Abstract
Background
Human immunodeficiencies characterized by hypomorphic mutations in critical developmental and signaling pathway genes allow for the dissection of the role of these genes in the development of the T-cell receptor (TCR) repertoire and the correlation of alterations of the TCR repertoire with diverse clinical phenotypes.
Objective
The presence of T cells in patients with Omenn syndrome (OS) and patients with atypical presentations of severe combined immunodeficiency gene mutations presents an opportunity to study the effects of the causal genes on TCR repertoires and provides a window into the clinical heterogeneity observed.
Methods
We performed deep sequencing of TCRβ complementarity-determining region 3 (CDR3) regions in subjects with a series of immune dysregulatory conditions caused by mutations in recombination activating gene 1/2 (RAG 1/2), IL-2 receptor γ (IL2RG), and ζ chain–associated protein kinase 70 (ZAP70); a patient with atypical DiGeorge syndrome; and healthy control subjects.
Results
We found that patients with OS had marked reductions in TCRβ diversity compared with control subjects, as expected. Patients with atypical presentations of RAG or IL2RG mutations associated with autoimmunity and granulomatous disease did not have altered overall diversity but instead had skewed V-J pairing and skewed CDR3 amino acid use. Although germline TCRs were more abundant and clonally expanded in patients with OS, nongermline sequences were expanded as well. TCRβ from patients with RAG mutations had less junctional diversity and smaller CDR3s than patients with OS caused by other gene mutations and healthy control subjects but relatively similar CDR3 amino acid use.
Conclusions
High-throughput TCR sequencing of rare immune disorders has demonstrated that quantitative TCR diversity can appear normal despite qualitative changes in repertoire and strongly suggests that in human subjects RAG enzymatic function might be necessary for normal CDR3 junctional diversity.
Keywords: T-cell receptor, recombination activating gene, Omenn syndrome, T-cell receptor sequencing
Genetic mutations associated with severe combined immunodeficiency (SCID) can be associated with a range of disease manifestations, from the absence of T cells to Omenn syndrome (OS) characterized by oligoclonal T-cell receptor (TCR) expansion, lymphocytosis, organomegaly, erythroderma, high serum IgE levels, and eosinophilia to diseases associated with autoimmunity and granulomatous disease.1–4 The heterogeneity can, in part, be explained by the degree of residual activity in recombination activating gene 1/2 (RAG1/2)1,5–7 or other SCID-related genes.8,9
The presence of T cells in patients with OS and other hypomorphic forms of SCID disorders presents an opportunity to study the effects of the underlying mutated genes or the consequences of poor thymic development on TCR repertoires by analyzing the overall diversity of the residual cells, as well as TCR characteristics, such as junctional diversity and amino acid use. It also provides a window into the clinical heterogeneity of this spectrum of disease.
We performed deep sequencing of TCRβ complementarity-determining region 3 (CDR3) regions in patients with OS, patients with hypomorphic forms of SCID, and healthy control subjects. We found that patients with OS with RAG mutations had less junctional diversity than control subjects and patients with OS of other causes. Patients with atypical clinical manifestations of mutations in SCID-associated genes had normal quantitative diversity but skewed V-J use and CDR3 amino acid composition compared with patients with OS or control subjects. Although TCR sequences without junctional diversity (germline sequences) were more abundant and clonally expanded in patients with OS, TCR sequences with junctional diversity (nongermline sequences) were expanded as well. The results suggest a role for RAG genes in CDR3 junctional diversity. It appears that aberrant TCR generation, but not numeric diversity, contributes to immune dysregulation in patients with certain hypomorphic forms of SCID.
METHODS
Patients
Five patients with the classical OS presentation, including infancy-onset hepatosplenomegaly, lymphadenopathy, erythroderma, and TCR oligoclonality, were included. Their mutations were W959X, R410W/R778W, del887G (protein negative), R559S/M435V, and R410Q/M435V in patients OS_RAG1 through OS_RAG5, respectively (see Table E1 in this article’s Online Repository at www.jacionline.org). Patient PB7 with OS caused by ζ chain–associated protein kinase 70 (ZAP70) deficiency has been described,10 as has patient 1228601 with RAG mutations causing autoimmunity and granulomata.4 Patient DIG with atypical complete DiGeorge anomaly had a rash and lymphadenopathy at 4 months of age, 18 months before thymus transplantation. The sample tested was obtained 14.3 months before thymus transplantation.11 Patient CMNL13687 with an IL-2 receptor γ (IL2RG) mutation has been described in part (Chinen et al,12 patient P1; De Ravin and Malech,13 noted on page 231 as an example of SCID-X1 with multiple viral infections). The patient’s mutation was in the poly-A tail of IL2RG. He presented in infancy with Pneumocystis jiroveci pneumonia that responded to antibiotic therapy. In his first 4 years of life, he underwent 4 attempts at transplantation with haploidentical lymphocyte-depleted parental bone marrow without ever achieving engraftment in any lineages. Despite failure to engraft, he survived on antibiotic/antiviral prophylaxis and regular infusions of intravenous gamma globulin alone. Although he had occasional viral infections and pneumonias, his main clinical problems were severe inflammatory bowel disease, severe eczema, and autoimmune alopecia. At 11 years of age, this patient was treated with ex vivo retrovirus gene therapy without marrow conditioning that resulted in only a low level of gene marking (approximately 3% to 6% in T lymphocytes and none persisting in other lineages), no restoration of measured immune functions, and little change in clinical status. Since providing the blood sample at a routine clinic visit that was used for the sequencing shown in the current report, he has received a matched unrelated cord blood hematopoietic stem cell transplant with conditioning resulting in full multilineage engraftment without any graft-versus-host disease that has led to complete resolution of the autoimmune symptoms, restoration of normal nutrition and growth, cessation of infections, and restoration of immune cell numbers and functions (see Table E2 in this article’s Online Repository at www.jacionline.org).
Patient and adult control samples were obtained with National Institute of Allergy and Infectious Diseases Institutional Review Board approval or Duke University Institutional Review Board approval, according to the guidelines of the local Medical Ethics Committee of the Erasmus MC, or under a standard Newcastle upon Tyne Hospitals’ Paediatric Bone marrow transplant research protocol for use of deidentified samples.
Flow cytometry and cell sorting
Flow cytometric cell sorting was performed on a 20-parameter FACSAria (BD Biosciences, San Jose, Calif) running FACSDiva software (version 6.1.3, BD Biosciences). B cells, T cells, and monocytes were discriminated by staining PBMCs with the following fluorescently labeled, mouse anti-human mAbs: anti-CD20 fluorescein isothiocyanate (in-house conjugated), anti-CD127 phycoerythrin (in-house conjugated), anti-CD3 H7APC (BD Biosciences), anti-CD14 Pacific Blue (BD Biosciences), anti-CD4 QD605 (Invitrogen, Carlsbad, Calif), anti-CD8 QD705 (Invitrogen), and ViViD LIVE/DEAD Fixable Dead Cell Stain (Invitrogen). Total CD4+ or CD8+ T cells were defined as CD3+, ViViD−, CD14− and CD4+ or CD8+, respectively.
TCRβ library construction for illumina sequencing
T cells from different subjects were sorted, with 708 to 2,000,000 cells (median, 126,000 cells) per patient directly sorted into collection tubes with RNAlater solution (Invitrogen). Cell lysis and mRNA extraction were performed according to the μMacs mRNA isolation kit (Miltenyi Biotec, Auburn, Calif). Each sample was eluted in 40 μL. Anchored RT-PCR was performed with a modified version of the SMARTer RACE cDNA amplification kit (Clontech Laboratories, Mountain View, Calif). The cDNA synthesis was performed in a total of 17.5 μL for 1.5 hour at 42°C by using 8 μL of the extracted mRNAwith final concentrations of the following reagents: Invitrogen synthesized 5′CDS Oligo dT primer, 0.685 μmol/L; 1× Clontech first-strand buffer; Clontech dithiothreitol, 1.14 mmol/L; Clontech dNTP Mix, 0.57 mmol/L; Clontech SMARTER oligo, 0.685 μmol/L; Invitrogen RNAseOUT Ribonuclease Inhibitor, 2.29 U/μL; and Invitrogen SuperScript II RT, 11.4 U/μL. The cDNA was cleaned with a modified version of the Agencourt AMPure XP kit (Beckman Coulter, Brea, Calif). Briefly, AMPure XP was added at a ratio of 1.8 volumes of the cDNA reaction. The binding was incubated for 5 minutes at room temperature. The AMPure Xp beads were separated with the Agencourt SPRIPlate magnet plate by incubating the cDNA-AMPure XP beads in the plate for 2 minutes. The cDNA reaction mix was removed, and the cDNA was resuspended in 30 μL of Invitrogen molecular grade water. TCRβ was amplified by using the KAPA Real Time Library Amplification kit (Kapa Biosystems, Woburn, Mass). Briefly, the total volume for the KAPA amplification reaction was 50 μL. We used the Clontech 5 Primer II A primer at a final concentration of 0.168 μmol/L as the 5′ primer and the TCRβ constant primer 5′-TGCTTCTGATGGCTCAAACACAGCGACCT-3′ at a final concentration of 0.2 μmol/L as the 3′ primer. The touchdown PCR consisted of 1 hold of 5 minutes at 95°C, 5 cycles of 20 seconds at 98°C and 1 minute at 72°C, and a variable number of cycles of 20 seconds at 98°C and 1 minute at 68°C until the amplification curve crossed the second standard, as described in the KAPA RT library amplification kit. The amplified samples were run in Invitrogen E-Gel SizeSelect 2%-gel system according to the manual. Twenty microliters of each library underwent a second PCR for addition of the Illumina adaptors (Illumina, San Diego, Calif). For the second PCR, we used the KAPA Real Time Library Amplification kit. We used two 5′ primers: the PE1 FCB ILL 1_2 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ at a final concentration of 0.2 μmol/L and the corresponding PE1 ILL Bar code 2_2 primer (see Table E3 in this article’s Online Repository at www.jacionline.org) at a final concentration of 0.02 μmol/L. The 3′ primer 5′-CAAGCAGAAGACGGCATACGAGATTGCTTCTGATGGCTCAAACACAGCGACCT-3′ was used at a final concentration of 0.2 μmol/L. The touch-up PCR conditions are as follow: 1 hold for 5 minutes at 95°C; 2 cycles of 20 seconds at 98°C, 30 seconds at 54°C, and 1 minute at 68°C; and 7 cycles of 20 seconds at 98°C and 1 minute at 68°C. The PCR product was cleaned by using the Agencourt AMPure XP kit, according to the protocol. The libraries were quantified with the Kapa Library quantification kit–Illumina, according to the manual.
Illumina sequencing and data analysis
The libraries were clustered in the Illumina Cbot according to the Illumina protocol at 8 pmol/L. Sequencing was performed in an Illumina HiSeq (Illumina) instrument using a modified protocol for 150 paired end reads. Lane 1 was dedicated to PhiX and used as a control lane for cross-talk matrix generation and phasing/prephasing. Read 1 was sequenced with the Illumina read 1 primer. For read 2, we added a mix of 13 TCRβ J genes’ primers to the Illumina primer read 2 (HP7) at a final concentration of 0.3 nmol/L each. The minimum coverage of the sampled populations was approximately 10 reads per input cell (see Table E1).
TCRβ annotation was performed by combining a custom Java program written in house and the National Center for Biotechnology Information’s BLAST+ program. Briefly, BLAST+ was used to identify the V and J germline genes of a TCRβ read. The CDR3 was then determined by finding the conserved cysteine at the 5′ end of the CDR3 and the conserved phenylalanine at the 3′ end of the CDR3. Unique TCRβ species, in which species is defined as a unique TCRβ V–CDR3 (nucleotide)–J combination, were then collapsed to determine the count for each species. For each unique species, the number of nucleotides within the CDR3 contributed by the germline V, J, and D genes was calculated, as was the number of nucleotide additions. The numbers of nucleotide additions were determined by taking the length of the CDR3 nucleotides and subtracting the number of nucleotides encoded by the V, J, and D genes. A measurement called the germline index was calculated by dividing the number of nucleotides in the CDR3 encoded by V, J, and D genes by the length of the CDR3, producing a value of between 0.0 and 1.0.
In an effort to identify potential sequencing errors, TCRβ species represented by a single sequencing read (ie, having a count of 1) were discarded. Additionally, the coverage was calculated for each sequenced sample in which coverage was defined as follows:
(Total count of annotated reads/Cell count used for library generation).
TCRβ species represented by fewer reads than half of the coverage were then discarded so that if a TCRβ species is represented by 4 reads and the coverage is 10×, that TCRβ species will be discarded.
PCR amplification error was addressed by identifying species in which the same Vand J genes are used and the CDR3 is of the same length but the CDR3s vary by only 1 nucleotide. The count of the species pair was then probed, and if the count of species A was found to be less than 5% of the count of species B, then species A would be discarded as the likely product of PCR amplification error. Replicates from 3 separate CD8+ T-cell memory populations from 2 different healthy donors were compared to determine the variability of sequences from sample run to sample run. Clonotype frequencies correlated with a P value of less than .0001 and an r2 median value of 0.85 (SD, 0.19).
Shannon entropy, species richness, and species evenness were calculated for each TCRβ repertoire by using the R package Vegan. Shannon entropy was defined by using the following equation:
where pi was the proportion of species i, and S was the number of species in a repertoire. Species richness was defined as the number of species found in a population, and species evenness was defined as the Shannon entropy divided by the log of the species richness.
Shannon entropy and species richness were normalized by calculating the maximum Shannon entropy and maximum species richness for each repertoire based on the cell count used for library generation. The calculated Shannon entropy and species richness were then divided by their respective maximums to return a number between 0.0 and 1.0.
For each repertoire, the average and SD were calculated for germline index, CDR3 length, and number of nucleotide additions. This was done twice. In one scenario all TCRβ species were given the same weight (unique sequence analysis), and in the other scenario the relative frequency of each clone within a repertoire was considered (total sequence analysis).
Custom Perl scripts were used to calculate the distribution of CDR3 length, V-J pairing percentage, and amino acid compositions of each CDR3 position, as well as whole CDR3s from all of the annotated TCRβ sequence reads in each subject and group. Each TCRβ percentage of each subject was used for clonally expanded analysis. All of the figures and various statistical analyses were performed with GraphPad Prism 6 (GraphPad Software, La Jolla, Calif) and computing environment R software; additional packages (reshape, ggplot2, and pheatmap) were taken from the Comprehensive R Archive Network. Unpaired t tests were carried out to demonstrate statistical significance for comparison of diversity indexes (*.01 < P < .05, **.001 < P < .01, and ***P < .001), V-J use diversity (*.01 < P < .05, **.001 < P < .01, and ***P < .001), and amino acid composition of each CDR3 position (P < .01). All of the TCRβ sequence reads from each subject in the same group were taken into account when comparing nucleotide addition in V(D)J junctional regions, CDR3 length, and germline index among groups. The Wilcoxon rank sum test (“wilcox.test” in R) was applied in comparison of clone sizes between germline and nongermline TCRβ sequences.
RESULTS
We performed deep sequencing on 5′ RACE PCR products of TCRβ CDR3s from sorted CD4+ and CD8+ T cells14 from (1) healthy control subjects; (2) 2 patients with mutations in SCID-causing genes (IL2RG and RAG) who had lymphopenia but did not have classic presentations of severe early immunodeficiency and who had autoimmunity, granulomata, or both over their lifetimes; (3) 4 patients with OS caused by RAG1/2 mutations; and (4) 2 patients with OS not caused by RAG mutations (1 with a ZAP70 mutation10 and 1 with atypical complete DiGeorge syndrome15,16), which phenocopies OS because of the near-total lack of thymic tissue (see Table E2). The residual RAG enzymatic activity in the patients with OS was substantially less than the RAG mutant activity from the patient with granuloma and autoimmunity.17 For individual samples, means of 27,849 to 76,431 and 4,835 to 109,599 unique CDR3 sequences were generated in CD4+ and CD8+ T cells from healthy control subjects, respectively, and 35 to 4,642 and 6 to 877 unique CDR3 sequences were generated in patients’ CD4+ and CD8+ total T cells. Overall coverage (total annotated TCRβ sequence reads per T cell) ranged from 9× to 136×, which can be attributed to differences in diversity (see Table E1).
Poor TCRβ CDR3 diversity in patients with OS
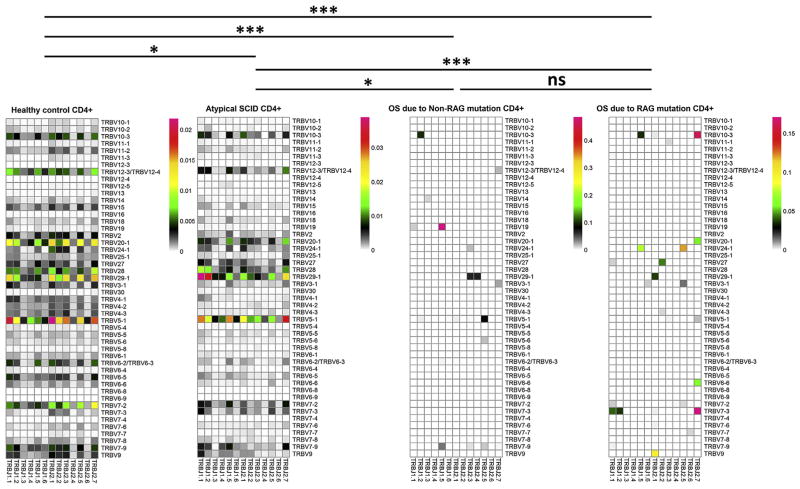
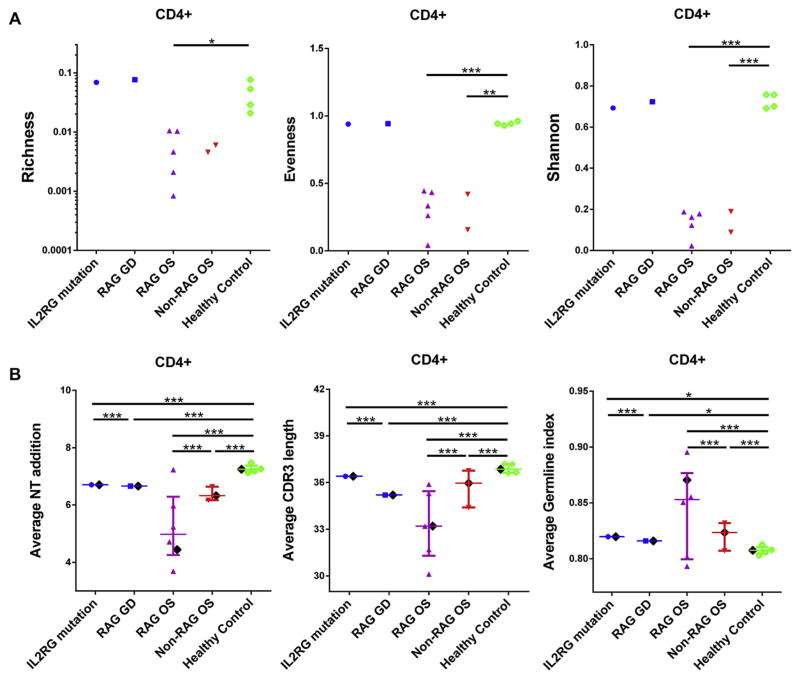
We applied measures of ecosystem diversity within the T-cell populations. Richness represents the numbers of unique TCRs in a given sample. The richness index was calculated by dividing the measured richness by the maximum richness (ie, the number of cells used to generate a library, where it is assumed that each cell could possibly contain a unique TCRβ). Evenness describes how evenly distributed the frequencies of each unique TCR are across the population measured. Shannon entropy is a measure of diversity that takes both richness and evenness into account. The Shannon entropy index was determined by dividing the measured Shannon entropy by the theoretical maximum Shannon entropy that is determined by assuming maximum evenness, and each cell used to generate a sequencing library could possibly contain a unique TCRβ.18 As expected, CD4+ and CD8+ T cells from patients with OS had significant reductions in V-J use (Fig 1 and see Figs E1–E3 in this article’s Online Repository at www.jacionline.org) and in richness, evenness, and Shannon entropy (Fig 2, A, and see Fig E4, A, in this article’s Online Repository at www.jacionline.org) compared with cells from healthy control subjects. Mutations in RAG or IL2RG causing autoimmunity and granulomata did not lead to a diminution of overall sequence diversity (Fig 2, A); however, V-J use was significantly skewed compared with normal values, arguing for preferential generation, expansion, or both of certain V-J segments (Fig 1 and see Figs E1–E3).
FIG 1.
Two-dimensional heat maps of TCRβ V-J use (percentage of different V-J pairing in total annotated TCRβ sequence reads from each group) for CD4+ T cells from healthy control subjects (n = 4), patients with atypical SCID (n = 2), patients with OS caused by non-RAG mutations (n = 2), and patients with OS caused by a RAG mutation (n = 5). The different Vβ genes are on the longitudinal ordinate, and the different Jβ genes are on the horizontal ordinate. P values of unpaired t tests are shown as follows: *.01 < P < .05, **.001 < P < .01, and ***P < .001. ns, Nonsignificant.
FIG 2.
Repertoire characteristics of CD4+ T cells from patients and healthy control subjects. A, Richness, evenness, and Shannon entropy (IL2RG mutation, n = 1; granulomatous disease caused by RAG mutation—RAG granulomatous disease, n = 1; OS caused by RAG mutations, n = 5; OS caused by non-RAG mutations, n = 2; and healthy control subjects, n = 4). B, Junctional nucleotide additions, CDR3 length, and germline index were calculated for total annotated TCRβ sequence reads from individual patients, as well as total annotated TCRβ sequence reads from all subjects in same group, and compared with each other. Patients with atypical SCID, patients with OS caused by RAG mutations, patients with OS caused by non-RAG mutations, and healthy control subjects are shown in blue, purple, red, and green, respectively. The black diamond represents the mean value calculated from pooled TCRβ sequences from all subjects of the same group. Medians with interquartile ranges are shown in each group. P values of unpaired t tests are shown as follows: *.01 < P < .05, **.001 < P < .01, and ***P < .001.
We also calculated CDR3 length and measured nucleotide addition in V(D)J junctional regions to qualitatively describe the TCR repertoires from CD4+ and CD8+ T-cell populations in each subject (see the Methods section). Total sequences from patients with OS caused by RAG1/2 mutations had fewer nucleotide additions in V(D)J junctional regions and smaller CDR3 lengths and were more likely to be germline than control sequences (Fig 2, B, and see Fig E5 in this article’s Online Repository at www.jacionline.org). TCR sequences from patients with OS caused by RAG mutations had significantly smaller CDR3s and less junctional diversity (defined by deviations of V-D or D-J sequences from the germline) compared with that from patients with OS with ZAP70 mutations or DiGeorge syndrome, who themselves had a slight diminution in junctional diversity compared with control subjects. Of note, the patient with granulomatous disease caused by a hypomorphic RAG mutation also showed slightly shorter TCRβ CDR3 lengths than the patients with IL2RG mutations or control subjects (Fig 2, B). Similar patterns were seen when analyzing unique TCRβ sequences (see Fig E6, A, in this article’s Online Repository at www.jacionline.org), although the differences in CDR3 characteristics between patients with OS caused by RAG and others were not as substantial, and in CD8+ T cells (see Fig E4, B; Fig E6, B, and Fig E7 in this article’s Online Repository at www.jacionline.org); however, the paucity of CD8+ T cells in patients with OS and the questionable status of CD8 T cells in patients with OS as bona fide class I–restricted cells precludes more rigorous analysis of this subset. Among the patients with OS caused by RAG mutations, residual enzymatic activity17 did not correlate with the intracohort variations in any of the measures of diversity.
TCRβ with germline sequences is more likely to be expanded peripherally but less so in patients with OS compared with control subjects
The markedly reduced initial numbers of T cells in patients with OS likely undergo massive lymphopenia-induced proliferation,19 which leads to the profound lymphocytosis that patients experience within the first few months of life. However, it is not known what specifically triggers this proliferation in some patients with SCID and not others, whether this proliferation of T cells is driven by TCR stimulation, and, if so, whether it is toward foreign antigens or self-antigens.20
To begin to describe the features of clonally expanded cells, we analyzed the relative expansion of rearrangements with and without junctional diversity and examined the amino acid use of patients’ CDR3s compared with those of control subjects.
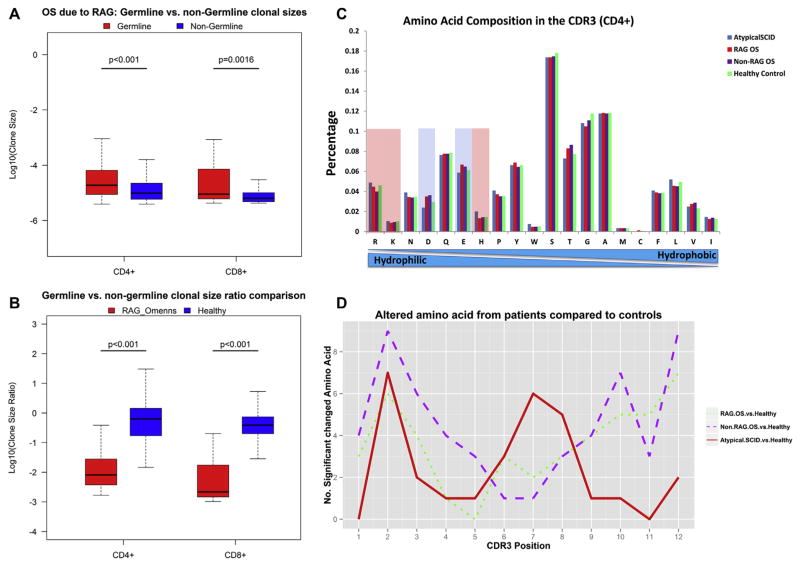
Unique sequences without junctional diversity (germline sequences) were more prevalent in the majority of patients with OS (see Fig E6, A), and those sequences were associated with larger average clone sizes than TCRβ sequences with junctional diversity (nongermline sequences) in patients with OS because of RAG1/2 mutations and healthy control subjects (Fig 3, A, and see Fig E8, A and F, in this article’s Online Repository at www.jacionline.org; P < .01, Wilcoxon rank sum test). This pattern was seen, to a lesser extent, in patients with non-RAG OS and the 2 patients with atypical manifestations of SCID gene mutations (see Fig E8, B–E; P > .01).
FIG 3.
Analysis of clone sizes of germline versus nongermline TCRβ sequences and amino acid composition of CDR3s in patients and control subjects. A, Box plots show clone sizes of CD4+ and CD8+ T cells with germline versus nongermline TCRβ sequences in patients with OS caused by RAG mutations. P values of Wilcoxon rank sum tests are shown. B, Box plots show the ratio of clone size of germline versus nongermline CD4+ and CD8+ T-cell TCRβ sequences compared with the ratio in healthy control subjects. P values of Wilcoxon rank sum tests are shown. C, Amino acid frequencies of the CDR3s for all unique CD4+ T-cell TCRβ sequences from patients with atypical SCID, patients with OS caused by RAG mutations, patients with OS caused by non-RAG mutations, and healthy control subjects (amino acids are ranked from hydrophilic to hydrophobic). Negatively charged amino acids are shown in light blue, and positively charged amino acids are shown in pink. D, Number of statistically significant changed amino acid frequencies in patient populations compared with healthy control subjects (P < .01) at each amino acid position of the CDR3 of TCRβ sequences that were 12 amino acids long.
The increased prevalence of unique germline sequences in patients with OS could be an indication that these CDR3s represent evolutionarily conserved segments that are more likely to survive positive selection in the context of only a small residual amount of thymopoiesis.
Of note, there was nonetheless a substantial population of expanded nongermline sequences in patients with OS caused by RAG1/2 mutations (Fig 3, B; P <.001). This expansion argues that the numeric diminution in thymic progenitors still allows for a substantial subset of randomly modified TCRβ sequences to be profoundly expanded in the periphery.
Clonal TCR expansion in patients with OS or the patients with atypical RAG or IL2RG mutations did not appear to be due to differences in total amino acid use in CDR3s because patients did not differ substantially from control subjects when comparing the hydrophobicity and charge of the residues found (Fig 3, C, and see Fig E9 in this article’s Online Repository at www.jacionline.org). Despite this lack of difference, it was still possible that residue use at individual positions might differ, and therefore we selected a CDR3 length (12 amino acids) that was present commonly in all samples to carefully compare amino acid use at each position (see Fig E10, A, in this article’s Online Repository at www.jacionline.org). Although the gross patterns of amino acid use in patients and control subjects at each position within their CDR3s was similar (see Fig E10), there were significant alterations in amino acid use rates in patients with OS toward the N-terminus and C-terminus of CDR3, which likely represents differential Vand J use in patients compared with healthy control subjects (Fig 3, D). Unlike the patients with OS, however, the 2 patients with atypical manifestations of SCID gene mutations had differential amino acid use centrally (Fig 3, D), arguing that in addition to the skewed V-J use (Fig 1 and see Fig E1), the peripheral population in these patients differs substantially from that in healthy control subjects and in a way not seen in the clonal expansion associated with classic OS. The differences observed between patients and healthy control subjects were not due to normal interindividual amino acid use variation, which is minimal in healthy control subjects (see Fig E11).
DISCUSSION
Deep sequencing has permitted a comprehensive analysis of the TCR repertoires of patients with rare immunodeficiencies that affect T-cell development but do not lead to the absence of T cells. In addition, it has shed light on what happens to the repertoire in different clinical scenarios of TCR development diseases and lymphopenia.
Our finding of smaller CDR3s and reduced junctional diversity in patients with marked peripheral expansion of T cells caused by RAG mutations compared with those with atypical complete DiGeorge syndrome or ZAP70 mutations suggests that normal RAG function itself is required for junctional diversity. Further support that this observation in T cells from patients is not likely due to disease-specific factors, such as cytokine- or microbe-driven peripheral activation, comes from studies of in vitro VDJ recombination in B cells reconstituted with mutant RAG, which was found to also lead to reduced CDR3 lengths.17 Although it has been observed that Rag and terminal deoxynucleotidyl transferase (TdT) activities tightly correlate during thymocyte developmental stages, the 2 genes appear to have discoordinated regulation during early-life thymopoiesis in mice.21 It is not known whether Rag (or, more precisely, variations in Rag function) could directly affect TdT function, junctional diversity, or both. These studies with human disease models have raised the possibility that the open conformation of V-D or D-J, which is maintained by RAG, is a rate-limiting step to N-nucleotide addition by TdT.22–24
The 2 patients with autoimmunity, granulomatous disease, or both caused by IL2RG or RAG mutations were lymphopenic; however, the diversity of their repertoire appeared quantitatively normal. Qualitative differences in V-J pairings and amino acid use at central CDR3 residues point toward a marked skewing of this repertoire. Whether such skewing is the result of a priori abnormalities in thymic rearrangements or a quantitative defect in central and peripheral tolerance remains to be determined. In addition to the qualitative alterations compared with healthy control subjects, the increased TCR repertoire diversity in these patients compared with that seen in patients with OS might explain the different clinical phenotype, with lower risk of severe infections and increased occurrence of autoimmunity in the former and more profound lymphocytosis and global inflammation in the latter. Our findings are in agreement with those of Lee et al,17 who also found that quantitative B-cell receptor diversity is maintained in a patient with granulomatous disease caused by hypomorphic RAG mutations but that skewed V-D-J use leads to a qualitative difference compared with that seen in healthy control subjects.
Finally, the presence of a substantial proportion of expanded TCRs with junctional diversity in addition to those without argues for strong degeneracy of TCR specificities, given that randomly rearranged and altered TCR junctions are capable of marked peripheral expansion, despite a substantial reduction in the number of possible rearrangements caused by the inborn error in TCR production. Further studies of these sequences might help identify the driving mechanism, specificity, or both of these expanded T cells.
Supplementary Material
Clinical implications: The results suggest a role for RAG genes in junctional diversity. It appears that aberrant TCR generation, but not numeric diversity, contributes to immune dysregulation in atypical presentations of SCID gene mutations.
Acknowledgments
Supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
We thank the patients and their families for participating in this study.
Abbreviations used
- CDR3
Complementarity-determining region 3
- IL2RG
IL-2 receptor γ
- OS
Omenn syndrome
- RAG
Recombination activating gene
- SCID
Severe combined immunodeficiency
- TCR
T-cell receptor
- TdT
Terminal deoxynucleotidyl transferase
- ZAP70
ζ chain–associated protein kinase 70
Footnotes
Disclosure of potential conflict of interest: M. van der Burg has received research support from ZonMW. I. Chinn has received research support from the National Institutes of Health. The rest of the authors declare that they have no relevant conflicts of interest.
References
- 1.Villa A, Sobacchi C, Notarangelo LD, Bozzi F, Abinun M, Abrahamsen TG, et al. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood. 2001;97:81–8. doi: 10.1182/blood.v97.1.81. [DOI] [PubMed] [Google Scholar]
- 2.Sobacchi C, Marrella V, Rucci F, Vezzoni P, Villa A. RAG-dependent primary immunodeficiencies. Hum Mutat. 2006;27:1174–84. doi: 10.1002/humu.20408. [DOI] [PubMed] [Google Scholar]
- 3.Niehues T, Perez-Becker R, Schuetz C. More than just SCID—the phenotypic range of combined immunodeficiencies associated with mutations in the recombinase activating genes (RAG) 1 and 2. Clin Immunol. 2010;135:183–92. doi: 10.1016/j.clim.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 4.De Ravin SS, Cowen EW, Zarember KA, Whiting-Theobald NL, Kuhns DB, Sandler NG, et al. Hypomorphic Rag mutations can cause destructive midline granulomatous disease. Blood. 2010;116:1263–71. doi: 10.1182/blood-2010-02-267583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villa A, Santagata S, Bozzi F, Giliani S, Frattini A, Imberti L, et al. Partial V(D)J recombination activity leads to Omenn syndrome. Cell. 1998;93:885–96. doi: 10.1016/s0092-8674(00)81448-8. [DOI] [PubMed] [Google Scholar]
- 6.Santagata S, Villa A, Sobacchi C, Cortes P, Vezzoni P. The genetic and biochemical basis of Omenn syndrome. Immunol Rev. 2000;178:64–74. doi: 10.1034/j.1600-065x.2000.17818.x. [DOI] [PubMed] [Google Scholar]
- 7.Wada T, Takei K, Kudo M, Shimura S, Kasahara Y, Koizumi S, et al. Characterization of immune function and analysis of RAG gene mutations in Omenn syndrome and related disorders. Clin Exp Immunol. 2000;119:148–55. doi: 10.1046/j.1365-2249.2000.01101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ege M, Ma Y, Manfras B, Kalwak K, Lu H, Lieber MR, et al. Omenn syndrome due to ARTEMIS mutations. Blood. 2005;105:4179–86. doi: 10.1182/blood-2004-12-4861. [DOI] [PubMed] [Google Scholar]
- 9.Marrella V, Maina V, Villa A. Omenn syndrome does not live by V(D)J recombination alone. Curr Opin Allergy Clin Immunol. 2011;11:525–31. doi: 10.1097/ACI.0b013e32834c311a. [DOI] [PubMed] [Google Scholar]
- 10.Turul T, Tezcan I, Artac H, de Bruin-Versteeg S, Barendregt BH, Reisli I, et al. Clinical heterogeneity can hamper the diagnosis of patients with ZAP70 deficiency. Eur J Pediatr. 2009;168:87–93. doi: 10.1007/s00431-008-0718-x. [DOI] [PubMed] [Google Scholar]
- 11.Chinn IK, Milner JD, Scheinberg P, Douek DC, Markert ML. Thymus transplantation restores the repertoires of Foxp3 and Foxp3 T cells in complete Digeorge anomaly. Clin Exp Immunol. 2013;173:140–9. doi: 10.1111/cei.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chinen J, Davis J, De Ravin SS, Hay BN, Hsu AP, Linton GF, et al. Gene therapy improves immune function in preadolescents with X-linked severe combined immunodeficiency. Blood. 2007;110:67–73. doi: 10.1182/blood-2006-11-058933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Ravin SS, Malech HL. Partially corrected X-linked severe combined immunodeficiency: long-term problems and treatment options. Immunol Res. 2009;43:223–42. doi: 10.1007/s12026-008-8073-6. [DOI] [PubMed] [Google Scholar]
- 14.Quigley MF, Almeida JR, Price DA, Douek DC. Unbiased molecular analysis of T cell receptor expression using template-switch anchored RT-PCR. Curr Protoc Immunol. 2011;Chapter 10(Unit10.33) doi: 10.1002/0471142735.im1033s94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markert ML, Alexieff MJ, Li J, Sarzotti M, Ozaki DA, Devlin BH, et al. Postnatal thymus transplantation with immunosuppression as treatment for DiGeorge syndrome. Blood. 2004;104:2574–81. doi: 10.1182/blood-2003-08-2984. [DOI] [PubMed] [Google Scholar]
- 16.Markert ML, Alexieff MJ, Li J, Sarzotti M, Ozaki DA, Devlin BH, et al. Complete DiGeorge syndrome: development of rash, lymphadenopathy, and oligoclonal T cells in 5 cases. J Allergy Clin Immunol. 2004;113:734–41. doi: 10.1016/j.jaci.2004.01.766. [DOI] [PubMed] [Google Scholar]
- 17.Lee YN, Frugoni F, Dobbs K, Walter JE, Giliani S, Gennery AR, et al. A systematic analysis of recombination activity and genotype-phenotype correlation in human recombination-activating gene 1 deficiency. J Allergy Clin Immunol. 2014 doi: 10.1016/j.jaci.2013.10.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naumova EN, Gorski J, Naumov YN. Two compensatory pathways maintain long-term stability and diversity in CD8 T cell memory repertoires. J Immunol. 2009;183:2851–8. doi: 10.4049/jimmunol.0900162. [DOI] [PubMed] [Google Scholar]
- 19.Khiong K, Murakami M, Kitabayashi C, Ueda N, Sawa S, Sakamoto A, et al. Homeostatically proliferating CD4 T cells are involved in the pathogenesis of an Omenn syndrome murine model. J Clin Invest. 2007;117:1270–81. doi: 10.1172/JCI30513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourgeois C, Stockinger B. T cell homeostasis in steady state and lymphopenic conditions. Immunol Lett. 2006;107:89–92. doi: 10.1016/j.imlet.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Bogue M, Gilfillan S, Benoist C, Mathis D. Regulation of N-region diversity in antigen receptors through thymocyte differentiation and thymus ontogeny. Proc Natl Acad Sci U S A. 1992;89:11011–5. doi: 10.1073/pnas.89.22.11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weigert M, Gatmaitan L, Loh E, Schilling J, Hood L. Rearrangement of genetic information may produce immunoglobulin diversity. Nature. 1978;276:785–90. doi: 10.1038/276785a0. [DOI] [PubMed] [Google Scholar]
- 23.Gauss GH, Lieber MR. Mechanistic constraints on diversity in human V(D)J recombination. Mol Cell Biol. 1996;16:258–69. doi: 10.1128/mcb.16.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komori T, Okada A, Stewart V, Alt FW. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–5. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.