Abstract
Cocaine-mediated repression of the histone methyltransferase (HMT) G9a has recently been implicated in transcriptional, morphological, and behavioral responses to chronic cocaine administration. Here, using a ribosomal affinity purification approach, we find that G9a repression by cocaine occurs in both Drd1 (striatonigral)- and Drd2 (striatopallidal)-expressing medium spiny neurons (MSNs). Conditional knockout and overexpression of G9a within these distinct cell types, however, reveals divergent behavioral phenotypes in response to repeated cocaine treatment. Our studies further indicate that such developmental deletion of G9a selectively in Drd2 neurons results in the unsilencing of transcriptional programs normally specific to striatonigral neurons, and the acquisition of Drd1-associated projection and electrophysiological properties. This partial striatopallidal to striatonigral ‘switching’ phenotype in mice indicates a novel role for G9a in contributing to neuronal subtype identity, and suggests a critical function for cell-type specific histone methylation patterns in the regulation of behavioral responses to environmental stimuli.
Chronic exposure to cocaine, or other substances of abuse, induces persistent changes in genome-wide transcriptional profiles in the brain’s reward circuitry, resulting in long-lasting behavioral abnormalities that characterize drug addiction1. Recent evidence suggests that cocaine-induced alterations in gene expression are mediated, in part, by direct regulation of the chromatin modifying machinery and associated transcription factor complexes2–4. Data now indicate a prominent role for cocaine-mediated reductions in expression and activity of G9a, a repressive HMT, and the euchromatic modification that it deposits, H3K9me2 (dimethylation of Lys9 of histone H3), in nucleus accumbens (NAc) in regulating transcriptional, structural, and behavioral plasticity in response to repeated drug treatment5–8. Repression of G9a has similarly been observed in postmortem NAc of human cocaine addicts (unpaired t-test, effect of addiction: t16 = 2.028, p ≤ 0.05; n = 9/group) (Fig. S1 and Supplementary Table 1), indicating a clinically relevant role for G9a and histone methylation in human addiction. To date, however, little is known about the cell-type specific activities of G9a in NAc in regulating physiological and behavioral responses to cocaine. Recently, through targeted deletions of G9a in either Drd1 or Drd2 medium spiny neurons (MSNs), Schaefer and colleagues demonstrated a role for this HMT in altering behavioral responsiveness to several pharmacological stimuli in a cell-type specific manner9, suggesting unique functions for G9a, and associated histone methylation states, in the regulation of distinct neuronal phenotypes.
Here, we aimed to better understand G9a’s role in regulating cocaine-induced behaviors through segregated analyses of G9a function in Drd1 vs. Drd2 MSNs using a combination of genetic, behavioral, and electrophysiological approaches. We found that G9a expression is reduced in both Drd1 and Drd2 cells by repeated cocaine exposure, however, opposing behavioral responses to cocaine treatment were observed in animals with conditional G9a knockout (KO) or adult G9a overexpression in Drd1 vs. Drd2 neurons. Genomic, anatomical, and electrophysiological characterizations of G9a revealed a previously uncharacterized role for this enzyme, whereby developmental loss of G9a selectively in Drd2 MSNs results in a partial striatopallidal to striatonigral ‘switching-like’ phenotype, with G9a KO Drd2 cells partly resembling Drd1 cells. Although G9a is a well-known mediator of tissue specific transcriptional programs10, our data implicate the enzyme as an important regulator of neuronal subtype identity in the adult central nervous system (CNS).
Results
Cocaine reduces G9a expression in Drd1- and Drd2 MSNs
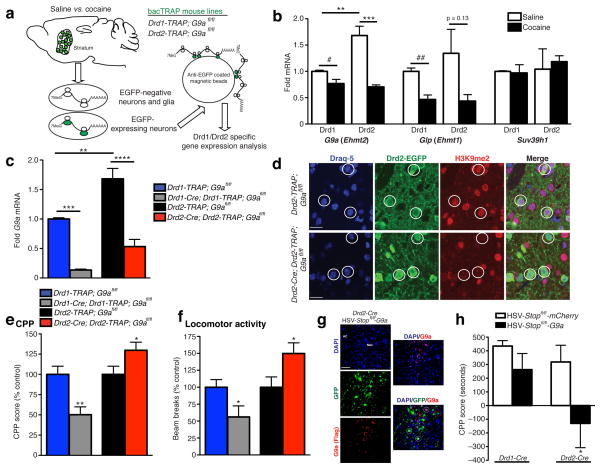
To assess G9a expression profiles selectively in Drd1 vs. Drd2 MSNs following repeated cocaine administration, we employed a cell-type specific translational profiling methodology (translating ribosome affinity purification (TRAP))11,12, which allows for affinity purification of EGFP-tagged polyribosomal complexes (and associated mRNAs) from adult neurons under the control of either Drd1a or Drd2 promoters (Drd1-TRAP; G9afl/fl or Drd2; G9afl/fl) (Fig. 1a).
Figure 1. Cocaine regulation of G9a in Drd1 and Drd2 striatal MSNs.
(a) Schematic outlining the TRAP approach for purifying cell-type specific polyribosomal associated mRNAs for Drd1 vs. Drd2 gene expression analysis. (b) G9a expression is significantly reduced in both Drd1 and Drd2 MSNs in striatum in response to repeated cocaine. Glp, G9a’s principal binding partner, displays similar regulation to G9a, however Suv39h1, an H3K9me3 HMT, was unaffected by cocaine treatment. (c) In comparison to wildtype littermate controls, G9a expression is significantly and selectively reduced in Drd1 and Drd2 MSNs following Cre-mediated recombination resulting in (d) selective loss of euchromatic H3K9me2 in these cells (scale bars for representative immunofluorescence images equal 20 μm). Such conditional KO of G9a in Drd1 vs. Drd2 MSNs results in distinct effects on both (e) cocaine reward (CPP) and (f) the locomotor activating effects of repeated cocaine exposure. (g) Representative images (scale bar equals 50 μm) demonstrating cell type specific overexpression of G9a (Flag tagged) in Drd2-Cre mice. Both HSV-Stopfl/fl-mCherry and G9a viruses express GFP irrespective of Cre expression, yet nuclear G9a (Flag) expression is only observed in a subpopulation of these MSNs (Drd2-Cre+). (h) Drd2, but not Drd1, specific G9a overexpression in adult NAc reduces cocaine CPP. Data are presented as averages ± SEM.
In accordance with previous findings5–7, quantitative polymerase chain reaction (qPCR) analyses indicate that G9a expression is significantly reduced in striatum following repeated cocaine. This reduction was predominantly observed in Drd2-expressing MSNs (two-way ANOVA, cell type x drug, drug: F1,8 = 37.05, p < 0.001; post hoc Bonferroni t-test, effect of drug Drd2 MSN: t8 = 6.955, p < 0.001, effect of cell type saline: t8 = 4.856, p < 0.01; n = 3/group), with a more modest reduction observed in Drd1 cells (unpaired t-test, effect of drug Drd1 MSN: t4 = 2.818, p < 0.05; n = 3/group) (Fig. 1b). G9a’s principal binding partner, G9a-like protein (Glp), which has also previously been demonstrated to be downregulated by repeated cocaine treatment6, was found to be robustly repressed in Drd1 MSNs, with a non-significant trend towards downregulation observed in Drd2 cells (unpaired t-test, effect of drug Drd1 MSN: t4 = 4.997, p < 0.01/effect of drug Drd2 MSN: t4 = 1.926, p = 0.13; n = 3/group) (Fig. 1b). Consistent with previous data5,7, the H3K9me3 HMT, Suv39h1, was not significantly regulated by repeated cocaine in either cell-type in striatum (Fig. 1b).
G9a oppositely regulates behaviors in D1 vs. D2 MSNs
Since cocaine-mediated reductions in G9a expression in NAc have previously been demonstrated to increase cocaine reward5, we next generated selective Drd1 and Drd2 G9a KO mice (Drd1-Cre; Drd1-TRAP; G9afl/fl or Drd2-TRAP; G9afl/fl) for direct examination of cell-type specific contributions of G9a to cocaine-mediated behaviors. G9a expression was significantly reduced following Cre-mediated recombination selectively in Drd1 or Drd2 cells (two-way ANOVA, cell type x genotype, genotype: F1,9 = 103.6, p < 0.0001; post hoc Bonferroni t-test, effect of genotype Drd1 MSN: t9 = 6.399, p < 0.001/Drd2 MSN: t9 = 7.951, p < 0.001; n = 3–4/group) (Fig. 1c), loss of which resulted in complete (100%) ablation of H3K9me2 in Cre-expressing striatal neurons (Fig. 1d). Loss of G9a was associated with non-significant trends for reduced expression of Glp in both cell types, with no effect on Suv39h1 (Fig. S2). Further work is needed to understand what controls the coordinated expression of G9a and Glp and how the balance between them influences neural gene expression.
Conditional KO of G9a in Drd2 MSNs phenocopied increased cocaine reward (unpaired t-test, effect of KO conditioned place preference (CPP): t16 = 2.090, p ≤ 0.05; n = 9/group) (Fig. 1e) and locomotor sensitization (LS) (unpaired t-test, effect of KO on LS: t16 = 2.264, p < 0.05; n = 9/group) (Fig. 1f) previously observed with global (i.e., both Drd1 and Drd2) KO of G9a in NAc5. In contrast, conditional G9a KO in Drd1 MSNs had the opposite effect on cocaine CPP and sensitization (unpaired t-test, effect of KO CPP: t16 = 3.482, p < 0.01; n = 8–10/group and LS: t16 = 2.211, p < 0.05; n = 9/group) (Fig. 1e–f) indicating functional differences for G9a between Drd1 and Drd2 neuronal subtypes. Lastly, to address the functional significance of G9a in adult MSNs, we generated conditional Stopfl/fl Herpes simplex viral (HSV) vectors to overexpress G9a selectively in Drd1- or Drd2 MSNs in NAc (Fig. 1g). Consistent with robust downregulation of G9a expression in Drd2 MSNs following repeated cocaine administration, as well as enhanced behavioral responses to the drug, in Drd2 developmental KOs, overexpression of G9a selectively in Drd2 cells reduced cocaine CPP (unpaired t-test, effect of Drd2 overexpression: t10 = 2.159, p ≤ 0.05, n = 5–7/group) (Fig. 1h), suggesting a prominent role for Drd2 G9a activity in regulating cocaine-mediated reward. Overexpression of G9a in Drd1 MSNs, however, did not affect cocaine CPP (unpaired t-test, effect of Drd1 overexpression: t11 = 1.294, p > 0.05, n = 6–7/group), further indicating different functions for G9a in these two inter-related cell types.
D2 KO promotes transcriptional unsilencing of D1 genes
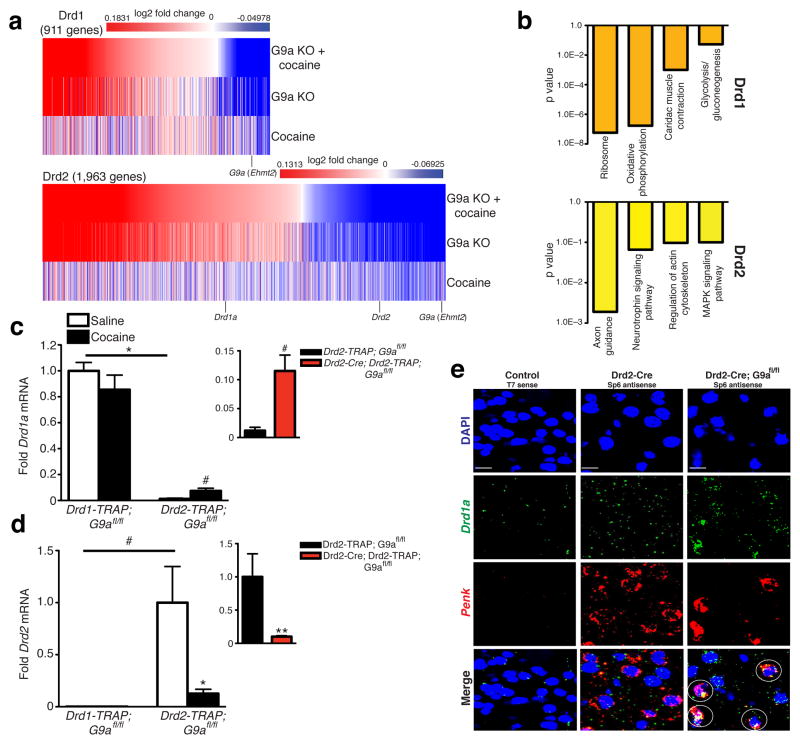
To better understand the contribution of G9a to the observed behavioral phenotypes, we next employed genome-wide analyses to monitor the impact of repeated cocaine, G9a KO, or cocaine plus G9a KO in Drd1 (Supplementary Table 2) vs. Drd2 (Supplementary Table 3) MSNs. Similar to previous findings11, we identified a limited set of genes in both cell types that display unique regulation by repeated cocaine (Drd1-253 genes and Drd2-239 genes). The effects of G9a KO, however, within these two cell types, both in the presence and absence of cocaine, were much more pronounced, with many of these genes exhibiting unique patterns of transcriptional unsilencing between Drd1 and Drd2 MSNs. The majority of these de-repression events were non-overlapping with genes displaying endogenous regulation by repeated cocaine in wildtype animals, likely indicating an indirect link between the effects of developmental G9a KO and adult cocaine exposure on the behavioral phenotypes observed in Fig. 1e–f.
Further analysis of gene expression profiles following G9a KO identified a distinct pattern of transcriptional de-repression in Drd2 neurons, whereby expression of Drd1a was upregulated, with concomitant reductions in Drd2 expression and enhanced expression of numerous genes previously identified as Drd1 enriched (heatmaps: two-way ANOVA, drug x genotype for both Drd1 and Drd2 MSNs independently, FDR 0.15) (Fig. 2a). Further bioinformatic analysis of genes displaying altered expression profiles between wildtype and G9a KO cells indicated highly relevant associations between previously identified Drd1 enriched and Drd1 associated cocaine-mediated pathways selectively in Drd2 neurons (e.g., regulation of the actin cytoskeleton, MAP kinase signaling, etc.) (Fig. 2b). Of particular interest was the identification of two genes previously demonstrated to play essential roles in the postnatal differentiation of striataonigral neurons, Ebf1 (early B-cell factor 1) and its associated binding partner Zfp521 (zinc finger protein 521)13,14. Both genes, which are enriched in wildtype Drd1 MSNs, were significantly upregulated following G9a KO in Drd2 neurons. Similar results were not observed for Drd1 cells lacking G9a. These data suggest that repression of G9a in Drd2 cells during development results in the unsilencing of a Drd1-like transcriptional program that influences an animal’s behavioral responsiveness to subsequent cocaine exposure in adulthood.
Figure 2. G9a KO alters the transcriptional identity of Drd2 MSNs in striatum.
(a) Comparative heatmaps displaying genes significantly regulated in Drd1 vs. Drd2 neurons by repeated cocaine, G9a KO, or repeated cocaine plus G9a KO. (b) Bioinformatics analysis (Database for Annotation, Visualization and Integrated Discovery (DAVID)) of gene expression pathways regulated by cocaine exposure ± G9a KO in Drd1 vs. Drd2 neurons demonstrating Drd2-selective enrichment for genes previously shown to be basally enriched or regulated by cocaine in Drd1 MSNs. Independent validations of cocaine’s and G9a’s impact on (c) Drd1a and (d) Drd2 expression in striatal MSNs reveals opposing patterns of regulation, whereby Drd1a is induced and Drd2 is repressed selectively in Drd2 MSNs following both repeated cocaine exposure and G9a KO (inset). (e) FISH analysis of Drd2-Cre vs. Drd2-Cre; G9afl/fl mice reveals increased colocalization of Drd1a (a Drd1 specific gene) and Penk (a Drd2 specific gene) in striatum of Drd2 G9a KOs in comparison to wildtype controls. White circles indicate DAPI+ cells co-expressing Drd1a and Penk, and red circles indicate DAPI+ cells expressing either Drd1a or Penk, but not both. T7 sense controls indicate gene specific labeling for both Drd1a and Penk probes. All images were acquired at 40X objective. Data are presented as averages ± SEM.
Following independent validations of both cocaine’s and G9a’s effects on Drd1a (two-way ANOVA, cell type x drug, cell type: F1,8 = 182.4, p < 0.0001; post hoc Bonferroni t-test, effect of cell type saline: t8 = 10.67, p < 0.0001/unpaired t-test, effect of cocaine Drd2 MSN: t4 = 2.945, p < 0.05/unpaired t-test, effect of genotype Drd2 MSN: t4 = 3.673, p < 0.05; n = 3/group) (Fig. 2c) and Drd2 (two-way ANOVA, cell type x drug, cell type: F1,8 = 10.32, p ≤ 0.01; unpaired t-test, effect of cell type saline: t4 = 2.875, p < 0.05, post hoc Bonferroni t-test, effect of drug: t8 = 3.531, p < 0.05/two-way ANOVA, cell type x genotype, genotype: F1,9 = 8.035, p < 0.05; post hoc Bonferroni t-test, effect of genotype: t9 = 3.884, p < 0.01; n = 3–4/group) (Fig. 2d) expression selectively in Drd2 neurons, we performed in situ hybridization to further examine Drd1a expression in Drd2 MSNs upon G9a KO. Consistent with data obtained from our bacTRAP arrays and subsequent qPCR analyses, G9a KO selectively in Drd2 MSNs increased the number of Penk+ cells (a marker for Drd2 MSNs) that are also Drd1a+ (Fig. 2e).
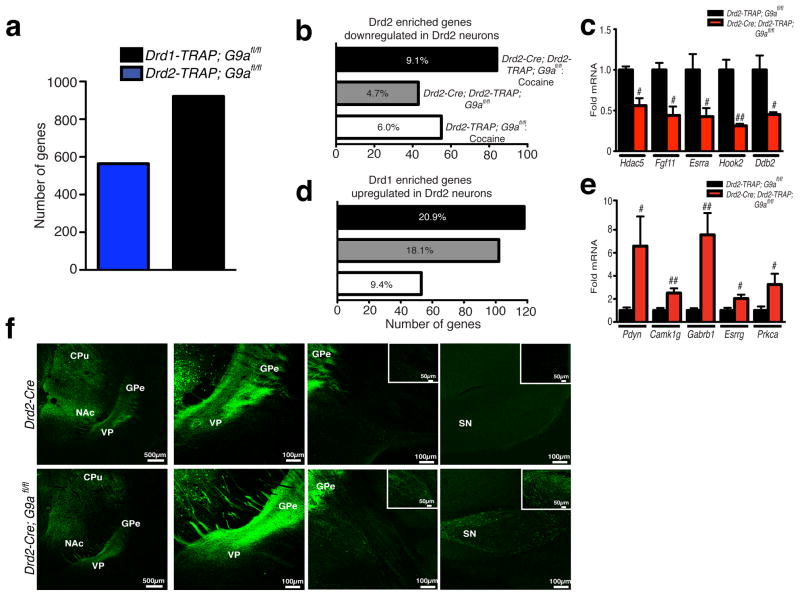
Next, compiling bioinformatic analyses from our bacTRAP arrays and from two independent data sets (bac-TRAP11 and fluorescence activated cell sorting (FACS)13), we found that genes across these lists overlap significantly. We statistically categorized genes as either Drd1 or Drd2 ‘enriched’ (pairwise comparison between Drd1 and Drd2 control saline, p < 0.005) in order to investigate whether a subset of Drd1 enriched genes are indeed unsilenced by cocaine, G9a KO or both. Numerous genes were significantly enriched in either Drd1 (564 genes, e.g., Drd1a, Eya1 and Pdyn) or Drd2 (921 genes, e.g., Drd2, Penk and Adora2a) MSNs (Fig. 3a and Supplementary Tables 4 and 5). We then identified a subset of these genes, which were validated as Drd2 enriched, that were significantly downregulated in Drd2 MSNs by either repeated cocaine (6.0%), G9a KO (4.7%), or both (9.1%) (p < 0.005) (Fig. 3b and Supplementary Tables 6–8), effects that were verified through independent qPCR analysis (unpaired t-test, effect of genotype: Hdac5-t4 = 4.382, p < 0.05; Fgf11-t4 = 4.019, p < 0.05; Esrra-t4 = 2.607, p ≤ 0.05; Hook2-t4 = 5.391, p < 0.01; Ddb2-t4 = 3.062, p < 0.05; n = 3/group) (Fig. 3c). Consistent with the notion that Drd2 MSNs experience a partial shift in transcriptional identity in response to developmental KO of G9a, subsequent analyses also demonstrated that a significant proportion of genes identified as Drd1 ‘enriched’ display significant upregulation in Drd2 cells in response to repeated cocaine (9.4%), G9a KO (18.1%), or both (20.9%) (p < 0.005) (Fig., 3d and Supplementary Tables 9–11), results that were later verified by qPCR (unpaired t-test, effect of genotype: Pdyn-t8 = 2.711, p < 0.05; Camk1g-t8 = 3.579, p < 0.01; Gabrb1-t8 = 4.338, p < 0.01; Esrrg-t7 = 2.669, p < 0.05; Prkca-t7 = 2.503, p < 0.05; n = 4–6/group) (Fig. 3e).
Figure 3. Selective loss of G9a in Drd2 MSNs results in the unsilencing of striatonigral specific gene expression profiles and abnormal afferent projections.
(a) Identification of Drd1 vs. Drd2 enriched gene expression profiles in striatum. (b) Numbers of Drd2 enriched genes displaying significant downregulation in Drd2 MSNs following repeated cocaine, G9a KO, or both. (c) Validation of Drd2 enriched genes downregulated by G9a KO selectively in Drd2 MSNs. (d) Numbers of Drd1 enriched genes displaying significant upregulation in Drd2 MSNs following repeated cocaine, G9a KO, or both. (e) Validation of Drd1 enriched genes upregulated by G9a KO selectively in Drd2 MSNs. Percentages indicate the number of Drd2 (b) or Drd1 (d) enriched genes displaying significant regulation by repeated cocaine, G9a KO, or both, divided by the total number of genes enriched within a given cell type. (f) Afferent projection terminal tracing demonstrates that selective loss of G9a in Drd2 neurons leads to abnormal ‘direct pathway’ projections to midbrain (substantia nigra) from Drd2 G9a KO MSNs in comparison to wildtype controls. Data are presented as averages ± SEM.
D2 KO leads to a partial D2 to D1 ‘switching’ phenotype
Given that conditional deletion of G9a enhances Drd1 specific gene expression profiles in Drd2 MSNs, we investigated whether G9a KO results in functional adaptations in anatomical projections or cellular physiology that may further explain the observed behavioral responses to cocaine. Drd2-Cre (control) and Drd2-Cre; G9afl/fl (Drd2 G9a KO) mice were stereotaxically injected intra-striatum with adeno-associated virus (AAV)-DIO-EYFP vectors containing a doubly floxed inverted EYPF marker. With this system, EYFP is exclusively expressed in Cre+ MSNs, allowing for cell-type specific tagging for use in anatomical tracing and slice physiology. First, to determine whether early postnatal deletion of G9a selectively in Drd2 MSNs alters the neurons’ projection profiles, wildtype and Drd2 G9a KO animals received successive intra-caudate putamen (CPu) and intra-NAc injections of AAV-DIO-EYFP, followed by projection labeling one month after infection. In wildtype animals, projection terminals were clearly identified in both the ventral pallidum (VP) and the globus pallidus externus (GPe), arising from the ventral and dorsal striatum, respectively. Consistent with previous literature, no labeling was observed beyond pallidal regions in wildtype Drd2-Cre mice. In Drd2 G9a KO animals, however, clear labeling of projection terminals was observed in substantia nigra, a midbrain region traditionally known to receive ‘direct pathway’ projections from Drd1, but not Drd2, MSNs originating in striatum (Fig. 3f). These data suggest that conditional KO of G9a in Drd2 MSNs leads to aberrant transcriptional programs that might influence the appropriate establishment of striatal circuitry.
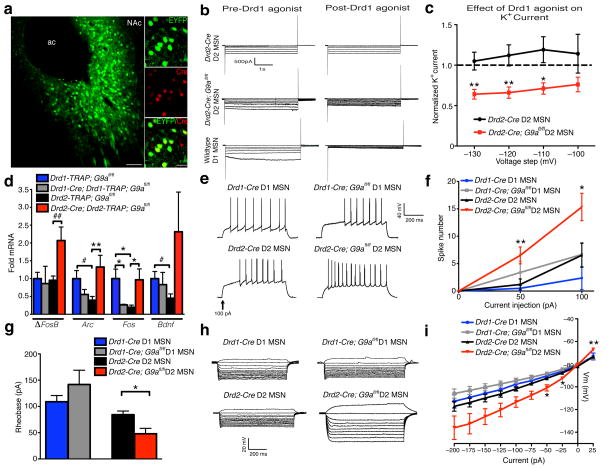
Next, to investigate whether G9a KO-induced expression of Drd1a in Drd2 MSNs results in functional Drd1 activity, AAV-DIO-EYFP infected NAc slices (Fig. 4a) from wildtype and Drd2 G9a KO animals were incubated in the presence or absence of the Drd1 agonist, dihydrexidine hydrochloride, and potassium (K+) currents were measured in voltage clamp. Consistent with published reports15, application of the D1 agonist decreased K+ currents at hyperpolarized potentials in wildtype Drd1 MSNs, with no evoked responses observed in control Drd2 MSNs (Fig. 4b). In G9a KO Drd2 neurons, however, D1 activation significantly reduced K+ currents, similar to effects observed in wildtype Drd1 MSNs (unpaired t-test, effect of D1 agonist, −130: t13 = 3.086, p < 0.01; −120: t13 = 3.077, p < 0.01; −110: t13 = 2.613, p < 0.05; n = 7–8/group) (Fig. 4b–c), demonstrating the presence of functional D1 receptors on G9a KO Drd2 neurons.
Figure 4. Ablation of G9a in Drd2 MSNs results in a functional Drd2-Drd1 ‘switching’ phenotype and enhanced cell excitability.
(a) Representative immunofluorescence image of AAV-DIO-EYFP expression in Drd2-Cre expressing MSNs in NAc (scale bars equal 100 μm and 20 μm respectively for low (left) vs. high (right) power images). (b) Representative traces of K+ channel mediated currents in voltage clamp in response to hyperpolarizing voltage steps for putative Drd1 vs. EYFP+ Drd2 MSNs ± G9a KO in the absence or presence of the D1 agonist dihydrexidine hydrochloride. (c) Quantification of normalized K+ current across multiple voltage steps indicates that D1 agonism attenuates K+ currents at hyperpolarized potentials in both G9a KO Drd2 MSNs and in Drd1 MSNs, but not in wildtype Drd2 neurons. (d) IEG analysis in Drd1 vs. Drd2 MSNs ± G9a KO demonstrates a selective upregulation of activity-dependent genes in Drd2 MSNs lacking G9a. (e) Representative traces of spike number in response to current injections over time as a measure of intrinsic neuronal excitability. Whole cell quantification of current-induced spike number in Drd1 vs. Drd2 MSNs ± G9a KO indicates that G9a ablation selectively from Drd2 neurons results in increased cellular excitability (f) and reduced threshold for the first spike (rheobase) (g). (h) Representative voltage traces in response to current injections. (i) Consistent with enhanced excitability of Drd2 neurons following G9a KO, increased membrane resistance in the I–V relationship was observed, with no effects seen in Drd1 MSNs, indicating increased excitability of G9a KO Drd2 neurons. Data are presented as averages ± SEM.
Recent work has established distinct roles for evoked excitability and postsynaptic responses between Drd1 and Drd2 MSNs in the regulation of cocaine-mediated behaviors16,17, in which increased Drd1, and decreased Drd2, MSN activity or excitatory synaptic function increases sensitivity to both the rewarding and locomotor activating effects of cocaine. Therefore, we examined directly if G9a plays a cell-type specific role in regulating the excitability of NAc MSNs. We began by profiling immediate early gene (IEG) expression, as a molecular marker of cellular activity, in Drd1 and Drd2 neurons in control vs. conditional G9a mutant mice. We found that several IEGs, which are enriched in Drd1 MSNs at baseline, are significantly induced in Drd2 neurons upon loss of G9a, with either opposite or insignificant changes observed in G9a KO Drd1 neurons (e.g., Arc (two-way ANOVA, cell type x genotype, interaction: F1,13 = 11.38, p < 0.01; post hoc Bonferroni t-test, effect of genotype: t13 = 3.538, p < 0.01/unpaired t-test, effect of cell type: t7 = 2.824, p < 0.05; n = 3–6/group) and Fos (two-way ANOVA, cell x genotype, interaction: F1,13 = 17.9, p ≤ 0.001; post hoc Bonferroni t-test, effect of genotype Drd1 MSN: t13 = 2.704, p < 0.05; Drd2 MSN: t13 = 3.356, p < 0.05/effect of cell type control: t13 = 3.192, p < 0.05; n = 3–6/group)). Also, consistent with previous data5, G9a KO from Drd2 MSNs increased basal expression of ΔFosB (unpaired t-test, effect of genotype Drd2 MSN: t8 = 3.339, p ≤ 0.01; n = 4–6/group), a splice product of the IEG FosB, which accumulates in striatum18, primarily in Drd1 cells19,20,27, in response to chronic cocaine exposure and has been shown to increase drug reward20 (Fig. 4d). Increased ΔFosB and other IEG expression in response to G9a KO exclusively in Drd2 MSNs further suggests a shift towards Drd1 transcriptional re-programming and altered cellular physiology.
Although observed increases in IEG expression in G9a KO Drd2 MSNs suggest increased activity, such changes are difficult to infer, since IEGs can be regulated by cellular signaling cascades in the absence of changes in neuronal firing. We thus directly investigated Drd1 and Drd2 MSN excitability in NAc after conditional G9a KO. We observed a dramatic enhancement in neuronal firing in response to current injections selectively in Drd2 neurons lacking G9a (unpaired t-test, effect of genotype Drd2 MSN, 50 pA: t29 = 2.846, p < 0.01, 100 pA: t28 = 2.655, p ≤ 0.01; n = 15–16/group), with no effect seen in G9a KO Drd1 MSNs (Fig. 4e–f). Consistent with increased excitability, Drd2 neurons lacking G9a also display a reduced threshold to induce the first spike (rheobase) in response to current injections (unpaired t-test, effect of genotype: t25 = 2.608, p ≤ 0.01; n = 8–16/group) (Fig. 4g) and increased membrane input resistance in the steady state current voltage (I–V) relationship (unpaired t-test, effect of genotype Drd2 MSN, −50 pA: t22 = 2.180, p < 0.05, −25 pA: t22 = 2.269, p < 0.05, 25 pA: t22 = 2.842, p < 0.01; n = 8–16/group) (Fig. 4h–i).
Discussion
Repeated cocaine exposure reduces G9a expression and activity in striatum, leading to persistent changes in gene expression, neuronal morphology, and behavior5. Here, we demonstrate that G9a expression is downregulated in both Drd1 and Drd2 MSNs, however, the functional impact of G9a ablation or overexpression on cocaine-induced behaviors segregates by neuronal subtype. Further investigations examining the consequences of developmental G9a KO in Drd1 vs. Drd2 neurons demonstrated a previously uncharacterized role for G9a in influencing striatal neuron subtype specification. Selective G9a KO in Drd2 expressing neurons induces a partial phenotypic switch: by downregulating certain striatopallidal and de-repressing certain striatonigral enriched genes, and by altering the axonal projection profiles and functional D1 receptor responsiveness in Drd2 neurons. This partial Drd2-Drd1 ‘switching’ phenotype is associated with enhanced sensitivity to the rewarding and locomotor activating effects of cocaine. Collectively, our results provide novel insight into the role of cell-type specific chromatin regulatory mechanisms in the development of behavioral plasticity to cocaine and reveal a role for G9a in controlling neuronal subtype identity during development.
G9a has previously been shown to suppress neuronal genes in non-neuronal cells through repressive complex associations with RE1 silencing transcription factor (REST) and Mediator, a multiprotein interface between RNA polymerase II and locus specific transcription factors21. To our knowledge, however, a role for G9a in cell-type specification within a given tissue has not yet been demonstrated. Here we provide direct evidence that G9a mediated silencing, likely coupled to its euchromatic HMT activity, not only controls tissue specific gene expression programs, but also the specification of closely related neuronal subtypes within the same tissue (e.g., Drd1 vs. Drd2 MSNs of striatum). Although Drd1 vs. Drd2 MSNs have previously been shown to differentially express numerous genes, many of which contribute to their cellular identities and distinct functional outputs11,13, the mechanisms controlling such distinct fates have remained unclear. The Drd2-to-Drd1 phenotypic switch mediated by G9a KO is obviously not complete: the changes observed here in transcriptional profiles and anatomical projections are partial. Moreover, while G9a KO in Drd2 MSNs increased their excitability, previous work has shown that Drd1 MSNs are less excitable than Drd2 MSNs22. Thus, G9a is likely only one of many chromatin regulatory proteins involved in establishing MSN cell-type specificity. Nevertheless, it is becoming increasingly clear that alterations in G9a, both developmentally and in the adult CNS, contribute significantly to neuronal function. It also must be noted that G9a may control cell function through mechanisms other than its HMT activity10. Moving forward, it will be important to further dissect the molecular mechanisms of G9a action in the CNS during periods of neurodevelopment and in adult brain in response to environmental stimuli.
Our findings suggest a disassociation between G9a’s roles during neurodevelopment in regards to mediating neuronal subtype specification, and its function in adult brain as a modulator of stimulus-induced gene expression. Although both developmental KO of G9a, and endogenous repression of G9a in adult brain following repeated cocaine, result in altered behavioral responses to the drug, it appears that the underlying molecular mechanisms are distinct. For example, repeated exposure to cocaine has been shown to increase dendritic spine density of NAc MSNs23–26; this phenomenon is thought to contribute to an animal’s increased behavioral responsiveness to cocaine and has been linked directly to suppression of G9a5. This response—both increased dendritic spines and suppression of G9a5—is mediated by ΔFosB, which forms a negative feedback loop with G9a during chronic cocaine exposure. Of intrigue is the recent observation that ΔFosB persistently accumulates, and thereby promotes increased dendritic spine density, exclusively in Drd1 MSNs in response to repeated cocaine19,27,28, suggesting a vital role for Drd1 specific G9a repression in the adult in the potentiation of cocaine-induced behaviors. That deletion of G9a from Drd1 MSNs during development results in an opposite behavioral phenotype likely reflects a developmental phenomenon.
Although G9a is also reduced in Drd2 MSNs in response to cocaine, the behavioral consequences of this effect in adult NAc are less clear. This is because the dramatic developmental abnormalities seen in Drd2 neurons upon loss of G9a make it difficult to infer anything about the function of this enzyme in the adult. On the other hand, the dramatic suppression of G9a in adult Drd2 MSNs in response to chronic cocaine, and the ability of G9a overexpression in Drd2 MSNs of adult animals to suppress cocaine reward, nevertheless suggest that G9a also regulates cocaine action via this cell type. The developmental effects of G9a KO in Drd2 MSNs raise the interesting question of whether similar abnormalities might be induced in adult neurons by cocaine. We believe that this is unlikely given the timeframe in which rodent studies have been conducted, however, it is difficult to truly gauge the impact of long-term repression of G9a by cocaine. For example, ~1 week of cocaine exposure downregulates G9a expression in striatum, an effect which persists for several days during withdrawal5, but this may not constitute enough time to permit deficits in neural cell-type specification seen upon developmental deletion of the gene. Therefore, future studies, involving cell-type specific KO of G9a in adult animals, will be needed to further uncover distinct functional roles of the enzyme in Drd1 vs. Drd2 MSNs in response to cocaine exposure.
Interestingly, chronic cocaine exposure has been shown to reduce the expression and functionality of Drd2 in striatum29, a phenomenon that is believed to contribute to increased drug reward and drug seeking behavior30. These findings have recently been supported by cell-type specific optogenetic dissections of Drd1 vs. Drd2 MSNs in mouse NAc: Lobo et. al. demonstrated that increasing the firing of Drd1 MSNs increases cocaine reward and locomotor sensitization, while activation of Drd2 MSNs exerts the opposite effects16. Our current data are in direct agreement with these previous studies, as we too observed cocaine-mediated alterations in dopamine receptor expression in striatum consistent with increased Drd1 tone: conditional G9a KO selectively from Drd2 neurons results in the unsilencing of Drd1 specific gene expression profiles and functional Drd1 activity in these cells, associated with increases in cellular excitability and behavioral responsiveness to cocaine, thus mimicking those effects seen via optogenetic stimulation of Drd1 MSNs.
In conclusion, our data demonstrate that G9a contributes to the specification of striatal MSN subtypes during development and that such actions have prominent effects on cocaine sensitivity. We also provide a genome-wide exploration of genes in Drd1 vs. Drd2 MSNs that show abnormal regulation in response to repeated cocaine administration as a consequence of the cell-type specific loss of G9a during this developmental period. Although a master regulator of gene expression and euchromatic silencing, G9a also appears to play an important role in the CNS in guiding fundamental aspects of neuronal subtype identity and, therefore function, in adult brain. Gaining a broader understanding of the physiological and behavioral consequences of chromatin dysregulation during early neurodevelopment will aid in the evolution of novel therapeutics aimed at treating addiction and other devastating psychiatric disorders.
Online Methods
Animals
Mice were housed at four to five per cage with a 12-hour light/dark cycle (lights on from 0700 to 1900 hours) at constant temperature (23°C) with ad libitum access to food and water. All animal protocols were approved by IACUC at UT Southwestern Medical Center, Mount Sinai, and The Rockefeller University.
Generation of Transgenic Mouse Lines
Drd1- and Drd2-Cre mice (EY262 and ER44, Gensat31, and Drd1-, and Drd2-TRAP mice11 were bred to G9afl/fl mice 32 to generate Drd1-Cre; G9afl/fl and Drd2-Cre; G9afl/fl for electrophysiological analysis, as well as Drd1-Cre; Drd1-TRAP; G9afl/fl, Drd1-TRAP; G9afl/fl, Drd2-Cre; Drd2-TRAP; G9afl/fl and Drd2-TRAP; G9afl/fl mice for cell-type specific ribosomal-associated RNA purification as previously described9. Drd2-Cre; Drd2-eGFP; G9afl/fl and Drd2-eGFP; G9afl/fl used for immunofluorescence studies were generated as previously described9. All mice were backcrossed to the C57Bl/6 background for >5 generations, and genotyping was performed as previously described.
Drugs
Cocaine-HCl was obtained from Sigma-Aldrich and used at a concentration of 20 mg/kg/i.p. Mice were immediately returned to their home cage after each injection, unless otherwise stated.
Quantifying Protein Expression from Human Postmortem Brain
Postmortem human brain tissue was obtained from the Quebec Suicide Brain Bank (IRB approval from Douglas Mental Health University Institute, Montreal, Quebec, Canada). Tissue preservation proceeded as described previously33. Once obtained, brains were placed on wet ice and transported to the Quebec Suicide Brain Bank facilities. Once dissected, tissues were flash frozen in 2-methylbutane at −40°C. All frozen tissues were kept separately in plastic bags at −80°C for long-term storage. Specific brain regions were later sectioned from frozen coronal slices.
This cohort was composed of one female and 17 male subjects, ranging in age between 18 and 66 years. All subjects died suddenly without protracted medical illness or extended agonal states. For each case, the cause of death was determined by the Quebec Coroner Office, and toxicological screens were conducted to obtain information on medication and illicit substance use at the time of death. The subject group consisted of 9 individuals who met the Structured Clinical Interview for DSM-IV (Diagnostic and Statistical Manual of Mental Disorders-IV) Axis I Disorders: Clinician Version (SCID-I) criteria for cocaine dependence. The control group comprised 9 subjects with no history of cocaine dependence and no major comorbid psychiatric diagnoses. Groups were matched for subject age, postmortem interval, and pH. For all subjects, psychological autopsies were performed, giving us access to detailed information on psychiatric and medical histories, as well as other relevant sociodemographic and clinical data (Supplementary Table 1).
To quantify G9a protein expression (Fig. S1), frozen NAc tissue was homogenized in 30 μL of homogenization buffer (320 mM sucrose, 5 mM Hepes buffer, 1% SDS, phosphatase inhibitor cocktails I and II (Sigma) and protease inhibitors (Roche) using an ultrasonic processor (Cole Parmer). Protein concentrations were determined using a DC protein assay (Bio-Rad), and 50 μg of protein was loaded onto 4–15% gradient Tris-HCl polyacrylamide gels for electrophoresis fractionation (Bio-Rad). Samples were next transferred to nitrocelluose membranes and blocked in Odyssey® blocking buffer (Li-Cor). Blocked membranes were incubated overnight at 4°C in primary antibody (1/500 G9a, Millipore 09-071) in Odyssey® blocking buffer. After washing with TBST, membranes were incubated with IRDye® secondary antibodies (1/5000 to 1/10000; Li-Cor) dissolved in Odyssey® blocking buffer for 1 hr at room temperature. Blots were then imaged with the Odyssey® Infrared Imaging system (Li-Cor) and were quantified by densitometry using ImageJ (NIH). The amount of protein loaded into each lane was normalized to levels of glyceraldehyde 3-phosphate dehydrogenase (1: 5000, Millipore MAB374).
TRAP, qPCR & Arrays
Polyribosome associated mRNAs from 2–5 month old, age and sex matched Drd1-Cre; Drd1-TRAP; G9afl/fl and Drd1-TRAP; G9afl/fl, Drd2-Cre; Drd2-TRAP; G9afl/fl and Drd2-TRAP; G9afl/fl mice (n = 2–4 mice/genotype/drug treatment, 2 hours after the last of eight repeated cocaine injections of 20 mg/kg/day) were obtained as previously described9,11,12. EGFP labeled ribosomes and associated mRNAs were immunoprecipitated using a mix of two monoclonal anti-GFP antibodies (50 μg of clones #19C8 and #19F7 for each IP, available at Sloan-Kettering Monoclonal Antibody Facility). Purified mRNA was amplified and processed for microarray and qPCR analysis using the Affymetrix two-cycle cDNA Synthesis kit (Affymetrix) as previously described11,12. Affymetrix Mouse Genome 430 2.0 arrays were used in all experiments. Information regarding the array design and features can be found at www.affymetrix.com. Mouse Genome 430 2.0 arrays were scanned using the GeneChip Scanner 3000 (Affymetrix) and globally scaled to 150 using the Affymetrix GeneChip Operating Software (GCOS v1.4).
cDNA was quantified by qPCR with SYBR Green. Each reaction was run in duplicate and analyzed using the ΔΔCt method as previously described34. Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) was used as a normalization control and was not found to be altered by any manipulations described throughout the manuscript. See supplementary Table 12 for mRNA primer sequences.
Behavior
Conditioned Placed Preference (CPP)
CPP was conducted as previously described5. Briefly, 8–12 week old Drd1-TRAP; G9afl/fl, Drd1-Cre; Drd1-TRAP; G9afl/fl, Drd2-TRAP and Drd2-Cre; Drd2-TRAP; G9afl/fl mice were placed into conditioning chambers consisting of three distinct environments. Mice displaying significant preference for either of the two conditioning chambers were excluded (<5% of all animals). Conditioning groups were subsequently balanced to adjust for chamber bias. On conditioning days (subsequent 4 days), animals were injected with either saline (days 1 and 3) or cocaine (10 mg/kg, i.p.) (days 2 and 4) and were then confined to one chamber in the afternoon for 30 minutes. On the test day, animals were placed back into the CPP apparatus without treatment for 20 minutes and evaluated for which side they preferred. Locomotor responses to cocaine were assessed by beam break analysis in the cocaine-paired chambers to ensure the effectiveness of drug treatment. For all groups, baseline locomotor responses to saline were assessed to ensure that locomotion was not affected by genotype (data not shown). For HSV CPP experiments (Fig. 1i), an accelerated conditioning paradigm was employed. Briefly, 48 hr after surgery, mice were injected with saline (day 1 morning, day 2 night) or cocaine (day 1 night, day 2 morning) during which they were confined to one chamber for 30 minutes as described. On day 3, CPP testing proceeded as stated previously. Behaviors were assessed during the animals’ light cycle.
Locomotor Activity
8–12 week old Drd1-TRAP; G9afl/fl, Drd1-Cre; Drd1-TRAP; G9afl/fl, Drd2-TRAP and Drd2-Cre; Drd2-TRAP; G9afl/fl mice injected once daily for 14 days with cocaine (20 mg/kg, i.p.) and immediately returned to their home cages. 1 week later, animals were habituated in locomotor boxes for 60 min and then monitored for locomotor activity after a single saline injection (60 minutes) using the Photobeam Activity System (San Diego Instruments). In accordance with previous findings9,35, all animals habituated normally and displayed similar responses to a saline injection. Next animals were injected with a low dose of cocaine (5 mg/kg, i.p.) and monitored for locomotor activity for 30 minutes. Note that all animals used in this study displayed enhancements in locomotor activity in response to cocaine above that of saline treated controls. Data are presented as a percent control of total beam breaks over the course of the 30 minute test comparing mutant mice to respective wildtype littermates. Locomotor behaviors were assessed during the animals’ light cycle.
TRAP Array Analyses
Affymetrix Probe Sets Summarization
Probe level intensity files (CEL) were imported into Affymetrix’s Expression Console (v1.1) program for analysis. The RMA algorithm was used for normalization and probe level summarization files (CHP) were then exported for further analysis. Only 1 of 30 files was filtered out, as it failed default quality thresholds. Two negative control files were also removed, leaving 27 files remaining across the three design factors.
Differential Analysis
Two-way ANOVAs among different genotypes and experimental treatments were performed for each cell type individually to identify genes displaying differential expression. For each gene, three F-tests were performed for genotype, treatment and their interaction. All F-test results were subjected to multiple testing corrections using the Benjamini-Hochberg procedure. FDR < 15% was used as a cutoff to select genes for the heatmaps displayed in Fig. 2.
Pairwise comparisons were performed between two groups of any combination of the three factors using student t-tests. A p-value of < 0.005 was used as a cutoff. This approach was first used to compare compiled data sets: 1) control-saline-Drd1 vs. control-saline-Drd2 from our bacTRAP arrays; 2) control-saline-Drd1 vs. control-saline-Drd2 from Heiman et al.11; and 3) Drd1 vs. Drd2 FACS arrays from Lobo et al.14 to identify cell-type specific gene expression enrichment under basal conditions. It was then used to compare the effects of cocaine treatment, G9a KO, or a combination of the two with respective cell-type specific saline controls. These comparisons were performed to identify genes displaying increased expression of Drd1 enriched genes or decreased expression of Drd2 enriched genes in Drd2 MSNs following different perturbations.
Immunohistochemistry
8- to 12-week-old sex- and age-matched Drd2-Cre; Drd2-eGFP; G9afl/fl, and their respective littermate controls were anesthetized with pentobarbital and transcardially perfused with 10 ml of phosphate-buffered saline (PBS) followed by 50 ml of 4% paraformaldehyde (PFA) in PBS. Brains were processed as previously described9 and coronally sectioned at either 12 μm thickness on a Cryostat (Fig. 1d) or 35 μm thickness on a microtome following cryoprotection in 30% sucrose (Fig. 1h). GFP (1:5000, Abcam ab6556 or 1:8000, Aves Labs, Inc. GFP-1020), Flag M2 (1:400, Sigma F1804) and H3K9me2 (1:500, Abcam ab1220) were visualized using indirect immunofluorescence analysis (Alexa Fluor 647/546/488 labeled goat anti-mouse/anti-rabbit IgGs (H+L) dilution 1:500, Invitrogen Corporation). Draq5 (Biostatus Limited) or DAPI (Life Technologies) were used to counterstain the nucleus and sections were mounted using Prolong Gold antifade (Invitrogen) before being imaged on a LSM 710 confocal microscope (Zeiss). For validation of H3K9me2 knockdown in Drd2 G9a KO MSNs, cells were counted from 5 independent images/genotype (control: 545 Draq-5+ cells and Mutant: 537 Draq-5+ cells). Data were recorded as the percentage of Drd2+ cells co-expressing H3K9me2 in wildtype vs. KO striatum.
Drd2-Cre and Drd2-Cre; G9afl/fl mice, which had received stereotaxic intra-NAc injections 1 month prior with adeno-associated virus (AAV)-DIO-EYFP, were sedated with a lethal dose of chloral hydrate and perfused with PBS followed by 4% PFA before being analyzed by single or double immunohistochemistry for GFP (EYFP) and Cre recombinase. Briefly, brains were post-fixed and then incubated at room temperature overnight in 30% sucrose in PBS before being coronally sectioned at 35 μm (for projection tracing, brains were sectioned in the sagittal plane on a cryostat (Leica)). Free-floating NAc sections were washed with PBS, blocked (3% normal donkey serum, 0.3% tritonX, PBS) for 1 hour, and later incubated with anti-GFP (to detect EYFP expression) (1:5000–8000, GFP 1020, Aves Labs, Inc) and anti-Cre recombinase (1:1000, Millipore MAB3120) antibodies in blocking solution. Following overnight incubation in primary antibody, NAc sections were rinsed 3 times for 10 minutes each with PBS, followed by incubation in 1:500 donkey anti-chicken Cy2 and 1:500 donkey anti-mouse Cy3 fluorescent-coupled secondary antibodies (Jackson ImmunoResearch) in blocking solution for 1 hour (for tracing studies: Alexa Fluor 488 labeled goat anti-chicken 1:1000). Sections were subsequently washed 3x with PBS. Finally, NAc sections were mounted onto superfrost plus slides, allowed to dry and were then dehydrated in ethanol and cleared with Citrisolv. Slides were coverslipped in DePex and sections were imaged on either LSM 710 or Olympus confocal microscopes. For tracing studies, NAc, dorsal striatum, VP, GPe, axons to midbrain and SN were imaged.
Viral Mediated Gene Transfer
Following ketamine (100 mg/kg)/xylazine (10 mg/kg) induced anesthesia, Drd1-Cre, Drd1-Cre; G9afl/fl, Drd2-Cre and Drd2-Cre; G9afl/fl mice were placed in stereotaxic instruments, and cranial surfaces were exposed. 33 gauge syringe needles were used to infuse 0.5 μl of virus bilaterally into NAc at a 10° angle (AP + 1.6; ML + 1.5; DV – 4.4) at a rate of 0.1 μl/min (for successive CPu injections, after NAc injections were complete, needles were raised +1.0 mm DV and virus was infused as previously indicated). Animals receiving AAV-DIO-EYFP injections were allowed to recover for 4 weeks following surgery. Animals receiving HSV-GFP-Stopfl/fl-mCherry/hG9a viruses were allowed to recover for 48 hours prior to CPP training. These times are consistent with periods of maximal viral-mediated transgene expression5,16.
Fluorescent in situ hybridization (FISH)
Briefly, PCR products generated using primers (IDT technology) with an attached Sp6 or T7 polymerase sequence were used for in vitro transcription of Drd1a-UTP-Digoxigenin or Penk-UTP-Biotin (Roche Diagnostics) labeled RNA probes. The following primer sequences were used:
Drd1a Forward T7-CTGTAATACGACTCACTATAGGGCAGATCGGGCATTTGGAGAGAT
Drd1a Reverse Sp6-GGGATTTAGGTGACACTATAGAATGACCAAGACAGCCACCAAG
Penk Forward T7-CTGTAATACGACTCACTATAGGGTTCCTGAGGCTTTGCACC
Penk Reverse Sp6-GGGATTTAGGTGACACTATAGAATCACTGCTGGAAAAGGGC
14 μm thick cryosectioned tissue was hybridized with riboprobes overnight, followed by incubation with peroxidase antibodies directed against digoxigenin (Roche) and biotin (Jackson Immunoresearch). Amplification of the peroxidase-based antibodies was performed with the TSA Plus Fluorescent Kit (Perkin Elmer) for 8 minutes during which time fluorescein was deposited on Drd1a-labeled cells and tetramethylrhodamine was deposited on Penk-labeled cells. Slides were mounted using Vectamount with DAPI (Vector Laboratories) and imaged using a Leica upright fluorescent microscope. Sense probes were used to control for non-specific labeling.
In Vitro Patch-Clamp Electrophysiology
Whole cell recordings were obtained from NAc shell MSNs in acute brain slices from Drd1-Cre, Drd1-Cre; G9afl/fl, Drd2-Cre and Drd2-Cre; G9afl/fl mice that had been stereotaxically injected intra-NAc with AAV-DIO-EYFP. To minimize stress and to obtain healthy NAc slices, mice were anesthetized and perfused immediately for 40–60 seconds with ice-cold aCSF (artificial cerebrospinal fluid), which contained 128 mM NaCl, 3 mM KCl, 1.25 mM NaH2PO4, 10 mM D-glucose, 24 mM NaHCO3, 2 mM CaCl2, and 2 mM MgCl2 (oxygenated with 95% O2 and 5% CO2, pH 7.4, 295–305 mOsm). Acute brain slices containing NAc MSNs were cut using a microslicer (DTK-1000, Ted Pella) in cold sucrose-aCSF, which was derived by fully replacing NaCl with 254 mM sucrose and saturated by 95% O2 and 5% CO2. Slices were maintained in a holding chamber with aCSF for 1 hour at 37°C. Patch pipettes (3–5 mΩ) for whole cell current-clamp and voltage-clamp recordings were filled with internal solution containing the following: 115 mM potassium gluconate, 20 mM KCl, 1.5 mM MgCl2, 10 mM phosphocreatine, 10 mM HEPES, 2 mM magnesium ATP, and 0.5 mM GTP (pH 7.2, 285 mOsm). Whole cell recordings were carried out using aCSF at 34°C (flow rate = 2.5 ml/min). For all recordings, responses were measured using the Multiclamp 700B amplifier and data acquisition was performed in pClamp 10 (Molecular Devices). Series resistance was monitored during experiments, and membrane currents and voltages were filtered at 3KHz (Bessel filter). To measure the intrinsic membrane properties of NAc MSNs, whole cell recordings were carried out in current clamp mode.
For measurements of neuronal excitability, current was injected in incremental steps (step 50 pA; range 0 to 100 pA). Measurements of input resistance (current-voltage response) were also performed with incremental current steps (steps 25 pA; range 50 to −200 pA). For all recordings, cells were held at −70 mV. To measure the effects of the D1 agonist (Dihydrexidine – DHX; 10uM) on potassium currents in Drd2-Cre G9a KO neurons, recordings were performed in voltage clamp mode (neurons held at −70 mV). To isolate potassium currents during whole-cell recordings, tetrodotoxin (TTX; 1 μM), cadmium chloride (CdCl2; 200 μM), kynurenic acid (1 mM), and picrotoxin (100 μM) were added to the bath aCSF solution in order to block sodium channels, calcium channels, glutamate receptors and GABAA receptors, respectively. Potassium current response to voltage steps (−40 to −130 mV) was measured in the absence and presence of the D1 agonist.
Statistics
For all qPCR analyses, two-way ANOVAs with Bonferroni multiple comparisons (*), or unpaired student’s t-tests (two-tailed) (#), were performed to determine significance. Student’s t-tests were used for other comparisons including behavioral experiments (mutant mice vs. wildtype littermates (*)) and all electrophysiological measurements (*). Microarray analyses were performed as described above. All values included in the figure legends represent mean ± SEM (*/#p ≤ 0.05; **/##p < 0.01, ***p < 0.001, ****p < 0.0001). Detailed statistical analyses are provided throughout the main text.
Sample Sizes
No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications from our laboratory. Furthermore, variance was found to be similar between groups being statistically compared.
Blinding
With the exception of qPCR and microarray analyses, all experiments were conducted with the experimenter blind to genotype.
Randomization
For all molecular, behavioral and electrophysiological experiments, animals were randomly assigned to groups (segregated by genotype). Furthermore, for all molecular experiments, tissue collected from treated animals was randomly pooled to provide ample tissue for biochemical procedures and to minimize variance across cohorts.
Normality Testing
Although normality was not formally tested for every experiment, observations of the data suggest that sample distributions do not deviate significantly from that of a normal distribution curve.
Supplementary Material
Supplementary Table 1: Human demographic data
Supplementary Table 2: List of genes significantly regulated by cocaine, G9a KO, or both in striatal Drd1 neurons (table.D1.pa0.15.xls)
Supplementary Table 3: List of genes significantly regulated by cocaine, G9a KO, or both in striatal Drd2 neurons (table.D2.pa0.15.xls)
Supplementary Table 4: List of genes significantly enriched basally in Drd1 neurons in striatum (Enrich.probesetList.D1.xls)
Supplementary Figure 1: G9a expression is reduced in NAc of human cocaine addicts
Supplementary Figure 2: Regulation of other HMTs by G9a KO in Drd1 and Drd2 MSNs.
Supplementary Table 5: List of genes significantly enriched basally in Drd2 neurons in striatum (Enrich.probesetList.D2.xls)
Supplementary Table 6: List of Drd2 enriched genes displaying significant downregulation in Drd2 neurons in response to cocaine (dat_D2like_CocOnly.xls)
Supplementary Table 7: List of Drd2 enriched genes displaying significant downregulation in Drd2 neurons in response to G9a KO (dat_D2like_ MutOnly.xls)
Supplementary Table 8: List of Drd2 enriched genes displaying significant downregulation in Drd2 neurons in response to cocaine plus G9a KO (dat_D2like_ CocMut.xls)
Supplementary Table 9: List of Drd1 enriched genes displaying significant upregulation in Drd2 neurons in response to cocaine (dat_D1like_CocOnly.xls)
Supplementary Table 10: List of Drd1 enriched genes displaying significant upregulation in Drd2 neurons in response to G9a KO (dat_D1like_ MutOnly.xls)
Supplementary Table 11: List of Drd1 enriched genes displaying significant upregulation in Drd2 neurons in response to cocaine plus G9a KO (dat_D1like_ CocMut.xls)
Supplementary Table 12: mRNA Primers (mRNA Primers.doc)
Acknowledgments
This work was supported by grants from the National Institute on Drug Abuse and National Institute of Mental Health, among others: P01DA08227 and P50MH96890 (E.J.N), MH092306 (D.C., M.H.H.) and J&J/IMHRO (M.H.H.), and DA025962 (A.S.). The following provided additional support for this work: NARSAD Young Investigator Award (18194) (A.S.) and the Seaver Foundation (A.S.).
Footnotes
Contributions
I.M. & E.J.N. conceived of the project. I.M., D.C., Y.L.H., M.H.H., A.S. and E.J.N designed the experiments. I.M., D.C., D.M.D., M.V.S., P.J.K., M.K.L., S.E.D., M.L.M., R.C.B., H.S., L.S., and A.S. collected and analyzed the data. G.T. and R.L.N. provided human tissue and essential viral vector reagents, respectively. I.M., A.S. and E.J.N. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests
Genome Database Accession # GSE54656
References
- 1.Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–98. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- 2.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nature reviews Neuroscience. 2011;12:623–37. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maze I, Nestler EJ. The epigenetic landscape of addiction. Annals of the New York Academy of Sciences. 2011;1216:99–113. doi: 10.1111/j.1749-6632.2010.05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogge GA, Wood MA. The role of histone acetylation in cocaine-induced neural plasticity and behavior. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2013;38:94–110. doi: 10.1038/npp.2012.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maze I, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–6. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Covington HE, 3rd, et al. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron. 2011;71:656–70. doi: 10.1016/j.neuron.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kennedy PJ, et al. Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nature Neuroscience. 2013;16:434–40. doi: 10.1038/nn.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun H, et al. Morphine epigenomically regulates behavior through alterations in histone H3 lysine 9 dimethylation in the nucleus accumbens. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:17454–64. doi: 10.1523/JNEUROSCI.1357-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaefer A, et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64:678–91. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes & development. 2011;25:781–8. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–48. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doyle JP, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–62. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–52. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 14.Lobo MK, Yeh C, Yang XW. Pivotal role of early B-cell factor 1 in development of striatonigral medium spiny neurons in the matrix compartment. Journal of neuroscience research. 2008;86:2134–46. doi: 10.1002/jnr.21666. [DOI] [PubMed] [Google Scholar]
- 15.Podda MV, Riccardi E, D’Ascenzo M, Azzena GB, Grassi C. Dopamine D1-like receptor activation depolarizes medium spiny neurons of the mouse nucleus accumbens by inhibiting inwardly rectifying K+ currents through a cAMP-dependent protein kinase A-independent mechanism. Neuroscience. 2010;167:678–90. doi: 10.1016/j.neuroscience.2010.02.075. [DOI] [PubMed] [Google Scholar]
- 16.Lobo MK, et al. Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science. 2010;330:385–90. doi: 10.1126/science.1188472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Park BH, Lee JH, Park SK, Kim JH. Cell type-specific alterations in the nucleus accumbens by repeated exposures to cocaine. Biological psychiatry. 2011;69:1026–34. doi: 10.1016/j.biopsych.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 18.Hope BT, et al. Induction of a long-lasting AP-1 complex composed of altered Fos-like proteins in brain by chronic cocaine and other chronic treatments. Neuron. 1994;13:1235–44. doi: 10.1016/0896-6273(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 19.Lee KW, et al. Cocaine-induced dendritic spine formation in D1 and D2 dopamine receptor-containing medium spiny neurons in nucleus accumbens. Proc Natl Acad Sci U S A. 2006;103:3399–404. doi: 10.1073/pnas.0511244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nestler EJ. Review. Transcriptional mechanisms of addiction: role of DeltaFosB. Philos Trans R Soc Lond B Biol Sci. 2008;363:3245–55. doi: 10.1098/rstb.2008.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding N, et al. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol Cell. 2008;31:347–59. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gertler TS, Chan CS, Surmeier DJ. Dichotomous anatomical properties of adult striatal medium spiny neurons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2008;28:10814–24. doi: 10.1523/JNEUROSCI.2660-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson TE, Kolb B. Persistent structural modifications in nucleus accumbens and prefrontal cortex neurons produced by previous experience with amphetamine. J Neurosci. 1997;17:8491–7. doi: 10.1523/JNEUROSCI.17-21-08491.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russo SJ, et al. Nuclear factor kappa B signaling regulates neuronal morphology and cocaine reward. J Neurosci. 2009;29:3529–37. doi: 10.1523/JNEUROSCI.6173-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaPlant Q, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–43. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dumitriu D, et al. Subregional, dendritic compartment, and spine subtype specificity in cocaine regulation of dendritic spines in the nucleus accumbens. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:6957–66. doi: 10.1523/JNEUROSCI.5718-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robison AJ, et al. Behavioral and structural responses to chronic cocaine require a feedforward loop involving DeltaFosB and calcium/calmodulin-dependent protein kinase II in the nucleus accumbens shell. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2013;33:4295–307. doi: 10.1523/JNEUROSCI.5192-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lobo MK, et al. DeltaFosB Induction in Striatal Medium Spiny Neuron Subtypes in Response to Chronic Pharmacological, Emotional, and Optogenetic Stimuli. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2013;33:18381–95. doi: 10.1523/JNEUROSCI.1875-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volkow ND, et al. Decreased dopamine D2 receptor availability is associated with reduced frontal metabolism in cocaine abusers. Synapse. 1993;14:169–77. doi: 10.1002/syn.890140210. [DOI] [PubMed] [Google Scholar]
- 30.Thanos PK, Michaelides M, Umegaki H, Volkow ND. D2R DNA transfer into the nucleus accumbens attenuates cocaine self-administration in rats. Synapse. 2008;62:481–6. doi: 10.1002/syn.20523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong S, et al. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2007;27:9817–23. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sampath SC, et al. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell. 2007;27:596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 33.Quirion R, et al. Human brain receptor autoradiography using whole hemisphere sections: a general method that minimizes tissue artefacts. Synapse. 1987;1:446–54. doi: 10.1002/syn.890010508. [DOI] [PubMed] [Google Scholar]
- 34.Tsankova NM, et al. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–25. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 35.Bateup HS, et al. Distinct subclasses of medium spiny neurons differentially regulate striatal motor behaviors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14845–50. doi: 10.1073/pnas.1009874107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Human demographic data
Supplementary Table 2: List of genes significantly regulated by cocaine, G9a KO, or both in striatal Drd1 neurons (table.D1.pa0.15.xls)
Supplementary Table 3: List of genes significantly regulated by cocaine, G9a KO, or both in striatal Drd2 neurons (table.D2.pa0.15.xls)
Supplementary Table 4: List of genes significantly enriched basally in Drd1 neurons in striatum (Enrich.probesetList.D1.xls)
Supplementary Figure 1: G9a expression is reduced in NAc of human cocaine addicts
Supplementary Figure 2: Regulation of other HMTs by G9a KO in Drd1 and Drd2 MSNs.
Supplementary Table 5: List of genes significantly enriched basally in Drd2 neurons in striatum (Enrich.probesetList.D2.xls)
Supplementary Table 6: List of Drd2 enriched genes displaying significant downregulation in Drd2 neurons in response to cocaine (dat_D2like_CocOnly.xls)
Supplementary Table 7: List of Drd2 enriched genes displaying significant downregulation in Drd2 neurons in response to G9a KO (dat_D2like_ MutOnly.xls)
Supplementary Table 8: List of Drd2 enriched genes displaying significant downregulation in Drd2 neurons in response to cocaine plus G9a KO (dat_D2like_ CocMut.xls)
Supplementary Table 9: List of Drd1 enriched genes displaying significant upregulation in Drd2 neurons in response to cocaine (dat_D1like_CocOnly.xls)
Supplementary Table 10: List of Drd1 enriched genes displaying significant upregulation in Drd2 neurons in response to G9a KO (dat_D1like_ MutOnly.xls)
Supplementary Table 11: List of Drd1 enriched genes displaying significant upregulation in Drd2 neurons in response to cocaine plus G9a KO (dat_D1like_ CocMut.xls)
Supplementary Table 12: mRNA Primers (mRNA Primers.doc)