Abstract
Zinc finger, BED-type containing 6 (ZBED6) is an important transcription factor in placental mammals, affecting development, cell proliferation and growth. In this study, we found that the expression of the ZBED6 and IGF2 were upregulated during C2C12 differentiation. The IGF2 expression levels were negatively associated with the methylation status in beef cattle (P < 0.05). A luciferase assay for the IGF2 intron 3 and P3 promoter showed that the mutant-type 439 A-SNP-pGL3 in driving reporter gene transcription is significantly higher than that of the wild-type 439 G-SNP-pGL3 construct (P < 0.05). An over-expression assay revealed that ZBED6 regulate IGF2 expression and promote myoblast differentiation. Furthermore, knockdown of ZBED6 led to IGF2 expression change in vitro. Taken together, these results suggest that ZBED6 inhibits IGF2 activity and expression via a G to A transition disrupts the interaction. Thus, we propose that ZBED6 plays a critical role in myogenic differentiation.
Most economically important traits of interest in cattle, such as growth, carcass, fatness and meat quality, have a multifactorial background and are controlled by environmental factors and an unknown number of quantitative trait locus (QTLs). These quantitative traits arise from interactions between two or more genes and their environment and can be mapped to their underlying genes via closely linked stretches of DNA1. IGF2 is a secreted peptide hormone that plays an important role in muscle tissues via both endocrine and local autocrine/paracrine mechanisms primarily by stimulating the IGF1 receptor (IGF1R). The muscle hypoplasia phenotype of IGF1R-null mice confirms the necessity of IGF signaling in muscle development2,3,4. A paternally expressed QTL affecting muscle growth, fat deposition and heart size in pigs maps to the insulin-like growth factor 2 (IGF2) region5,6.
Gene mapping studies revealed a single nucleotide transition from intron 3-G3072A of the IGF2 as being responsible for much of the difference in body composition between lean pigs and wild boars7. This single nucleotide substitution is located in a CpG site surrounded by a 16 bp evolutionarily conserved region. It is abrogates a binding site for a repressor and leads to a 3-fold greater postnatal expression of IGF2 mRNA in skeletal muscle7. This quantitative trait nucleotide (QTN) is one of the rare examples in which a single base substitution underlying a complex trait has been identified and the mechanism of action is partially understood.
Zinc finger, BED-type containing 6 (ZBED6) is a novel transcription factor that was identified and shown to act as a repressor of IGF2 transcription in skeletal muscle myogenesis and development8. ZBED6 as a domesticated DNA transposon, unique to placental mammals, located in intron 1 of a “host” gene called ZC3H11A8,11. ZBED6 has a single exon comprising more than 900 codons and two DNA-binding BED domains9. ZBED6 is specific for placental mammals and derived from a domesticated DNA transposon10. In addition, ZBED6 is a novel transcription factor that appears to have evolved an essential function in the common ancestor of all placental mammals11. The ZBED6 gene is exclusive to placental mammals and highly conserved among species across 26 placental mammals according to the available genome sequence data8. The functional characterization of ZBED6 shows that it has a broad tissue distribution and may affect the expression of approximately 2,500 putative downstream targets, many of which have essential biological functions in placental mammals8.
In this study, we aimed to elucidate the DNA methylation profiles in the differentially methylated region (DMR) of the intron 3 of the IGF2 gene in nine tissues and organs and their relationships to mRNA expression patterns of fetal and adult using two-tail samples of Chinese Qincuan beef cattle breed with different growth performance. we also investigated the effect of ZBED6 over-expression and knockdown on myoblasts differentiation and IGF2 expression with the goal to provide novel insight on the function of ZBED6 as well as molecular mechanisms underlying myoblasts differentiation and muscle development. Thus, we hypothesized that ZBED6 and IGF2 are directly associated with muscle development or that ZBED6 regulates IGF2 transcription in myoblasts. This hypothesis could provide an insight into the transcriptional regulation of the ZBED6 gene and the potential biological roles of its gene product.
Results
No single nucleotide polymorphism (SNP) in bovine IGF2 intron 3 by DNA sequencing
Previous published results show that a single nucleotide substitution in intron 3-G3072A of IGF2 in pigs7. We amplified and sequenced the intron 3 of IGF2 from 200 DNA samples selected randomly from the study four cattle populations. Compared with the published gene sequence from NCBI, there were no SNP identified in these animals (Figure 1).
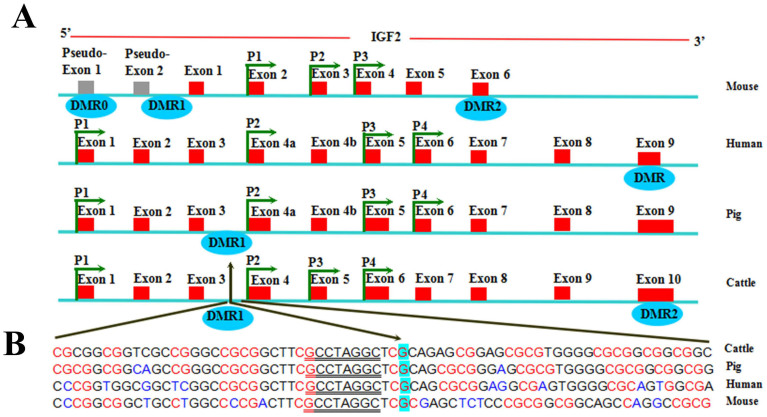
Figure 1. Comparative representation of molecular structure of the IGF2 gene in four different mammalian species.
(A) Numbered red rectangles represent exons, promoter regions are indicated as P1–P4 and intragenic differentially methylated regions as DMR below a rectangle. The mouse IGF2 gene consists of six exons and two additional pseudo-exons28,29. The human30 and pig31,32 IGF2 gene consists of ten exons. The bovine IGF2 gene consists of ten exons, transcription of the gene is initiated from four promoters (P1–P4) and transcripts are alternatively spliced33,34,35,36. Exon 10 contains a DMR that is hypermethylated on the paternal allele27,37. (B) The QTN is indicated with an arrow and the surrounding sequence is shown for four mammals (CpGs are highlighted in red; SNPs are highlighted in blue; a palindromic sequence is underlined).
IGF2 expression was downregulated during fetal and adult stages
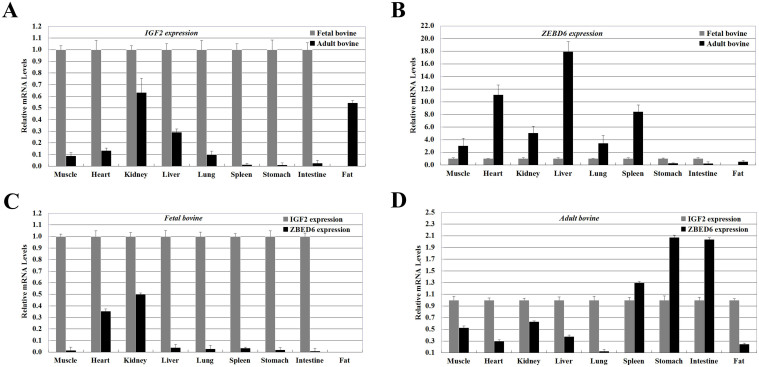
To detect the tissue distribution of bovine ZBED6 and IGF2 mRNA, qPCR was carried out with cDNA from nine cattle tissues and organs (longissimus dorsi muscle, heart, kidney, liver, lung, spleen, stomach, intestine, and fat tissue). As shown in Figure 2, the qPCR analysis showed that IGF2 (Figure 2A) and ZBED6 (Figure 2B), has a broad tissue distribution in cattle tissues and organs, which is consistent with its role as the IGF2 repressor in all examined tissues and organs from six individuals (three individuals per stage). The relative expression levels of IGF2 gene were decreased in all tissues from day 90 fetuses (fetal bovine, FB) to 24-month-old adult bovines (adult bovine, AB), respectively. The results indicated that the bovine ZBED6 mRNA level was relatively lower than IGF2 mRNA in five examined tissues and organs except spleen, stomach, and intestine in fetal and adult bovine groups (Figure 2C and 2D).
Figure 2. Expression pattern analysis of ZBED6 and IGF2 in nine bovine tissues and organs.
IGF2 (A) and ZBED6 (B) mRNA expression were normalized to the geometric mean of the suitable housekeeping genes (GAPDH and ACTINB) and expressed relative to gene expression in the fetal bovine group. IGF2 (C and D) and ZBED6 (C and D) mRNA expression were normalized to the geometric mean of the suitable housekeeping genes (GAPDH and ACTINB) and expressed relative to IGF2 mRNA expression in fetal and adult bovine groups. Error bars represent standard error of the mean (SEM). Each column value represents the means ± SEM of three replicates (n = 3).
IGF2 methylation status was upregulated during fetal and adult stages
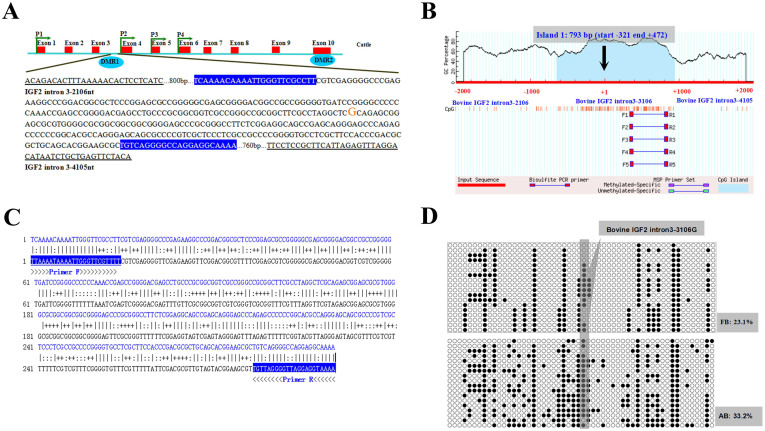
The CpG islands located in the intragenic IGF2 DMR (Figure 3A and 3B) was predicted using online MethPrimer software14 and amplified by PCR. The identities of all amplified PCR products were verified by sequencing. The methylation patterns of the CpG islands were determined using bisulfite-assisted sequencing. The DNA methylation pattern of the intragenic IGF2 DMR in the bovine muscle tissues analysed by BSP are shown in Figure 3. There were 54 CpG sites (Figure 3C) were identified in all of the clones. On the basis of the methylation pattern of each of the clones, we found that at least 30 alleles were different for each bovine group. Methylation data from BSP sequencing were analysed by computing the percentage of methylated CpGs of the total number of CpGs using QUMA software. BSP amplified sequences of DMR (366 bp) have 54 CpGs, and we sequenced 18 clones in each sample. Hence, there was the total 972 CpGs of IGF2 DMR in muscle of fetal bovine (FB) group, the percentage of methylated CpGs is 225/972 = 23.1%. Statistical results showed that the 24-month-old adult bovine (AB) group animals had significantly higher DNA methylation levels in muscle (33.2%) than the day 90 fetal bovine (FB) group animals(Figure 3D).
Figure 3. Molecular structure and differentially methylated region (DMR), BSP amplified nucleotide sequences and DNA methylation patterns of IGF2 DMR1.
(A) Molecular structure of the bovine IGF2 gene. Intron 3 and exon 10 contains two differentially methylated regions (DMRs). A 2000-bp fragment centred around intron 3–3106 (intron 3–3072 mutation on pig IGF2) was considered to methylation analysis. (B) Schematic representation of the proximal promoter region (IGF2 intron 3 −2106 to −4105 base pairs) of the bovine IGF2 gene (modified output of MethPrimer program) to predict regions of high GC content. Dashed lines indicate the GC percentage as represented on the y-axis and the x-axis denotes the bp position on the IGF2 gene intron 3 −2106 to −4105 base pairs untranslated region. Arrows indicate the intron 3–3106 mutation. Coordinates are given in relation to the intron 3–3106 mutation site (shown as +1); vertical lines indicate relative positions of CpG dinucleotides; solid lines depict location of the PCR primers. The 793 bp CpG islands are evident in the intron 3 of the gene, with GC content of 62.50%. (C) Nucleotide sequences for a 366 bp IGF2 DMR (total 54 CpG site) fragment (upper strands) and its bisulfite-converted version (lower strands). Primer sequences are underlined. (D) DNA methylation patterns of muscle tissue in the fetal bovine (FB: 23.1%) group and adult bovine (AB: 33.2%) group analyzed by BSP. Each line represents one individual bacterial clone, and each circle one single CpG dinucleotide. Open circles show unmethylated CpG's and black circles methylated CpG's. The gray box show the 28th CpG site of the intron 3–3106 mutation site by BSP analyses.
ZBED6 and IGF2 were upregulated during C2C12 differentiation
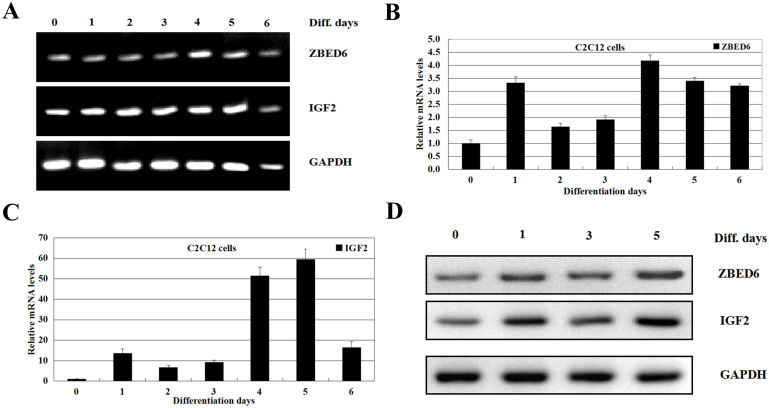
To examine the role of ZBED6 in myogenesis, we first assessed ZBED6 and IGF2 genes expression in the mouse satellite cell line C2C12 upon differentiation. C2C12 myoblasts grown to 100% confluence were induced to differentiate by serum withdrawal (2% horse serum). Typically, cell fusion and formation of small myotubes were evident 2 days after induction, and myotubes were fully formed in 3 days. We analysed the expression of the ZBED6 and IGF2 genes expressing by semi-qPCR during C2C12 differentiation. We found that the ZBED6 and IGF2 genes were upregulated (Figure 4A). To further test the expression change of the ZBED6 and IGF2 genes during C2C12 differentiation, qRT-PCR was performed. After 1 day of differentiation the mRNA levels of both ZBED6 and IGF2 increased more than threefold, and they stayed elevated throughout the differentiation from day 1 to day 6 during myogenic differentiation in C2C12 cells (Figure 4B and 4C). The protein levels of ZBED6 and IGF2 also increased during C2C12 differentiation, as detected by western blotting (Figure 4D). This correlation of differentiation and upregulation of ZBED6 suggested that ZBED6 might be involved in myogenic differentiation of C2C12 cells.
Figure 4. The expression of the ZBED6 and IGF2 genes are upregulation during C2C12 myoblast differentiation.
(A) Semi-qPCR was performed to determine the endogenous expression level of ZBED6, and IGF2 and GAPDH and ACTINB using mRNA isolated from C2C12 cells. Data are representative of three to four independent experiments performed in duplicate (n = 3 or 4). (B and C) C2C12 cells myoblasts were induced to differentiate at 100%. Total RNA was isolated from the differentiating cells on various days as indicated (differentiation days) and subjected to analysis by qRT-PCR to determine the relative levels of ZBED6 (B) and IGF2 (C) genes. The values were normalized to GAPDH and ACTINB mRNA expression level and the value of day 0 was set to 1. Error bars represent standard error of the mean (SEM). The data in (B) and (C) are mean ± SEM from three to four independent experiments (n = 3 or 4). (D) Crude proteins were extracted from the treated cells and then subjected to Western blot with antibodies against ZBED6 and IGF2, GAPDH was used as a control for equal protein loading. Data are representative of three to four independent experiments performed in duplicate (n = 3 or n = 4). The data revealed that the protein levels of ZBED6 and IGF2 also increased during C2C12 differentiation.
ZBED6 repressed transcriptional activity of IGF2 in C2C12
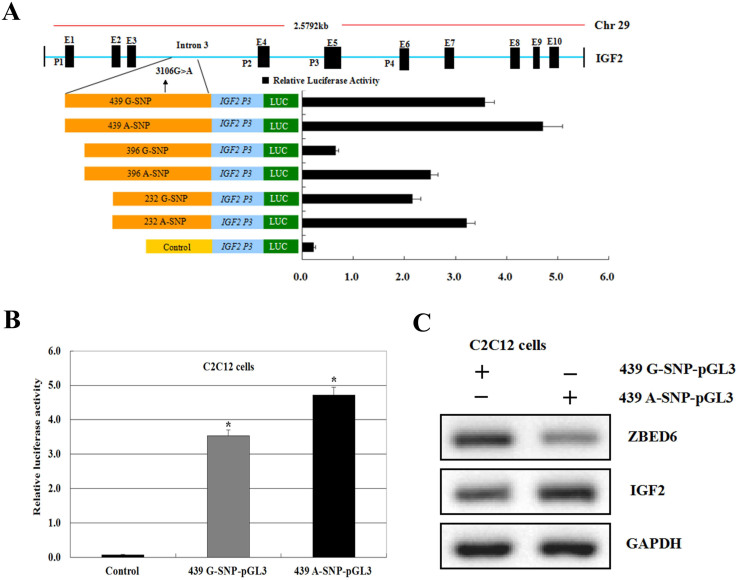
We next sought to determine the minimal region required for promoter activity within the upstream bovine IGF2 gene sequences. A series of deletion constructs from the intron 3 region of this gene were generated via PCR-based approaches (Figure 5A). These deletion constructs were transiently transfected into C2C12 cells, and luciferase activity was detected. The results showed that six deletion constructs 439G-SNP-pGL3 to 232 A-SNP-pGL3 displayed elevated promoter activity relative to control (Figure 5A and 5B). They indicated a potential transcription factor located the wild-type 439G-SNP-pGL3 and the mutant-type 439A-SNP-pGL3 because this region was identified and shown to a novel transcription factor ZBED6 act as a repressor of IGF2 transcription in skeletal muscle myogenesis and development8.
Figure 5. Recombinant vectors used to assay the sequence activity of IGF2 gene intron 3.
(A) Luciferase activity of IGF2 (Black bars) intron 3 sequence 3106G>A mutant constructs in C2C12 cells. The location and size of each fragment is indicated to the left of each bar relative to the intron 3 start codon. IGF2 promoter activity modulated by genetic polymorphisms (intron 3–3106G>A). Luciferase activity were transfected with recombinant plasmids containing the intron 3–3106 G-SNP and −3106 A-SNP and the bovine IGF2 P3 promoter in cell lines. Results from pGL3-Basic plasmid are given as a negative control. (B) Comparison of luciferase activity levels of IGF2 in C2C12 cell lines between transfected with pcDNA3.1+ (control), pcDNA3.1+-ZBED6. The blank expression vector (pcDNA3.1+ and pGL3-Basic) was used to maintain equivalent amounts of DNA. Gray solid bar show luciferase activities from cell lines transfected with the wild type haplotype IGF2 439G-SNP-pGL3 construct containing the wild-type allele. Black solid bar represent luciferase activities from cell lines transfected with the mutant haplotype 439A-SNP-pGL3 construct containing the mutant allele. Data are mean ± SEM of normalized luciferase activity. (Values represent the mean ± SEM of three duplications.) *P < 0.05. (C) Crude proteins were extracted from the treated cells and then subjected to Western blot with antibodies against ZBED6 and IGF2, GAPDH was used as a control for equal protein loading. Data are representative of at least three independent experiments performed in duplicate (n = 3 or n = 4).
Moreover, deletion construct mutant-type 439A-SNP-pGL3 had significantly elevated promoter activity relative to others (Figure 5A and 5B), the protein level of IGF2 also increased during C2C12 differentiation, as detected by western blotting (Figure 5C), leading us to conclude that the core region of the basal promoter of bovine IGF2 is located within the −3083 to −3544 sequence, which is a region in the vicinity of the intron 3 sequence 3106G>A mutant (intron 3–3072 mutation on pig IGF2).
ZBED6 overexpression inhibited IGF2 gene expression
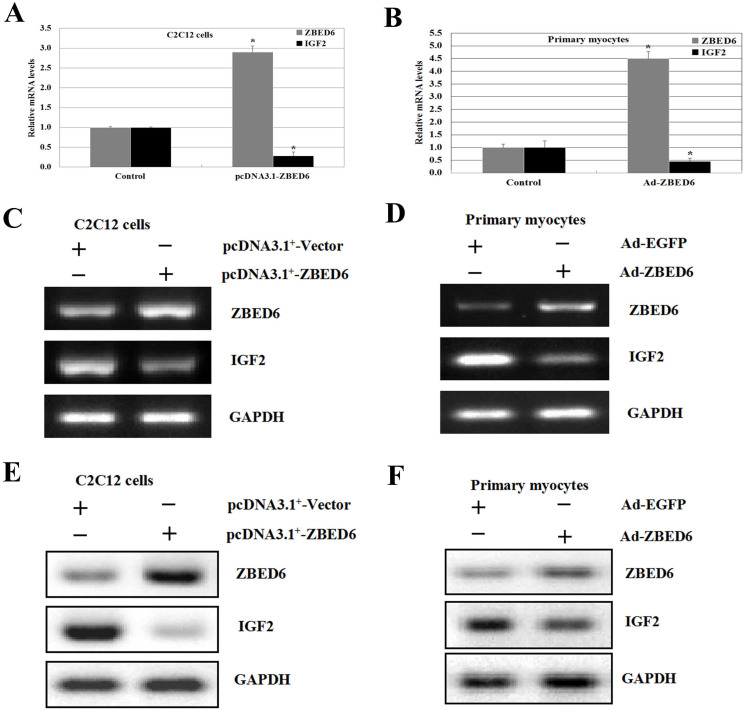
To further investigate the inhibition of IGF2 in myoblasts treated with pcDNA3.1+-ZBED6, C2C12 myoblasts were examined for ZBED6 expression after differentiation media (DMEM, 2% FBS) with or without pcDNA3.1+-ZBED6 treatment. ZBED6 mRNA and protein expression at 48 hrs after transduction, with pcDNA3.1+-ZBED6 treatment, showed a significant increase in ZBED6 expression compared with control myoblasts (Figure 6A, 6C, and 6E). By contrast, IGF2 expression was not increased in the pcDNA3.1+-ZBED6-treated C2C12 myoblasts cultured in differentiation media (Figure 6A, 6C, and 6E).
Figure 6. ZBED6 mediated IGF2 expression in C2C12 cell lines and bovine primary myocyte cells.
(A) Comparison of mRNA expression levels of bovine ZBED6 and IGF2 in C2C12 cell lines between transfected with pcDNA3.1+ (control), pcDNA3.1+-ZBED6. A blank expression vector (pcDNA3.1+) was used to maintain equivalent amounts of DNA. (B) The qPCR analysis of ZBED6 and IGF2 mRNA levels in bovine primary myocyte cells transfected with Ad-EGFP (control) and Ad-ZBED6. A blank expression vector (Ad-EGFP) was used to maintain equivalent amounts of DNA. The mRNA expression was normalized to the geometric mean of the suitable housekeeping genes (GAPDH and ACTINB) and expressed relative to gene expression in the control group. Error bars represent standard error of the mean (SEM). Each column value represents the mean ± SEM of at least three independent experiments (n = 3 or 4). *P < 0.05. (C and D) Semi-qPCR was performed to determine the endogenous expression level of ZBED6 and IGF2 using mRNA isolated from C2C12 cell lines and bovine primary myocyte cells. Data are representative of at least three independent experiments performed in duplicate (n = 3 or 4). (E and F) Crude proteins were extracted from the treated cells and then subjected to Western blot with antibodies against ZBED6 and IGF2, GAPDH was used as a control for equal protein loading. Data are representative of at least three independent experiments performed in duplicate (n = 3 or 4).
We next examined if Ad-ZBED6 treatment can down-regulate IGF2 expression when it is added to the bovine primary myoblast after Ad-ZBED6 expression. To this end, primary myoblast were cultured and Ad-ZBED6 was added when cell confluence reached approximately 60–70%. 48 hrs after transduction, ZBED6 and IGF2 expression were analyzed by qPCR, semi-qPCR, and western blot analyses. As can be seen in Figure 6B, 6D, and 6F, without Ad-ZBED6 treatment, IGF2 is upregulation. When Ad-ZBED6 was added to primary myoblast, as seen previously, IGF2 expression was not increased in bovine primary myoblast.
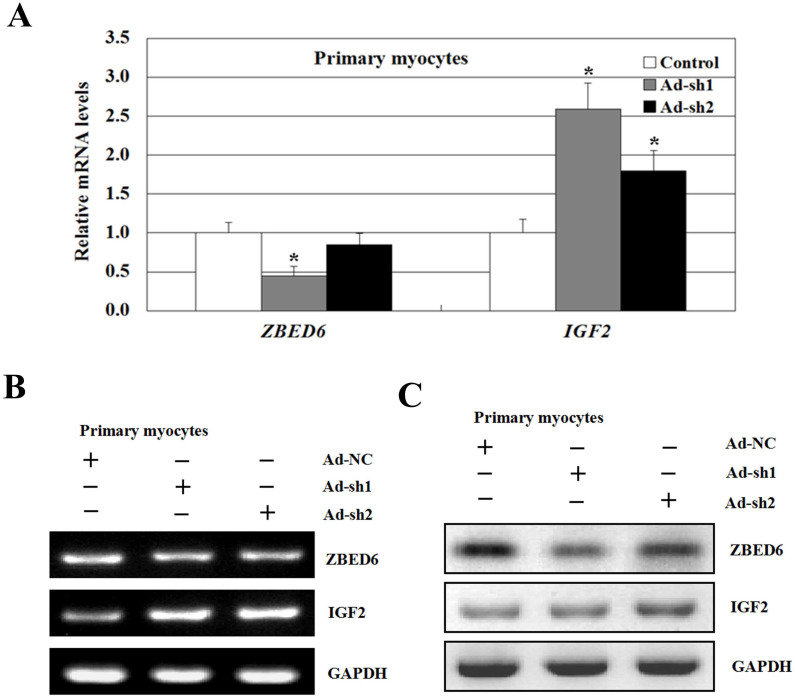
ZBED6 knockdown increases IGF2 gene expression in bovine primary myoblast
Bovine primary myoblast were transduced with the shRNA-ZBED6 or a shRNA-control for 48 hrs. RNA was then extracted, and the expression levels of ZBED6 and IGF2 were analyzed in ZBED6 knockdown primary myoblast (Figure 7A and 7B). These findings suggest ZBED6 negatively regulates IGF2 through increase expression of IGF2 gene. Western blot analysis also revealed that IGF2 protein was strongly repressed by ZBED6 (Figure 7C). These results indicated that ZBED6 may repress the expression of IGF2 through interacting with IGF2 during differentiation of primary myoblast.
Figure 7. ZBED6 and IGF2 expression change after ZBED6 knockdown in bovine primary myocyte cells.
(A) Comparison of mRNA expression levels of bovine ZBED6 and IGF2 between transfected with Ad-sh906 and Ad-sh2819 (transduced with two adenoviruses at 300 multiplicity of infection for 48 hrs) in decreasing ZBED6 expression in bovine primary myocyte cells. A blank expression vector (Ad-NC) was used to maintain equivalent amounts of DNA. The mRNA expression was normalized to the geometric mean of the suitable housekeeping genes (GAPDH and ACTINB) and expressed relative to gene expression in the control group. Error bars represent standard error of the mean (SEM). Each column value represents the mean ± SEM of at least three independent experiments (n = 3 or 4). *P < 0.05. (B) Semi-qCR was performed to determine the endogenous expression level of ZBED6 and IGF2 using mRNA isolated from bovine primary myocyte cells. Data are representative of at least three independent experiments performed in duplicate (n = 3 or 4). (C) Crude proteins were extracted from the treated cells and then subjected to Western blot with antibodies against ZBED6 and IGF2, GAPDH was used as a control for equal protein loading. Data are representative of three independent experiments performed in duplicate (n = 3). The data revealed that Ad-sh1906 had the highest knockdown of ZBED6 transcript and protein.
Discussion
Because the IGF2 mutation discovered in pigs is associated with enhanced muscle mass and reduced backfat, with the mechanism involving the release of postnatal ZBED6-mediated transcriptional repression of IGF2 in skeletal muscle7,8, we hypothesized that ZBED6 may also play a role in regulating myoblast differentiation. In this study, we evaluated the effects of DNA methylation, expression, overexpression and knockdown ZBED6 mRNA on C2C12 myoblasts and bovine primary myoblast proliferation and differentiation.
Despite the importance of ZBED6 in muscle development, little is known about its regulation. As described above, an in vitro study indicated that compared with 439-A-SNP-pGL3, 439-G-SNP-pGL3 has lower activity in driving reporter gene transcription. The bovine ZBED6 and IGF2 mRNA expression in nine different tissues was compared between full-sibling cattle (share the same sire and dam) of the fetal bovine (n = 3) and the adult bovine (n = 3). The results indicated that the bovine ZBED6 mRNA levels were upregulation in adult bovine than in fetal bovine in nine different tissues; however, the adult bovine exhibited a lower IGF2 mRNA level than in fetal bovine (Figure 2A and 2B). Specifically, in fetal stage, the IGF2 had the higher expression levels than ZBED6 in the eight tissues except fat; In adult stage, the IGF2 had the higher expression levels than ZBED6 in the five tissues except spleen, stomach, and intestine, respectively (Figure 2C and 2D). This result suggested that IGF2 acts as a repressor in the adult bovine individuals and that the ZBED6 lead to down-regulation of IGF2 expression in the skeletal muscle of cattle.
DNA methylation play an important role in the regulation of gene expression, and the proper DNA methylation is essential for normal gene function19,20. DNA methylation has been popularly investigated due to its heritable epigenetic modifications of the genome and has been implicated in the regulation of most cellular processes. It has a large impact on the regulation of gene expression and is critical in establishing patterns of gene repression during development21. ZBED6 targets can be released from repression by epigenetic activation. This is implied by the finding that EMSA using an oligonucleotide with a methylated CpG site was not bound by ZBED67. The binding of the repressor to its target site represses transcription from at least four IGF2 promoters spread over a 4-kb region. Furthermore, the repressor binds its target site only when it is unmethylated7.
In the present study, we examined the mRNA expression and DNA methylation levels of imprinted gene IGF2 in two key stages of myogenesis and muscle maturation to determine whether epigenetic modification of imprinted genes was responsible for fetal and adult growth and development. The relative gene expression levels of nine tissues between fetal bovine and adult bovines, whereas the adult bovine group exhibited a significantly lower mRNA level compared to controls (FB group). However, the adult bovine group exhibited a significantly higher DNA methylation level than the fetal bovine group. There are several other surrounding CpG that show greater differences in methylation between fetal and adult bovines. These results indicate that IGF2 expression levels were negatively associated with the methylation status of the IGF2 DMR during the two developmental stages.
To assess whether the haplotypes affect bovine IGF2 gene transcription, the 439 bp fragment spanning the bovine IGF2 intron 3 region from −3083 to −3544 (relative to intron 3) was cloned and linked to a luciferase reporter construct. Two constructs differing only at intron 3–3106 were obtained, both of which harbor the identified haplotypes, namely the wild-type haplotype 439-G-SNP-pGL3 and the mutant-type haplotype 439-A-SNP-pGL3. The promoter activity of these haplotypes was assessed by transient transfection of the individual reporter constructs into C2C12 cells. As shown in Figure 5B, the activity of 439-A-SNP-pGL3 in driving reporter gene transcription is significantly higher than that of the 439-G-SNP-pGL3 construct (P < 0.01). Of the two haplotypes identified in C2C12 cells, 439-A-SNP-pGL3 has the highest activity, and 439-G-SNP-pGL3 has the lowest activity.
By generating a series of deletion constructs of the bovine IGF2 intron 3 region, the optimal promoter was defined to be the 439 bp (−3083 to −3544) region upstream of the initiation codon. Sequence analysis of this region showed that it lacked a TATA box, which is known to recruit transcriptional machinery needed for efficient expression of a gene22. Recent bioinformatics studies have suggested that the majority of mammalian gene promoters lack a “TATA box”, have multiple TSSs and are highly GC-rich23,24,25,26. Our sequence analysis demonstrated that the IGF2 intron 3 region had a significantly high GC content (62.50%) in the one CpG islands. (Figure 3B).
Previous experiments have suggested that ZBED6 is an intronless gene located in the first intron of ZC3H11A, and the ZBED6 gene is co-transcribed with ZC3H11A by using a common promoter8. When the ZC3H11A first intron containing ZBED6 is present in the transcript, only ZBED6 protein is translated because it contains the first translated codon and a termination codon. Transcripts of the ZC3H11A gene will be translated into proteins only when the first intron is spliced out8,11. In our previous study, rapid amplification of 5′ cDNA ends (5′-RACE) analysis revealed two transcription start sites (TSS) for the bovine ZBED6 starting within exon 1 of the ZC3H11A gene (TSS-1) and upstream of the translation start codon of the ZBED6 gene (TSS-2)27. Conventionally, transcriptional initiation is believed to occur from a single focused “TSS”22. Therefore, the presence of two distinct transcriptional start sites suggests that ZBED6 transcription may be regulated by “non-traditional” mechanisms.
Despite the importance of ZBED6 in muscle development, little is known about its regulation. To identify the possible TFBS in the bovine ZBED6 promoter, the promoter sequence using web-based software showed that there were many potential factors; several binding sites for transcription factors were predicted for the region containing the myocyte enhancer factor-2 (MEF2), POU domain, class 1, transcription factor 1 (PIT1) and MYOD. The presence of all of the described transcription factor binding sites in the bovine ZBED6 promoter indicates that it is under a high level of transcriptional control by several transcription factors27. Future research should address the interaction between all of these sites to provide a better understanding of ZBED6 regulation and the myogenic process.
We carried out the deletion mutant analysis to demonstrate the effect of the transcription factors on the activity of the promoter. The results of a dual-luciferase reporter assay (Figure 5) showed that the full-length promoter fragment (439-G-SNP-pGL3) has a strong promoter activity (firefly reporter luciferase levels in relation to control renilla luciferase level), indicating that the obtained promoter region was correct. The other promoter fragments had promoter activity similar to or lower than the full-length promoter, showing that these regions contain transcription factor binding sites mediating negative regulation. The ZBED6 gene is located in the first intron of zinc finger CCCH type containing 11A (ZC3H11A), but the ZBED6 protein has no significant sequence similarity to the ZC3H11A protein. Chromatin immunoprecipitation (ChIP) followed by high-throughput DNA sequencing (ChIP-Seq) using C2C12 cells revealed approximately 2,500 ZBED6 binding genomic regions bound by ZBED6, and the deduced consensus motif gave a perfect match with the established binding site in IGF2, and approximately 1,200 genes had at least one putative ZBED6 binding site occurring within 5 kb of the TSS in C2C12 cells. The gene ontology analysis results suggest that ZBED6 is an important regulator of development, cell proliferation and growth8.
To further confirm the effect of ZBED6 overexpression on the regulation of IGF2 expression, we used semi-qPCR, qPCR and western blot to detect the amount of ZBED6 and IGF2 mRNA and protein expression in the C2C12 cells and bovine primary myoblast from overexpressed pcDNA3.1+-ZBED6 and Ad-ZBED6. As shown in Figure 6, the expression of IGF2 mRNA and protein expression were significantly down-regulated by ZBED6 overexpression cells compared with the vector control.
The results suggest that overexpression of ZBED6 significantly inhibited the expression level of the IGF2 gene, the IGF2 mRNA expression level was significantly downregulated in C2C12 cells and bovine primary myoblast. This result is similar to the increased IGF2 expression in skeletal muscle of pigs carrying the mutation at the ZBED6 target site in IGF2 gene7. ZBED6 acts as a repressor of IGF2 transcription in skeletal muscle myogenesis and development8. These studies suggested that ZBED6 was a major regulator of IGF2 expression in skeletal muscle but a repressor of the constitutive expression of IGF2.
In summary, ZBED6 and IGF2 expression were upregulated from day 0 to day 6 at the mRNA level in differentiating C2C12 cells. The overexpression and knockdown of ZBED6 led to IGF2 expression change in C2C12 cells in vitro. We have identified a biological function for ZBED6 in C2C12 and bovine primary myoblast and found that ZBED6 play a regulatory role in IGF2 gene transcription by binding in the core promoter region between 3083 bp and 3544 bp of the bovine IGF2 intron 3. Together, our results suggest that ZBED6 directly represses transcription of IGF2 during myogenesis. These results give new insight into the biological function and regulatory mechanism of the ZBED6 gene.
Methods
Ethics statement
All experiments were performed by the Regulations for the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, 2004) and approved by the Institutional Animal Care and Use Committee (College of Animal Science and Technology, Northwest A&F University, China). Fetal and adult Chinese Qinchuan beef cattle were obtained from Shannxi Kingbull Animal Husbandry Co., Ltd (Baoji, China). Adult animals were allowed access to feed and water ad libitum under normal conditions and humanely sacrificed as necessary to ameliorate suffering.
DNA sequencing
To explore the genetic variation of bovine IGF2 gene, we sequenced four pooled DNA samples (Pool-Seq, total = 200) from four cattle breeds in China: Nanyang cattle (NY), Qinchuan cattle (QC), Jiaxian cattle (JX) and Chinese Holstein (CH). The pooled DNA samples consisted of 50 DNA samples selected randomly from each cattle breed. NY, QC and JX are three important breeds for beef production in China, whereas CH is a dairy breed. These four breeds are the main breeds of China, and they are reared in the provinces of Henan and Shaanxi. Calves were weaned at six months of age on average and raised from weaning to slaughter on a diet of corn and corn silage. The animals of each breed were selected to be unrelated for at least three generations with the aim of having diverse lineages within each breed.
Genomic DNA was isolated from 2% heparin-treated blood samples and stored at −80°C following standard procedures12. The content of DNA was estimated spectrophotometrically, and the genomic DNA was then diluted to 50–100 ng/μL. All DNA samples were stored at −20°C for subsequent analysis.
Primers used to amplify the bovine IGF2 gene (NCBI: EU518675.1, AC_000186.1) were designed from a published gene sequence. Primers, fragment sizes and annealing temperature (AT) are provided in Table S1.
Tissue and RNA isolation
Samples of nine tissues and organs (longissimus dorsi muscle, heart, kidney, liver, lung, spleen, stomach, intestine, and fat tissue) from nine individuals (three individuals per stage) were harvested for RNA isolated within 10 min after slaughter at three key stages of myogenesis and muscle maturation: 90 days at embryo (fetal bovine, FB), and 24-month-old (adult bovine, AB).
An analysis of gene expression patterns in six individuals from full-sibling Chinese Qinchuan cattle (i.e., in which all six animals have the same sire and dam) was conducted. The LDM sample was collected from castrated full-sibling individuals (three fetal bovine animals and three adult bovine animals) raised by Shannxi Kingbull Animal Husbandry Co., Ltd. (Baoji City, China). The specimens used in this study were collected immediately after slaughter and used for gene expression analysis. Muscle specimens for gene expression analysis were sampled and snap frozen in liquid nitrogen. Total RNA was isolated from 80–100 mg of tissue samples from QC, and RNA from samples was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. All samples were subjected to reverse transcription using a cDNA high capacity kit (Invitrogen, Carlsbad, CA, USA).
C2C12 and bovine primary myoblast culture and differentiation
Mouse myoblast cell line C2C12 was purchased from American Type Culture Collection. C2C12 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin (final concentration) and 100 μg/mL streptomycin. Cells were cultured in a 5% CO2 humidified incubator at 37°C.
The C2C12 cells were seeded in six-well plates. After approximately 12–16 h, when cell confluence reached approximately 60%–70%, the differentiation of C2C12 myoblasts into myotubes was induced by the addition of differentiation medium (DMEM containing 2% horse serum instead of 10% FBS). Starting at the beginning of differentiation, the C2C12 cells were cultured in six-well plates and harvested for RNA extraction at 0, 1, 2, 3, 4, 5 and 6 days.
Bovine myoblast cultures were derived and enriched from Musculus semitendinosus of 120-day fetuses resulting in cultures of at least 90% myoblast purity. Purified bovine primary myoblast cultures were grown prior to assay in minimal essential medium (MEM; Invitrogen) containing 10% FBS for 24 h before trypsinization and seeding onto coverslips.
Total RNA samples were extracted using TRIZOL (Invitrogen, USA) according to the manufacturer's instructions. The cDNA samples were obtained by reverse transcription from 1 mg RNA using oligo (dT) and reverse transcriptase. All samples were subjected to were subjected to reverse transcription using a cDNA high capacity kit (Invitrogen, Carlsbad, CA, USA).
Semi-quantitative reverse transcription-polymerase chain peaction (semi - qPCR) and quantitative real - time PCR (qPCR)
The semi - qPCR (Reverse Transcription-Polymerase Chain Reaction) was performed in a volume of 10 mL containing 1 μL of 10× PCR buffer (plus Mg 2+), 100 μmol/L of each dNTP, 0.5 μmol/L of each PCR primer, 1.0 U Taq DNA polymerase (Takara, Japan) and 0.5 μL of cDNA template. The primers were designed and synthesized as shown in Table S1. The PCR reaction was 5 min at 95°C followed by 27 cycles of 30 s at 95°C, 30 s at the specific melting temperatures of each primer pair, 30 s at 72°C and a final extension time of 5 min at 72°C.
The qPCR analysis using SYBR Green PCR Master Mix (Takara, Dalian, China) was performed on a CFX96™ Real-Time PCR Detection System (Bio-Rad, USA). Data were normalized to the geometric mean of data from GAPDH and ACTINB, which were used as endogenous control genes. All primers for validation were designed to cross exon-exon junctions and are shown in Supplementary Table S1. Relative expression levels of objective mRNAs were calculated using the ΔΔCt method13.
Bisulphite sequencing polymerase chain reaction (BSP)
Genomic DNA was extracted following the standard procedures using TIANamp Genomic DNA Kit (Tiangen, Beijing, China). DNA treatment with sodium bisulphite was performed using the EZ DNA Methylation Kit (Zymo Research, USA) according to the manufacturer's protocol, except that the conversion temperature was changed to 55°C. The modified DNA samples were diluted in 10 μL of distilled water and should be immediately used in BSP or stored at −80°C until PCR amplification. Three separate bisulphite modification treatments were performed for each DNA sample.
The BSP primers were designed by the online MethPrimer software14. The sequences of the PCR primers used for amplifying the targeted products are shown in Figure 4A. The details of BSP amplified nucleotide sequences of IGF2 DMR are shown in Figure 3 and 4. We used hot start DNA polymerase (Zymo Taq ™ Premix, Zymo Research, USA) for BSP. PCR was performed in 50 μL of reaction volume, containing 200 ng/50 μL genomic DNA, 0.3 to 1 μM of each primer, Zymo Taq ™ Premix 25 μL. The PCR was performed with a DNA Engine Thermal Cycler (Bio-Rad, USA) using the following program: 10 min at 95°C, followed by 45 cycles of denaturation for 30 s at 94°C, annealing for 40 s at 50.4°C and extension for 30 s at 72°C, with a final extension at 72°C for 7 min. The PCR products were gel purified using Gel Purification Kit (Sangon, Shanghai, China). The purified fragments were subcloned into the pGEM T-easy vector (Promega, Madison, WI, USA). Different positive clones for each subject were randomly selected for sequencing (Sangon, Shanghai, China). Three independent amplification experiments were performed for IGF2 gene in each sample. We sequenced 6 clones from each independent set of amplification and cloning; hence, there were 18 clones for the IGF2 DMR in each sample. The final sequence results were processed by online QUMA software15.
Adenovirus expression vector
The adenovirus expression vector of ZBED6 (Ad-ZBED6) was constructed to study its function in C2C12 cells and bovine primary myoblast. The full-length DNA sequence of the ZBED6 gene was amplified by polymerase chain reaction (PCR) as a template, while the Kpn I and Xba I cleavage points were introduced and then ligated with adenovirus shuttle plasmid pAdTrack-CMV by T4 DNA ligase after double digestion of Kpn I and Xba I, which generates the complementary sticky ends with pAdTrack-CMV. The recombinant plasmid was transformed into E. Coli DH5a and extracted for the double digestion test and following studies. After PmeI digestion, the linearized plasmid was in homologous recombination with the skeleton plasmid pAdEasy-1 in BJ5183 (pAdEasy-1 present in Escherichia coli BJ5183), while the positive clones were selected for culture, plasmid extraction, and PacI digestion. After the recombinant adenoviral plasmid was digested by Pac I, the larger fragment was recovered using a gel extraction kit and transfected into HEK293A cells by a liposome. After 8 or 10 days, the recombinant adenovirus was collected by repeated freezing and thawing of the HEK293 cells and infected HEK293 cells; 2 or 3 days later, the cells showed the cytopathic effect (CPE) phenomenon and the virus was collected.
Adenovirus-delivered RNAi
Expression of shRNA to knock down a gene has become a valuable alternative approach to study gene function. It has the advantage of being a much faster and cost-effective method than generating conditional knock-out mice16. All short hairpin (sh)RNAs used in this study were constructed in pENTR/CMV-GFP/U6. The shRNA sequences were designed using the WI siRNA Selection Program (http://sirna.wi.mit.edu/home.php) and BLOCK-iT RNAi Designer (http://rnaidesigner.invitrogen.com/rnaiexpress/) using the bovine ZBED6 gene sequence (NCBI: AC_000173.1). We selected the highest ranked shRNA sequences. Meanwhile, those sequences were selected and synthetized at acommercial facility (Invitrogen,Shanghai,China) with BamH I and Xho I restriction sites suitable for the cloning process (see Table S3). Lastly, four shRNA were generated by heat treatment annealing and constructed into pENTR/CMV-GFP/U6-shRNA. The CDS of ZBED6 was subcloned into the pDsRed1-N1 plasmid vector between the EcoR I and Kpn I restriction sites to generate pDsRed1-N1-ZBED6.
shRNA expression cassettes with an EGFP reporter gene in the pENTR vector were switched into an adenoviral vector (pAd/PL-DEST) using the Gateway technique (Invitrogen, USA) to generate pAd-shRNA vectors. Pac I linearized adenoviral plasmids were 8 to 10 days after transfection, the recombinant virus was collected and subjected to two rounds of amplification in 293A cells. The viral titers were determined in transduced 293A cells through GFP expression as previously described17,18.
Bovine primary myocyte cells at 70–80% confluence were transduced with adenovirus supernatant at a multiplicity of infection (MOI) of 300. The medium was replaced with fresh medium 6 h later. The shRNA negative control adenovirus (Ad-NC) was used as a control. Cells were harvested 48 hrs after transduction.
Construction of promoter haplotypes of IGF2 intron 3
The genomic DNAs were extracted from the bovine muscle tissue using a phenol extraction method12. Six different primers (listed in Table S2) designed according to the sequence of the bovine IGF2 gene were used in the PCR with bovine genomic DNA as a template to obtain the complete bovine IGF2 promoter (439G-SNP-pGL3) and the truncated promoter fragments of IGF2 intron 3.
The constructs contained 439 bp, 396 bp, and 232 bp from IGF2 intron 3 (nucleotides 3083–3108), followed by the IGF2 P3 promoter (nucleotides −427 to +327 relative to the exon 6)6,7,8,9 and a luciferase reporter. A DNA fragment that covers the polymorphic sites (intron 3–3106 G>A) of the intron 3 region of the bovine ZBED6 gene was amplified by PCR with PrimeSTAR HS DNA polymerase (2.5 U/μL) (Takara, Dalian, China) and ligated into the pGL3-basic vector (Promega, Madison, WI, USA). The primer used for constructing vectors contain the Kpn I (forward primer) and Bgl II (reverse primer) restriction sites, is indicated by the lowercase letters in Table S2. After being digested by restriction enzymes Kpn I and Bgl II, PCR products were separately inserted into the pGL3-base vector (Promega, Madison, WI, USA), which contains the luciferase reporter gene, according to the manufacturer's instructions.
Construction of the pcDNA3.1+-ZBED6 expression vector
The primer pairs (Table S1) designed according to the GenBank sequence database were used in PCR to obtain the CDS sequence of the bovine ZBED6 gene. The restriction enzymes Kpn I (forward primer) and Xba I (reverse primer) were used to digest the amplified bovine ZBED6 CDS and pcDNA3.1+ vector (Invitrogen, Carlsbad, CA, USA). T4 DNA ligase (Takara, Dalian, China) was used to link the products of digestion. Standard molecular cloning techniques12 were then used to obtain the pcDNA3.1+- ZBED6 vectors.
Plasmid transfection and dual luciferase reporter assay
When cells reached 90% confluence, the proliferation medium was removed, and the cells were rinsed with phosphate-buffered saline and treated for 2 min with 0.25% trypsin to detach cells. Cells were collected, centrifuged and diluted in a proliferation medium prepared with DMEM without penicillin/streptomycin (pen/strep) and split onto two gelatin-coated 24-well plates at a density of ~105 cells per well. The following day, cells were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Briefly, for each well, 2.0 μl of Lipofectamine 2000 and 0.8 μg of DNA were mixed in 100 μl of FBS-free and pen/strep-free Opti-MEM I medium (Promega, Madison, WI, USA) for 20 min. To normalize the transfection efficiency, the pRL-TK plasmid vector (Promega, Madison, WI, USA), which carries a renilla luciferase gene, was co-transfected with the reporter construct as described above. The experiments were performed in three replicates for each construct.
The relative activities of the full-length promoter and the truncated promoter fragments of the bovine IGF2 gene were analyzed using the Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA) in accordance with the manufacturer's protocol. Cells were harvested 24 h post-transfection, and firefly and renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) and a BHP9504 Fluorescent Analytic Instrument (Hamamatsu, Japan). The firefly luciferase activities were normalized by the renilla luciferase activities in each well. The data given in the results section are the average of three replicates. Data are expressed as the mean ± standard error from two independent experiments.
Western blotting
Adenovirus-treated or untreated C2C12 cells were extracted using lysis buffer (10 mM Tris-HCl, pH 7.5, 1% Triton X-100, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 10 g/ml aprotinin, and 10 g/mL leupeptin) on ice. Protein samples (20 g) were separated on 10% SDS-PAGE and electroblotted to a nitrocellulose membrane. After being blocked with 5% fat-free milk in TTBS (20 mM Tris-HCl, pH 7.6, 150 mM NaCl, 0.1% Tween 20) for 1 h, the membranes were incubated with primary antibodies overnight at 4 uC. After washing three times with Tris-buffered saline/0.1% Tween 20 (TBST) at room temperature, the membranes were hybridized with secondary antibody for 1 h at room temperature, and again washed three times with TBST. The targeted proteins were detected using the Gel Doc XR System as per the instructions of the manufacturer.
Author Contributions
Conceived and designed the experiments: Y.Z.H. and H.C. Performed the experiments: Y.Z.H., L.Z.Z., X.S.L., M.X.L. and Y.J.S. Analyzed the data: Y.Z.H. Contributed reagents/materials/analysis tools: H.C., X.Y.L., C.Z.L. and C.L.Z. Wrote the paper: Y.Z.H. Provided editorial suggestions and revisions: H.C., X.Z. And C.J.L. All authors have read and approved the final manuscript.
Supplementary Material
Dataset 1
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant No. 31272408 and 30972080), Agricultural Science and Technology Innovation Projects of Shaanxi Province (Grant No. 2012NKC01-13), Program of National Beef Cattle and Yak Industrial Technology System (Grant No. CARS-38) and National 863 Program of China (Grant No. 2013AA102505).
References
- Andersson L. Genetic dissection of phenotypic diversity in farm animals. Nat Rev Genet. 2, 130–138 (2001). [DOI] [PubMed] [Google Scholar]
- Jones J. I. & Clemmons D. R. (1995) Insulin - like growth factors and their binding proteins: biological actions. Endocr Rev. 16, 3–34. [DOI] [PubMed] [Google Scholar]
- Nakae J., Kido Y. & Accili D. Distinct and overlapping functions of insulin and IGF - I receptors. Endocr Rev. 22, 818–835 (2001). [DOI] [PubMed] [Google Scholar]
- Liu J. P., Baker J., Perkins A. S., Robertson E. J. & Efstratiadis A. Mice carrying null mutations of the genes encoding insulin- like growth factor I (Igf - 1) and type 1 IGF receptor (Igf1r). Cell 75, 59–72 (1993). [PubMed] [Google Scholar]
- Jeon J. T. et al. A paternally expressed QTL affecting skeletal and cardiac muscle mass in pig s maps to the IGF2 locus. Nat Genet. 21, 157–158 (1999). [DOI] [PubMed] [Google Scholar]
- Nezer C. et al. An imprinted QTL with major effect on muscle mass and fat deposition maps to the IGF2 locus in pigs. Nat Genet. 21, 155–156 (1999). [DOI] [PubMed] [Google Scholar]
- Van Laere A. S. et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 425, 832–826 (2003). [DOI] [PubMed] [Google Scholar]
- Markljung E. et al. ZBED6, a Novel Transcription Factor Derived from a Domesticated DNA Transposon Regulates IGF2 Expression and Muscle Growth. PLOS Biology 7, e1000256. 10.1371/journal.pbio10 00256 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A., Ghazal A., Andersson G., Andersson L. & Jern P. ZBED evolution: repeated utilization of DNA transposons as regulators of diverse host functions. PLoS One 8, e59940 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L. Studying phenotypic evolution in domestic animals: a walk in the footsteps of Charles Darwin. Cold Spring Harb Symp Quant Biol. 74, 319–325 (2009). [DOI] [PubMed] [Google Scholar]
- Andersson L. et al. ZBED6: The birth of a new transcription factor in the common ancestor of placental mammals. Transcription 1, 144–148 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J. & Russell D. W. (Translated by Huang PT) (2002). Molecular Cloning A Laboratory Manual. Science, Beijing.
- Livak K. J. & Schmitteg T. D. Analysis of relative gene expression data using Quantitative Real-Time PCR and the 2[-delta delta c(t)) method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Li L. C. & Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 18, 1427–1431 (2002). [DOI] [PubMed] [Google Scholar]
- Kumaki Y., Oda M. & Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 36, W170–175 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witting S. R., Brown M., Saxena R., Nabinger S. & Morral N. Helper-dependent adenovirus - mediated short hairpin RNA expression in the liver activates the interferon response. J Biol Chem. 283, 2120–2128 (2008). [DOI] [PubMed] [Google Scholar]
- Luo J. et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nature Protocols. 2, 1236–1247 (2007). [DOI] [PubMed] [Google Scholar]
- Ostapchuk P. & Hearing P. Control of adenovirus packaging. J Biol Chem. 96, 25–35 (2005). [DOI] [PubMed] [Google Scholar]
- Morgan H. D., Santos F., Green K., Dean W. & Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 14, R47–58 (2005). [DOI] [PubMed] [Google Scholar]
- Jones P. A. & Takai D. The role of DNA methylation in mammalian epigenetics. Science 293, 1068–1070 (2001). [DOI] [PubMed] [Google Scholar]
- Cedar H. & Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 10, 295–304 (2009). [DOI] [PubMed] [Google Scholar]
- Bjornsdottir G. & Myers L. C. Minimal components of the RNA polymerase II transcription apparatus determine the consensus TATA box. Nucleic Acids Res. 36, 2906–2916 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A. et al. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet 8, 424–436 (2007). [DOI] [PubMed] [Google Scholar]
- Bajic V. B. et al. Mice and men: their promoter properties. PLoS Genet 2, e54 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald P. C., Shlyakhtenko A., Mir A. A. & Vinson C. Clustering of DNA sequences in human promoters. Genome Res 14, 1562–1574 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anish R., Hossain M. B., Jacobson R. H. & Takada S. Characterization of transcription from TATA-less promoters: identification of a new core promoter element XCPE2 and analysis of factor requirements. PLoS ONE 4, e5103 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. Z. et al. 5′- Regulatory Region and Two Coding Region Polymorphisms Modulate Promoter Activity and Gene Expression of the Growth Suppressor Gene ZBED6 in Cattle. PLoS One 8, e79744 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotwein P. & Hall L. J. Evolution of insulin-like growth factor II: characterization of the mouse IGF-II gene and identification of two pseudo-exons. DNA Cell Biol. 9, 725–735 (1990). [DOI] [PubMed] [Google Scholar]
- Tran V. G. et al. H19 antisense RNA can up-regulate Igf2 transcription by activation of a novel promoter in mouse myoblasts. PLoS One 7, e37923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pagter-Holthuizen P. et al. Differential expression of the human insulin-like growth factor II gene: characterization of the IGF-II mRNAs and an mRNA encoding a putative IGF-II-associated protein. Biochim. Biophys. Acta 950, 282–295 (1988). [DOI] [PubMed] [Google Scholar]
- Amarger V. et al. Comparative sequence analysis of the INS-IGF2-H19 gene cluster in pigs. Mamm. Genome 13, 388–398 (2002). [DOI] [PubMed] [Google Scholar]
- Van Laere A. S. et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 425, 832–836 (2003). [DOI] [PubMed] [Google Scholar]
- Brown W. M., Dziegielewska K. M., Foreman R. C. & Saunders N. R. The nucleotide and deduced amino acid sequences of insulin-like growth factor II cDNAs from adult bovine and fetal sheep liver. Nucleic Acids Res. 18, 4614 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall J. J. & Schmutz S. M. Linkage mapping of IGF2 on cattle chromosome 29. Anim. Genet. 34, 313 (2003). [DOI] [PubMed] [Google Scholar]
- Gebert C. et al. DNA methylation in the IGF2 intragenic DMR is re-established in a sex-specific manner in bovine blastocysts after somatic cloning. Genomics 94, 63–69 (2009). [DOI] [PubMed] [Google Scholar]
- Holm P., Booth P. J., Schmidt M. H., Greve T. & Callesen H. High bovine blastocysts development in a static in vitro production system using SOFaa medium supplemented with sodium citrate and myo-inositol with or without serumproteins. Theriogenology 52, 683–700 (1999). [DOI] [PubMed] [Google Scholar]
- Han D. W. et al. Methylation status of putative differentially methylated regions of porcine IGF2 and H19. Mol. Reprod. Dev. 75, 777–784 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset 1