serandour et al., 2011). Members of the FoxA, D, E, and O families have been shown to have these properties (Lalmansingh et al., 2012; Zaret et al., 2008).
One of the first and most well-characterized examples of pioneer factor activity is the regulation of the albumin gene Alb1 during endoderm differentiation. Alb1 is normally activated by liver-inducing signals (Gualdi et al., 1996; Jung et al., 1999), and the Alb1 enhancer is highly methylated and silent in young embryos and in embryonic stem cells (Zaret et al., 2008). However a single CpG in the silent Alb1 enhancer is already bound by Foxd3 in ES cells (Xu et al., 2009). As endoderm begins to differentiate, Foxd3 is replaced by Foxa class proteins, and GATA factors such as Gata6 (Bossard and Zaret, 1998, 2000), although the Alb1 locus is still silent. However, this occupancy of the silent locus by Foxa factors is necessary for liver induction (Lee et al., 2005). Upon exposure to liver-inducing signals (Wandzioch and Zaret, 2009), lineage-specifc transcription factors are recruited to the Alb1 enhancer and transcription commences.
Is there a role for pioneer factors in segregating cell fates during the differentiation of the neural plate border region? As described above, Foxd3, an early marker of neural ectoderm and a subsequent marker of neural crest, can act as a pioneer factor in liver differentiation (Xu et al., 2009; Zaret et al., 2008). Zebrafish foxi1 shares several properties of pioneer factors (Yan et al., 2006) – for example, foxi1, like mouse Foxa1, has been shown to remain bound to DNA during mitosis and may thus act as an epigenetic mark (Yan et al., 2006; Zaret et al., 2008). In addition, Gata2 and Gata3 are expressed early in the differentiation of non-neural ectoderm (reviewed in Grocott et al., 2012) and can regulate the expression of pre-placodal markers and competence to respond to pre-placodal inducing signals (Bhat et al., 2013; Kwon et al., 2010; Pieper et al., 2012). Although this is purely circumstantial evidence, it is nevertheless possible that a number of the transcription factors described throughout this review can act in two ways – firstly by regulating the expression of other genes with which they are co-expressed (e.g. Bhat et al., 2013), but also as pioneer factors, remaining bound to the chromatin associated with neural crest- or placode-specific genes and preparing them to become activated upon receipt of appropriate inducing signals. The ability of transcription factors to act as pioneer factors has implications for the construction and interpretation of gene regulatory networks for neural crest and placodes (Betancur et al., 2010; Bhat et al., 2013; Grocott et al., 2012; Sauka-Spengler and Bronner-Fraser, 2008), as pioneer factors would bind and occupy DNA regulatory sequences of lineage-specific genes well before transcription of those genes occurred.
CONCLUSION
The key developmental problem of the neural plate border is how to generate very different cell types – neural crest and placodes – from a spatially restricted population of cells receiving very similar levels of multiple inducing signals. In this review, we propose that small local changes in signaling levels can be amplified and stabilized by a series of repressive and positive interactions between transcription factors. These initially specify broad domains of neural or non-neural ectoderm, but then activate subsequent transcription factors that iteratively refine the border. The physical mechanisms by which these progenitor domains segregate are still unclear and will be a fruitful avenue for further investigation. We suggest that some of the transcription factors expressed early in the differentiation of the neural plate border may act as pioneer factors and remain expressed and bound to lineage-specific gene enhancers. The gradual recruitment of additional transcription factors to these enhancer elements may provide competence for the segregated progenitor populations to ultimately differentiate into neural crest and placode derivatives.
Highlights.
The neural plate border forms from an interaction between neural plate and epidermis
The border region will generate neural crest and craniofacial placodes
The identity of cells in the border is determined by different levels of FGF, Wnt and BMP signals.
The segregation of different cell fates is achieved by positive and negative feedback
Acknowledgments
We wish to thank Marianne Bronner and the Fondation Les Treilles for organizing the meeting that led to this review. They fostered a wonderfully congenial environment that to many exchanges of ideas between participants, and some of those ideas ultimately found their way into the text. A.K.G.’s travel to Les Treilles was supported by DC004675.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Andrew K. Groves, Email: akgroves@bcm.edu.
Carole LaBonne, Email: clabonne@northwestern.edu.
References
- Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, Bitner-Glindzicz M, Francis M, Lacombe D, Vigneron J, Charachon R, Boven K, Bedbeder P, Van Regemorter N, Weissenbach J, Petit C. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet. 1997;15:157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- Ahmed M, Wong EY, Sun J, Xu J, Wang F, Xu PX. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev Cell. 2012a;22:377–390. doi: 10.1016/j.devcel.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M, Xu J, Xu PX. EYA1 and SIX1 drive the neuronal developmental program in cooperation with the SWI/SNF chromatin-remodeling complex and SOX2 in the mammalian inner ear. Development. 2012b;139:1965–1977. doi: 10.1242/dev.071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens K, Schlosser G. Tissues and signals involved in the induction of placodal Six1 expression in Xenopus laevis. Dev Biol. 2005;288:40–59. doi: 10.1016/j.ydbio.2005.07.022. [DOI] [PubMed] [Google Scholar]
- Albazerchi A, Stern CD. A role for the hypoblast (AVE) in the initiation of neural induction, independent of its ability to position the primitive streak. Dev Biol. 2007;301:489–503. doi: 10.1016/j.ydbio.2006.08.057. [DOI] [PubMed] [Google Scholar]
- Aoki Y, Saint-Germain N, Gyda M, Magner-Fink E, Lee YH, Credidio C, Saint-Jeannet JP. Sox10 regulates the development of neural crest-derived melanocytes in Xenopus. Dev Biol. 2003;259:19–33. doi: 10.1016/s0012-1606(03)00161-1. [DOI] [PubMed] [Google Scholar]
- Arduini BL, Brivanlou AH. Modulation of FOXD3 activity in human embryonic stem cells directs pluripotency and paraxial mesoderm fates. Stem Cells. 2012;30:2188–2198. doi: 10.1002/stem.1200. [DOI] [PubMed] [Google Scholar]
- Arresta E, Bernardini S, Gargioli C, Filoni S, Cannata SM. Lens-forming competence in the epidermis of Xenopus laevis during development. J Exp Zool A Comp Exp Biol. 2005;303:1–12. doi: 10.1002/jez.a.138. [DOI] [PubMed] [Google Scholar]
- Bailey AP, Streit A. Sensory organs: making and breaking the pre-placodal region. Curr Top Dev Biol. 2006;72:167–204. doi: 10.1016/S0070-2153(05)72003-2. [DOI] [PubMed] [Google Scholar]
- Baker CV, Bronner-Fraser M. Vertebrate cranial placodes I. Embryonic induction. Dev Biol. 2001;232:1–61. doi: 10.1006/dbio.2001.0156. [DOI] [PubMed] [Google Scholar]
- Baker CV, Stark MR, Marcelle C, Bronner-Fraser M. Competence, specification and induction of Pax-3 in the trigeminal placode. Development. 1999;126:147–156. doi: 10.1242/dev.126.1.147. [DOI] [PubMed] [Google Scholar]
- Bally-Cuif L, Gulisano M, Broccoli V, Boncinelli E. c-otx2 is expressed in two different phases of gastrulation and is sensitive to retinoic acid treatment in chick embryo. Mech Dev. 1995;49:49–63. doi: 10.1016/0925-4773(94)00301-3. [DOI] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M, Garcia-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- Basch ML, Selleck MA, Bronner-Fraser M. Timing and competence of neural crest formation. Dev Neurosci. 2000;22:217–227. doi: 10.1159/000017444. [DOI] [PubMed] [Google Scholar]
- Beanan MJ, Sargent TD. Regulation and function of Dlx3 in vertebrate development. Dev Dyn. 2000;218:545–553. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1026>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Bellefroid EJ, Kobbe A, Gruss P, Pieler T, Gurdon JB, Papalopulu N. Xiro3 encodes a Xenopus homolog of the Drosophila Iroquois genes and functions in neural specification. EMBO J. 1998;17:191–203. doi: 10.1093/emboj/17.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessarab DA, Chong SW, Korzh V. Expression of zebrafish six1 during sensory organ development and myogenesis. Dev Dyn. 2004;230:781–786. doi: 10.1002/dvdy.20093. [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu Rev Cell Dev Biol. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat N, Kwon HJ, Riley BB. A gene network that coordinates preplacodal competence and neural crest specification in zebrafish. Dev Biol. 2013;373:107–117. doi: 10.1016/j.ydbio.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Bailey AP, Bronner-Fraser M, Streit A. Segregation of lens and olfactory precursors from a common territory: cell sorting and reciprocity of Dlx5 and Pax6 expression. Dev Biol. 2004;271:403–414. doi: 10.1016/j.ydbio.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Bronner-Fraser M. Hierarchy of regulatory events in sensory placode development. Curr Opin Genet Dev. 2004;14:520–526. doi: 10.1016/j.gde.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Bronner-Fraser M. Competence, specification and commitment to an olfactory placode fate. Development. 2008;135:4165–4177. doi: 10.1242/dev.026633. [DOI] [PubMed] [Google Scholar]
- Bilican B, Fiore-Heriche C, Compston A, Allen ND, Chandran S. Induction of Olig2 precursors by FGF involves BMP signalling blockade at the Smad level. PLoS One. 2008;3:e2863. doi: 10.1371/journal.pone.0002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonstein L, Elias S, Frank D. Paraxial-fated mesoderm is required for neural crest induction in Xenopus embryos. Dev Biol. 1998;193:156–168. doi: 10.1006/dbio.1997.8795. [DOI] [PubMed] [Google Scholar]
- Bossard P, Zaret KS. GATA transcription factors as potentiators of gut endoderm differentiation. Development. 1998;125:4909–4917. doi: 10.1242/dev.125.24.4909. [DOI] [PubMed] [Google Scholar]
- Bossard P, Zaret KS. Repressive and restrictive mesodermal interactions with gut endoderm: possible relation to Meckel’s Diverticulum. Development. 2000;127:4915–4923. doi: 10.1242/dev.127.22.4915. [DOI] [PubMed] [Google Scholar]
- Bricaud O, Collazo A. The transcription factor six1 inhibits neuronal and promotes hair cell fate in the developing zebrafish (Danio rerio) inner ear. J Neurosci. 2006;26:10438–10451. doi: 10.1523/JNEUROSCI.1025-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner ME, LeDouarin NM. Development and evolution of the neural crest: an overview. Dev Biol. 2012;366:2–9. doi: 10.1016/j.ydbio.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown ST, Wang J, Groves AK. Dlx gene expression during chick inner ear development. J Comp Neurol. 2005;483:48–65. doi: 10.1002/cne.20418. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Pandur PD, Kenyon KL, Pignoni F, Moody SA. Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development. 2004;131:5871–5881. doi: 10.1242/dev.01516. [DOI] [PubMed] [Google Scholar]
- Cabrita MA, Christofori G. Sprouty proteins, masterminds of receptor tyrosine kinase signaling. Angiogenesis. 2008;11:53–62. doi: 10.1007/s10456-008-9089-1. [DOI] [PubMed] [Google Scholar]
- Chapman SC, Brown R, Lees L, Schoenwolf GC, Lumsden A. Expression analysis of chick Wnt and frizzled genes and selected inhibitors in early chick patterning. Dev Dyn. 2004;229:668–676. doi: 10.1002/dvdy.10491. [DOI] [PubMed] [Google Scholar]
- Chen B, Kim EH, Xu PX. Initiation of olfactory placode development and neurogenesis is blocked in mice lacking both Six1 and Six4. Dev Biol. 2009;326:75–85. doi: 10.1016/j.ydbio.2008.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Streit A. Induction of the inner ear: stepwise specification of otic fate from multipotent progenitors. Hear Res. 2013;297:3–12. doi: 10.1016/j.heares.2012.11.018. [DOI] [PubMed] [Google Scholar]
- Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- Cheyette BN, Green PJ, Martin K, Garren H, Hartenstein V, Zipursky SL. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- Cho GS, Choi SC, Han JK. BMP signal attenuates FGF pathway in anteroposterior neural patterning. Biochem Biophys Res Commun. 2013;434:509–515. doi: 10.1016/j.bbrc.2013.03.105. [DOI] [PubMed] [Google Scholar]
- Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Adv Cancer Res. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- Christophorou NAD, Bailey AP, Hanson S, Streit A. Activation of Six1 target genes is required for sensory placode formation. Dev Biol. 2009;336:327–336. doi: 10.1016/j.ydbio.2009.09.025. [DOI] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson ME, Selleck MA, McMahon AP, Bronner-Fraser M. Dorsalization of the neural tube by the non-neural ectoderm. Development. 1995;121:2099–2106. doi: 10.1242/dev.121.7.2099. [DOI] [PubMed] [Google Scholar]
- Donner AL, Maas RL. Conservation and non-conservation of genetic pathways in eye specification. Int J Dev Biol. 2004;48:743–753. doi: 10.1387/ijdb.041877ad. [DOI] [PubMed] [Google Scholar]
- Esterberg R, Fritz A. dlx3b/4b are required for the formation of the preplacodal region and otic placode through local modulation of BMP activity. Dev Biol. 2009;325:189–199. doi: 10.1016/j.ydbio.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve P, Bovolenta P. cSix4, a member of the six gene family of transcription factors, is expressed during placode and somite development. Mech Dev. 1999;85:161–165. doi: 10.1016/s0925-4773(99)00079-9. [DOI] [PubMed] [Google Scholar]
- Ezin AM, Fraser SE, Bronner-Fraser M. Fate map and morphogenesis of presumptive neural crest and dorsal neural tube. Dev Biol. 2009;330:221–236. doi: 10.1016/j.ydbio.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fainsod A, Steinbeisser H, De Robertis EM. On the function of BMP-4 in patterning the marginal zone of the Xenopus embryo. EMBO J. 1994;13:5015–5025. doi: 10.1002/j.1460-2075.1994.tb06830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feledy JA, Beanan MJ, Sandoval JJ, Goodrich JS, Lim JH, Matsuo-Takasaki M, Sato SM, Sargent TD. Inhibitory patterning of the anterior neural plate in Xenopus by homeodomain factors Dlx3 and Msx1. Dev Biol. 1999a;212:455–464. doi: 10.1006/dbio.1999.9374. [DOI] [PubMed] [Google Scholar]
- Feledy JA, Morasso MI, Jang SI, Sargent TD. Transcriptional activation by the homeodomain protein distal-less 3. Nucleic Acids Res. 1999b;27:764–770. doi: 10.1093/nar/27.3.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Garre P, Rodriguez-Gallardo L, Gallego-Diaz V, Alvarez IS, Puelles L. Fate map of the chicken neural plate at stage 4. Development. 2002;129:2807–2822. doi: 10.1242/dev.129.12.2807. [DOI] [PubMed] [Google Scholar]
- Furlong EE. Integrating transcriptional and signalling networks during muscle development. Curr Opin Genet Dev. 2004;14:343–350. doi: 10.1016/j.gde.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Gallagher BC, Henry JJ, Grainger RM. Inductive processes leading to inner ear formation during Xenopus development. Dev Biol. 1996;175:95–9107. doi: 10.1006/dbio.1996.0098. [DOI] [PubMed] [Google Scholar]
- Garcia-Castro MI, Marcelle C, Bronner-Fraser M. Ectodermal Wnt function as a neural crest inducer. Science. 2002;297:848–851. doi: 10.1126/science.1070824. [DOI] [PubMed] [Google Scholar]
- Gasparini F, Degasperi V, Shimeld SM, Burighel P, Manni L. Evolutionary conservation of the placodal transcriptional network during sexual and asexual development in chordates. Dev Dyn. 2013 doi: 10.1002/dvdy.23957. [DOI] [PubMed] [Google Scholar]
- Gillespie LL, Paterno GD, Slack JM. Analysis of competence: receptors for fibroblast growth factor in early Xenopus embryos. Development. 1989;106:203–208. doi: 10.1242/dev.106.1.203. [DOI] [PubMed] [Google Scholar]
- Glavic A, Gomez-Skarmeta JL, Mayor R. The homeoprotein Xiro1 is required for midbrain-hindbrain boundary formation. Development. 2002;129:1609–1621. doi: 10.1242/dev.129.7.1609. [DOI] [PubMed] [Google Scholar]
- Glavic A, Maris Honore S, Gloria Feijoo C, Bastidas F, Allende ML, Mayor R. Role of BMP signaling and the homeoprotein Iroquois in the specification of the cranial placodal field. Dev Biol. 2004;272:89–8103. doi: 10.1016/j.ydbio.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Gomez-Skarmeta JL, Glavic A, de la Calle-Mustienes E, Modolell J, Mayor R. Xiro, a Xenopus homolog of the Drosophila Iroquois complex genes, controls development at the neural plate. EMBO J. 1998;17:181–190. doi: 10.1093/emboj/17.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goriely A, Diez del Corral R, Storey KG. c-Irx2 expression reveals an early subdivision of the neural plate in the chick embryo. Mech Dev. 1999;87:203–206. doi: 10.1016/s0925-4773(99)00149-5. [DOI] [PubMed] [Google Scholar]
- Graham A, Shimeld SM. The origin and evolution of the ectodermal placodes. J Anat. 2013;222:32–40. doi: 10.1111/j.1469-7580.2012.01506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grocott T, Tambalo M, Streit A. The peripheral sensory nervous system in the vertebrate head: a gene regulatory perspective. Dev Biol. 2012;370:3–23. doi: 10.1016/j.ydbio.2012.06.028. [DOI] [PubMed] [Google Scholar]
- Groves AK, Bronner-Fraser M. Competence, specification and commitment in otic placode induction. Development. 2000;127:3489–3499. doi: 10.1242/dev.127.16.3489. [DOI] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- Halfon MS, Carmena A, Gisselbrecht S, Sackerson CM, Jimenez F, Baylies MK, Michelson AM. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell. 2000;103:63–74. doi: 10.1016/s0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- Hans S, Christison J, Liu D, Westerfield M. Fgf-dependent otic induction requires competence provided by Foxi1 and Dlx3b. BMC Dev Biol. 2007;7:5. doi: 10.1186/1471-213X-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson IM. Mammalian homologues of the Drosophila eye specification genes. Semin Cell Dev Biol. 2001;12:475–484. doi: 10.1006/scdb.2001.0271. [DOI] [PubMed] [Google Scholar]
- Hatada Y, Stern CD. A fate map of the epiblast of the early chick embryo. Development. 1994;120:2879–2889. doi: 10.1242/dev.120.10.2879. [DOI] [PubMed] [Google Scholar]
- Hoffman TL, Javier AL, Campeau SA, Knight RD, Schilling TF. Tfap2 transcription factors in zebrafish neural crest development and ectodermal evolution. J Exp Zool B Mol Dev Evol. 2007;308:679–691. doi: 10.1002/jez.b.21189. [DOI] [PubMed] [Google Scholar]
- Hong CS, Saint-Jeannet JP. The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol Biol Cell. 2007;18:2192–2202. doi: 10.1091/mbc.E06-11-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CS, Saint-Jeannet JP. Sox proteins and neural crest development. Semin Cell Dev Biol. 2005;16:694–703. doi: 10.1016/j.semcdb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Ille F, Atanasoski S, Falk S, Ittner LM, Marki D, Buchmann-Moller S, Wurdak H, Suter U, Taketo MM, Sommer L. Wnt/BMP signal integration regulates the balance between proliferation and differentiation of neuroepithelial cells in the dorsal spinal cord. Dev Biol. 2007;304:394–408. doi: 10.1016/j.ydbio.2006.12.045. [DOI] [PubMed] [Google Scholar]
- Inui M, Montagner M, Ben-Zvi D, Martello G, Soligo S, Manfrin A, Aragona M, Enzo E, Zacchigna L, Zanconato F, Azzolin L, Dupont S, Cordenonsi M, Piccolo S. Self-regulation of the head-inducing properties of the Spemann organizer. Proc Natl Acad Sci U S A. 2012;109:15354–15359. doi: 10.1073/pnas.1203000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara T, Sato S, Ikeda K, Yajima H, Kawakami K. Multiple evolutionarily conserved enhancers control expression of Eya1. Dev Dyn. 2008;237:3142–3156. doi: 10.1002/dvdy.21716. [DOI] [PubMed] [Google Scholar]
- Itasaki N, Hoppler S. Crosstalk between Wnt and bone morphogenic protein signaling: a turbulent relationship. Dev Dyn. 2010;239:16–33. doi: 10.1002/dvdy.22009. [DOI] [PubMed] [Google Scholar]
- Jayasena CS, Ohyama T, Segil N, Groves AK. Notch signaling augments the canonical Wnt pathway to specify the size of the otic placode. Development. 2008;135:2251–2261. doi: 10.1242/dev.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Cook SA, Erway LC, Matthews AN, Sanford LP, Paradies NE, Friedman RA. Inner ear and kidney anomalies caused by IAP insertion in an intron of the Eya1 gene in a mouse model of BOR syndrome. Hum Mol Genet. 1999;8:645–653. doi: 10.1093/hmg/8.4.645. [DOI] [PubMed] [Google Scholar]
- Jung J, Zheng M, Goldfarb M, Zaret KS. Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science. 1999;284:1998–2003. doi: 10.1126/science.284.5422.1998. [DOI] [PubMed] [Google Scholar]
- Kaji T, Artinger KB. dlx3b and dlx4b function in the development of Rohon-Beard sensory neurons and trigeminal placode in the zebrafish neurula. Dev Biol. 2004;276:523–540. doi: 10.1016/j.ydbio.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Sato S, Ozaki H, Ikeda K. Six family genes--structure and function as transcription factors and their roles in development. Bioessays. 2000;22:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Khudyakov J, Bronner-Fraser M. Comprehensive spatiotemporal analysis of early chick neural crest network genes. Dev Dyn. 2009;238:716–723. doi: 10.1002/dvdy.21881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kil SH, Streit A, Brown ST, Agrawal N, Collazo A, Zile MH, Groves AK. Distinct roles for hindbrain and paraxial mesoderm in the induction and patterning of the inner ear revealed by a study of vitamin-A-deficient quail. Dev Biol. 2005;285:252–271. doi: 10.1016/j.ydbio.2005.05.044. [DOI] [PubMed] [Google Scholar]
- Knight RD, Nair S, Nelson SS, Afshar A, Javidan Y, Geisler R, Rauch GJ, Schilling TF. lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development. 2003;130:5755–5768. doi: 10.1242/dev.00575. [DOI] [PubMed] [Google Scholar]
- Kobayashi D, Kobayashi M, Matsumoto K, Ogura T, Nakafuku M, Shimamura K. Early subdivisions in the neural plate define distinct competence for inductive signals. Development. 2002;129:83–93. doi: 10.1242/dev.129.1.83. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Osanai H, Kawakami K, Yamamoto M. Expression of three zebrafish Six4 genes in the cranial sensory placodes and the developing somites. Mech Dev. 2000;98:151–155. doi: 10.1016/s0925-4773(00)00451-2. [DOI] [PubMed] [Google Scholar]
- Kobberup S, Schmerr M, Dang ML, Nyeng P, Jensen JN, MacDonald RJ, Jensen J. Conditional control of the differentiation competence of pancreatic endocrine and ductal cells by Fgf10. Mech Dev. 2010;127:220–234. doi: 10.1016/j.mod.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Lupton JR, Lai SL, Miller MR, Doe CQ. Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell. 2013;152:97–108. doi: 10.1016/j.cell.2012.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Doe CQ. Temporal fate specification and neural progenitor cometence during development. Nat Rev Neurosci. 2013;14:823–836. doi: 10.1038/nrn3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y, Ikeda K, Iwakura Y, Kawakami K. Six1 and Six4 promote survival of sensory neurons during early trigeminal gangliogenesis. Brain Res. 2006;1116:93–9102. doi: 10.1016/j.brainres.2006.07.103. [DOI] [PubMed] [Google Scholar]
- Köster RW, Kuhnlein RP, Wittbrodt J. Ectopic Sox3 activity elicits sensory placode formation. Mech Dev. 2000;95:175–187. doi: 10.1016/s0925-4773(00)00356-7. [DOI] [PubMed] [Google Scholar]
- Kozlowski DJ, Murakami T, Ho RK, Weinberg ES. Regional cell movement and tissue patterning in the zebrafish embryo revealed by fate mapping with caged fluorescein. Biochem Cell Biol. 1997;75:551–562. [PubMed] [Google Scholar]
- Kozlowski DJ, Whitfield TT, Hukriede NA, Lam WK, Weinberg ES. The zebrafish dog-eared mutation disrupts eya1, a gene required for cell survival and differentiation in the inner ear and lateral line. Dev Biol. 2005;277:27–41. doi: 10.1016/j.ydbio.2004.08.033. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Salic AN, Evans LM, Kirschner MW. Geminin, a neuralizing molecule that demarcates the future neural plate at the onset of gastrulation. Development. 1998;125:3247–3258. doi: 10.1242/dev.125.16.3247. [DOI] [PubMed] [Google Scholar]
- Kumar JP. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell Mol Life Sci. 2009;66:565–583. doi: 10.1007/s00018-008-8335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HJ, Bhat N, Sweet EM, Cornell RA, Riley BB. Identification of early requirements for preplacodal ectoderm and sensory organ development. PLoS Genet. 2010:6. doi: 10.1371/journal.pgen.1001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Neural crest induction in Xenopus: evidence for a two-signal model. Development. 1998;125:2403–2414. doi: 10.1242/dev.125.13.2403. [DOI] [PubMed] [Google Scholar]
- Labosky PA, Kaestner KH. The winged helix transcription factor Hfh2 is expressed in neural crest and spinal cord during mouse development. Mech Dev. 1998;76:185–190. doi: 10.1016/s0925-4773(98)00105-1. [DOI] [PubMed] [Google Scholar]
- Laclef C, Hamard G, Demignon J, Souil E, Houbron C, Maire P. Altered myogenesis in Six1-deficient mice. Development. 2003;130:2239–2252. doi: 10.1242/dev.00440. [DOI] [PubMed] [Google Scholar]
- Lagutin O, Zhu CC, Furuta Y, Rowitch DH, McMahon AP, Oliver G. Six3 promotes the formation of ectopic optic vesicle-like structures in mouse embryos. Dev Dyn. 2001;221:342–349. doi: 10.1002/dvdy.1148. [DOI] [PubMed] [Google Scholar]
- Lalmansingh AS, Karmakar S, Jin Y, Nagaich AK. Multiple modes of chromatin remodeling by Forkhead box proteins. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbagrm.2012.02.018. [DOI] [PubMed] [Google Scholar]
- Le Douarin N, Kalcheim C. The neural crest. 2. Cambridge University Press; Cambridge, UK; New York, NY, USA: 1999. [Google Scholar]
- Lee CS, Sund NJ, Behr R, Herrera PL, Kaestner KH. Foxa2 is required for the differentiation of pancreatic alpha-cells. Dev Biol. 2005;278:484–495. doi: 10.1016/j.ydbio.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Lee HH, Frasch M. Nuclear integration of positive Dpp signals, antagonistic Wg inputs and mesodermal competence factors during Drosophila visceral mesoderm induction. Development. 2005;132:1429–1442. doi: 10.1242/dev.01687. [DOI] [PubMed] [Google Scholar]
- Lee YH, Aoki Y, Hong CS, Saint-Germain N, Credidio C, Saint-Jeannet JP. Early requirement of the transcriptional activator Sox9 for neural crest specification in Xenopus. Dev Biol. 2004;275:93–103. doi: 10.1016/j.ydbio.2004.07.036. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Brivanlou AH. Proposal of a model of mammalian neural induction. Dev Biol. 2007;308:247–256. doi: 10.1016/j.ydbio.2007.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cornell RA. Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos. Dev Biol. 2007;304:338–354. doi: 10.1016/j.ydbio.2006.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, Rosenfeld MG. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- Lillevali K, Haugas M, Matilainen T, Pussinen C, Karis A, Salminen M. Gata3 is required for early morphogenesis and Fgf10 expression during otic development. Mech Dev. 2006;123:415–429. doi: 10.1016/j.mod.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Lim CY, Reversade B, Knowles BB, Solter D. Optimal histone H3 to linker histone H1 chromatin ratio is vital for mesodermal competence in Xenopus. Development. 2013;140:853–860. doi: 10.1242/dev.086611. [DOI] [PubMed] [Google Scholar]
- Linker C, De Almeida I, Papanayotou C, Stower M, Sabado V, Ghorani E, Streit A, Mayor R, Stern CD. Cell communication with the neural plate is required for induction of neural markers by BMP inhibition: evidence for homeogenetic induction and implications for Xenopus animal cap and chick explant assays. Dev Biol. 2009;327:478–486. doi: 10.1016/j.ydbio.2008.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsiou A, Hanson S, Streit A. A balance of FGF, BMP and WNT signalling positions the future placode territory in the head. Development. 2005;132:4051–4062. doi: 10.1242/dev.01964. [DOI] [PubMed] [Google Scholar]
- Liu C, Goswami M, Talley J, Chesser-Martinez PL, Lou CH, Sater AK. TAK1 promotes BMP4/Smad1 signaling via inhibition of erk MAPK: a new link in the FGF/BMP regulatory network. Differentiation. 2012;83:210–219. doi: 10.1016/j.diff.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Liu W, Lagutin OV, Mende M, Streit A, Oliver G. Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J. 2006;25:5383–5395. doi: 10.1038/sj.emboj.7601398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Matsuo-Takasaki M, Lim JH, Sargent TD. Differential regulation of Dlx gene expression by a BMP morphogenetic gradient. Int J Dev Biol. 2001a;45:681–684. [PubMed] [Google Scholar]
- Luo T, Matsuo-Takasaki M, Sargent TD. Distinct roles for Distal-less genes Dlx3 and Dlx5 in regulating ectodermal development in Xenopus. Mol Reprod Dev. 2001b;60:331–337. doi: 10.1002/mrd.1095. [DOI] [PubMed] [Google Scholar]
- Marchant L, Linker C, Mayor R. Inhibition of mesoderm formation by follistatin. Dev Genes Evol. 1998;208:157–160. doi: 10.1007/s004270050167. [DOI] [PubMed] [Google Scholar]
- Martin K, Groves AK. Competence of cranial ectoderm to respond to Fgf signaling suggests a two-step model of otic placode induction. Development. 2006;133:877–887. doi: 10.1242/dev.02267. [DOI] [PubMed] [Google Scholar]
- Matsuo-Takasaki M, Matsumura M, Sasai Y. An essential role of Xenopus Foxi1a for ventral specification of the cephalic ectoderm during gastrulation. Development. 2005;132:3885–3894. doi: 10.1242/dev.01959. [DOI] [PubMed] [Google Scholar]
- Mayor R, Guerrero N, Martinez C. Role of FGF and noggin in neural crest induction. Dev Biol. 1997;189:1–12. doi: 10.1006/dbio.1997.8634. [DOI] [PubMed] [Google Scholar]
- Mayor R, Morgan R, Sargent MG. Induction of the prospective neural crest of Xenopus. Development. 1995;121:767–777. doi: 10.1242/dev.121.3.767. [DOI] [PubMed] [Google Scholar]
- McCabe KL, Bronner-Fraser M. Molecular and tissue interactions governing induction of cranial ectodermal placodes. Dev Biol. 2009;332:189–195. doi: 10.1016/j.ydbio.2009.05.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLarren KW, Litsiou A, Streit A. DLX5 positions the neural crest and preplacode region at the border of the neural plate. Dev Biol. 2003;259:34–47. doi: 10.1016/s0012-1606(03)00177-5. [DOI] [PubMed] [Google Scholar]
- Milet C, Maczkowiak F, Roche DD, Monsoro-Burq AH. Pax3 and Zic1 drive induction and differentiation of multipotent, migratory, and functional neural crest in Xenopus embryos. Proc Natl Acad Sci U S A. 2013;110:5528–5533. doi: 10.1073/pnas.1219124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milet C, Monsoro-Burq AH. Neural crest induction at the neural plate border in vertebrates. Dev Biol. 2012;366:22–33. doi: 10.1016/j.ydbio.2012.01.013. [DOI] [PubMed] [Google Scholar]
- Mishima N, Tomarev S. Chicken Eyes absent 2 gene: isolation and expression pattern during development. Int J Dev Biol. 1998;42:1109–1115. [PubMed] [Google Scholar]
- Monsoro-Burq AH, Fletcher RB, Harland RM. Neural crest induction by paraxial mesoderm in Xenopus embryos requires FGF signals. Development. 2003;130:3111–3124. doi: 10.1242/dev.00531. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq AH, Wang E, Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev Cell. 2005;8:167–178. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq AH, Duprez D, Watanabe Y, Bontoux M, Vincent C, Brickell P, Le Douarin N. The role of bone morphogenetic proteins in vertebral development. Development. 1996;122:3607–3616. doi: 10.1242/dev.122.11.3607. [DOI] [PubMed] [Google Scholar]
- Murdoch B, DelConte C, Garcia-Castro MI. Embryonic Pax7-expressing progenitors contribute multiple cell types to the postnatal olfactory epithelium. J Neurosci. 2010;30:9523–9532. doi: 10.1523/JNEUROSCI.0867-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch B, DelConte C, Garcia-Castro MI. Pax7 lineage contributions to the mammalian neural crest. PLoS One. 2012;7:e41089. doi: 10.1371/journal.pone.0041089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogita J, Isogai E, Sudo H, Sakiyama S, Nakagawara A, Koseki H. Expression of the Dan gene during chicken embryonic development. Mech Dev. 2001;109:363–365. doi: 10.1016/s0925-4773(01)00522-6. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Expression of mouse Foxi class genes in early craniofacial development. Dev Dyn. 2004;231:640–646. doi: 10.1002/dvdy.20160. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Groves AK, Martin K. The first steps towards hearing: mechanisms of otic placode induction. Int J Dev Biol. 2007;51:463–472. doi: 10.1387/ijdb.072320to. [DOI] [PubMed] [Google Scholar]
- Ozaki H, Nakamura K, Funahashi J, Ikeda K, Yamada G, Tokano H, Okamura HO, Kitamura K, Muto S, Kotaki H, Sudo K, Horai R, Iwakura Y, Kawakami K. Six1 controls patterning of the mouse otic vesicle. Development. 2004;131:551–562. doi: 10.1242/dev.00943. [DOI] [PubMed] [Google Scholar]
- Pandur PD, Moody SA. Xenopus Six1 gene is expressed in neurogenic cranial placodes and maintained in the differentiating lateral lines. Mech Dev. 2000;96:253–257. doi: 10.1016/s0925-4773(00)00396-8. [DOI] [PubMed] [Google Scholar]
- Papalopulu N, Kintner C. Xenopus Distal-less related homeobox genes are expressed in the developing forebrain and are induced by planar signals. Development. 1993;117:961–975. doi: 10.1242/dev.117.3.961. [DOI] [PubMed] [Google Scholar]
- Papanayotou C, Mey A, Birot A-M, Saka Y, Boast S, Smith JC, Samarut J, Stern CD. A mechanism regulating the onset of Sox2 expression in the embryonic neural plate. PLoS Biol. 2008:6. doi: 10.1371/journal.pbio.0060002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu KS, Mardon G. Genetic control of retinal specification and determination in Drosophila. Int J Dev Biol. 2004;48:913–924. doi: 10.1387/ijdb.041875kp. [DOI] [PubMed] [Google Scholar]
- Pera E, Stein S, Kessel M. Ectodermal patterning in the avian embryo: epidermis versus neural plate. Development. 1999;126:63–73. doi: 10.1242/dev.126.1.63. [DOI] [PubMed] [Google Scholar]
- Pera EM, Ikeda A, Eivers E, De Robertis EM. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev. 2003;17:3023–3028. doi: 10.1101/gad.1153603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips BT, Kwon HJ, Melton C, Houghtaling P, Fritz A, Riley BB. Zebrafish msxB, msxC and msxE function together to refine the neural-nonneural border and regulate cranial placodes and neural crest development. Dev Biol. 2006;294:376–390. doi: 10.1016/j.ydbio.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Pieper M, Ahrens K, Rink E, Peter A, Schlosser G. Differential distribution of competence for panplacodal and neural crest induction to non-neural and neural ectoderm. Development. 2012;139:1175–1187. doi: 10.1242/dev.074468. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997a;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Zipursky SL. Identification of genes required for Drosophila eye development using a phenotypic enhancer-trap. Proc Natl Acad Sci U S A. 1997b;94:9220–9225. doi: 10.1073/pnas.94.17.9220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad MS, Sauka-Spengler T, LaBonne C. Induction of the neural crest state: control of stem cell attributes by gene regulatory, post-transcriptional and epigenetic interactions. Dev Biol. 2012;366:10–21. doi: 10.1016/j.ydbio.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell P, Oliver G, Mardon G, Donner AL, Maas RL. Pax6-dependence of Six3, Eya1 and Dach1 expression during lens and nasal placode induction. Gene Expr Patterns. 2005;6:110–118. doi: 10.1016/j.modgep.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Wysocka J. Epigenomics of human embryonic stem cells and induced pluripotent stem cells: insights into pluripotency and implications for disease. Genome Med. 2011;3:36. doi: 10.1186/gm252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram EV, Meshorer E. Transcriptional competence in pluripotency. Genes Dev. 2009;23:2793–2798. doi: 10.1101/gad.1881609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- Retting KN, Song B, Yoon BS, Lyons KM. BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development. 2009;136:1093–1104. doi: 10.1242/dev.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex M, Orme A, Uwanogho D, Tointon K, Wigmore PM, Sharpe PT, Scotting PJ. Dynamic expression of chicken Sox2 and Sox3 genes in ectoderm induced to form neural tissue. Dev Dyn. 1997;209:323–332. doi: 10.1002/(SICI)1097-0177(199707)209:3<323::AID-AJA7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Robledo RF, Lufkin T. Dlx5 and Dlx6 homeobox genes are required for specification of the mammalian vestibular apparatus. Genesis. 2006;44:425–437. doi: 10.1002/dvg.20233. [DOI] [PubMed] [Google Scholar]
- Rodriguez Esteban C, Capdevila J, Economides AN, Pascual J, Ortiz A, Izpisua Belmonte JC. The novel Cer-like protein Caronte mediates the establishment of embryonic left-right asymmetry. Nature. 1999;401:243–251. doi: 10.1038/45738. [DOI] [PubMed] [Google Scholar]
- Rogers CD, Ferzli GS, Casey ES. The response of early neural genes to FGF signaling or inhibition of BMP indicate the absence of a conserved neural induction module. BMC Dev Biol. 2011;11:74. doi: 10.1186/1471-213X-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CD, Moody SA, Casey ES. Neural induction and factors that stabilize a neural fate. Birth Defects Res C Embryo Today. 2009;87:249–262. doi: 10.1002/bdrc.20157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf RG, Xu PX, Silvius D, Otto EA, Beekmann F, Muerb UT, Kumar S, Neuhaus TJ, Kemper MJ, Raymond RM, Brophy PD, Berkman J, Gattas M, Hyland V, Ruf EM, Schwartz C, Chang EH, Smith RJH, Stratakis CA, Weil D, Petit C, Hildebrandt F. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc Natl Acad Sci U S A. 2004;101:8090–8095. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai N, Mizuseki K, Sasai Y. Requirement of FoxD3-class signaling for neural crest determination in Xenopus. Development. 2001;128:2525–2536. doi: 10.1242/dev.128.13.2525. [DOI] [PubMed] [Google Scholar]
- Sato S, Ikeda K, Shioi G, Nakao K, Yajima H, Kawakami K. Regulation of Six1 expression by evolutionarily conserved enhancers in tetrapods. Dev Biol. 2012;368:95–108. doi: 10.1016/j.ydbio.2012.05.023. [DOI] [PubMed] [Google Scholar]
- Sato S, Ikeda K, Shioi G, Ochi H, Ogino H, Yajima H, Kawakami K. Conserved expression of mouse Six1 in the pre-placodal region (PPR) and identification of an enhancer for the rostral PPR. Dev Biol. 2010;344:158–171. doi: 10.1016/j.ydbio.2010.04.029. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Evolutionary origins of vertebrate placodes: insights from developmental studies and from comparisons with other deuterostomes. J Exp Zool B Mol Dev Evol. 2005;304:347–399. doi: 10.1002/jez.b.21055. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Induction and specification of cranial placodes. Dev Biol. 2006;294:303–351. doi: 10.1016/j.ydbio.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Schlosser G. How old genes make a new head: redeployment of Six and Eya genes during the evolution of vertebrate cranial placodes. Integr Comp Biol. 2007;47:343–359. doi: 10.1093/icb/icm031. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Do vertebrate neural crest and cranial placodes have a common evolutionary origin? Bioessays. 2008;30:659–672. doi: 10.1002/bies.20775. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Making senses development of vertebrate cranial placodes. Int Rev Cell Mol Biol. 2010;283:129–234. doi: 10.1016/S1937-6448(10)83004-7. [DOI] [PubMed] [Google Scholar]
- Schubert FR, Mootoosamy RC, Walters EH, Graham A, Tumiotto L, Munsterberg AE, Lumsden A, Dietrich S. Wnt6 marks sites of epithelial transformations in the chick embryo. Mech Dev. 2002;114:143–148. doi: 10.1016/s0925-4773(02)00039-4. [DOI] [PubMed] [Google Scholar]
- Serandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Metivier R, Salbert G, Eeckhoute J. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 2011;21:555–565. doi: 10.1101/gr.111534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serikaku MA, O’Tousa JE. sine oculis is a homeobox gene required for Drosophila visual system development. Genetics. 1994;138:1137–1150. doi: 10.1093/genetics/138.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng G, Stern CD. Gata2 and Gata3: novel markers for early embryonic polarity and for non-neural ectoderm in the chick embryo. Mech Dev. 1999;87:213–216. doi: 10.1016/s0925-4773(99)00150-1. [DOI] [PubMed] [Google Scholar]
- Shin D, Lee Y, Poss KD, Stainier DY. Restriction of hepatic competence by Fgf signaling. Development. 2011;138:1339–1348. doi: 10.1242/dev.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skromne I, Stern CD. Interactions between Wnt and Vg1 signalling pathways initiate primitive streak formation in the chick embryo. Development. 2001;128:2915–2927. doi: 10.1242/dev.128.15.2915. [DOI] [PubMed] [Google Scholar]
- Smale ST. Pioneer factors in embryonic stem cells and differentiation. Curr Opin Genet Dev. 2010;20:519–526. doi: 10.1016/j.gde.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon KS, Fritz A. Concerted action of two dlx paralogs in sensory placode formation. Development. 2002;129:3127–3136. doi: 10.1242/dev.129.13.3127. [DOI] [PubMed] [Google Scholar]
- Squarzoni P, Parveen F, Zanetti L, Ristoratore F, Spagnuolo A. FGF/MAPK/Ets signaling renders pigment cell precursors competent to respond to Wnt signal by directly controlling Ci-Tcf transcription. Development. 2011;138:1421–1432. doi: 10.1242/dev.057323. [DOI] [PubMed] [Google Scholar]
- Steiner AB, Engleka MJ, Lu Q, Piwarzyk EC, Yaklichkin S, Lefebvre JL, Walters JW, Pineda-Salgado L, Labosky PA, Kessler DS. FoxD3 regulation of Nodal in the Spemann organizer is essential for Xenopus dorsal mesoderm development. Development. 2006;133:4827–4838. doi: 10.1242/dev.02663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern CD, Downs KM. The hypoblast (visceral endoderm): an evo-devo perspective. Development. 2012;139:1059–1069. doi: 10.1242/dev.070730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steventon B, Araya C, Linker C, Kuriyama S, Mayor R. Differential requirements of BMP and Wnt signalling during gastrulation and neurulation define two steps in neural crest induction. Development. 2009;136:771–779. doi: 10.1242/dev.029017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RA, Arduini BL, Berghmans S, George RE, Kanki JP, Henion PD, Look AT. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev Biol. 2006;292:174–188. doi: 10.1016/j.ydbio.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Streit A. Origin of the vertebrate inner ear: evolution and induction of the otic placode. J Anat. 2001;199:99–103. doi: 10.1046/j.1469-7580.2001.19910099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A. Extensive cell movements accompany formation of the otic placode. Dev Biol. 2002;249:237–254. doi: 10.1006/dbio.2002.0739. [DOI] [PubMed] [Google Scholar]
- Streit A. The preplacodal region: an ectodermal domain with multipotential progenitors that contribute to sense organs and cranial sensory ganglia. Int J Dev Biol. 2007;51:447–461. doi: 10.1387/ijdb.072327as. [DOI] [PubMed] [Google Scholar]
- Streit A, Berliner AJ, Papanayotou C, Sirulnik A, Stern CD. Initiation of neural induction by FGF signalling before gastrulation. Nature. 2000;406:74–78. doi: 10.1038/35017617. [DOI] [PubMed] [Google Scholar]
- Streit A, Lee KJ, Woo I, Roberts C, Jessell TM, Stern CD. Chordin regulates primitive streak development and the stability of induced neural cells, but is not sufficient for neural induction in the chick embryo. Development. 1998;125:507–519. doi: 10.1242/dev.125.3.507. [DOI] [PubMed] [Google Scholar]
- Streit A, Sockanathan S, Perez L, Rex M, Scotting PJ, Sharpe PT, Lovell-Badge R, Stern CD. Preventing the loss of competence for neural induction: HGF/SF, L5 and Sox-2. Development. 1997;124:1191–1202. doi: 10.1242/dev.124.6.1191. [DOI] [PubMed] [Google Scholar]
- Streit A, Stern CD. Establishment and maintenance of the border of the neural plate in the chick: involvement of FGF and BMP activity. Mech Dev. 1999;82:51–66. doi: 10.1016/s0925-4773(99)00013-1. [DOI] [PubMed] [Google Scholar]
- Stuhlmiller TJ, Garcia-Castro MI. Current perspectives of the signaling pathways directing neural crest induction. Cell Mol Life Sci. 2012a;69:3715–3737. doi: 10.1007/s00018-012-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhlmiller TJ, Garcia-Castro MI. FGF/MAPK signaling is required in the gastrula epiblast for avian neural crest induction. Development. 2012b;139:289–300. doi: 10.1242/dev.070276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E, Steventon B, Scrapra E, Garcia S, Trepat X, Streit A, Mayor R. Chase-and-run between adjacent cell populations promotes directional collective migration. Nat Cell Biol. 2013;15:763–772. doi: 10.1038/ncb2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh-Finkel L, Zeisel A, Brodt-Ivenshitz M, Shamai A, Yao Z, Seger R, Domany E, Tzahor E. BMP-mediated inhibition of FGF signaling promotes cardiomyocyte differentiation of anterior heart field progenitors. Development. 2010;137:2989–3000. doi: 10.1242/dev.051649. [DOI] [PubMed] [Google Scholar]
- Tompers DM, Foreman RK, Wang Q, Kumanova M, Labosky PA. Foxd3 is required in the trophoblast progenitor cell lineage of the mouse embryo. Dev Biol. 2005;285:126–137. doi: 10.1016/j.ydbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Tran KD, Miller MR, Doe CQ. Recombineering Hunchback identifies two conserved domains required to maintain neuroblast competence and specify early-born neuronal identity. Development. 2010;137:1421–1430. doi: 10.1242/dev.048678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribulo C, Aybar MJ, Nguyen VH, Mullins MC, Mayor R. Regulation of Msx genes by a Bmp gradient is essential for neural crest specification. Development. 2003;130:6441–6452. doi: 10.1242/dev.00878. [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev Cell. 2003;4:509–519. doi: 10.1016/s1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- Villanueva S, Glavic A, Ruiz P, Mayor R. Posteriorization by FGF, Wnt, and retinoic acid is required for neural crest induction. Dev Biol. 2002;241:289–301. doi: 10.1006/dbio.2001.0485. [DOI] [PubMed] [Google Scholar]
- Wandzioch E, Zaret KS. Dynamic signaling network for the specification of embryonic pancreas and liver progenitors. Science. 2009;324:1707–1710. doi: 10.1126/science.1174497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawersik S, Maas RL. Vertebrate eye development as modeled in Drosophila. Hum Mol Genet. 2000;9:917–925. doi: 10.1093/hmg/9.6.917. [DOI] [PubMed] [Google Scholar]
- Wayne S, Robertson NG, DeClau F, Chen N, Verhoeven K, Prasad S, Tranebjarg L, Morton CC, Ryan AF, Van Camp G, Smith RJ. Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Hum Mol Genet. 2001;10:195–200. doi: 10.1093/hmg/10.3.195. [DOI] [PubMed] [Google Scholar]
- Wilson SI, Edlund T. Neural induction: toward a unifying mechanism. Nat Neurosci. 2001;4(Suppl):1161–1168. doi: 10.1038/nn747. [DOI] [PubMed] [Google Scholar]
- Wilson SI, Rydstrom A, Trimborn T, Willert K, Nusse R, Jessell TM, Edlund T. The status of Wnt signalling regulates neural and epidermal fates in the chick embryo. Nature. 2001;411:325–330. doi: 10.1038/35077115. [DOI] [PubMed] [Google Scholar]
- Winchester CL, Ferrier RK, Sermoni A, Clark BJ, Johnson KJ. Characterization of the expression of DMPK and SIX5 in the human eye and implications for pathogenesis in myotonic dystrophy. Hum Mol Genet. 1999;8:481–492. doi: 10.1093/hmg/8.3.481. [DOI] [PubMed] [Google Scholar]
- Woda JM, Pastagia J, Mercola M, Artinger KB. Dlx proteins position the neural plate border and determine adjacent cell fates. Development. 2003;130:331–342. doi: 10.1242/dev.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Dude CM, Baker CVH. Fine-grained fate maps for the ophthalmic and maxillomandibular trigeminal placodes in the chick embryo. Dev Biol. 2008;317:174–186. doi: 10.1016/j.ydbio.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- Yan J, Xu L, Crawford G, Wang Z, Burgess SM. The forkhead transcription factor FoxI1 remains bound to condensed mitotic chromosomes and stably remodels chromatin structure. Mol Cell Biol. 2006;26:155–168. doi: 10.1128/MCB.26.1.155-168.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Watts J, Xu J, Wandzioch E, Smale ST, Sekiya T. Pioneer factors, genetic competence, and inductive signaling: programming liver and pancreas progenitors from the endoderm. Cold Spring Harb Symp Quant Biol. 2008;73:119–126. doi: 10.1101/sqb.2008.73.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Knosp BM, Maconochie M, Friedman RA, Smith RJH. A comparative study of Eya1 and Eya4 protein function and its implication in branchio-oto-renal syndrome and DFNA10. J Assoc Res Otolaryngol. 2004;5:295–304. doi: 10.1007/s10162-004-4044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Huang L, Wei ZB, Silvius D, Tang B, Xu PX. The role of Six1 in mammalian auditory system development. Development. 2003;130:3989–4000. doi: 10.1242/dev.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CC, Dyer MA, Uchikawa M, Kondoh H, Lagutin OV, Oliver G. Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development. 2002;129:2835–2849. doi: 10.1242/dev.129.12.2835. [DOI] [PubMed] [Google Scholar]
- Zou D, Silvius D, Fritzsch B, Xu PX. Eya1 and Six1 are essential for early steps of sensory neurogenesis in mammalian cranial placodes. Development. 2004;131:5561–5572. doi: 10.1242/dev.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D, Silvius D, Rodrigo-Blomqvist S, Enerback S, Xu PX. Eya1 regulates the growth of otic epithelium and interacts with Pax2 during the development of all sensory areas in the inner ear. Dev Biol. 2006;298:430–441. doi: 10.1016/j.ydbio.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]