Abstract
Background and Aims
To examine effects of equol, the soy phytoestrogen metabolite, on gene expression in the monkey iliac artery.
Methods and Results
A high fat/high cholesterol diet was fed to eight ovariectomized cynomolgus monkeys for 6.5 years. After biopsy of the left iliac artery, the animals were randomized to two treatment groups for 8 months; the treatment groups were equol (23.7 mg/100 g diet, n=4) and vehicle (n=4). The right iliac artery was removed at necropsy. Gene expression in the iliac arteries in response to equol was determined by DNA microarray. Comparison of atherosclerotic lesions and plasma lipids at pre- versus post-equol treatment time points and in vehicle versus equol treatment groups did not identify any significant differences. Despite the lack of effect of equol on these parameters, 59 genes were down-regulated and 279 were up-regulated in response to equol. Comparison of these data to previous work identified 10 genes regulated in opposite directions by equol compared to presence of atherosclerosis plaque (Menopause 2011;18:1087–1095) and 55 genes differentially expressed in the same direction in response to both equol and estradiol (Eyster et al., Menopause 2013; in press).
Conclusions
Similar responses of genes to both equol and estradiol may reflect the extent to which equol serves as a natural selective estrogen receptor modulator in the arteries. Opposite responses of 10 genes to equol versus the presence of atherosclerosis implicates those genes in the potential protective effects of equol in arteries.
Keywords: equol, soy, gene expression, DNA microarray, atherosclerosis, artery
Introduction
Equol is a metabolite of the soy isoflavone, daidzein, produced by the action of gastrointestinal bacteria [1]. Equol is a ligand for estrogen receptors (ER) with greater potency at ER than daidzein [1]. Many claims have been made regarding the cardiovascular health benefits of soy isoflavones, and by extension, equol [2]. Studies in laboratory animals support the atheroprotective effects of dietary soy components [3, 4]. However, scientific evidence in human studies in support of these claims has been variable [2, 5]. One contributor to the variability of soy isoflavones in humans is that as many as 30–50% of individuals are not significant producers of equol from daidzein because they lack the appropriate intestinal flora [6]. This heterogeneity leads to substantial variability in the circulating concentrations of equol and daidzein and difficulties in analysis and interpretation of results.
The effects of estrogen therapy on cardiovascular diseases are controversial. When the blood vessels are healthy, estrogen appears to protect them from the development of atherosclerotic plaque [7]. However, if atherosclerosis is well-established, estrogen appears to not be beneficial and may actually increase the risk of clinical events [8, 9]. The exact mechanisms by which estrogen influences cardiovascular disease are not entirely clear, thus the evidence that daidzein, genistein, and equol can influence estrogen receptor activity does not clarify their mechanism of action in the cardiovascular system. However, these soy/soy-derived compounds have greater affinity for ERβ than for ERα [10]. Thus, these compounds may act as natural selective estrogen receptor modulators (SERMs) [11].
Similarities in the cardiovascular system of human and cynomolgus monkeys make this animal model an ideal system for the study of the effects of equol [12]. Cynomolgus monkeys develop atherosclerosis when placed on a North American-type atherogenic diet [3, 12], and the effects of estrogen and soy isoflavones on the cardiovascular system and the development of atherosclerosis have been studied extensively in cynomolgus monkeys [3, 12].
The current study was undertaken to examine global gene expression patterns in the iliac arteries of cynomolgus monkeys in response to treatment with a synthetic racemic mixture of S- and R-equol in order to identify potential equol specific effects in the absence of other isoflavone components and to avoid problems associated with variable equol conversion rates. The extent of atherosclerosis in the iliac arteries has been shown to be directly correlated with that of the coronary arteries in macaques [3]; therefore, iliac arteries were used as proxies for the coronary artery in this study. Moreover, the use of the iliac arteries permitted a longitudinal assessment of pre-versus post-equol treatment.
Methods
Animals and Study Design
Eight adult female cynomolgus macaques (Macaca fascicularis), age 20 years or older, were used for this study. The animals were surgically menopausal with ovariectomy 4–6 years prior to the study. They had been housed in stable social groups of 3–4 animals each and had consumed various semi-purified high fat/high cholesterol diets for 6.5 years upon entering this study. The diet used for this study was formulated to mimic a typical North American diet and contained 0.20 mg cholesterol/Calorie of diet, 29.4% fat, and 19.8% protein from animal sources (casein/lactalbumin). The monkeys received approximately 120 kcal/kg body weight of the diet once daily for 8 months.
The left common iliac artery was biopsied to obtain pretreatment arterial tissue as described [4, 13]. Equol was added to the diet as a supplement at 23.7 mg equol/100 grams of diet (n=4); control animals received the same diet without equol (n=4). The equol supplement contained a 96.0% pure racemic mixture of S- and R-equol enantiomers in a 1:1 ratio and was provided by Solae, a division of Dupont (St. Louis, MO, USA). The dose of equol was designed to mimic the amount of isoflavones consumed by women in several clinical trials [2] and then scaled to account for metabolic differences between women and monkeys. The dietary supplement was the only source of equol in the diet; the diet contained no soy isoflavones. After 8 months on the North American diet with equol or control treatment, the monkeys were euthanized and the post-treatment common iliac artery was collected and processed as described [14]. Briefly, the iliac artery was opened longitudinally, laid flat, and divided into 3 equal segments. One segment was fixed in paraformaldehyde, embedded in paraffin, sectioned, and stained with Verhoeff-van Gieson’s stain for histologic assessment of atherosclerotic plaque intimal area. As previously described [3, 14, 15], digital images were captured from each arterial section and morphometric measurements were made to assess the extent (defined as cross-sectional area in mm2) of iliac artery plaque [3, 15]. A second segment of iliac artery was placed in RNAlater (Sigma R-0901) and stored at −70° until us ed for extraction of total RNA. The third section was frozen and archived. At the initiation of the study (baseline) and after 3, 6, and 8 months of treatment, blood samples were obtained for the measurement of lipids and lipoproteins as described [4, 14].
All animal procedures were carried out at Wake Forest University whose facilities and laboratory animal program are fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care. All procedures using animals conformed to State and Federal laws and were conducted in compliance with standards of the U.S. Department of Health and Human Services, and guidelines established by the Wake Forest University Animal Care and Use Committee (ACUC).
Analysis of gene expression
Total RNA was isolated from segments of iliac artery using a method designed to maximize RNA extraction from small tissue samples [13, 16]. Arterial segments ranging in size from 2.94–13.64 mg were minced in 600 μl TRI reagent (Molecular Research Center, Cincinnati, OH) and homogenized in a 2 ml tube with a 7 mm probe on a Polytron homogenizer (Kinematica, Luzern, CH). The probe was then rinsed in 400 μl of fresh TRI reagent to recover residual sample from the probe. The 2 aliquots of TRI reagent were pooled. Bromochloropropane (200 μl) and 3 M sodium acetate (60 μl) were added and the samples were centrifuged (5 min at 8,000g) to effect phase separation. The aqueous layer contraining RNA was then purified on a silica gel membrane spin column (RNeasy, Qiagen, Valencia, CA) per company instructions [16]. Gene expression signatures were analyzed using CodeLink Whole Human Genome Bioarrays (Applied Microarrays, Tempe, AZ) to compare pretreatment versus posttreatment gene expression for both equol and vehicle treated groups as described [13]. Differential expression of SET domain, bifurcated 2 (SETDB2) was identified by DNA microarray and confirmed by two-step real time reverse transcription-polymerase chain reaction (RT-PCR) as described [13]. Primers and probe were obtained from Applied Biosystems (Life Technologies; Hs00230475_m1). Data from real time PCR reactions were analyzed by qBase software as described [13].
Statistical analysis
Data are expressed as the mean ± standard error of the mean (SE), the experimental number (n) was 4 per group, and the p value was set at 0.05. Statistical analysis of DNA microarray data utilized GeneSpring GX 7.0 software (Agilent, Santa Clara, CA). Multiple testing correction used the Benjamini and Hochberg False Discovery Rate set at 0.05. Real time RT-PCR data were analyzed by paired t-test. Sizes of atherosclerotic lesions were compared by ANOVA. Differences in plasma lipid variables between equol and vehicle treated monkeys at baseline, 3, 6, and 8 months were analyzed by repeated measures ANOVA (JMP V-8, SAS, Cary, NC).
Results
Data were analyzed for statistically different expression in pretreatment versus post-equol treatment monkey iliac arteries. Gene expression data for equol treatment were corrected for vehicle control data. Transcribed sequences with no known function, genes for which the fold expression values in response to equol were less than 2.0-fold different from pretreatment values, and genes for which the average relative expression values were near the limits of detection of the assay were excluded from further analysis. Application of these criteria identified differential expression of 338 genes in the monkey iliac arteries in response to equol (Table 1). Of those genes, 59 were down-regulated by equol and 279 were up-regulated. The presence of atherosclerosis causes substantial changes in gene expression [13]. 76 of the genes that responded to equol were regulated similarly by atherosclerosis (Table 1), whereas 10 showed expression in the opposite direction to that of atherosclerosis (Table 2). Fifty-five genes showed differential expression in the same direction (increase or decrease) in response to both equol and estradiol [14] (Table 1).
Table 1.
Differentially expressed genes in the iliac arteries of ovariectomized cynomolgus macaques treated with equol for 8 months. Gene expression values are shown for pretreatment baseline data (PreTrt; n=4) and for post equol treatment (PostTrt; n=4). Fold expression data (Fold) compare posttreatment to pretreatment values for each gene (PostTrt/PreTrt). The GenBank accession number (GenBank ACCN#) and p value for statistical significance (p val) are also shown.
| Genbank ACCN # | Gene Name | PreTrt | PostTrt | Fold | p val |
|---|---|---|---|---|---|
| Lipid/Fatty acid biosynthesis/metabolism | |||||
| NM_012260 | 2-hydroxyphytanoyl-CoA lyase (HPCL2) | 1.11 | 3.06 | 2.8 | 0.027 |
| NM_020676 | abhydrolase domain containing 6 (ABHD6) | 0.89 | 2.04 | 2.3 | 0.010 |
| NM_030925 | calcium binding protein 39-like (CAB39L) | 6.04 | 2.83 | 0.5 | 0.006 |
| NM_001752 | catalase (CAT) | 0.40 | 1.21 | 3.0 | 0.017 |
| NM_000896 | cytochrome P450, family 4, subfamily F, polypeptide 3 (CYP4F3) | 4.56 | 9.39 | 2.1 | 0.003 |
| NM_003676 | degenerative spermatocyte homolog, lipid desaturase (DEGS) | 2.96 | 6.41 | 2.2 | 0.030 |
| NM_016245 | dehydrogenase/reductase (SDR family) member 8 (DHRS8) | 32.11 | 15.13 | 0.5 | 0.000 |
| NM_001398 | enoyl Coenzyme A hydratase 1, peroxisomal (ECH1) | 0.82 | 1.77 | 2.2 | 0.024 |
| NM_002395 | malic enzyme 1, NADP(+)-dependent, cytosolic (ME1) | 0.35 | 1.25 | 3.6 | 0.027 |
| NM_015922 | NAD(P) dependent steroid dehydrogenase-like (NSDHL) | 0.71 | 2.42 | 3.4 | 0.021 |
| NM_003846 | peroxisomal biogenesis factor 11B (PEX11B) | 0.95 | 2.79 | 2.9 | 0.025 |
| NM_003630 | peroxisomal biogenesis factor 3 (PEX3) | 2.13 | 4.40 | 2.1 | 0.033 |
| NM_018441 | peroxisomal trans-2-enoyl-CoA reductase (PECR)b | 1.80 | 6.11 | 3.4 | 0.048 |
| NM_153613 | lysophosphatidylcholine acyltransferase 4 | 0.52 | 1.10 | 2.1 | 0.043 |
| NM_016048 | isochorismatase domain containing 1 (ISOC1) | 0.58 | 1.66 | 2.9 | 0.013 |
| Glucose metabolism | |||||
| NM_000188 | hexokinase 1 (HK1)b | 3.48 | 8.61 | 2.5 | 0.036 |
| NM_002300 | lactate dehydrogenase B (LDHB) | 25.88 | 54.82 | 2.1 | 0.031 |
| NM_021965 | phosphoglucomutase 5 (PGM5) | 0.40 | 1.14 | 2.9 | 0.042 |
| NM_005609 | phosphorylase, glycogen; muscle (PYGM) | 1.86 | 7.48 | 4.0 | 0.028 |
| ATP synthesis | |||||
| NM_000143 | fumarate hydratase (FH) | 6.04 | 16.44 | 2.7 | 0.028 |
| NM_002080 | glutamic-oxaloacetic transaminase 2, mitochondrial (GOT2) | 3.16 | 9.66 | 3.1 | 0.031 |
| NM_005896 | isocitrate dehydrogenase 1 (NADP+), soluble (IDH1) | 1.77 | 3.95 | 2.2 | 0.039 |
| NM_002168 | isocitrate dehydrogenase 2 (NADP+), mitochondrial (IDH2) | 0.42 | 1.28 | 3.0 | 0.001 |
| NM_005000 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa (NDUFA5) b | 27.84 | 13.27 | 0.5 | 0.011 |
| NM_003002 | succinate dehydrogenase complex, subunit D (SDHD) | 17.66 | 35.44 | 2.0 | 0.011 |
| Protein metabolism | |||||
| NM_183050 | branched chain keto acid dehydrogenase E1, beta polypeptide (BCKDHB) | 1.74 | 3.67 | 2.1 | 0.017 |
| NM_152740 | 3-hydroxyisobutyrate dehydrogenase (HIBADH)b | 2.11 | 5.98 | 2.8 | 0.021 |
| General Enzymes/Metabolism | |||||
| NM_138340 | abhydrolase domain containing 3 (ABHD3)b | 0.68 | 1.86 | 2.7 | 0.047 |
| NM_018641 | carbohydrate (chondroitin 4) sulfotransferase 12 (CHST12)a | 3.39 | 7.48 | 2.2 | 0.028 |
| NM_020682 | Cyt19 protein (CYT19) | 1.69 | 3.65 | 2.2 | 0.007 |
| NM_032857 | lactamase, beta (LACTB)b | 57.55 | 29.57 | 0.5 | 0.050 |
| NM_022493 | nuclear prelamin A recognition factor-like (NARFL) | 0.48 | 1.48 | 3.1 | 0.003 |
| NM_033452 | tripartite motif-containing 47 (TRIM47)a,b | 2.07 | 5.79 | 2.8 | 0.020 |
| NM_017590 | ubiquitous tetratricopeptide containing protein RoXaN (RoXaN) | 3.15 | 6.75 | 2.1 | 0.001 |
| Signal transduction | |||||
| NM_002922 | regulator of G-protein signalling 1 (RGS1) | 17.34 | 3.04 | 0.2 | 0.006 |
| NM_003390 | WEE1 homolog (WEE1)a | 4.67 | 1.76 | 0.4 | 0.005 |
| NM_007314 | v-abl Abelson murine leukemia viral oncogene homolog 2 (ABL2) | 2.31 | 0.88 | 0.4 | 0.011 |
| NM_002928 | regulator of G-protein signalling 16 (RGS16) | 77.98 | 32.00 | 0.4 | 0.007 |
| AK124904 | similar to Rho/Rac guanine nucleotide exchange factora | 4.11 | 1.72 | 0.4 | 0.002 |
| NM_152422 | protein tyrosine phosphatase domain containing 1 (PTPDC1) | 1.00 | 0.43 | 0.4 | 0.032 |
| NM_006622 | polo-like kinase 2 (Drosophila) (PLK2) | 11.41 | 5.36 | 0.5 | 0.002 |
| NM_006224 | phosphotidylinositol transfer protein (PITPN) | 1.49 | 0.73 | 0.5 | 0.019 |
| NM_015093 | mitogen-activated protein kinase kinase kinase 7 interacting protein 2 (MAP3K7IP2)a | 17.52 | 8.63 | 0.5 | 0.031 |
| NM_014836 | Rho-related BTB domain containing 1 (RHOBTB1) | 50.87 | 25.28 | 0.5 | 0.004 |
| NM_022456 | RAB3A interacting protein (rabin3) (RAB3IP) | 1.93 | 0.97 | 0.5 | 0.025 |
| NM_003706 | phospholipase A2, group IVC (PLA2G4C) | 15.29 | 7.69 | 0.5 | 0.000 |
| NM_014225 | protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), alpha (PPP2R1A)a | 14.80 | 28.40 | 1.9 | 0.001 |
| NM_198938 | prostaglandin E synthase 2 (PTGES2) | 0.57 | 1.13 | 2.0 | 0.028 |
| NM_031417 | MAP/microtubule affinity-regulating kinase 4 (MARK4) | 0.49 | 0.98 | 2.0 | 0.000 |
| NM_016656 | Ras-related GTP binding B (RRAGB) | 1.17 | 2.35 | 2.0 | 0.040 |
| NM_004162 | RAB5A, member RAS oncogene family (RAB5A) | 1.68 | 3.38 | 2.0 | 0.009 |
| NM_015915 | spastic paraplegia 3A (autosomal dominant) (SPG3A) | 2.09 | 4.22 | 2.0 | 0.013 |
| NM_006290 | tumor necrosis factor, alpha-induced protein 3 (TNFAIP3) | 1.61 | 3.28 | 2.0 | 0.025 |
| NM_016573 | Gem-interacting protein (GMIP) | 0.53 | 1.08 | 2.0 | 0.045 |
| NM_007182 | Ras association (RalGDS/AF-6) domain family 1 (RASSF1)b | 17.18 | 35.09 | 2.0 | 0.034 |
| U70667 | Fas-ligand associated factor 1 | 16.80 | 34.35 | 2.0 | 0.045 |
| NM_001343 | disabled homolog 2, mitogen-responsive phosphoprotein (DAB2) | 2.56 | 5.26 | 2.1 | 0.027 |
| NM_206835 | TNF receptor-associated factor 7 (TRAF7)b | 5.40 | 11.10 | 2.1 | 0.028 |
| NM_014602 | phosphoinositide-3-kinase, regulatory subunit 4, p150 (PIK3R4) | 1.75 | 3.63 | 2.1 | 0.029 |
| NM_052902 | serine/threonine kinase 11 interacting protein (STK11IP) | 2.08 | 4.38 | 2.1 | 0.020 |
| NM_005219 | diaphanous homolog 1 (DIAPH1) | 5.41 | 11.53 | 2.1 | 0.017 |
| NM_015937 | phosphatidylinositol glycan, class T (PIGT) | 6.61 | 14.23 | 2.2 | 0.008 |
| NM_020983 | adenylate cyclase 6 (ADCY6) | 3.31 | 7.18 | 2.2 | 0.008 |
| NM_012478 | WW domain binding protein 2 (WBP2)a,b | 7.10 | 15.45 | 2.2 | 0.029 |
| NM_003022 | SH3 domain binding glutamic acid-rich protein like (SH3BGRL) | 32.43 | 71.01 | 2.2 | 0.038 |
| NM_005486 | target of myb1-like 1 (TOM1L1) | 0.71 | 1.56 | 2.2 | 0.026 |
| NM_000115 | endothelin receptor type B (EDNRB) | 1.18 | 2.61 | 2.2 | 0.010 |
| N22508 | similar to tumor necrosis factor receptor 2a | 3.29 | 7.43 | 2.3 | 0.006 |
| NM_013314 | B-cell linker (BLNK) | 5.13 | 11.59 | 2.3 | 0.000 |
| NM_002480 | protein phosphatase 1, regulatory (inhibitor) subunit 12A (PPP1R12A) | 13.27 | 30.19 | 2.3 | 0.046 |
| NM_013994 | discoidin domain receptor family, member 1 (DDR1) | 0.76 | 1.73 | 2.3 | 0.001 |
| NM_145245 | similar to ecotropic viral integration site 5; Neuroblastoma stage 4S genea | 3.15 | 7.19 | 2.3 | 0.038 |
| NM_000678 | adrenergic, alpha-1D-, receptor (ADRA1D) | 0.46 | 1.05 | 2.3 | 0.042 |
| NM_015609 | putative MAPK activating protein PM20, PM21 | 1.20 | 2.74 | 2.3 | 0.000 |
| NM_002227 | Janus kinase 1 (a protein tyrosine kinase) (JAK1) | 1.56 | 3.57 | 2.3 | 0.018 |
| NM_016151 | thousand and one amino acid protein kinase (TAO1)a | 6.10 | 14.17 | 2.3 | 0.000 |
| NM_002336 | low density lipoprotein receptor-related protein 6 (LRP6)a | 12.20 | 28.48 | 2.3 | 0.013 |
| NM_014376 | cytoplasmic FMR1 interacting protein 2 (CYFIP2) | 0.91 | 2.15 | 2.4 | 0.013 |
| NM_001242 | tumor necrosis factor receptor superfamily, member 7 (TNFRSF7)b | 0.82 | 1.96 | 2.4 | 0.032 |
| NM_182759 | TAFA3 protein (TAFA3)a | 3.48 | 8.58 | 2.5 | 0.006 |
| NM_198964 | parathyroid hormone-like hormone (PTHLH)a | 11.91 | 29.65 | 2.5 | 0.001 |
| NM_021044 | desert hedgehog homolog (DHH) | 0.66 | 1.67 | 2.5 | 0.011 |
| NM_000801 | FK506 binding protein 1A, 12kDa (FKBP1A) | 4.60 | 11.75 | 2.6 | 0.043 |
| NM_007369 | G protein-coupled receptor 161 (GPR161)a | 5.37 | 13.79 | 2.6 | 0.002 |
| NM_198452 | Similar to calcium/calmodulin-dependent protein kinase 1, beta | 1.49 | 3.85 | 2.6 | 0.005 |
| NM_004383 | c-src tyrosine kinase (CSK) | 1.90 | 4.97 | 2.6 | 0.021 |
| NM_007178 | serine/threonine kinase receptor associated protein (STRAP)b | 2.76 | 7.26 | 2.6 | 0.036 |
| NM_018010 | estrogen-related receptor beta like 1 (ESRRBL1)b | 0.87 | 2.30 | 2.6 | 0.037 |
| NM_024832 | Ras and Rab interactor 3 (RIN3)a | 1.05 | 2.80 | 2.7 | 0.033 |
| NM_020421 | aarF domain containing kinase 1 (ADCK1) | 0.48 | 1.28 | 2.7 | 0.040 |
| NM_181784 | sprouty-related, EVH1 domain containing 2 (SPRED2) | 1.06 | 2.84 | 2.7 | 0.039 |
| NM_003405 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta (YWHAH)b | 5.74 | 15.56 | 2.7 | 0.027 |
| NM_014220 | transmembrane 4 superfamily member 1 (TM4SF1) | 2.01 | 5.61 | 2.8 | 0.050 |
| NM_012302 | latrophilin 2 (LPHN2) | 1.72 | 4.85 | 2.8 | 0.039 |
| NM_016639 | tumor necrosis factor receptor superfamily, member 12A (TNFRSF12A)b | 10.41 | 29.44 | 2.8 | 0.027 |
| NM_002836 | protein tyrosine phosphatase, receptor type, A (PTPRA)b | 2.83 | 8.12 | 2.9 | 0.017 |
| NM_016478 | nuclear interacting partner of anaplastic lymphoma kinase (ALK) (NIPA) | 0.58 | 1.73 | 3.0 | 0.018 |
| NM_015662 | selective LIM binding factor, rat homolog (SLB) | 0.59 | 1.77 | 3.0 | 0.007 |
| NM_004838 | homer homolog 3 (HOMER3)a | 0.50 | 1.60 | 3.2 | 0.048 |
| NM_016586 | MAP3K12 binding inhibitory protein 1 (MBIP) | 1.84 | 5.95 | 3.2 | 0.005 |
| NM_017790 | regulator of G-protein signalling 3 (RGS3)a,b | 1.66 | 5.59 | 3.4 | 0.015 |
| NM_005493 | RAN binding protein 9 (RANBP9) | 0.32 | 1.09 | 3.4 | 0.023 |
| NM_003239 | transforming growth factor, beta 3 (TGFB3) | 0.30 | 1.08 | 3.6 | 0.005 |
| NM_005737 | ADP-ribosylation factor-like 7 (ARL7)a | 0.30 | 1.15 | 3.8 | 0.028 |
| NM_006271 | S100 calcium binding protein A1 (S100A1) | 1.18 | 4.74 | 4.0 | 0.012 |
| NM_006374 | serine/threonine kinase 25 (STK25)b | 2.28 | 9.30 | 4.1 | 0.048 |
| NM_002755 | mitogen-activated protein kinase kinase 1 (MAP2K1)b | 1.84 | 7.53 | 4.1 | 0.016 |
| NM_005539 | inositol polyphosphate-5-phosphatase, 40kDa (INPP5A) | 3.77 | 15.80 | 4.2 | 0.016 |
| NM_030798 | Williams-Beuren syndrome chromosome region 16 (WBSCR16)a | 0.23 | 1.00 | 4.3 | 0.014 |
| NM_212492 | G protein pathway suppressor 1 (GPS1) | 1.29 | 8.00 | 6.2 | 0.017 |
| Transcriptional regulation | |||||
| NM_199072 | I-mfa domain-containing protein (HIC)a | 11.74 | 2.58 | 0.2 | 0.001 |
| NM_001806 | CCAAT/enhancer binding protein (C/EBP), gamma (CEBPG) | 13.79 | 4.21 | 0.3 | 0.024 |
| NM_014112 | 1trichorhinophalangeal syndrome I (TRPS1)a | 29.14 | 12.57 | 0.4 | 0.001 |
| NM_005437 | nuclear receptor coactivator 4 (NCOA4) | 16.28 | 7.85 | 0.5 | 0.014 |
| BC033086 | transcription factor 19 (SC1) | 8.41 | 4.06 | 0.5 | 0.018 |
| NM_005069 | single-minded homolog 2 (SIM2)b | 19.17 | 37.60 | 2.0 | 0.038 |
| NM_018433 | jumonji domain containing 1A (JMJD1A) | 133.85 | 262.61 | 2.0 | 0.042 |
| NM_003419 | zinc finger protein 345 (ZNF345) | 0.94 | 1.86 | 2.0 | 0.047 |
| NM_003743 | nuclear receptor coactivator 1 (NCOA1) | 1.21 | 2.44 | 2.0 | 0.008 |
| NM_173539 | zinc finger protein 596 (ZNF596) | 0.78 | 1.64 | 2.1 | 0.014 |
| NM_017761 | proline-rich nuclear receptor coactivator 2 (PNRC2) | 5.05 | 10.84 | 2.1 | 0.007 |
| NM_001517 | general transcription factor IIH, polypeptide 4, 52kDa (GTF2H4) | 2.47 | 5.40 | 2.2 | 0.004 |
| NM_002017 | Friend leukemia virus integration 1 (FLI1) | 0.96 | 2.28 | 2.4 | 0.019 |
| M94046 | zinc finger protein (MAZ) mRNAa | 10.92 | 26.50 | 2.4 | 0.006 |
| NM_001206 | basic transcription element binding protein 1 (BTEB1)a | 0.79 | 1.95 | 2.5 | 0.019 |
| NM_021994 | zinc finger protein (C2H2 type) 277 (ZNF277)b | 1.12 | 2.82 | 2.5 | 0.025 |
| NM_013260 | transcriptional regulator protein (HCNGP) | 1.04 | 2.70 | 2.6 | 0.005 |
| NM_014423 | ALL1 fused gene from 5q31 (AF5Q31)b | 2.41 | 6.33 | 2.6 | 0.032 |
| NM_016535 | zinc finger protein 581 (ZNF581)a,b | 2.35 | 6.44 | 2.7 | 0.009 |
| NM_001164 | amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) (APBB1) | 0.92 | 2.61 | 2.8 | 0.011 |
| NM_003457 | zinc finger protein 207 (ZNF207) | 5.51 | 15.79 | 2.9 | 0.008 |
| NM_002398 | Meis1, myeloid ecotropic viral integration site 1 homolog (MEIS1) | 1.29 | 3.70 | 2.9 | 0.031 |
| NM_030767 | AT-hook transcription factor (AKNA) | 0.98 | 3.07 | 3.1 | 0.009 |
| NM_003575 | zinc finger protein 282 (ZNF282) | 0.45 | 1.45 | 3.2 | 0.017 |
| NM_004118 | forkhead-like 18 (Drosophila) (FKHL18)a | 3.74 | 12.10 | 3.2 | 0.041 |
| NM_014067 | LRP16 protein (LRP16) | 0.33 | 1.10 | 3.3 | 0.001 |
| NM_003418 | zinc finger protein 9 (a cellular retroviral nucleic acid binding protein) (ZNF9) | 7.95 | 28.17 | 3.5 | 0.029 |
| NM_005610 | retinoblastoma binding protein 4 (RBBP4) | 2.98 | 11.36 | 3.8 | 0.004 |
| NM_020338 | retinoic acid induced 17 (RAI17), mRNA | 0.66 | 2.53 | 3.8 | 0.019 |
| NM_019102 | homeo box A5 (HOXA5) | 0.73 | 2.80 | 3.8 | 0.031 |
| NM_021078 | GCN5 general control of amino-acid synthesis 5-like 2 (yeast) (GCN5L2)b | 1.31 | 5.13 | 3.9 | 0.014 |
| NM_014583 | LIM and cysteine-rich domains 1 (LMCD1) | 0.83 | 3.37 | 4.1 | 0.039 |
| U38904 | zinc finger protein C2H2-25 mRNA | 0.35 | 1.44 | 4.1 | 0.032 |
| NM_002167 | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein (ID3)a | 2.17 | 10.63 | 4.9 | 0.024 |
| Nucleic acid synthesis and regulation | |||||
| NM_004396 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (DDX5)a | 3.57 | 1.34 | 0.4 | 0.031 |
| NM_006284 | TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa (TAF10) | 1.95 | 0.74 | 0.4 | 0.001 |
| NM_031915 | SET domain, bifurcated 2 (SETDB2)b | 43.73 | 17.33 | 0.4 | 0.019 |
| NM_015450 | protection of telomeres 1 (POT1)a | 12.54 | 5.47 | 0.4 | 0.013 |
| NM_139281 | WD repeat domain 36 (WDR36) | 4.28 | 1.94 | 0.5 | 0.002 |
| NM_145715 | tigger transposable element derived 2 (TIGD2)b | 8.72 | 4.07 | 0.5 | 0.017 |
| NM_019070 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 (DDX49) | 13.89 | 6.92 | 0.5 | 0.001 |
| NM_000553 | Werner syndrome (WRN) | 3.59 | 1.79 | 0.5 | 0.000 |
| NM_006469 | influenza virus NS1A binding protein (IVNS1ABP) | 18.57 | 9.56 | 0.5 | 0.008 |
| NM_005777 | RNA binding motif protein 6 (RBM6) | 2.98 | 5.89 | 2.0 | 0.038 |
| NM_024096 | XTP3-transactivated protein A (XTP3TPA)b | 3.64 | 7.20 | 2.0 | 0.033 |
| NM_015117 | zinc finger CCCH type domain containing 3 (ZC3HDC3)a | 1.67 | 3.34 | 2.0 | 0.000 |
| NM_018119 | polymerase (RNA) III (DNA directed) polypeptide E (80kD) (POLR3E)b | 0.75 | 1.52 | 2.0 | 0.011 |
| NM_012232 | polymerase I and transcript release factor (PTRF)a | 4.03 | 8.34 | 2.1 | 0.006 |
| NM_020385 | XPMC2 prevents mitotic catastrophe 2 homolog (XPMC2H) | 0.79 | 1.64 | 2.1 | 0.002 |
| NM_014706 | squamous cell carcinoma antigen recognised by T cells 3 (SART3) | 1.40 | 2.91 | 2.1 | 0.001 |
| NM_001499 | GLE1 RNA export mediator-like (yeast) (GLE1L)b | 0.80 | 1.68 | 2.1 | 0.025 |
| NM_003883 | histone deacetylase 3 (HDAC3)b | 3.57 | 7.51 | 2.1 | 0.036 |
| NM_016732 | RNA binding protein (hnRNP-associated with lethal yellow) (RALY) | 4.27 | 9.02 | 2.1 | 0.014 |
| NM_005915 | MCM6 minichromosome maintenance deficient 6 (MCM6) | 1.47 | 3.18 | 2.2 | 0.025 |
| NM_024844 | pericentrin 1 (PCNT1) | 2.45 | 5.48 | 2.2 | 0.008 |
| NM_004504 | HIV-1 Rev binding protein (HRB) | 1.08 | 2.42 | 2.2 | 0.044 |
| NM_014596 | zinc ribbon domain containing, 1 (ZNRD1)b | 1.55 | 3.56 | 2.3 | 0.000 |
| NM_022874 | survival of motor neuron 1, telomeric (SMN1) | 8.55 | 20.03 | 2.3 | 0.041 |
| NM_006828 | activating signal cointegrator 1 complex 3 (ASCC3) (helicase HELIC1)b | 0.77 | 1.82 | 2.4 | 0.024 |
| NM_032361 | THO complex 3 (THOC3) | 1.97 | 4.66 | 2.4 | 0.016 |
| NM_006999 | polymerase (DNA directed) sigma (POLS)b | 0.90 | 2.29 | 2.5 | 0.034 |
| NM_006924 | splicing factor, arginine/serine-rich 1 (splicing factor 2) (SFRS1) | 2.89 | 7.92 | 2.7 | 0.013 |
| NM_031266 | heterogeneous nuclear ribonucleoprotein A/B (HNRPAB)b | 2.86 | 8.36 | 2.9 | 0.018 |
| L29065 | DNA-binding protein A gene | 2.90 | 8.97 | 3.1 | 0.040 |
| NM_004593 | splicing factor, arginine/serine-rich 10 (transformer 2 homolog) (SFRS10)b | 2.71 | 8.61 | 3.2 | 0.018 |
| NM_006391 | importin 7 (IPO7)b | 1.36 | 4.51 | 3.3 | 0.007 |
| NM_031492 | hypothetical protein similar to RNA-binding protein lark (MGC10871) | 1.17 | 3.92 | 3.4 | 0.029 |
| NM_004247 | U5 snRNP-specific protein, 116 kD (U5-116KD) | 0.56 | 1.93 | 3.4 | 0.001 |
| NM_002695 | polymerase (RNA) II (DNA directed) polypeptide E, 25kDa (POLR2E)a | 0.39 | 1.36 | 3.5 | 0.017 |
| NM_014740 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 48 (DDX48) | 4.97 | 19.13 | 3.8 | 0.022 |
| NM_002694 | polymerase (RNA) II (DNA directed) polypeptide C, 33kDa (POLR2C) | 1.81 | 6.99 | 3.9 | 0.007 |
| NM_201224 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 (DDX47)b | 0.86 | 3.91 | 4.5 | 0.016 |
| NM_006026 | H1 histone family, member X (H1FX)b | 1.23 | 7.78 | 6.3 | 0.026 |
| NM_004964 | histone deacetylase 1 (HDAC1) | 0.28 | 1.84 | 6.6 | 0.005 |
| NM_001126 | adenylosuccinate synthase (ADSS)b | 1.13 | 2.81 | 2.5 | 0.010 |
| NM_003875 | guanine monophosphate synthetase (GMPS) | 1.63 | 3.47 | 2.1 | 0.010 |
| NM_004075 | cryptochrome 1 (photolyase-like) (CRY1) | 0.77 | 1.70 | 2.2 | 0.021 |
| NM_175886 | phosphoribosyl pyrophosphate synthetase 1-like 1 (PRPS1L1) | 1.04 | 2.74 | 2.6 | 0.018 |
| Defense/Immune system | |||||
| NM_002982 | chemokine (C-C motif) ligand 2 (CCL2)b | 5.21 | 1.29 | 0.2 | 0.022 |
| NM_024711 | immune associated nucleotide 2 (hIAN2)b | 37.14 | 17.51 | 0.5 | 0.032 |
| NM_000585 | interleukin 15 (IL15) | 2.99 | 1.42 | 0.5 | 0.001 |
| NM_000538 | regulatory factor X-associated protein (RFXAP) | 1.29 | 2.53 | 2.0 | 0.026 |
| NM_006688 | complement component 1, q subcomponent-like 1 (C1QL1) | 3.14 | 6.34 | 2.0 | 0.009 |
| NM_009588 | lymphotoxin beta (TNF superfamily, member 3) (LTB)a | 1.57 | 3.24 | 2.1 | 0.010 |
| NM_018844 | B-cell receptor-associated protein 29 (BCAP29)b | 3.13 | 7.35 | 2.3 | 0.040 |
| NM_197974 | butyrophilin, subfamily 3, member A3 (BTN3A3)b | 2.36 | 5.55 | 2.4 | 0.044 |
| NM_021160 | HLA-B associated transcript 5 (BAT5)b | 3.14 | 9.37 | 3.0 | 0.012 |
| NM_006858 | interleukin 1 receptor-like 1 ligand (IL1RL1LG) | 0.43 | 1.35 | 3.1 | 0.009 |
| NM_030789 | histocompatibility (minor) 13 (HM13)b | 1.95 | 6.72 | 3.4 | 0.007 |
| NM_005335 | hematopoietic cell-specific Lyn substrate 1 (HCLS1) | 0.33 | 1.24 | 3.8 | 0.015 |
| Proteases and their regulators | |||||
| NM_030660 | Machado-Joseph disease (ataxin 3) (MJD) | 2.77 | 1.03 | 0.4 | 0.003 |
| NM_032802 | putative intramembrane cleaving protease (SPPL2A) | 7.61 | 3.57 | 0.5 | 0.003 |
| NM_006036 | putative prolyl oligopeptidase | 3.35 | 1.66 | 0.5 | 0.003 |
| NM_016022 | likely ortholog of C elegans anterior pharynx defective 1A (APH-1A) | 0.94 | 1.85 | 2.0 | 0.010 |
| NM_006215 | serine (or cysteine) proteinase inhibitor, A4 (antitrypsin) (SERPINA4) | 1.31 | 2.77 | 2.1 | 0.004 |
| NM_013379 | dipeptidylpeptidase 7 (DPP7)a,b | 9.54 | 22.95 | 2.4 | 0.047 |
| NM_139159 | dipeptidylpeptidase 9 (DPP9)a | 1.21 | 3.09 | 2.6 | 0.005 |
| NM_004279 | peptidase (mitochondrial processing) beta (PMPCB) | 8.87 | 23.35 | 2.6 | 0.039 |
| NM_005468 | N-acetylated alpha-linked acidic dipeptidase-like 1 (NAALADL1) | 0.96 | 2.83 | 2.9 | 0.003 |
| NM_007173 | protease, serine, 23 (SPUVE) | 2.45 | 7.87 | 3.2 | 0.048 |
| NM_016134 | 2plasma glutamate carboxypeptidase (PGCP)b | 0.86 | 3.63 | 4.2 | 0.015 |
| Cell cycle/cell fate | |||||
| AF220656 | apoptosis-associated nuclear protein PHLDA1 (PHLDA1) | 2.67 | 1.02 | 0.4 | 0.005 |
| NM_006716 | activator of S phase kinase (ASK)a | 7.94 | 3.51 | 0.4 | 0.013 |
| NM_022476 | fused toes homolog (mouse) (FTS) | 2.38 | 5.25 | 2.2 | 0.024 |
| NM_005190 | cyclin C (CCNC) | 0.61 | 1.42 | 2.3 | 0.049 |
| NM_024094 | defective in sister chromatid cohesion homolog 1 (MGC5528) | 33.98 | 82.09 | 2.4 | 0.019 |
| NM_020812 | dedicator of cytokinesis 6 (DOCK6) | 1.71 | 4.24 | 2.5 | 0.042 |
| NM_194271 | ring finger protein 34 (RNF34) | 1.52 | 3.81 | 2.5 | 0.004 |
| NM_017747 | ankyrin repeat and KH domain containing 1 (ANKHD1) | 1.50 | 3.79 | 2.5 | 0.027 |
| NM_032038 | spinster-like (SPINL)b | 2.60 | 6.81 | 2.6 | 0.030 |
| NM_006283 | transforming, acidic coiled-coil containing protein 1 (TACC1) | 1.81 | 4.91 | 2.7 | 0.011 |
| NM_002455 | metaxin 1 (MTX1)b | 2.06 | 5.87 | 2.8 | 0.017 |
| NM_004642 | CDK2-associated protein 1 (CDK2AP1)b | 3.62 | 11.18 | 3.1 | 0.016 |
| NM_016238 | anaphase promoting complex subunit 7 (ANAPC7) | 1.92 | 6.13 | 3.2 | 0.011 |
| NM_004765 | B-cell CLL/lymphoma 7C (BCL7C)a | 2.69 | 8.96 | 3.3 | 0.001 |
| NM_004632 | death associated protein 3 (DAP3)b | 1.58 | 5.58 | 3.5 | 0.018 |
| NM_005426 | tumor protein p53 binding protein, 2 (TP53BP2) | 1.59 | 6.08 | 3.8 | 0.004 |
| NM_001760 | cyclin D3 (CCND3)b | 1.76 | 8.95 | 5.1 | 0.014 |
| Adhesion | |||||
| NM_016174 | cerebral endothelial cell adhesion molecule 1 (CEECAM1)a | 7.68 | 15.94 | 2.1 | 0.009 |
| NM_005245 | FAT tumor suppressor homolog 1 (FAT) | 0.96 | 2.07 | 2.2 | 0.002 |
| NM_003872 | neuropilin 2 (NRP2) | 1.96 | 4.25 | 2.2 | 0.009 |
| NM_006670 | trophoblast glycoprotein (TPBG) | 1.59 | 3.46 | 2.2 | 0.007 |
| NM_021181 | SLAM family member 7 (SLAMF7)a | 3.46 | 8.27 | 2.4 | 0.015 |
| NM_032457 | BH-protocadherin (brain-heart) (PCDH7)b | 20.88 | 53.94 | 2.6 | 0.042 |
| NM_004148 | ninjurin 1 (NINJ1)b | 2.26 | 7.14 | 3.2 | 0.026 |
| NM_001423 | epithelial membrane protein 1 (EMP1) | 0.44 | 1.75 | 4.0 | 0.021 |
| NM_005529 | heparan sulfate proteoglycan 2 (perlecan) (HSPG2)a | 1.18 | 8.27 | 7.0 | 0.007 |
| Extracellular matrix | |||||
| NM_032048 | elastin microfibril interfacer 2 (EMILIN2)b | 11.80 | 5.58 | 0.5 | 0.012 |
| NM_000088 | collagen, type I, alpha 1 (COL1A1)a | 72.58 | 138.33 | 1.9 | 0.032 |
| NM_002023 | fibromodulin (FMOD) | 1.48 | 3.14 | 2.1 | 0.006 |
| NM_000095 | cartilage oligomeric matrix protein (COMP)b | 6.21 | 13.88 | 2.2 | 0.021 |
| NM_130444 | collagen, type XVIII, alpha 1 (COL18A1) | 1.37 | 3.16 | 2.3 | 0.005 |
| NM_022664 | extracellular matrix protein 1 (ECM1)a | 0.34 | 1.21 | 3.6 | 0.040 |
| NM_199235 | collectin sub-family member 11 (COLEC11)a | 1.13 | 4.07 | 3.6 | 0.007 |
| Chaperonins | |||||
| NM_024610 | HSPB (heat shock 27kDa) associated protein 1 (HSPBAP1) | 18.15 | 8.28 | 0.5 | 0.019 |
| NM_006431 | chaperonin containing TCP1, subunit 2 (beta) (CCT2)a | 52.85 | 24.46 | 0.5 | 0.001 |
| NM_012111 | AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (AHSA1)b | 4.21 | 8.56 | 2.0 | 0.041 |
| NM_006221 | protein (peptidyl-prolyl cis/trans isomerase) NIMA-interacting 1 (PIN1) | 2.66 | 5.89 | 2.2 | 0.048 |
| NM_006112 | peptidylprolyl isomerase E isoform 1; cyclophilin 33b | 3.51 | 12.41 | 3.5 | 0.034 |
| NM_144617 | heat shock protein, alpha-crystallin-related, B6 (HSPB6)a | 0.19 | 1.11 | 5.8 | 0.016 |
| Oxidation-reduction | |||||
| NM_000096 | ceruloplasmin (ferroxidase) (CP)b | 2.70 | 9.56 | 3.5 | 0.010 |
| NM_014080 | dual oxidase 2 (DUOX2)a | 6.30 | 18.92 | 3.0 | 0.002 |
| NM_015913 | endoplasmic reticulum thioredoxin superfamily member, 18 kDa (TLP19)b | 1.17 | 3.31 | 2.8 | 0.037 |
| NM_016275 | selenoprotein T (SELT)b | 2.40 | 8.77 | 3.7 | 0.024 |
| NM_000178 | glutathione synthetase (GSS) | 0.87 | 2.11 | 2.4 | 0.005 |
| Cytoskeleton | |||||
| NM_001376 | dynein, cytoplasmic, heavy polypeptide 1 (DNCH1) | 45.02 | 22.20 | 0.5 | 0.003 |
| AA912262 | similar to thymosin beta-4 | 21.92 | 10.96 | 0.5 | 0.007 |
| NM_014900 | COBL-like 1 (COBLL1) | 4.83 | 9.91 | 2.1 | 0.034 |
| NM_032608 | myosin XVIIIB (MYO18B) | 2.06 | 4.37 | 2.1 | 0.026 |
| NM_015033 | formin binding protein 1 (FNBP1)a | 19.47 | 41.60 | 2.1 | 0.019 |
| NM_001978 | erythrocyte membrane protein band 49 (dematin) (EPB49)b | 4.48 | 9.64 | 2.2 | 0.023 |
| NM_014000 | vinculin (VCL) | 8.07 | 23.87 | 3.0 | 0.011 |
| NM_021738 | supervillin (SVIL) | 1.75 | 5.30 | 3.0 | 0.010 |
| NM_053024 | profilin 2 (PFN2) | 1.15 | 3.58 | 3.1 | 0.006 |
| NM_000366 | tropomyosin 1 (alpha) (TPM1) | 4.22 | 15.86 | 3.8 | 0.017 |
| NM_006400 | dynactin 2 (p50) (DCTN2)b | 1.69 | 7.74 | 4.6 | 0.029 |
| Organelle function | |||||
| NM_001011 | ribosomal protein S7 (RPS7)a | 76.57 | 29.38 | 0.4 | 0.030 |
| NM_014445 | stress-associated endoplasmic reticulum protein 1 (SERP1) | 21.88 | 8.61 | 0.4 | 0.003 |
| AK057656 | similar to mitochondrial processing peptidase beta subunita | 14.41 | 6.46 | 0.4 | 0.016 |
| NM_001028 | ribosomal protein S25 (RPS25) | 15.99 | 7.64 | 0.5 | 0.018 |
| NM_013237 | px19-like protein (PX19) | 26.54 | 13.02 | 0.5 | 0.001 |
| NM_014713 | lysosomal-associated protein transmembrane 4 alpha (LAPTM4A) | 64.63 | 133.42 | 2.1 | 0.027 |
| NM_006855 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 (KDELR3) | 2.67 | 5.94 | 2.2 | 0.027 |
| NM_002902 | reticulocalbin 2, EF-hand calcium binding domain (RCN2) | 4.68 | 10.61 | 2.3 | 0.044 |
| NM_006351 | translocase of inner mitochondrial membrane 44 homolog (TIMM44) | 1.29 | 2.95 | 2.3 | 0.004 |
| NM_002901 | reticulocalbin 1, EF-hand calcium binding domain (RCN1) | 3.31 | 7.64 | 2.3 | 0.030 |
| NM_032478 | mitochondrial ribosomal protein L38 (MRPL38)a | 4.24 | 11.15 | 2.6 | 0.002 |
| NM_172251 | mitochondrial ribosomal protein L54 (MRPL54) | 0.32 | 1.01 | 3.2 | 0.002 |
| Protein processing | |||||
| NM_016480 | poly(A) binding protein interacting protein 2 (PAIP2) | 50.05 | 98.55 | 2.0 | 0.015 |
| NM_052870 | sorting nexin associated golgi protein 1 (SNAG1)b | 0.92 | 1.82 | 2.0 | 0.021 |
| NM_182551 | lysocardiolipin acyltransferase 1 | 0.85 | 1.69 | 2.0 | 0.011 |
| NM_007230 | mannosidase, alpha, class 1B, member 1 (MAN1B1)b | 0.89 | 2.44 | 2.7 | 0.034 |
| NM_002676 | phosphomannomutase 1 (PMM1)b | 0.88 | 2.44 | 2.8 | 0.004 |
| NM_000434 | sialidase 1 (lysosomal sialidase) (NEU1)b | 0.79 | 3.77 | 4.8 | 0.009 |
| NM_003032 | sialyltransferase 1 (beta-galactoside alpha-2,6-sialyltransferase) (SIAT1) | 1.92 | 3.87 | 2.0 | 0.011 |
| NM_005216 | dolichyl-diphosphooligosaccharide-protein glycosyltransferase (DDOST)b | 6.91 | 20.35 | 2.9 | 0.031 |
| NM_003863 | dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit (DPM2)b | 2.74 | 11.14 | 4.1 | 0.008 |
| NM_005500 | SUMO-1 activating enzyme subunit 1 (SAE1) | 0.84 | 2.30 | 2.7 | 0.009 |
| NM_012214 | mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isoenzyme A (MGAT4A) | 6.14 | 2.92 | 0.5 | 0.003 |
| NM_020156 | core 1 UDP-galactose:N-acetylgalactosamine-alpha-R beta 1,3-galactosyltransferase (C1GALT1) | 8.35 | 4.22 | 0.5 | 0.022 |
| NM_002047 | glycyl-tRNA synthetase (GARS) | 13.70 | 6.61 | 0.5 | 0.000 |
| NM_001751 | cysteinyl-tRNA synthetase (CARS) | 0.27 | 1.00 | 3.7 | 0.002 |
| NM_004184 | tryptophanyl-tRNA synthetase (WARS) | 1.15 | 2.45 | 2.1 | 0.003 |
| Vesicle transport | |||||
| NM_018261 | SEC3-like 1 (SEC3L1) | 6.17 | 2.72 | 0.4 | 0.000 |
| NM_015386 | component of oligomeric golgi complex 4 (COG4) | 1.75 | 3.55 | 2.0 | 0.044 |
| NM_007277 | SEC6-like 1 (S cerevisiae) (SEC6L1)b | 2.80 | 6.87 | 2.5 | 0.045 |
| NM_138567 | synaptotagmin VIII (SYT8)a | 0.44 | 1.17 | 2.7 | 0.048 |
| NM_198398 | serologically defined breast cancer antigen 84 (SDBCAG84)b | 7.49 | 24.60 | 3.3 | 0.040 |
| NM_004781 | vesicle-associated membrane protein 3 (cellubrevin) (VAMP3) | 2.65 | 10.01 | 3.8 | 0.045 |
| Ubiquitin system and related | |||||
| NM_016406 | Ufm1-conjugating enzyme 1 (Ufc1) | 3.20 | 1.38 | 0.4 | 0.000 |
| NM_006313 | ubiquitin specific protease 15 (USP15) | 4.99 | 2.41 | 0.5 | 0.001 |
| NM_014412 | Siah-interacting protein (SIP) | 17.39 | 8.70 | 0.5 | 0.011 |
| NM_006156 | neural precursor cell expressed, developmentally down-regulated 8 (NEDD8) | 13.02 | 6.66 | 0.5 | 0.016 |
| NM_006913 | ring finger protein 5 (RNF5)b | 7.32 | 14.39 | 2.0 | 0.006 |
| NM_018438 | F-box protein 6 (FBXO6)b | 2.76 | 5.76 | 2.1 | 0.033 |
| NM_003939 | beta-transducin repeat containing (BTRC) | 0.92 | 1.93 | 2.1 | 0.049 |
| NM_020892 | deltex homolog 2 (DTX2) | 0.75 | 1.62 | 2.2 | 0.025 |
| NM_012308 | F-box and leucine-rich repeat protein 11 (FBXL11)a | 8.93 | 21.07 | 2.4 | 0.016 |
| NM_022039 | split hand/foot malformation (ectrodactyly) type 3 (SHFM3)b | 2.88 | 7.38 | 2.6 | 0.040 |
| NM_016129 | COP9 constitutive photomorphogenic homolog subunit 4 (COPS4) | 4.44 | 13.16 | 3.0 | 0.035 |
| NM_006837 | COP9 constitutive photomorphogenic homolog subunit 5 (COPS5)b | 1.89 | 7.74 | 4.1 | 0.022 |
| NM_173647 | ring finger protein 149 (RNF149)b | 154.37 | 68.53 | 0.4 | 0.021 |
| Channels/transporters | |||||
| NM_001679 | ATPase, Na+/K+ transporting, beta 3 polypeptide (ATP1B3) | 3.00 | 1.25 | 0.4 | 0.039 |
| NM_003562 | solute carrier family 25 (oxoglutarate carrier), member 11 (SLC25A11) | 2.12 | 4.19 | 2.0 | 0.004 |
| NM_015945 | solute carrier family 35, member C2 (SLC35C2) | 1.60 | 3.18 | 2.0 | 0.009 |
| NM_014437 | solute carrier family 39 (zinc transporter), member 1 (SLC39A1) | 1.53 | 3.10 | 2.0 | 0.037 |
| NM_001038 | sodium channel, nonvoltage-gated 1 alpha (SCNN1A) | 0.59 | 1.21 | 2.1 | 0.030 |
| NM_022003 | FXYD domain containing ion transport regulator 6 (FXYD6) | 0.91 | 1.90 | 2.1 | 0.029 |
| NM_004732 | potassium voltage-gated channel, shaker-related subfamily, beta member 3 (KCNAB3) | 4.04 | 8.72 | 2.2 | 0.041 |
| NM_001695 | ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C, isoform 1 (ATP6V1C1)b | 2.56 | 5.59 | 2.2 | 0.029 |
| NM_001293 | chloride channel, nucleotide-sensitive, 1A (CLNS1A) | 11.66 | 25.98 | 2.2 | 0.034 |
| NM_133496 | solute carrier family 30 (zinc transporter), member 7 (SLC30A7)b | 0.99 | 2.28 | 2.3 | 0.040 |
| NM_005689 | ATP-binding cassette, sub-family B (MDR/TAP), member 6 (ABCB6) | 0.56 | 1.29 | 2.3 | 0.015 |
| NM_018344 | solute carrier family 29 (nucleoside transporters), member 3 (SLC29A3) | 4.20 | 9.70 | 2.3 | 0.001 |
| NM_015638 | transient receptor potential cation channel, subfamily C, member 4 associated protein (TRPC4AP)b | 0.87 | 2.15 | 2.5 | 0.035 |
| NM_004955 | solute carrier family 29 (nucleoside transporters), member 1 (SLC29A1) | 2.27 | 6.16 | 2.7 | 0.023 |
| NM_000387 | solute carrier family 25, member 20 (SLC25A20) | 0.53 | 1.49 | 2.8 | 0.020 |
| NM_017458 | major vault protein (MVP)a | 1.07 | 3.18 | 3.0 | 0.045 |
| NM_199037 | sodium channel, voltage-gated, type I, beta (SCN1B)a | 2.17 | 6.53 | 3.0 | 0.008 |
| NM_005765 | ATPase, H+ transporting, lysosomal accessory protein 2 (ATP6AP2) | 0.39 | 1.27 | 3.3 | 0.022 |
| NM_004317 | arsA arsenite transporter, ATP-binding, homolog 1 (ASNA1)a | 0.31 | 1.01 | 3.3 | 0.013 |
| NM_016016 | solute carrier family 25 member 39 (SLC25A39)b | 1.37 | 4.90 | 3.6 | 0.014 |
| Unknown function | |||||
| NM_018103 | leucine rich repeat containing 5 (LRRC5)b | 40.70 | 15.24 | 0.4 | 0.026 |
| AK124720 | similar to paraneoplastic antigen MA1 (PNMA1) | 4.37 | 8.60 | 2.0 | 0.004 |
| NM_001294 | cleft lip and palate associated transmembrane protein 1 (CLPTM1) | 2.89 | 5.76 | 2.0 | 0.004 |
| NM_014254 | transmembrane protein 5 (TMEM5) | 4.66 | 10.19 | 2.2 | 0.042 |
| NM_014575 | schwannomin interacting protein 1 (SCHIP1) | 2.12 | 5.11 | 2.4 | 0.046 |
| NM_138373 | myeloid-associated differentiation marker (MYADM) | 0.35 | 1.02 | 2.9 | 0.015 |
| NM_152285 | arrestin domain containing 1 (ARRDC1)a,b | 0.62 | 1.81 | 2.9 | 0.020 |
Table 2.
Genes in the iliac arteries of ovariectomized cynomolgus macaques that were regulated in opposite directions in response to equol treatment compared to the presence of atherosclerosis. Ten genes were up-regulated by equol but down-regulated in the presence of atherosclerosis or vice versa. Data are expressed as fold expression; posttreatment versus pretreatment values for equol (Fold Equol; PostTrt/PreTrt) and presence versus absence of atherosclerosis [13] (Fold Athero; presence/absence). The GenBank accession number (GenBank ACCN#) and gene ontology for each gene are also shown.
| Genbank ACCN # | Gene Name | Fold Equol | Fold Athero | Gene Ontology |
|---|---|---|---|---|
| NM_005069 | single-minded homolog 2 (SIM2) | 2.0 | 0.4 | Transcriptional regulation |
| NM_032457 | BH-protocadherin (brain-heart) (PCDH7) | 2.6 | 0.3 | Adhesion |
| NM_001978 | erythrocyte membrane protein band 49 (dematin) (EPB49) | 2.2 | 0.4 | Cytoskeleton |
| NM_024711 | immune associated nucleotide 2 (hIAN2) | 0.5 | 2.0 | Defense/Immune system |
| NM_032048 | elastin microfibril interfacer 2 (EMILIN2) | 0.5 | 2.4 | Extracellular matrix |
| NM_173647 | ring finger protein 149 (RNF149) | 0.4 | 2.3 | Ubiquitin system and related |
| NM_032857 | lactamase, beta (LACTB) | 0.5 | 3.1 | Catabolic/Metabolism |
| NM_031915 | SET domain, bifurcated 2 (SETDB2) | 0.4 | 2.3 | Nucleic acid regulation |
| NM_145715 | tigger transposable element derived 2 (TIGD2) | 0.5 | 2.3 | Nucleic acid regulation |
| NM_018103 | leucine rich repeat containing 5 (LRRC5) | 0.4 | 2.4 | Unknown function |
The size of atherosclerotic lesions in the iliac vessels of monkeys prior to initiation of treatment with equol averaged 0.51±0.19 mm2 with a range of 0.113–1.003 mm2. After 8 months of treatment with equol, the average plaque size was 0.748±0.22 mm2 with a range of 0.171–1.188 mm2. In control animals, the size of atherosclerotic plaque lesions averaged 0.36±0.21 mm2 with a range of 0–0.873 mm2 prior to initiation of vehicle treatment, and 0.47±0.27 mm2 with a range of 0.025–1.217 mm2 after the 8 month treatment period. Comparison of pre- versus post-treatment and vehicle versus equol treatment did not identify any significant differences among lesion sizes. Plasma lipids were assessed at baseline and at 3, 6, and 8 months of the study. No significant differences in plasma lipids in response to equol treatment were identified (Table 3).
Table 3.
Plasma lipids and body weight (BW, kg) in equol treated (n=4) ovariectomized cynomolgus macaques at baseline and at 3, 6, and 8 months (m) of treatment. Abbreviations: total plasma cholesterol (TPC, mmol/L), triglycerides (TG, mmol/L), high density lipoprotein cholesterol (HDLC, mmol/L), low density lipoprotein plus very low density lipoprotein cholesterol (LDL+VLDL, mmol/L).
| BW (kg) | TPC | TG | HDLC | LDL+VLDL | TPC/HDLC | |
|---|---|---|---|---|---|---|
| Baseline | 2.85±0.18 | 428.75±21.5 | 32.25±1.7 | 37±8.7 | 391.75±15.3 | 13.29±2.5 |
| 3 m | 3.04±0.21 | 340.5±28.6 | 35.25±0.5 | 38.3±7.7 | 302.25±21.1 | 9.59±1.3 |
| 6 m | 3.02±0.19 | 337.5±34.2 | 29.5±1.2 | 33±9.4 | 304.50±38.4 | 13.30±4.5 |
| 8 m | 3.01±0.17 | 372.25±25.9 | 34.75±2.6 | 23±6.6 | 349.25±28.5 | 23.04±9.2 |
SET domain, bifurcated 2 (SETDB2) was identified by DNA microarray as a down-regulated gene in response to equol (Figure 1A). Real time RT-PCR confirmed down-regulation of SETDB2 by equol (Figure 1B).
Figure 1.
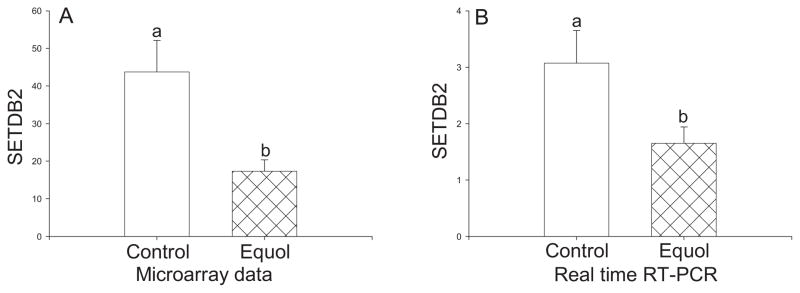
Differential expression of SETDB2 in the iliac arteries of cynomolgus monkeys treated with equol for 8 months. Panel A: Data from DNA microarray analysis, n=4/group. Panel B: Data from real time RT-PCR analysis. Data are expressed as the mean ± SEM. Statistical analysis utilized Student’s t-test. Bars with different letter superscripts denote that the data for those groups are significantly different from each other, p<0.05.
The data set for these DNA microarrays has been deposited in the NCBI Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo) and can be accessed through GEO Series accession numbers GSE37186 and GSE26326 (GSM646184, GSM646185, GSM6946190-5).
Discussion
Of the genes in the iliac arteries that responded to equol in this study, 16% also responded to estradiol [14] but 84% did not. As equol has been shown to activate the ERβ receptor [1], these data support the concept that equol acts as a natural SERM [11]. In contrast to previous findings with soy protein and isoflavones in the cynomolgus monkey [4, 12], equol did not alter plasma lipids or lipoproteins. However, blood lipids and lipoproteins do not appear to be the sole source of positive effects of estrogens in the vasculature [15]. Thus, the gene expression effects of equol make it a potential SERM candidate for modulation of vascular biology. Moreover, soy products have also been reported to exhibit antihypertensive effects [17]. Blood pressure was not measured in the current study, but antihypertensive activity remains a potential protective mechanism of equol in the cardiovascular system. Further research to define the extent of estrogen agonist activity of equol in the arteries is warranted.
Arteries from control and equol-treated animals contained established atherosclerotic plaque prior to initiation of equol treatment. It is likely that the profound gene expression profile differences due to the presence of atherosclerosis [13, 18, 19] at least partially masked the effects of equol on gene expression. Indeed, 86 genes appeared on the differential gene expression lists for both presence of atherosclerosis [13] and response to equol. Of those 86 genes, 76 showed the same directional changes in expression. The pattern of expression in individual arteries suggests that those changes may have been due to the presence of atherosclerotic plaque and further suggest that treatment with equol was unable to reverse the effects of atherosclerosis on expression of those genes. In contrast, the expression of ten genes was regulated in opposite directions in response to equol versus atherosclerosis. These genes may represent points of potentially atheroprotective effects of equol in the arteries. One of these genes, SETDB2, was down-regulated in response to equol but up-regulated in the presence of atherosclerosis [13]. SETDB2 (aliases CLLD8, KMT1F) is a histone 3 lysine 9 (H3K9) methyltransferase with a methyl-CpG-binding domain (MBD). H3K9 methylation is associated with chromatin condensation and transcriptional inactivation. SETDB2 may have a role in chromatin condensation/decondensation as described by Falandry and coworkers [20], or it may have a more directed role in the regulation of specific genes as described by Xu and coworkers [21], or both. In the iliac arteries, the presence of atherosclerosis increased the expression of SETDB2 [13], whereas treatment with equol resulted in a decrease in SETDB2 expression. Therefore, we can speculate that the presence of atherosclerosis would lead to chromatin condensation and repression of the expression of specific genes, whereas treatment with equol would allow chromatin decondensation and up-regulation of specific genes. This is a novel finding as the potential for equol to counter the effects of atherosclerosis on arterial gene expression at the level of histone/chromatin regulation has not previously been reported. However, the effects of regulation of the expression of this gene on arterial function remain to be determined.
Equol activates other members of the steroid receptor superfamily of nuclear receptors in addition to the estrogen receptor. For example, equol can activate peroxisome proliferator activated receptor gamma (PPARγ) [22]. A protective role for PPARγ has been identified in atherosclerosis [23]. We identified up-regulation of 7 genes associated with peroxisome synthesis and function in the monkey iliac arteries in response to equol (CAT, ECH1, HPCL2, ISOC1, PECR, PEX3, and PEX11B) which suggests that equol effectively activated PPARs in the iliac arteries. Another nuclear receptor, the pregnane X receptor (PXR, alias NR1I2) can also be activated by equol [24]. Recent evidence links PXR activation to energy metabolism [25]. Whether PPAR or PXR plays a role in the regulation of gene expression in the iliac arteries in response to equol requires further investigation.
Summary and Conclusions
The differential expression of genes in the arteries in response to equol may reflect agonism at estrogen receptors, PPARγ, or PXR. The 10 genes for which equol treatment was able to overcome the effects of atherosclerosis on expression represent potential targets for protective effects of equol in the arteries.
Acknowledgments
This work was supported by PO1 HL45666 (TBC), RO1 AG28641 (TCR), and RO1 AG27847 (SEA). The Genomics Core facility at the University of South Dakota is supported by NIH INBRE 2 P20 RR016479. The racemic mixture of equol was a generous donation from Solae, a division of Dupont, St. Louis, MO.
These sponsors had no role in the study design, data collection, analysis or interpretation, in the writing of the manuscript, or in the decision to submit the manuscript for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Muthyala RS, Ju YH, Sheng S, Williams LD, Doerge DR, Katzenellenbogen BS, et al. Equol, a natural estrogenic metabolite from soy isoflavones: convenient preparation and resolution of R- and S-equols and their differing binding and biological activity through estrogen receptors alpha and beta. Bioorg Medicin Chem. 2004;12:1559–1567. doi: 10.1016/bmc.2003.11.035. [DOI] [PubMed] [Google Scholar]
- 2.North American Menopause Society. The role of soy isoflavones in menopausal health: report of The North American Menopause Society/Wulf H. Utian Translational Science Symposium in Chicago, IL (October 2010) Menopause. 2011;18:732–753. doi: 10.1097/gme.0b013e31821fc8e0. [DOI] [PubMed] [Google Scholar]
- 3.Clarkson TB, Anthony MS, Morgan TM. Inhibition of postmenopausal atherosclerosis progression: a comparison of the effects of conjugated equine estrogens and soy phytoestrogens. J Clin Endocrinol Metab. 2001;86:41–47. doi: 10.1210/jc.86.1.41. [DOI] [PubMed] [Google Scholar]
- 4.Walker SE, Register TC, Appt SE, Adams MR, Clarkson TB, Chen H, et al. Plasma lipid-dependent and –independent effects of dietary soy protein and social status on atherogenesis in premenopausal monkeys: implications for postmenopausal atherosclerosis burden. Menopause. 2008a;15:950–957. doi: 10.1097/gme.0b013e3181612cef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodis HN, Mack WJ, Kono N, Azen SP, Shoupe D, Hwang-Levine J, et al. for the Women’s Isoflavone Soy Health Research Group. Isoflavone soy protein supplementation and atherosclerosis progression in healthy postmenopausal women. A randomized controlled trial. Stroke. 2011;42:3168–3175. doi: 10.1161/STROKEAHA.111.620831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassidy A, Brown JE, Hawdon A, Faughnan MS, King LJ, Millward J, et al. Factors affecting the bioavailability of soy isoflavones in humans after ingestion of physiologically relevant levels from different soy foods. J Nutr. 2006;136:45–51. doi: 10.1093/jn/136.1.45. [DOI] [PubMed] [Google Scholar]
- 7.Miller VM, Duckles SP. Vascular actions of estrogens: functional implications. Pharmacol Rev. 2008;60:210–241. doi: 10.1124/pr.107.08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarkson TB. Estrogen effects on arteries vary with stage of reproductive life and extent of subclinical atherosclerosis progression. Menopause. 2007;14:373–384. doi: 10.1097/GME.0b013e31803c764d. [DOI] [PubMed] [Google Scholar]
- 9.Dubey RK, Imthurn B, Barton M, Jackson EK. Vascular consequences of menopause and hormone therapy: importance of timing of treatment and type of estrogen. Cardiovasc Res. 2005;66:295–306. doi: 10.1016/j.cardiores.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Jackson RL, Greiwe JS, Schwen RJ. Emerging evidence of the health benefits of S-equol, an estrogen receptor β agonist. Nutr Rev. 2011;69:432–448. doi: 10.1111/j.1753-4887.2011.00400.x. [DOI] [PubMed] [Google Scholar]
- 11.Brzezinski A, Debi A. Phytoestrogens: the “natural” selective estrogen receptor modulators? Eur J Obstet Gynecol Reprod Biol. 1999;85:47–51. doi: 10.1016/50301-2115(98)00281-4. [DOI] [PubMed] [Google Scholar]
- 12.Shelton KA, Clarkson TB, Kaplan JR. Nonhuman primate models of atherosclerosis. In: Abee CR, Mansfield K, Tardif SD, Morris T, editors. Nonhuman Primates in Biomedical Research. 2. San Diego, CA: Elsevier; 2012. pp. 385–411. [Google Scholar]
- 13.Eyster KM, Appt S, Mark-Kappeler CJ, Chalpe A, Register T, Clarkson TB. Gene expression signatures differ with extent of atherosclerosis in monkey iliac artery. Menopause. 2011;18:1087–1095. doi: 10.1097/gme.0b013e3182163fea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eyster KM, Appt S, Chalpe A, Mark-Kappeler CJ, Register TC, Clarkson TB. Effects of estradiol on transcriptional profiles in atherosclerotic iliac arteries in ovariectomized cynomolgus macaques. Menopause. 2013 doi: 10.1097/GME.0b013e31829367c0. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarkson TB, Ethun KF, Chen H, Golden D, Floyd E, Appt SE. Effects of bazedoxifene alone and with conjugated equine estrogens on coronary and peripheral artery atherosclerosis in postmenopausal monkeys. Menopause. 2013;20:274–281. doi: 10.1097/GME.0b013e318271e59b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eyster KM, Brannian JD. Gene expression profiling in the aging ovary. In: Methods in Molecular Biology: Molecular Endocrinology. 2009;590:71–89. doi: 10.1007/978-1-60327-378-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eyster KM, Breitkopf NP, Martin DS. Antihypertensive activity of soy and soy-derived compounds. In: Ahmad A, editor. Soy: Nutrition, Consumption and Health. Nova Scientific Publishers; Hauppauge, NY: 2012. pp. 79–104. [Google Scholar]
- 18.Lutgens E, Faber B, Schapira K, Evelo CT, van Haaften R, Heeneman S, et al. Gene profiling in atherosclerosis reveals a key role for small inducible cytokines. Validation using a novel monocyte chemoattractant protein monoclonal antibody. Circulation. 2005;111:3443–3452. doi: 10.1161/CIRCULATIONAHA.104.510073. [DOI] [PubMed] [Google Scholar]
- 19.Bijnens AP, Lutgens E, Ayoubi T, Kuiper J, Horrevoets AJ, Daemen MJ. Genome-wide expression studies of atherosclerosis. Critical issues in methodology, analysis, interpretation of transcriptomics data. Arterioscler Thromb Vasc Biol. 2006;26:1226–1235. doi: 10.1161/ATV.0000219289.06529.f1. [DOI] [PubMed] [Google Scholar]
- 20.Falandry C, Fourel G, Galy V, Ristriani T, Horard B, Bensimon E, Salles G, Gilson E, Magdinier F. CLLD8/KMT1F is a lysine methyltransferase that is important for chromosome segregation. J Biol Chem. 2010;285:20234–20241. doi: 10.1074/jbc.M109.052399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu P-F, Zhu K-Y, Jin Y, Chen Y, Sun X-J, Deng M, et al. Setdb2 restricts dorsal organizer territory and regulates left-right asymmetry through suppressing fgf8 activity. Proc Natl Acad Sci USA. 2010;107:2521–2526. doi: 10.1073/pnas.0914396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho KW, Lee O-H, Banz WJ, Moustaid-Moussa N, Shay NF, Kim Y-C. Daidzein and the daidzein metabolite, equol, enhance adipocyte differentiation and PPARγ transcriptional activity. J Nutr Biochem. 2010;21:841–847. doi: 10.1016/jnutbio.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 23.Wang N, Yin R, Liu Y, Mao G, Xi F. Role of peroxisome proliferator-activated receptor-γ in atherosclerosis. Circ J. 2011;75:528–535. doi: 10.1253/circj.CJ-11-0060. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Ross-Viola JS, Shay NF, Moore DD, Ricketts M-L. Human CYP3A4 and murine Cyp3A11 are regulated by equol and genistein via the pregnane X receptor in a species-specific manner. J Nutr. 2009;139:898–904. doi: 10.3945/jn.108.103572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ihunnah CA, Jiang M, Xie W. Nuclear receptor PXR, transcriptional circuits and metabolic relevance. Biochim Biophys Acta. 2011;1812:956–963. doi: 10.1016/j.bbadis.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]


