Abstract
Background
Public Health England (PHE) holds a collection of Shigella flexneri Type strains isolated between 1949 and 1972 representing 15 established serotypes and one provisional type, E1037. In this study, the genomes of all 16 PHE Type strains were sequenced using the Illumina HiSeq platform. The relationship between core genome phylogeny and serotype was examined.
Results
The most common target gene for the detection of Shigella species in clinical PCR assays, ipaH, was detected in all genomes. The type-specific target genes were correctly identified in each genome sequence. In contrast to the S. flexneri in serotype 5 strain described by Sun et al. (2012), the two PHE serotype 5 Type strains possessed an additional oac gene and were differentiated by the presence (serotype 5b) or absence (serotype 5a) of gtrX. The somatic antigen structure and phylogenetic relationship were broadly congruent for strains expressing serotype specific antigens III, IV and V, but not for those expressing I and II. The whole genome phylogenies of the 15 isolates sequenced showed that the serotype 6 Type Strain was phylogenetically distinct from the other S. flexneri serotypes sequenced. The provisional serotype E1037 fell within the serotype 4 clade, being most closely related to the Serotype 4a Type Strain.
Conclusions
The S. flexneri genome sequences were used to evaluate phylogenetic relationships between Type strains and validate genotypic and phenotypic assays. The analysis confirmed that the PHE S. flexneri Type strains are phenotypically and genotypically distinct. Novel variants will continue to be added to this archive.
Keywords: Shigella flexneri type strains, Next generation sequencing technology, Molecular serotyping
Background
Shigella flexneri is the predominant cause of shigellosis in the developing world [1], making appropriate subtyping tools for tracking S. flexneri epidemiology vital to global public health. The S. flexneri serotyping scheme differentiates isolates serologically based on the expression of the major type specific somatic antigen (I-VI) and common group factor antigens (3,4 designated Y and 7,8 designated X) [2]. The common group factor antigens account for the complex intra-serotype relationships. Currently, there are 15 established serotypes. Traditional S. flexneri serotyping is performed by slide agglutination using antiserum raised in rabbits against type specific and group factor antigens. Recently, Sun et al.[3] published a multiplex PCR approach for molecular serotyping of S. flexneri. This method differentiates the 15 accepted serotypes based on known differences in (i) their gtr genes encoding the type specific antigens I, II, IV, and V, group factor antigen 7,8 (X) and 1c (gtrI, gtrII, gtrIV, gtrV, gtrX, and gtrIC) (ii) the oac gene that mediates O-acetylation modification in serotypes 1b, 3a, 3b, and 4b and (iii) the wzx 6 for detection of serotype 6.
Public Health England (PHE) holds an historic collection of 16 S. flexneri Type strains isolated between 1949 and 1972. Strains belonging to this set have been used to produce standardised antiserum for the phenotypic serotyping scheme at PHE for over 60 years. To increase the utility of this collection, we report the draft whole genome sequences of the 16 PHE S. flexneri Type strains in order to facilitate a greater understanding of how whole genome phylogenies compare to typing data generated from diagnostic and molecular serotyping targets.
Methods
Bacterial strains
The 16 strains of S. flexneri analysed in this study are shown in Table 1. Strains used in this study were serotyped by slide agglutination using both commercially available monovalent antisera (Denka Seiken, Japan) and monoclonal antibody reagents (Reagensia AB, Sweden) and in-house antisera raised in rabbits [4] to all type specific somatic antigens and the group factor antigens. All strains were tested using the PCR serotyping assay described by Sun et al. [3].
Table 1.
Comparison of the phenotypic and genotypic serotyping
|
Phenotypic results |
PCR results |
Genome sequencing results |
||||
|---|---|---|---|---|---|---|
| Agglutination reactions | Phenotypic serotype [[2]] | Genes detected using PCR scheme [[3]]. | PCR serotype [[3]] | Serotype derived from genome sequence according to PCR scheme [[3]] | Number of SNPs different in the core regions compared with reference strain | Proportion of reference mapped to |
| Type I |
1a |
gtrI |
1a |
1a |
2549 |
0.88 |
| Type I + Group 6 |
1b |
gtrI + oac |
1b |
1b |
2552 |
0.87 |
| MASF 1c |
1c |
gtrI + gtrC |
1c |
1c |
2661 |
0.88 |
| Type II + Y |
2a |
gtrII |
2a |
2a |
731 |
0.91 |
| Type II + X |
2b |
gtrII + gtrX |
2b |
2b |
2650 |
0.89 |
| Type III + X |
3a |
gtrIII + gtrX |
3a |
3a |
6947 |
0.86 |
| Type III + Y |
3b |
gtrIII |
3b |
3b |
6976 |
0.85 |
| Type III + Y |
3c |
gtrIII |
3c Not included in the PCR scheme |
Not included in the PCR scheme |
7030 |
0.85 |
| Type IV + Y |
4a |
gtrIV |
4a |
4a |
2595 |
0.87 |
| Type IV + Group 6 |
4b |
gtrIV + oac |
4b |
4b |
2528 |
0.87 |
| Type V + Y |
5a |
gtrV + oac |
5a Not included in the PCR scheme |
Not included in the PCR scheme |
4388 |
0.89 |
| Type V + X |
5b |
gtrV + oac + gtrX |
5b Not included in the PCR scheme |
Not included in the PCR scheme |
4294 |
0.88 |
| Type VI |
6 |
wzx6 |
6 |
6 |
47787 |
0.74 |
| MASF IV-1 E1037 |
E1037 |
gtrIV + lpt-O |
MASF IV-1 E1037 |
4a with Ipt-O (opt) gene |
3333 |
0.88 |
| X |
X |
gtrX |
X |
X |
6937 |
0.84 |
| Y | Y | No amplification | Y | Y | 752 | 0.92 |
Genome sequencing and analysis
Genomic DNA was isolated from an overnight culture using the Wizard kit (Promega, Madison, Wisconsin, USA) and was sequenced at the Wellcome Trust Sanger Institute (WTSI) and PHE. Paired end libraries where each pair was 100 bp in length were generated on the Illumina Hiseq 2500 instrument (San Diego, California, USA). Resulting FASTQ reads were processed using Trimmomatic v0.27 [5] to remove bases with a PHRED score of less than 30 and read length less than 50 bp after quality trimming. High quality reads were then mapped to the reference strain, S. flexneri serotype 2a strain 2457 T (AE014073.1) [6], using BWA v0.6.2 and Single Nucleotide Polymorphisms called using GATK v2.5.2 in Unified Genotyper mode [7]. Positions in the reference genome where GATK mapping quality was below 30 and genotyping quality was below 50 in any strain were excluded from further analysis. Single Nucleotide Polymorphisms (SNPs) were defined as the sub-set of high quality positions (MQ > 30, GQ > 50) where the base identified varied from the reference position. De novo assembly was performed using Velvet v1.2.3 [8] with K-mer selected using VelvetK (Table 2) (http://www.vicbioinformatics.com/software.velvetk.shtml). A maximum likelihood phylogenetic tree was drawn using MEGA v5.1 with 500 bootstraps based on an alignment of 10632 SNPs called against the S. flexneri serotype 2a strain 2457 T reference genome.
Table 2.
Genome statistics for the S. flexneri genomes sequenced in this study
| Serotype | Number of high quality mapped reads | Average coverage of S. flexneri 2457 T reference | Kmer used in Velvet assembly | N50 | Number of contigs | De novo assembly genome size |
|---|---|---|---|---|---|---|
| 1a |
5593126 |
121.5896957 |
79 |
31660 |
526 |
4369471 |
| 1b |
5332110 |
115.9154348 |
79 |
31344 |
387 |
4176066 |
| 1c |
5593956 |
121.6077391 |
81 |
30176 |
425 |
4255254 |
| 2a |
5541229 |
120.4615 |
79 |
33398 |
417 |
4262526 |
| 2b |
4869600 |
105.8608696 |
77 |
32299 |
421 |
4328906 |
| 3a |
4590499 |
99.79345652 |
77 |
33146 |
510 |
4601423 |
| 3b |
4803610 |
104.4263043 |
79 |
33159 |
450 |
4494495 |
| 3c |
5604902 |
121.8456957 |
81 |
33478 |
451 |
4495552 |
| 4a |
5328467 |
115.8362391 |
81 |
33309 |
413 |
4267213 |
| 4b |
5106835 |
111.0181522 |
79 |
34439 |
391 |
4188134 |
| 5a |
5461876 |
118.7364348 |
79 |
29622 |
443 |
4362146 |
| 5b |
5250432 |
114.1398261 |
79 |
29416 |
459 |
4354058 |
| 6 |
6350211 |
138.0480652 |
85 |
20677 |
614 |
4401927 |
| E1037 |
15797791 |
343.4302391 |
93 |
36513 |
378 |
4350496 |
| X |
17982247 |
390.918413 |
93 |
33586 |
414 |
4324701 |
| Y | 20943063 | 455.2839783 | 95 | 29717 | 447 | 4491024 |
Genomic data deposition
Wellcome Trust Sanger Institute sequence data is available in the Short Read Archive under the following accession numbers (serotype): ERS088060 (1a); ERS088061 (1b); ERS088062 (1c); ERS088063 (2a); ERS088064 (2b); ERS088065 (3a); ERS088066 (3b); ERS088067 (3c); ERS088068 (4a); ERS088069 (4b); ERS088071 (5a); ERS088072 (5b); ERS088073 (6); ERS088074 (X); ERS088075 (Y); ERS088076 (E1037).
Findings
Mapping of the sequencing reads to the 4.6 Mbp S. flexneri serotype 2a strain 2457 T reference genome resulted in 99–455 times coverage, with between 731 and 47787 SNPs compared to the reference genome (Table 1). De novo assembly resulted in an average N50 of 31621 with an average of 447 contigs (Table 2).
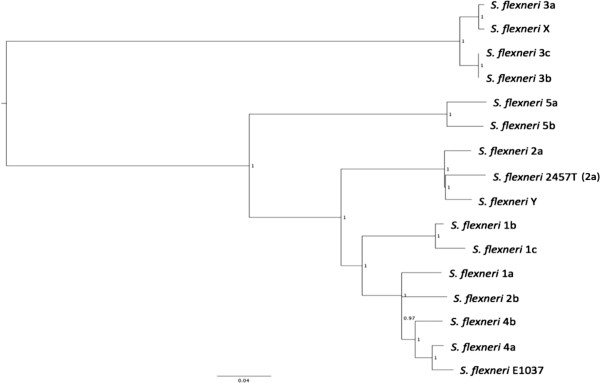
The phylogenetic relationships of the S. flexneri Type strains showed the somatic antigen structure and phylogenetic relationships were broadly congruent for strains expressing type specific antigens III, IV and V, but not I and II (Figure 1). In addition, serotype 3a was more closely related to the serotype X isolate than isolates expressing serotypes 3b and 3c. Serotype 3c was phylogenetically closely related to serotype 3b but differed phenotypically as it failed to agglutinate with the 3,4 (y) group factor antigen. Serotype 3c is not longer included in the current serotyping scheme [3] as it is very rarely identified (nine isolates submitted to GBRU since 2004).
Figure 1.
Midpoint-rooted phylogenetic tree of S. flexneri type strains based on 10631 variant positions in the core genome, node labels are bootstrap values based on 500 bootstraps.S. flexneri serotype 6 is very distantly related to the S. flexneri strains described in this study (Table 1) and is therefore excluded from the tree.
It has long been reported that the somatic O antigen of S. flexneri serotype 6 differs considerably from that of the other S. flexneri serotypes and that strains of S. flexneri serotype 6 resemble strains of S. boydii immunochemically [9]. Consistent with previous studies and phenotypic information, serotype 6 formed an out group from the other S. flexneri serotypes sequences (data not shown) [10] being more closely related to Shigella boydii CDC 3083–94 (GenBank: CP001063.1); differing by 47 787 SNPs from S. flexneri 2a (Table 1) and approximately 7300 SNPs from S. boydii CDC 3083–94 (data not shown).
In 1972, colleagues in our laboratory reported a provisional new serotype, designated E1037, frequently submitted to PHE between 2004 and 2013 (276 isolates submitted to GBRU since 2004). Phylogenetically, E1037 is closely related to Serotype 4a (Figure 1). Other groups have supported the extension of the accepted classification scheme to include this novel type [11,12].
The presence of key diagnostic and molecular serotyping genes was also determined. We confirmed the presence of the ipaH gene (the target gene for the detection of Shigella species in diagnostic PCR assays) in all the PHE Type strains. It was not possible to de novo assemble the complete ipaH gene in any strain analysed here due to the presence of multiple homologues of ipaH in the genome. However, all 16 genomes showed the presence of the entire length of ipaH by either BLAST comparison of multiple contigs or mapping to the S. flexneri 2a 2457 T reference genome.
The molecular serotyping detailed in Sun et al. [3] correlated with the phenotypic data for all isolates tested (Table 1). The provisional type, E1037, was the only Type Strain to contain a copy of the plasmid-mediated seroconverting Ipt-O (opt) gene [12]. In contrast to the serotype 5 strain described by Sun et al. (2012) [3], both PHE serotype 5 Type strains encoded an additional oac gene which was intact according to de novo assembly and the presence of the oac gene was confirmed by PCR [3]. The 5a and 5b serotypes were differentiable by the presence (serotype 5b) or absence (serotype 5a) of gtrX (Table 1).
Future directions
The PHE S. flexneri Type strain data set has been used in the validation and evaluation of genotypic and phenotypic assays and has facilitated the study of phylogenetic relationships within this species during outbreak investigations (unpublished observations). Analysis of the genome sequences, in conjunction with the phenotypic serotyping data, provided new insights into this historic strain set. Comparisons with the PCR serotyping scheme highlighted the need to add novel variants [13] in order to maintain a comprehensive collection of relevant Type strains.
Abbreviations
SNPs: Single nucleotide polymorphisms.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AG and CJ performed serology and DNA extraction. DW carried out the DNA sequencing. PA, KB, NRT and TJD analyzed the sequencing data. PA, KB, TJD and CJ contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Philip M Ashton, Email: philip.ashton@phe.gov.uk.
Kate S Baker, Email: kb14@sanger.ac.uk.
Amy Gentle, Email: amy_gentle_15@hotmail.co.uk.
David J Wooldridge, Email: david.wooldridge@phe.gov.uk.
Nicholas R Thomson, Email: nrt@sanger.ac.uk.
Timothy J Dallman, Email: tim.dallman@phe.gov.uk.
Claire Jenkins, Email: claire.jenkins@phe.gov.uk.
Acknowledgements
The authors would like to acknowledge the support of Kathie Grant, Catherine Arnold, Jonathan Green and PHE NGS Implementation Group.
References
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M. et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case–control study. Lancet. 2013;382(9888):209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- Ewing WH, Carpenter KP. Recommended designations for the subserotypes of Shigella flexneri. Int J Syst Bacteriol. 1966;16:145–149. doi: 10.1099/00207713-16-2-145. [DOI] [Google Scholar]
- Sun Q, Lan R, Wang Y, Zhao A, Zhang S, Wang J, Wang Y, Xia S, Jin D, Cui Z, Zhao H, Li Z, Ye C, Zhang S, Jing H, Xu J. Development of a multiplex PCR assay targeting O-antigen modification genes for molecular serotyping of Shigella flexneri. J Clin Microbiol. 2011;49:3766–3770. doi: 10.1128/JCM.01259-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross RJ, Rowe B. In: The Virulence of Escherichia coli: Reviews and Methods 1985; (Special Publication of the Society for General Microbiology no. 13): 345–360. Sussman M, editor. London: Academic Press; Serotyping of Escherichia coli. [Google Scholar]
- Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40(Web Server issue):W622-7. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Goldberg MB, Burland V, Venkatesan MM, Deng W, Fournier G, Mayhew GF, Plunkett G 3rd, Rose DJ, Darling A, Mau B, Perna NT, Payne SM, Runyen-Janecky LJ, Zhou S, Schwartz DC, Blattner FR. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457 T. Infect Immun. 2003;71:2775–2786. doi: 10.1128/IAI.71.5.2775-2786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slopek S, Mulczyk M. Concerning the classification of Shigella flexneri 6 bacilli. Arch Immunol Ther Exp (Warsz) 1967;15:600–603. [PubMed] [Google Scholar]
- Yang J, Nie H, Chen L, Zhang X, Yang F, Xu X, Zhu Y, Yu J, Jin Q. Revisiting the molecular evolutionary history of Shigella spp. J Mol Evol. 2007;64:71–79. doi: 10.1007/s00239-006-0052-8. [DOI] [PubMed] [Google Scholar]
- Pryamukhina NS, Khomenko NA. Suggestion to supplement Shigella flexneri classification scheme with the subserovar Shigella flexneri 4c: phenotypic characteristics of strains. J Clin Microbiol. 1988;26:1147–1149. doi: 10.1128/jcm.26.6.1147-1149.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Knirel YA, Lan R, Wang J, Senchenkova SN, Jin D, Shashkov AS, Xia S, Perepelov AV, Chen Q, Wang Y, Wang H, Xu J. A novel plasmid-encoded serotype conversion mechanism through addition of phosphoethanolamine to the O-antigen of Shigella flexneri. PLoS One. 2012;7:e46095. doi: 10.1371/journal.pone.0046095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Lan R, Wang J, Xia S, Wang Y, Wang Y, Jin D, Yu B, Knirel YA, Xu J. Identification and characterization of a novel Shigella flexneri serotype Yv in China. PLoS One. 2013;8:e70238. doi: 10.1371/journal.pone.0070238. [DOI] [PMC free article] [PubMed] [Google Scholar]