Abstract
Short interfering RNA (siRNA) has been an important laboratory tool in the last two decades and has allowed researchers to better understand the functions of non-protein-coding genes through RNA interference (RNAi). Although RNAi holds great promise for this purpose as well as for treatment of many diseases, efforts at using siRNA have been hampered by the difficulty of safely and effectively introducing it into cells of interest, both in vitro and in vivo. To overcome this challenge, many biomaterials and nanoparticles (NPs) have been developed and optimized for siRNA delivery, often taking cues from the DNA delivery field, although different barriers exist for these two types of molecules. In this review, we discuss general properties of biomaterials and nanoparticles that are necessary for effective nucleic acid delivery. We also discuss specific examples of bioengineered materials, including lipid-based NPs, polymeric NPs, inorganic NPs, and RNA-based NPs, which clearly illustrate the problems and successes in siRNA delivery.
Introduction
Since its discovery in 1998,1 the RNA interference (RNAi) gene silencing pathway has been a focus of many major areas of research. Chief among these are the natural mechanism of viral defense in plants and insects,2–4 the discovery of endogenous protein function by reducing its production,5 and the treatment of diseases that are caused by overproduction of a specific gene.6, 7 The last of these applications in particular requires safe and effective methods for siRNA delivery in order to maximize the clinical potential of RNAi.
The natural RNAi pathway begins with double-stranded RNA (dsRNA) that is cleaved by the protein Dicer into ~21-base pair dsRNA known as short interfering RNA (siRNA). The antisense strand of siRNA is incorporated into the RNA-induced silencing complex (RISC), which binds and subsequently cleaves, in a catalytic fashion, strands of mRNA complementary to the siRNA. This prevents protein production and results in sequence-specific gene knockdown (for review, see Hannon8). Early methods to non-virally induce RNAi included direct injection of dsRNA9–11 or mechanical agitation of cells in the presence of dsRNA.12 However, these methods are not clinically translatable, and introduction of dsRNA longer than 30 base pairs has been shown to induce an interferon (IFN) response.13 As a result, delivery of siRNA to circumvent IFN is more frequently employed.
Although very effective, viral methods for nucleic acid delivery have been associated with immunogenicity and tumorigenicity.14 Non-viral nucleic acid delivery systems are traditionally less effective15 but can be designed to avoid issues typical of viruses. Several types of materials have been used for delivery of nucleic acids and of siRNA in particular. Because many early efforts at siRNA delivery used materials that were already well-studied in the context of DNA delivery, we first discuss properties of these materials that make them suitable for nucleic acid delivery in general and then describe their utility for overcoming barriers to siRNA delivery (Figure 1) in particular.
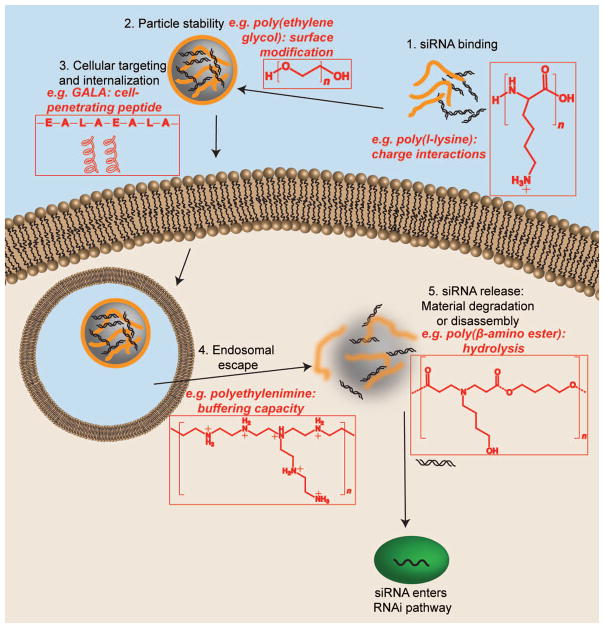
Figure 1.
siRNA faces several barriers during intracellular delivery. Representative biomaterials that are able to overcome these barriers are shown above, along with the particular strategy employed by that material.
General properties of nucleic acid delivery nanoparticles
Nucleic acid binding or encapsulation ability
The biomaterial transfection agent must have sufficient ability to bind to or encapsulate the nucleic acid. Because nucleic acids are negatively charged, positively charged biomaterials are commonly used. Poly(L-lysine) (PLL)’s ability to bind DNA was discovered through studies of DNA-histone binding and conformation,16 and it was later explored for delivery of exogenous DNA.17, 18 PLL, however, lacks the ability to escape the endosome, an essential step in intracellular delivery through the endocytic internalization route.19, 20 Other biomaterials such as liposomes are able to encapsulate nucleic acids within their aqueous interior. These artificial vesicles can be made with tight control over physical properties by changing the amphiphilic lipids that compose them as well as by varying fabrication methods.21–23 Nucleic acid binding or encapsulation is generally a necessary, but not sufficient, biomaterial attribute to prevent degradation of the nucleic acid and to facilitate the entry of such highly negatively charged molecules into the cell.
Shielding
Liposomes, as well as other types of nanoparticles (NPs), often suffer from quick clearance from the circulation by cells of the mononuclear phagocyte system (MPS).24, 25 Chemical structure of specific materials aside, most NPs for nucleic acid delivery rely at least partially on ionic interactions for cellular uptake and carry charge on their surface. Although electrostatic repulsion contributes to NP colloidal stability in aqueous suspension, physiological salt24, 25 and serum26 conditions are often enough to coat the NPs non-specifically with proteins, and cause aggregation, leading to decreased delivery efficiency and increased clearance by MPS cells. One way to overcome this potential problem is to coat the NP surface with a shielding molecule, commonly poly(ethylene glycol) (PEG), whose hydrated structure can prevent non-specific interactions with biomolecules.27 This strategy, dubbed “PEGylation,” has been employed in a number of gene delivery systems, including NPs based on chitosan,28 gelatin,29 cationic polymers,30 lipids,31 and metallic NPs.32
It should be noted that PEGylation has also been found to prevent desirable interactions, such as those leading to intracellular delivery to target cells. As a result, researchers have explored the use of cleavable PEG chains on NP surfaces to increase circulation time but also allow effective transfection.33
Targeting and cellular internalization
A number of methods can be used to promote uptake of nucleic acids by cells of interest. For example, taking advantage of the properties of the lipid bilayer of cell membranes, cationic lipids can facilitate cellular uptake by interacting with anionic cell surface molecules.34–36 Similarly, other cationic materials, such as polymers, also show high uptake into cells compared to neutral or anionic materials due to interactions with the relatively negative cell surface.37 Unfortunately, these interactions with NPs and the cell membrane can also contribute to cytotoxicity.38
Various chemical moieties and biological ligands can also be incorporated into the material or into the NP as an alternative or additional method of increasing cellular internalization. For instance, the amphipathic peptide GALA (30 residue polypeptide containing 4 Glu-Ala-Leu-Ala repeats) forms an alpha-helical structure that promotes interaction with lipid bilayers,39 as does the KALA peptide (30 residue polypeptide containing 3 Lys-Ala-Leu-Ala repeats),40 which has the additional advantage of being positively-charged at neutral or low pH for binding to nucleic acids. The transition from random coil to alpha helix, and therefore the ability to disrupt membranes, is pH-dependent for both of these peptides, making them potentially useful specifically for environmentally triggered membrane fusion.41, 42 Other cell-penetrating peptides (CPPs) used for this purpose include the trans-activating transcriptional activator (TAT) peptide derived from the human immunodeficiency virus (HIV).43, 44 This strategy can be combined with a disassembly approach by synthesizing a high molecular-weight form of TAT peptide, linked with disulfide bridges, that can be degraded by bioreduction to reduce potential cytotoxicity.45
In addition to general, non-specific cellular uptake, it is often desirable to be able to deliver siRNA to a specific cell or tissue type. Ligands such as sugars, aptamers, peptides, and proteins for cell- or tissue-specific cell surface proteins can also be coated on46, 47 or conjugated to48, 49 a biomaterial or NP to enhance uptake and delivery in a targeted manner.
Endosomal Escape
For successful intracellular delivery of nucleic acids, it is critical that that the NP be able to reach the cytoplasm safely and efficiently. A common method of overcoming the endosomal escape barrier is the proton sponge mechanism, in which a reversibly protonated material is believed to act as a buffer. This not only protects the cargo from acidification of the endosomal compartment but also causes a water influx that leads to endosome lysis and release of the cargo into the cytosol. This mechanism was proposed to explain the effectiveness of polyethylenimine (PEI) for gene transfer, as PEI contains reversibly protonated tertiary amines,50 and while its validity has not been incontrovertibly proven and has been challenged,51 it nonetheless remains the most widely believed hypothesis.52, 53 Other materials like PLL can be modified to contain titratable amines, such as by substituting histidine or arginine for lysine residues,54, 55 leading to improved transfection. Other materials like poly(amidoamine) (PAA) dendrimers are also reversibly protonated at physiologically relevant pH.56 However, while these materials increased DNA delivery efficacy, some, like PAAs, bind siRNA less tightly, compromising their ability to act as siRNA delivery agents without additional modifications.57
Other methods for endosomal escape include hydrophobic biomaterials that can fuse with the membrane, such as dioleoylphosphatitylethanolamine (DOPE) in lipid-based particles.58, 59 Polyanionic biomaterials have also been employed to promote membrane destabilization, often in a pH-dependent manner that allows triggered endosomal escape when NPs are in certain environments.60 Membrane-disruptive peptides, such as GALA and KALA, can also be conjugated to or non-covalently associated with NPs to cause endosomal escape.18, 41, 42 KALA in particular maintains α-helical character even at low pH (4.5), thus making it capable of membrane disruption and leakage even in lower pH environments comparable to late endosomes. KALA was shown to cause nearly 100% leakage of endosomal contents over pH range 4.5–8.40
Degradability and nucleic acid release
Release of a NP into the cytoplasm after endosomal escape is not necessarily sufficient for biological effect. It was shown that DNA plasmids must unbind sufficiently from a delivery vehicle for transfection to occur.61, 62 However, while DNA must be trafficked to the nucleus, siRNA must be released in the cytoplasm, its principal site of action.63 A number of factors can affect material degradation and cargo release rate; for example, poly(beta-amino ester)s (PBAE)s are hydrolytically degradable,64 with their degradation and complex disassociation rate dependent on the local pH and the polymer conformation.65 Because hydrolytic degradation of free PBAEs in solution takes place in hours at pH 7 at 37°C, these nanoparticles can diffuse or be convected to cells and release their cargo shortly thereafter. Moreover, the polymers showed slowed degradation at the lower pH (5–6) found in endosomal compartments, which could provide some protection for nucleic acids until after endosomal escape to the cytoplasm.
Aside from hydrolysis, as is characteristic of PBAEs and other polyesters, another mode of degradation useful in siRNA delivery is bioreduction due to disulfide linkages (for review, see Son et al.66). The latter mechanism takes advantage of the reducing cytoplasmic environment to cause quick, environmentally-triggered degradation and siRNA release into the correct cellular compartment.
NPs that do not have a mechanism to stimulate siRNA release may cause low knockdown. Gold NPs, for example, without an siRNA release mechanism may achieve lower knockdown or require higher siRNA doses67 versus similar delivery vehicles that contain an efficient release mechanism.68
Biocompatibility
It is critical that materials administered to a patient be biocompatible. Although cationic lipids have been studied often for nucleic acid delivery, their disruption of the membrane, while an advantage for cellular uptake, can also cause excessive cytotoxicity.69 Biomaterial degradation is one mechanism of increasing biocompatibility. Degradation of a biomaterial can reduce cytotoxicity, as molecular weight of cationic polymers has been positively correlated with cytotoxicity.70 For instance, poly[alpha-(4-aminobutyl)-l-glycolic acid] (PAGA), a hydrolytically degradable PLL analogue containing ester bonds in place of the amide bond of the polypeptide, showed not only increased transfection efficacy but also negligible toxicity in vitro compared to unmodified PLL.71 A hydrolytically degradable form of branched PEI showed a similar increase in biocompatibility72 compared to the typically high cytotoxicity of unmodified PEI.73, 74
Stability
Although many of the above biomaterials have been explored for siRNA delivery, and most continue to be investigated for this purpose, several challenges remain to be overcome. Many of these biomaterials and NPs were initially developed for DNA delivery. Aside from differences in the site of action of these two nucleic acids, there are also biophysical differences between them. Although both are composed of similar anionic bases, siRNA molecules are much smaller in size than DNA plasmids. In addition, dsRNA is stiffer than dsDNA,75, 76 and segments as short as siRNA act as rigid rods. This has important implications for nucleic acid binding properties77, 78: there is less multivalency in siRNA than in DNA when interacting with a cationic polymer because of fewer binding sites per molecule. In addition, the stiff RNA molecule, which is expected to condense very little, may not be able to bend to the conformations ideal for high affinity binding and encapsulation, and this can lead to weak stability of siRNA-containing NPs.
In addition to low binding affinity, instability of the RNA cargo itself is a major problem in many of the delivery systems currently investigated. siRNA is more prone to enzymatic degradation than DNA, though less so than single-stranded RNA. Therefore, protection from the extracellular and endosomal environments is necessary for successful delivery. Stability is a challenge for lipid-based materials as well since lipid-based colloids often exhibit low stability.79
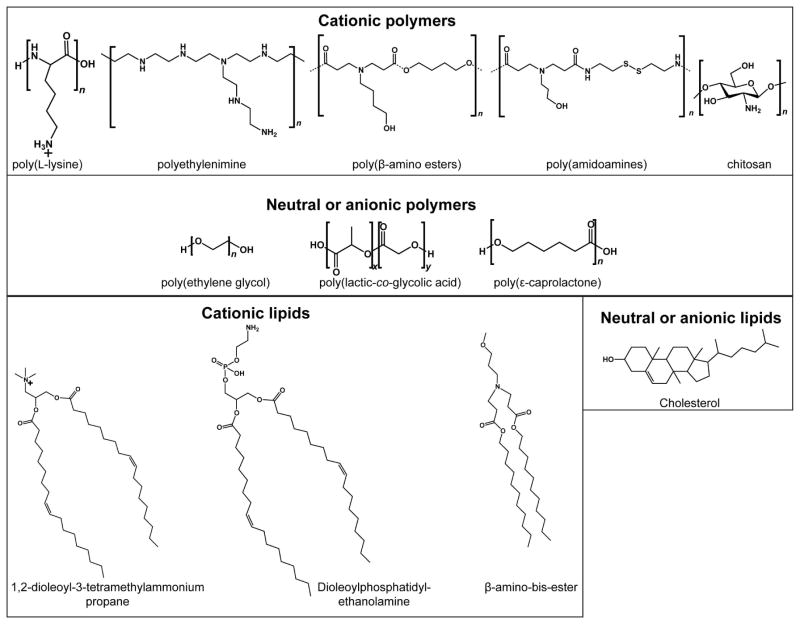
Examples of broad classes of biomaterials used for siRNA delivery are presented in Figure 2. Below and in Table 1, we will discuss illustrative examples of next-generation materials that address many of the challenges listed here, including siRNA binding and protection, particle stability, cellular internalization and targeting, material biocompatibility and efficacy, endosomal escape and intracellular targeting, and siRNA release.
Figure 2.
Examples of classes of polymeric and lipidic biomaterials used for siRNA delivery.
Table 1.
Types of biomaterials used for siRNA delivery and the challenges they pose.
| Biomaterial type | Major Challenges | Strategies to overcome | Examples |
|---|---|---|---|
| Lipid-based nanoparticles | Off-target immunogenicity | Surface-coating with non-immunogenic materials | DOTAP-based nanoparticles surface-modified with hyaluronic acid and PEG84 |
| Toxicity | Low lipid-to- siRNA ratio | Amine-containing lipidoid synthesized from 12-C acrylamides was complexed with siRNA at different ratios to find maximum siRNA loading efficiency87 | |
| Empirical testing of different compounds | Screening of a combinatorial library of synthetic lipidoids and test for toxicity and efficacy86 | ||
| Cellular uptake | Fusogenic lipids | Addition of cholesterol to liposome86–89 | |
| Lipid structure engineered to promote fusogenic phase transition92 | |||
| Other lipid interaction with membrane | Screening of lipid polar headgroups with different size, charge, and acid-dissociation constant94 | ||
| Liposome instability | Surface modification | Incorporation of PEG into liposome formulation86, 87, 93 | |
| Polymeric nanoparticles | Endosomal escape | Proton-sponge effect (buffering capacity) | Reversibly protonated tertiary amines (e.g. PEI,50 PAA,56 PBAE64) |
| Fusogenic peptides | PLL conjugated to pH-sensitive endosomolytic peptide101 | ||
| PLGA-b-PLL nanoparticles non-covalently coated with endosomolytic peptide122 | |||
| Membrane-penetrating peptide KALA crosslinked into an siRNA-binding polymer119 | |||
| siRNA binding | High polymer cationic character | Primary polymer blended or doped with polycation (e.g. chitosan121) | |
| Addition of a small molecule with primary amines57, 105, 111 | |||
| Copolymerization with polycation122 | |||
| Crosslinked particles | Oxidation of thiol-functionalized polymers to form disulfides throughout nanoparticle105, 106 | ||
| Toxicity | Degradation | Hydrolytically-cleavable ester moieties (e.g. PBAEs)7, 115–117 | |
| Bioreducible disulfide bridges57, 108–111, 113, 115, 117, 119 | |||
| Cellular targeting | Tissue-specific ligands | Conjugation of ligand to polymer106 | |
| Non-covalent coating of nanoparticle surface with ligand122 | |||
| Inorganic NPs | Long-term efficacy | Controlled release | Porous silica particles that can be loaded with siRNA-containing NPs140 |
| Toxicity | Gold-based NPs | Gold NPs coated with PEI and siRNA using LbL process67, 68 | |
| Gold NPs functionalized with PEG and siRNA, then coated with PBAEs68 | |||
| Gold NPs functionalized with polyvalent siRNA138 and made into spherical nucleic acids139 | |||
| Particle tracking | Quantum dot- based NPs | Quantum dots covalently modified with siRNA, PEG, and a tumor homing peptide148 | |
| Engineered RNA-based NPs | Particle instability | Multimeric siRNA | siRNA strands with “sticky” overhangs that reversibly concatamerize77, 107, 150, 152 |
| RNAi microsponges containing densely packed, multimeric siRNA157 | |||
| Interferon response | Control over RNA size | Disulfide-linked multimeric siRNA77, 156 | |
| siRNA strands with “sticky” overhangs77, 156 | |||
| Loss of siRNA biological activity | Labile, long, or crosslinked linkages | Disulfide linkages107, 150, 152 or long, non-labile linkages150 | |
| Polyvalent nucleic acid nanostructures159 |
Lipid-based nanoparticles
Lipid-based materials are the most widely-used biomaterials for nanoparticulate siRNA delivery. Of over 20 siRNA phase I clinical trials, nearly half use NPs as the delivery vehicle, and almost all of these are lipid-based.80 Many of the leading commercially available reagents are lipid-based, including Lipofectamine™ 2000,81 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP),82 RNAifect,82 and TransIT-TKO and TransIT-siQuest.83 It has become clear, however, that careful controls are necessary for the successful interpretation of results, as siRNA in combination with cationic lipids can cause off-target effects. For example, an in vivo study in mice showed that intravenous injection of naked siRNA had no measurable effect; however, injection of liposomal siRNA NPs based on 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP), a commonly used cationic lipid for nucleic acid delivery, induced a potent IFN response, including the upregulation of downstream molecule STAT1 (signal transducer and activator of transcription 1).82 Although the greatest effect was seen with DOTAP/siRNA complexes, including several different siRNA sequences, DOTAP alone did cause an IFN and STAT1 response, emphasizing the importance of careful design of the material used for lipid-based NPs to avoid potentially adverse side effects as well as careful interpretation of results. Some researchers have focused on overcoming this problem in particular; for example, Chono et al.84 used the additional components of hyaluronic acid, a polysaccharide with low immunogenicity, PEG, and a targeting ligand, and found that their liposome elicited no significant immunotoxicity, measured by cytokine expression, within their therapeutic range. Other strategies have been employed to reduce the off-target immunological response to siRNAs, particularly when delivered within liposomes; for instance, 2′-O-methyl-modified siRNAs were designed and delivered in stable nucleic acid lipid particles (SNALPs)85 with less unwanted immune stimulation observed.
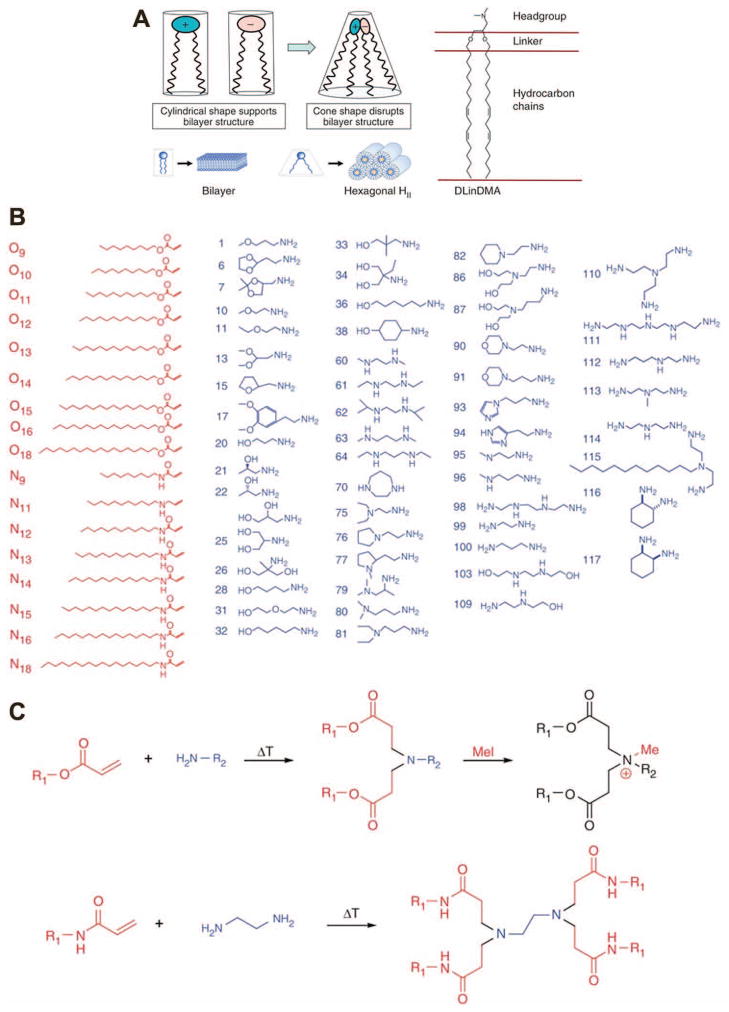
To bypass problematic toxicity and particle instability and to maximize siRNA delivery and gene knockdown, several strategies can be employed. Akinc et al. used a combinatorial library to link acrylate esters or acrylamides containing 9- to 18-carbon hydrocarbon tails via one of many different amine-containing small molecules,86 thereby creating a wide range of lipidoid materials with slightly varying structure (Figure 3). These lipidoid molecules were mixed with cholesterol and a PEG-lipid (polyethylene glycol conjugated to a lipid moiety for incorporation into liposomes via hydrophobic interactions) and loaded with siRNA. Using this method, the authors optimized nanocomplex stability and efficacy in vitro, then used top materials to achieve >90% knockdown in vivo after two daily intravenous injections of 2 mg siRNA/kg mouse. A follow-up study showed increased efficacy of their material to nearly 100% when the siRNA dose was increased to 10 mg/kg, though high toxicity associated with cationic lipids necessitated a lower lipid concentration to be tolerable. The authors improved the biocompatibility by maximizing siRNA loading, thereby reducing the total lipid content delivered, and optimized the PEG chain length to improve in vivo particle stability.87
Figure 3.
Various approaches have been taken to lipid-based siRNA delivery. While some groups use rational design and focus on specific delivery aspects, such as lipid polymorphism leading to membrane fusion (A), others have employed high-throughput methods to screen through a wide array of different molecules to empirically determine the best structures (B–C). Reprinted by permission from Macmillan Publishers Ltd: Nature Biotechnology,86, 94 copyright (2010) (A) and copyright (2008) (B–C).
Lipid material optimization and selection has also been done to examine very specific aspects of transfection. For example, one reason for the use of cholesterol in liposome formulations is to increase fusion with the cell membrane for internalization,88, 89 in addition to affecting the fluidity or rigidity of the liposome bilayer. Other lipids have also been found to promote fusion of the liposome with the endosomal membrane because their structure requires less energy to transition to a non-lamellar phase, forming tubular structures with hydrophobic tails exposed and promoting siRNA release into the cytoplasm.90, 91 While many groups have incorporated fusogenic lipids into their NP formulations and some trends had been implicated in increasing fusogenicity, Heyes et al. specifically studied the effect of saturation and chain length on fusogenic phase transition as well as on transfection efficacy.92 They and others have investigated siRNA delivery from SNALPs by selecting the best fusogenic lipid for the endosomal escape step. Semple et al. used in vivo screening in mice to develop siRNA-containing SNALPs that could be administered i.v. to non-human primates and cause 80% knockdown of a target gene.93
The in vivo studies described, while showing high efficacy, required ~1 mg/kg of siRNA to achieve knockdown. By using a similar combinatorial method to that mentioned above, Love et al. identified a linker molecule, N-(2-(4-(2-aminoethyl)piperazin-1-yl)ethyl)ethane-1,2-diamine, that, after reaction with the 12-carbon epoxide 1,2-epoxydodecane, formed lipid-based NPs that were primarily taken up by macropinocytosis rather than endocytosis, as with their previously-studied materials.95 They were able to achieve one to two orders of magnitude better efficiency (0.03–0.3 mg/kg dose) in non-human primates with up to ~75% knockdown at the lowest dose, possibly because their internalization route allowed their particles to avoid the endosome. The authors note that lower siRNA doses, while causing the same initial effect, had lower knockdown duration compared to high siRNA doses. This high nucleic acid efficiency allowed the authors to reduce the total amount of lipidoid material needed for therapeutic benefit, lowering potential toxicity, and also allowed them to knock down several genes in a single combined dose.
In addition to nanomaterial composition, siRNA delivery efficacy in vivo can also depend heavily on the route of administration. Leconet et al., using commercially available reagents (the lipid-based Invivofectamine 2.0 and the polymeric JET-PEI), achieved transient knockdown in diabetic mice, with a single injection of siRNA/lipid-based carrier able to knock down a target gene for at least seven days in their system.96 In particular, they found that pancreatic immune cells were transfected with much greater efficacy after intraperitoneal injection compared with intravenous injection. All of these various factors, therefore--specific siRNA sequence and chemical modification, material composition, dose of siRNA and/or material, additives into the liposome, route of cellular entry, and route of in vivo administration--can each have enormous effects on RNAi mediated by lipid-based nanocarriers.
The ability of lipid-based materials to stably encapsulate siRNA has led to their prolific use in ongoing clinical trials relative to other siRNA delivery technologies (for review, see Forbes and Peppas80). In particular, the decreased immunogenicity and enhanced in vivo stability and shielding promoted by the PEG layers surrounding SNALPs has enabled multiple clinical trials involving SNALPs. Drug TKM-080301 sponsored by Tekmira Pharmaceuticals Corporation and Alnylam Pharmaceuticals is a SNALP containing siRNA targeting polo-like kinase-1 (PLK-1). This drug has completed a Phase I trial administered via hepatic arterial infusion for primary or secondary liver cancer, and is currently undergoing an additional Phase I trial targeting solid tumors via intravenous infusion.97, 98 SNALP-based siRNA drug ALN-TTR01, also sponsored by Alnylam Pharmaceuticals, has completed a Phase I trial using siRNA targeting transthyretin in an effort to treat Transthyretin-Mediated Amyloidosis.99 Cationic lipid nanoparticle-based siRNA drug Atu027 sponsored by Silence Therapeutics AG has completed a dose-escalation, Phase I clinical trial to assess toxicity.100 These and other lipid-based siRNA delivery technologies represent the class of material nearest to reaching the clinic for siRNA delivery.
Polymeric nanoparticles
Although lipid-based nanoparticles have been historically more widely used for nucleic acid delivery, polymer-based nanoparticles are very promising newer class of materials for the delivery of nucleic acids and are an active subject of ongoing research. Because many of the polymers used for siRNA delivery were originally investigated as DNA delivery materials, many of them have gone through iterations of modification as various hurdles specific to siRNA delivery were discovered. Key classes of materials are described below, along with examples of the bottlenecks encountered and the strategies used to sidestep them.
Poly(l-lysine)- and polyethylenimine-based materials
PLL and PEI, as mentioned above, were early candidate materials for siRNA delivery, with PEI still used frequently in research today. Derivatives of both polymers continue to be developed and optimized and are the basis of many of the polymeric systems being studied for siRNA delivery. Because PLL/nucleic acid complexes can be taken up by cells but cannot efficiently escape the endosome, Meyer et al. developed PLL- and PEI- based materials by conjugating the polycation to an endosomolytic mellitin peptide sequence modified with pH-labile protecting groups (dimethylmaleic anhydride), which restrict the lytic activity of the peptide until they are cleaved at acidic pH.101 However, because the peptide was negatively charged and was necessary in high number for effective endosomal escape, this covalent modification destabilized the polymer/siRNA complex, precluding the formation of sufficiently small NPs for cell uptake. As a solution for this, the authors grafted PEG to the polycation before modification with the peptide, which increased the stability of the complex and caused gene knockdown in vitro in human neuroblastoma cells.
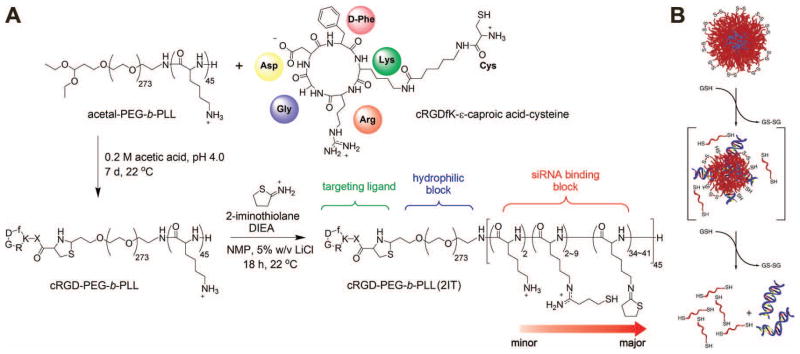
Miyata et al. used a previously-developed102, 103 PEG-b-PLL copolymer to deliver nucleic acids.104 By modifying some of the lysine sidechains with thiol-containing functional groups, they could form micelles via electrostatic interactions with anionic DNA and then oxidize the complexes, forming disulfide bridges to stabilize the particles.104 This method was also effective for in vitro siRNA delivery to Huh7 human hepatoma cells, with ~80% knockdown seen after delivery of 100 nM siRNA to Huh7 cells in the presence of serum. Because of weaker binding seen in siRNA polyplexes compared with DNA polyplexes, the authors further modified their polymer with 2-iminothiolane to increase the cationic character and thus strengthen siRNA binding.105 Because of the difference in redox potential between the cytoplasm and extracellular environment, this system carries with it the advantage of quick siRNA release once in the desired cellular compartment. Further improvements on this system included the conjugation of cyclic RGD peptide to the PEG block, thereby providing a method to target tissues in vivo (Figure 4a).106 These siRNA-containing micelles accumulated in tumor tissue and the surrounding vasculature in a subcutaneous HeLa tumor model. Delivery of 24 μg siRNA per mouse (~0.5–1 mg/kg) against vascular endothelial growth factor (VEGF) and VEGF receptor 2 (VEGFR-2) two times every four days for 12 days showed ~50% decreased in measured VEGF mRNA levels in the tumor and significantly slowed tumor growth (~60–70% smaller tumor volume after 12 days compared to control) without apparent toxic side effects on the animal. Similar strategies have also been employed with PEI, using PEGylated siRNA against VEGF to form PEI-based micelles that caused VEGF knockdown in an in vivo PC-3 prostate cancer model.107
Figure 4.
Widely-used approaches to improving polymeric siRNA delivery include conjugation to PEG and targeting ligands (A) and introduction of disulfide bridges for controlled cytoplasmic release (B). Part A reprinted with permission from Christie, R. J.; Matsumoto, Y.; Miyata, K.; Nomoto, T.; Fukushima, S.; Osada, K.; Halnaut, J.; Pittella, F.; Kim, H. J.; Nishiyama, N.; Kataoka, K.: Targeted Polymeric Micelles for siRNA Treatment of Experimental Cancer by Intravenous Injection. ACS Nano 2012, 6, 5174–5189.106 Copyright (2012) American Chemical Society. Part B reprinted from Journal of Controlled Release, 150/2, van der Aa et al, Optimization of poly(amido amine)s as vectors for siRNA delivery, 177–186,112 Copyright (2011), with permission from Elsevier.
With toxicity as a major problem limiting the use of PEI, forms of degradable PEI have been studied extensively for siRNA as well as DNA delivery. Breunig et al. linked low-molecular weight linear PEI segments with disulfide bridges to form a bioreducible PEI.108 Combining the efficacy of high-molecular PEI with the biocompatibility of shorter polymer chains, they delivered 100 nM siRNA in vitro to Chinese hamster ovary cells (CHO-K1) in serum-free medium and were able to achieve in vitro RNAi of green fluorescent protein (GFP) expression (~50% knockdown) that was comparable in efficacy to that of high-molecular weight branched PEI (bPEI) while being less cytotoxic. This design carried with it the additional benefit of targeted siRNA release into the cytoplasm, as disulfide bridges would be expected to be reduced to thiols.
Other classes of polymers
Also capitalizing on bioreducible linkages for targeted release, Jeong et al. described the use of a bioreducible PAA, designated SS-PAEI [poly(amido ethylenimine)], for siRNA delivery.109 Having found previously that SS-PAEI had higher buffering capacity than PEI,110 they further demonstrated that their bioreducible polymer could cause significant knockdown of vascular endothelial growth factor (VEGF) expression from human prostate cancer cells in vitro with 30 nM siRNA delivered in serum-free medium. Their polymer was more effective (up to ~80% less mRNA expression) and less toxic than the linear PEI used as a comparison. Because there was not a significant increase in cellular uptake of SS-PAEI/siRNA particles compared to PEI/siRNA, the authors concluded that intracellular events, likely the disulfide reduction step, was the main reason for the increased efficacy. The siRNA binding efficiency of SS-PAEI is lower than that of PEI; therefore, SS-PAEI was copolymerized with a polymer block containing ethylene diamine.57, 111 The increased amine content and therefore increased positive charge caused tighter siRNA binding and, at optimized ratios, better in vitro knockdown efficiency in human head and neck carcinoma cells (UM-SCC-14C) and non-small-cell lung carcinoma (H1299).
Further optimization of this system included PEGylation by adding PEG-amine into the SS-PAEI synthesis, then mixing PEG-containing SS-PAEI with PEG-free SS-PAEI in order to improve particle stability in the presence of blood components like serum.113 However, while particles with higher PEG content were more stable against aggregation in serum or salt, they were also more easily dissociated in the presence of heparin, suggesting less stable binding interactions between siRNA and highly PEGylated polymers. Toxicity in erythrocytes as well as H1299 cells decreased with increasing PEG content, although this came at the cost of strongly compromised knockdown efficacy as well.
Similar to PAAs are poly(ester amine)s (PEAs) or poly(beta-amino ester)s (PBAEs), which are hydrolytically degradable through their ester groups. Initially studied primarily for DNA delivery, some early attempts to use PBAEs for siRNA delivery fell short without additionally conjugating siRNA to solid particles.114 As with other cationic polymers, this class of materials faced obstacles in polymer-siRNA binding efficiency and intracellular release. By increasing the ratio of PBAE to siRNA (100:1 or greater PBAE:nucleic acid ratio by weight, compared to ~50:1 for DNA delivery), better particle formation and siRNA complexation was achieved, resulting in successful knockdown (~70%) in human umbilical vein endothelial cells when 60 nM siRNA was delivered in medium with 2% serum.115
Other groups were able to show ~70% in vitro knockdown in hepatoma cells in serum-free medium with lower PBAE:siRNA weight ratios when using PBAEs of similar chemical structure.116 Interestingly, the PBAE molecular weight reported was higher than that of the previous two studies cited and this may have enabled the increased efficacy at lower PBAE:siRNA weight ratios. Trends in PBAE structure with siRNA delivery efficacy have since been further examined,65 confirming the general trend of increasing knockdown efficacy with increasing PBAE molecular weight, as well as other polymer properties like hydrophobicity. This study also showed that the siRNA dose could be decreased by a factor of twelve (as low as 5 nM) without negatively affecting the knockdown efficacy (~90% knockdown in serum-free medium) as long as the total polymer concentration was not altered. Transfection in 10% serum-containing medium was lower (~70%), which the authors believed was due to some destabilization of the nanoparticles in the presence of serum proteins. In particular, this was observed with the top PBAEs in the study, most of which contained a primary amine terminal functional group that could be cleaved in a reducing environment. By utilizing disulfide bonds, this study and others117, 118 have found bioreducible linkages can be a key attribute for successful non-viral siRNA delivery. In another study, poly(ethylene oxide) (PEO)-b-PBAE NPs were made for siRNA delivery in combination with PEO-b-poly(ε-caprolactone) (PCL) NPs loaded with paclitaxel.7 This study showed successful knockdown of MDR-1, a cause of multiple drug resistance in cancer cells, and consequently increased efficacy of a chemotherapeutic in vitro in SKOV3 ovarian cancer cells.
The combination of PEGylation with disulfide linkages has also been applied to polypeptides, including KALA.119 Mok et al. conjugated PEG to siRNA via a disulfide bridge. Cysteine-containing KALA was then self-crosslinked to form a high-molecular weight, but bioreducible polymer, which electrostatically complexed with siRNA-PEG. This system was able to achieve nearly 50% gene knockdown in MDA-MB-435 melanoma cells, cultured in vitro in the presence of 10% serum with siRNA concentration of ~60 nM.
Woodrow et al. had an interesting finding while investigating the effectiveness of poly(lactic-co-glycolic acid) (PLGA) NPs at siRNA-mediated silencing in vivo.120 Although a positively-charged component, spermidine, was included in the encapsulation formulation, it was used as an excipient rather than as a primary delivery agent. The PLGA/siRNA NPs themselves were small enough for cellular internalization, and siRNA released over time from the particles could be isolated and delivered separately without loss of function. Using these particles, the authors delivered siRNA across the vaginal mucosal barrier and effected knockdown in cells in the vaginal tract. A major advantage of this system is the biocompatibility of PLGA and related polyesters, which have been used in a number of FDA-approved devices. In addition, being solid particles, this delivery system does not suffer from unstable binding interactions as do many polymeric electrostatic complexes. Researchers are interested in adding cationic materials, such as chitosan or the above spermidine, as part of PLGA-based NPs to increase siRNA loading into anionic PLGA NPs and increase knockdown efficacy.121
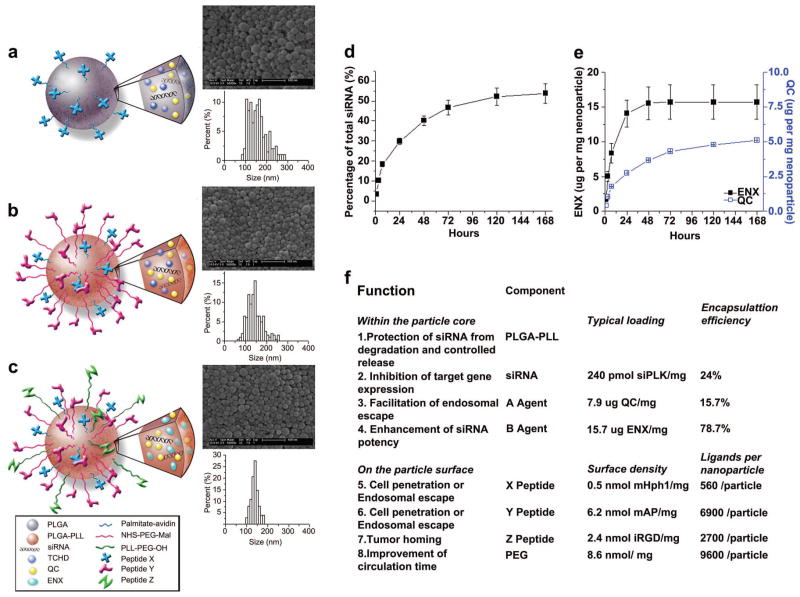
A study by Zhou et al.122 provides an example of how several components can be combined in a NP design to incorporate many different functions and properties. The authors used as its base material a PEG-grafted PLGA-b-PLL block copolymer previously developed for fabrication of surface-functionalized NPs.123, 124 DNA or siRNA was loaded into the NPs during synthesis, along with palmitoyl-avidin, allowing biotinylated peptides to be conjugated to the NP surface. In this way, the authors were able to design NPs containing polycations for nucleic acid complexation; PLGA as a biocompatible matrix; PEG to improve stability and circulation time; two encapsulated drugs to enhance delivery efficacy and siRNA processing; and three peptides to promote cellular uptake, endosomal escape, and tissue homing (Figure 5). While this type of drug delivery vehicle is more complex than some of the other examples described here, it provides flexibility in formulation and design details and was effective in vitro for ~91% knockdown in A549 lung cancer cells in culture medium.
Figure 5.
PLGA-b-PLL-g-PEG NPs contain siRNA, two drugs, and three ligands for targeting, cell penetration, and trafficking. Reprinted from Biomaterials, 33/2, Zhou et al, Octa-functional PLGA nanoparticles for targeted and efficient siRNA delivery to tumors, 177–186,122 Copyright (2012), with permission from Elsevier.
Other elegant polymer designs for siRNA delivery, including a multifunctional, cyclodextrin-based vehicle, have had successful siRNA delivery preclinical studies and have moved forward to clinical testing.125 This formulation, denoted CALAA-01, is a self-assembled complex of siRNA with a multi-component delivery vehicle, consisting of (i) a cyclodextrin-containing polymer, which also contains positive charges for siRNA binding; (ii) adamantane-conjugated PEG, with PEG added for particle stability and adamantine used to bind to cyclodextrin; and (iii) an adamantine-PEG-transferrin conjugate for targeting. This polymeric siRNA formulation, designed for good in vivo stability and site-specific delivery, is currently in a phase I trial.
Inorganic NPs
Calcium Phosphate
Calcium phosphate (CaP) crystals were originally developed for DNA delivery and were fabricated by highly saturated solutions of DNA and CaP that resulted in spontaneous coprecipitation.126–128 In an effort to limit the rapid and often difficult-to-control growth of CaP crystals, Kakisawa et al. developed an inorganic-organic NP hybrid for DNA delivery in which polyaspartate segments of polyaspartate-PEG block copolymers could adsorb onto the expanding crystal surface, thereby limiting continued growth.129 The PEG segments impart biocompatibility and particle stability.
This NP formulation was eventually used for in vitro siRNA delivery to human embryonic kidney 293 cells and was able to achieve roughly 50% gene knockdown using a 125 nM siRNA dose in serum-containing media.130 However, these particles required chloroquine for efficient endosomal escape, so the same researchers replaced polyaspartate with poly(methacrylic acid) (PMA), as PMA becomes more hydrophobic in pH 4–6 and could disrupt the endosomal membrane. To assess the siRNA delivery of this CaP-PMA-PEG system to 293 cells in a more physiologically relevant way in vitro, the group incubated all siRNA transfection agents in serum-containing media for 30 min prior to transfection. They were able to show significantly lower toxicity than Lipofectamine™ 2000 and a significantly higher gene knockdown of 65% versus RNAifect™ at 42%.130
siRNA loading into CaP based NPs was improved five-fold versus previously described systems by Zhang et al. via the covalent linkage of PEG to siRNA before precipitation with CaP. These NPs achieved roughly 60% gene knockdown in HeLa cells with 100 nM siRNA in serum-containing media.131
Gold
Gold-based nanoparticles (AuNPs) are attractive drug delivery candidates due to their low cytotoxicity,132 easily tunable physical properties,133 and readily functionalizable surface chemistry.134–136 Elbakry et al. used AuNPs and the ionic layer-by-layer (LbL)137 surface modification process for siRNA delivery. These researchers took advantage of sulfur-gold binding to coat AuNPs with 11-mercaptoundecanoic acid (MUA), then used electrostatic interactions to coat the particles with a layer of PEI, followed by siRNA and another layer of PEI. In vitro delivery to CHO-K1 cells using 290 nM siRNA in serum-containing media resulted in approximately 80% knockdown.67 However, this particular NP formulation required a high siRNA dose most likely because it included no siRNA release mechanism. Cytoplasmically-targeted siRNA release was achieved by Lee et al. in a AuNP formulation in which siRNA was attached to AuNPs via a PEG linker and bioreducible disulfide bonds, then coated with PBAEs. This formulation achieved near-complete gene knockdown in HeLa cells when a 90 nM dose of siRNA was delivered in serum-containing media.68 Giljohann et al. have also demonstrated transfection agent-free delivery of siRNA through the use of AuNPs densely functionalized with RNA oligonucleotides.138 In this study, ~100 nM of RNA duplex (3 nM AuNPs) caused 73% knockdown in HeLa cells. In an intriguing recent development, spherical nucleic acids, consisting of 13 nm AuNP cores and 30 siRNAs per particle densely functionalized around the surfaces, were able to penetrate skin and cause functional knockdown in hairless mice following topical treatment in vivo.139
Silica
As NP-based siRNA therapeutics are developed for clinical use, an important obstacle will be to create optimal dosing regimens, ideally those that minimize dosing frequency. Multistage release vectors were developed to address this issue by using nanosized lipid-based siRNA carriers loaded into micron sized, porous silicon particles.140 In an in vivo study of orthotopic mouse ovarian carcinoma, this systemic delivery system extended gene knockdown from a few days to more than three weeks and effectively reduced angiogenesis, cell proliferation, and tumor burden. Another formulation was developed in which porous silicon nanoparticles were loaded with the chemotherapeutic doxorubicin and siRNA targeting a drug efflux transporter, and then coated with siRNA. In vitro delivery of these particles to multidrug-resistant KB-V1 cells showed knockdown of the drug transporter, resulting in increased intracellular and intranuclear doxorubicin levels.141
Quantum Dots
Quantum dots (QDs) are inorganic semiconductor nanoparticles and can be used as fluorophores. QDs with easily tunable emission properties, such as CdSe/ZnS nanoparticles, are promising candidates for imaging purposes in that they are brighter,142, 143 less susceptible to photobleaching,144 are easier to detect amongst in vivo background fluorescence versus most fluorescent dyes,142 and are able to be made biocompatible enough to be used with cells.145, 146 Chen et al. first codelivered siRNA and QDs in order to trace the delivered siRNA in vitro.147 This concept was extended to make QDs part of the delivery system itself by Derfus et al., in which QDs were covalently modified with PEG, siRNA, and a tumor homing peptide (F3) to deliver siRNA to HeLa cells.148 They were able to show that cell uptake was facilitated by F3 conjugation, and the NPs achieved approximately 30% gene knockdown in serum conditions with a 50 nM dose of siRNA. Although the NPs could not improve upon the knockdown achieved by Lipofectamine™ 2000, this delivery system showed an exciting proof-of-concept for a method to deliver and specifically track siRNA molecules using QDs.
Engineered RNA-based NPs
As gene knockdown via direct delivery of siRNA is a transient, dose-dependent process, siRNA loading into the delivery system is critical for complete and long-lasting gene suppression. Delivery systems in which siRNA is conjugated directly to a biomaterial or in which engineered RNA is the delivery material itself present exciting strategies to maximize loading. An example of increased loading was examined earlier in this review when siRNA was directly conjugated to PEG and coprecipitated with CaP to improve siRNA loading five-fold.131 Additionally, particle stability can be enhanced with the use of multimeric siRNAs, a class of material created by either covalently or noncovalently linking siRNA molecules. These multimeric siRNAs promote multivalent biomaterial-nucleic acid interactions similar to those seen with plasmid DNA, resulting in improved NP stability with RNA. Strategies for modifying siRNA while maintaining its biological activity are discussed below.
siRNA Conjugates
Important considerations for chemically modifying siRNA include assuring that the modification will not interfere with RNAi,149 and, for modifications where multiple siRNAs are attached to each other, that the long chain of RNA will not induce an IFN response.13 Singh et al. examined the former issue using the PEGylated siRNA-QD conjugates described earlier in this review. The authors found that decreasing the length of the siRNA-QD linker resulted in less RNAi, while lengthening this linker promoted more RNAi. They also found that the site of conjugation on the siRNA molecule, 5′ versus 3′, sense versus antisense, did not seem to effect RNAi.150 These results confirmed what an earlier study had concluded with conjugation to magnetic NPs.151
Another investigation into PEG-siRNA conjugation utilized a disulfide linker and formed NPs using PEG-siRNA and PEI. These NPs were able to achieve 80% and 96.5% gene knockdown in serum and serum-free conditions, respectively, in human prostate carcinoma PC-3 cells using a 100 nM siRNA dose.152 The same material in an in vivo study using subcutaneous PC-3 tumors and IV injection of NPs with 1.5 nmol VEGF-targeting siRNA showed 86 ± 4% decreased VEGF expression, resulting in a 78 ± 9% reduction in microvessel formation and a ten-fold decrease in tumor volume.107
An alternative siRNA conjugation strategy involves conjugating the 3′ end of the sense strand of siRNA to cholesterol. Such a system can cause 50% knockdown in HeLa cells with a ~200 nM siRNA dose without using any additional transfection agent or nanoparticle.153 Cholesterol-siRNA, but not unmodified siRNA, has improved pharmacokinetic properties and can mediate knockdown in mice following i.v. administration. Other lipophilic modifications to siRNA can also enhance in vivo knockdown.154
Multimeric siRNA
While most strategies focus on tuning vector material properties to make DNA delivery vehicles work for siRNA delivery, another approach is to adjust the siRNA itself to make it physically more like DNA. Multimeric siRNA allows for increased multivalent interactions in addition to lending the geometric flexibility that is favorable for higher affinity binding. An interesting method to form multimeric siRNA was introduced by Bolcato-Bellemin et al. in which they synthesized siRNA strands with 5–8 bp overhangs to yield “sticky siRNA ends” that would reversibly concatemerize to form long repeats of siRNA.155 When delivered with linear PEI to A549 human lung carcinoma cells in serum-free conditions, these NPs achieved 80% gene knockdown with 50 nM siRNA and 70% knockdown with as little as 20 nM siRNA. The authors also showed that this material triggered no IFN response. This concept was later expanded upon by covalently linking siRNA strands with either a bioreducible or non-degrading linkage.156 When complexed with linear PEI, both the cleavable and non-cleavable siRNA NPs achieved near complete gene knockdown in PC3 cells with a 90 nM siRNA dose in serum-free conditions. However, when using rapid amplification of cDNA ends to look for the specific cleaved target mRNA, the authors only found a significant concentration of the expected fragment in the samples treated with the disulfide-linked siRNA.
A unique method to create an siRNA-based material was recently suggested by Lee et al.157 The group employed rolling circle transcription158 to make connected repeats of hairpin RNAs, which then crystallized into growing sheets to eventually form microsponges. From these microsponges, micron-sized, spherical particles could be pinched off and coated with bPEI to compact them into roughly 200 nm sized particles. These particles achieved such incredibly high siRNA loading that the authors were able to show comparable knockdown to commercially available siRNA delivery systems using three orders of magnitude fewer particles. In vitro delivery of these particles to T22 cells achieved nearly 60% knockdown with 100 nM siRNA, and an in vivo intratumoral injection to T22 cells showed significant gene knockdown at 4 days post-transfection.
An additional class of three-dimensional nucleic acid-only structures for RNA delivery are polyvalent nucleic acid nanostructures. These have been synthesized by Chad Mirkin and his research group by constructing intra-crosslinked spherical nucleic acids with an inorganic nanoparticle core and then subsequent dissolution of this core.159 These polyvalent nucleic acid nanostructures share many of the characteristics of spherical nucleic acids including being able to cause comparable efficient gene knockdown.
Conclusion and outlook
Despite its clinical potential, use of siRNA as a therapeutic has been hampered by a lack of effective and safe methods of delivery. Many different materials have been explored in the laboratory, with some, mostly lipid-based, having been translated to the clinic in early-phase trials. It is important to note, however, that both the siRNA molecule itself and also the delivery vehicle can have off-target effects that may confound results and affect the translation of this technology.
As each biomaterial is developed, challenges in siRNA delivery are illuminated, aiding in the design of improved carrier NPs. New biomaterials have now been developed to fit the chemistry, biophysical structure, and biological function of siRNA. Many research groups are exploiting the benefits of increased siRNA binding affinity, for example, by increasing the positive charge of a biomaterial or by synthesizing multimeric siRNA molecules, to improve stability and delivery efficacy. Similarly, other researchers are investigating triggered release properties, such as disulfide bonds to cause siRNA release upon entering the cytoplasm, to improve delivery efficacy and reduce cytotoxicity. These next-generation materials have shown promise in the laboratory both in vitro and in vivo. As better ways for siRNA delivery are discovered, the wide range of therapeutic benefits to using siRNA will more fully come to light.
Acknowledgments
This work was supported in part by the NIH (1R01EB016721). KLK thanks the NIH Cancer Nanotechnology Training Center (R25CA153952) at the JHU Institute for Nanobiotechnology for fellowship support and SYT thanks the National Science Foundation for fellowship support.
References
- 1.Fire A, Xu SQ, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez-Vargas I, Travanty EA, Keene KM, Franz AWE, Beaty BJ, Blair CD, Olson KE. RNA interference, arthropod-borne viruses, and mosquitoes. Virus Res. 2004;102:65–74. doi: 10.1016/j.virusres.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 4.Voinnet O. Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- 5.Kuwabara PE, Coulson A. RNAi--prospects for a general technique for determining gene function. Parasitol Today. 2000;16:347–349. doi: 10.1016/s0169-4758(00)01677-x. [DOI] [PubMed] [Google Scholar]
- 6.Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: Current status and prospective. Int J Cancer. 2007;120:953–960. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- 7.Yadav S, van Vlerken LE, Little SR, Amiji MM. Evaluations of combination MDR-1 gene silencing and paclitaxel administration in biodegradable polymeric nanoparticle formulations to overcome multidrug resistance in cancer cells. Cancer Chemother Pharmacol. 2009;63:711–722. doi: 10.1007/s00280-008-0790-y. [DOI] [PubMed] [Google Scholar]
- 8.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 9.Boxem M, Srinivasan DG, van den Heuvel S. The Caenorhabditis elegans gene ncc-1 encodes a cdc2-related kinase required for M phase in meiotic and mitotic cell divisions, but not for S phase. Development. 1999;126:2227–2239. doi: 10.1242/dev.126.10.2227. [DOI] [PubMed] [Google Scholar]
- 10.Longman D, Johnstone IL, Caceres JF. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 2000;19:1625–1637. doi: 10.1093/emboj/19.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Svoboda P, Stain P, Hayashi H, Schultz RM. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development. 2000;127:4147–4156. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- 12.Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stark GR, Kerr IM, Williams BRG, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 14.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 15.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 16.Li HJ, Chang C, Weiskopf M. Helix-Coil Transition in Nucleoprotein-Chromatin Structure. Biochemistry. 1973;12:1763–1772. doi: 10.1021/bi00733a016. [DOI] [PubMed] [Google Scholar]
- 17.Wu GY, Wu CH. Receptor-mediated in vitro gene transformation by a soluble DNA carrier system. J Biol Chem. 1987;262:4429–4432. [PubMed] [Google Scholar]
- 18.Wagner E, Plank C, Zatloukal K, Cotten M, Birnstiel ML. Influenza-Virus Hemagglutinin-Ha-2 N-Terminal Fusogenic Peptides Augment Gene-Transfer by Transferrin Polylysine DNA Complexes - toward a Synthetic Virus-Like Gene-Transfer Vehicle. Proc Natl Acad Sci. 1992;89:7934–7938. doi: 10.1073/pnas.89.17.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curiel DT, Agarwal S, Wagner E, Cotten M. Adenovirus Enhancement of Transferrin Polylysine-Mediated Gene Delivery. Proc Natl Acad Sci. 1991;88:8850–8854. doi: 10.1073/pnas.88.19.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Midoux P, Monsigny M. Efficient gene transfer by histidylated polylysine pDNA complexes. Bioconjugate Chem. 1999;10:406–411. doi: 10.1021/bc9801070. [DOI] [PubMed] [Google Scholar]
- 21.Vadiei K, Lopez-Berestein G, Perez-Soler R, Luke DR. In vitro evaluation of liposomal cyclosporine. Int J Pharm. 1989;57:133–138. [Google Scholar]
- 22.Scherphof GL, Dijkstra J, Spanjer HH, Derksen JT, Roerdink FH. Uptake and intracellular processing of targeted and nontargeted liposomes by rat Kupffer cells in vivo and in vitro. Ann N Y Acad Sci. 1985;446:368–384. doi: 10.1111/j.1749-6632.1985.tb18414.x. [DOI] [PubMed] [Google Scholar]
- 23.Alving CR, Steck EA, Chapman WL, Jr, Waits VB, Hendricks LD, Swartz GM, Jr, Hanson WL. Therapy of leishmaniasis: superior efficacies of liposome-encapsulated drugs. Proc Natl Acad Sci. 1978;75:2959–2963. doi: 10.1073/pnas.75.6.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wightman L, Kircheis R, Rossler V, Carotta S, Ruzicka R, Kursa M, Wagner E. Different behavior of branched and linear polyethylenimine for gene delivery in vitro and in vivo. J Gene Med. 2001;3:362–372. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 25.Ogris M, Steinlein P, Kursa M, Mechtler K, Kircheis R, Wagner E. The size of DNA/transferrin-PEI complexes is an important factor for gene expression in cultured cells. Gene Ther. 1998;5:1425–1433. doi: 10.1038/sj.gt.3300745. [DOI] [PubMed] [Google Scholar]
- 26.Ward CM, Read ML, Seymour LW. Systemic circulation of poly(L-lysine)/DNA vectors is influenced by polycation molecular weight and type of DNA: differential circulation in mice and rats and the implications for human gene therapy. Blood. 2001;97:2221–2229. doi: 10.1182/blood.v97.8.2221. [DOI] [PubMed] [Google Scholar]
- 27.Ogris M, Brunner S, Schuller S, Kircheis R, Wagner E. PEGylated DNA/transferrin-PEI complexes: reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Ther. 1999;6:595–605. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]
- 28.Mao HQ, Roy K, Troung-Le VL, Janes KA, Lin KY, Wang Y, August JT, Leong KW. Chitosan-DNA nanoparticles as gene carriers: synthesis, characterization and transfection efficiency. J Control Release. 2001;70:399–421. doi: 10.1016/s0168-3659(00)00361-8. [DOI] [PubMed] [Google Scholar]
- 29.Kaul G, Amiji M. Tumor-targeted gene delivery using poly(ethylene glycol)-modified gelatin nanoparticles: In vitro and in vivo studies. Pharmaceut Res. 2005;22:951–961. doi: 10.1007/s11095-005-4590-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suk JS, Suh J, Choy K, Lai SK, Fu J, Hanes J. Gene delivery to differentiated neurotypic cells with RGD and HIV Tat peptide functionalized polymeric nanoparticles. Biomaterials. 2006;27:5143–5150. doi: 10.1016/j.biomaterials.2006.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatakeyama H, Akita H, Kogure K, Oishi M, Nagasaki Y, Kihira Y, Ueno M, Kobayashi H, Kikuchi H, Harashima H. Development of a novel systemic gene delivery system for cancer therapy with a tumor-specific cleavable PEG-lipid. Gene Ther. 2007;14:68–77. doi: 10.1038/sj.gt.3302843. [DOI] [PubMed] [Google Scholar]
- 32.Kawano T, Yamagata M, Takahashi H, Niidome Y, Yamada S, Katayama Y, Niidome T. Stabilizing of plasmid DNA in vivo by PEG-modified cationic gold nanoparticles and the gene expression assisted with electrical pulses. J Control Release. 2006;111:382–389. doi: 10.1016/j.jconrel.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 33.Meyer M, Wagner E. pH-responsive shielding of non-viral gene vectors. Expert Opin Drug Deliv. 2006;3:563–571. doi: 10.1517/17425247.3.5.563. [DOI] [PubMed] [Google Scholar]
- 34.Hafez I, Maurer N, Cullis P. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8:1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 35.Xu Y, Szoka FC., Jr Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 1996;35:5616–5623. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- 36.Zelphati O, Szoka FC., Jr Mechanism of oligonucleotide release from cationic liposomes. Proc Natl Acad Sci. 1996;93:11493–11498. doi: 10.1073/pnas.93.21.11493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verma A, Stellacci F. Effect of Surface Properties on Nanoparticle-Cell Interactions. Small. 2010;6:12–21. doi: 10.1002/smll.200901158. [DOI] [PubMed] [Google Scholar]
- 38.Lockman PR, Koziara JM, Mumper RJ, Allen DD. Nanoparticle surface charges alter blood-brain barrier integrity and permeability. J Drug Target. 2004;12:635–641. doi: 10.1080/10611860400015936. [DOI] [PubMed] [Google Scholar]
- 39.Subbarao NK, Parente RA, Szoka FC, Jr, Nadasdi L, Pongracz K. The pH-dependent bilayer destabilization by an amphipathic peptide. Biochemistry. 1987;26:2964–2972. doi: 10.1021/bi00385a002. [DOI] [PubMed] [Google Scholar]
- 40.Wyman TB, Nicol F, Zelphati O, Scaria P, Plank C, Szoka FC., Jr Design, synthesis, and characterization of a cationic peptide that binds to nucleic acids and permeabilizes bilayers. Biochemistry. 1997;36:3008–3017. doi: 10.1021/bi9618474. [DOI] [PubMed] [Google Scholar]
- 41.Niidome T, Ohmori N, Ichinose A, Wada A, Mihara H, Hirayama T, Aoyagi H. Binding of Cationic Alpha-Helical Peptides to Plasmid DNA and Their Gene Transfer Abilities into Cells. J Biol Chem. 1997;272:15307–15312. doi: 10.1074/jbc.272.24.15307. [DOI] [PubMed] [Google Scholar]
- 42.Plank C, Oberhauser B, Mechtler K, Koch C, Wagner E. The Influence of Endosome-Disruptive Peptides on Gene-Transfer Using Synthetic Virus-Like Gene-Transfer Systems. J Biol Chem. 1994;269:12918–12924. [PubMed] [Google Scholar]
- 43.Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 44.Green M, Loewenstein PM. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell. 1988;55:1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 45.Manickam DS, Bisht HS, Wan L, Mao GZ, Oupicky D. Influence of TAT-peptide polymerization on properties and transfection activity of TAT/DNA polyplexes. J Control Release. 2005;102:293–306. doi: 10.1016/j.jconrel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 46.Shmueli RB, Anderson DG, Green JJ. Electrostatic surface modifications to improve gene delivery. Expert Opin Drug Del. 2010;7:535–550. doi: 10.1517/17425241003603653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris TJ, Green JJ, Fung PW, Langer R, Anderson DG, Bhatia SN. Tissue-specific gene delivery via nanoparticle coating. Biomaterials. 2010;31:998–1006. doi: 10.1016/j.biomaterials.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng CJ, Saltzman WM. Enhanced siRNA delivery into cells by exploiting the synergy between targeting ligands and cell-penetrating peptides. Biomaterials. 2011;32:6194–6203. doi: 10.1016/j.biomaterials.2011.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou J, Neff CP, Swiderski P, Li H, Smith DD, Aboellail T, Remling-Mulder L, Akkina R, Rossi JJ. Functional in vivo delivery of multiplexed anti-HIV-1 siRNAs via a chemically synthesized aptamer with a sticky bridge. Mol Ther. 2013;21:192–200. doi: 10.1038/mt.2012.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boussif O, Lezoualch F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and in-Vivo - Polyethylenimine. Proc Natl Acad Sci. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benjaminsen RV, Mattebjerg MA, Henriksen JR, Moghimi SM, Andresen TL. The Possible “Proton Sponge” Effect of Polyethylenimine (PEI) Does Not Include Change in Lysosomal pH. Mol Ther. 2013;21:149–157. doi: 10.1038/mt.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sonawane ND, Szoka FC, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 53.Nel AE, Madler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Understanding biophysicochemical interactions at the nano-bio interface. Nature Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 54.Okuda T, Sugiyama A, Niidome T, Aoyagi H. Characters of dendritic poly(L-lysine) analogues with the terminal lysines replaced with arginines and histidines as gene carriers in vitro. Biomaterials. 2004;25:537–544. doi: 10.1016/s0142-9612(03)00542-8. [DOI] [PubMed] [Google Scholar]
- 55.Benns JM, Choi JS, Mahato RI, Park JS, Kim SW. pH-sensitive cationic polymer gene delivery vehicle: N-Ac-poly(L-histidine)-graft-poly(L-lysine) comb shaped polymer. Bioconjugate Chem. 2000;11:637–645. doi: 10.1021/bc0000177. [DOI] [PubMed] [Google Scholar]
- 56.Haensler J, Szoka FC., Jr Polyamidoamine cascade polymers mediate efficient transfection of cells in culture. Bioconjugate Chem. 1993;4:372–379. doi: 10.1021/bc00023a012. [DOI] [PubMed] [Google Scholar]
- 57.Vader P, van der Aa LJ, Engbersen JFJ, Storm G, Schiffelers RM. Disulfide-Based Poly(amido amine)s for siRNA Delivery: Effects of Structure on siRNA Complexation, Cellular Uptake, Gene Silencing and Toxicity. Pharmaceut Res. 2011;28:1013–1022. doi: 10.1007/s11095-010-0344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Felgner JH, Kumar R, Sridhar CN, Wheeler CJ, Tsai YJ, Border R, Ramsey P, Martin M, Felgner PL. Enhanced Gene Delivery and Mechanism Studies with a Novel Series of Cationic Lipid Formulations. J Biol Chem. 1994;269:2550–2561. [PubMed] [Google Scholar]
- 59.El Ouahabi A, Thiry M, Pector V, Fuks R, Ruysschaert JM, Vandenbranden M. The role of endosome destabilizing activity in the gene transfer process mediated by cationic lipids. FEBS Lett. 1997;414:187–192. doi: 10.1016/s0014-5793(97)00973-3. [DOI] [PubMed] [Google Scholar]
- 60.Drummond DC, Zignani M, Leroux JC. Current status of pH-sensitive liposomes in drug delivery. Prog Lipid Res. 2000;39:409–460. doi: 10.1016/s0163-7827(00)00011-4. [DOI] [PubMed] [Google Scholar]
- 61.Gary DJ, Puri N, Won YY. Polymer-based siRNA delivery: perspectives on the fundamental and phenomenological distinctions from polymer-based DNA delivery. J Control Release. 2007;121:64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 62.Luo D, Saltzman WM. Synthetic DNA delivery systems. Nat Biotechnol. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 63.Kawasaki H, Taira K. Short hairpin type of dsRNAs that are controlled by tRNAVal promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res. 2003;31:700–707. doi: 10.1093/nar/gkg158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lynn DM, Langer R. Degradable poly (beta-amino esters): synthesis, characterization, and self-assembly with plasmid DNA. J Am Chem Soc. 2000;122:10761–10768. [Google Scholar]
- 65.Tzeng SY, Green JJ. Subtle changes to polymer structure and degradation mechanism enable highly effective nanoparticles for siRNA and DNA delivery to human brain cancer. Adv Healthcare Mater. 2013;2:467. doi: 10.1002/adhm.201200257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Son S, Namgung R, Kim J, Singha K, Kim WJ. Bioreducible Polymers for Gene Silencing and Delivery. Accounts of Chemical Research. 2012:45. doi: 10.1021/ar200248u. [DOI] [PubMed] [Google Scholar]
- 67.Elbakry A, Zaky A, Liebl R, Rachel R, Goepferich A, Breunig M. Layer-by-layer assembled gold nanoparticles for siRNA delivery. Nano Lett. 2009;9:2059–2064. doi: 10.1021/nl9003865. [DOI] [PubMed] [Google Scholar]
- 68.Lee JS, Green JJ, Love KT, Sunshine J, Langer R, Anderson DG. Gold, poly (beta-amino ester) nanoparticles for small interfering RNA delivery. Nano Lett. 2009;9:2402–2406. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spagnou S, Miller AD, Keller M. Lipidic carriers of siRNA: differences in the formulation, cellular uptake, and delivery with plasmid DNA. Biochemistry. 2004;43:13348–13356. doi: 10.1021/bi048950a. [DOI] [PubMed] [Google Scholar]
- 70.Hill IR, Garnett MC, Bignotti F, Davis SS. In vitro cytotoxicity of poly(amidoamine)s: relevance to DNA delivery. Biochim Biophys Acta. 1999;1427:161–174. doi: 10.1016/s0304-4165(99)00021-5. [DOI] [PubMed] [Google Scholar]
- 71.Lim YB, Han SO, Kong HU, Lee Y, Park JS, Jeong B, Kim SW. Biodegradable polyester, poly[alpha-(4 aminobutyl)-L-glycolic acid], as a non-toxic gene carrier. Pharmaceut Res. 2000;17:811–816. doi: 10.1023/a:1007552007765. [DOI] [PubMed] [Google Scholar]
- 72.Forrest ML, Koerber JT, Pack DW. A degradable polyethylenimine derivative with low toxicity for highly efficient gene delivery. Bioconjugate Chem. 2003;14:934–940. doi: 10.1021/bc034014g. [DOI] [PubMed] [Google Scholar]
- 73.Sutton D, Kim SJ, Shuai XT, Leskov K, Marques JT, Williams BRG, Boothman DA, Gao JM. Efficient suppression of secretory clusterin levels by polymer-siRNA nanocomplexes enhances ionizing radiation lethality in human MCF-7 breast cancer cells in vitro. Int J Nanomedicine. 2006;1:155–162. doi: 10.2147/nano.2006.1.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grayson ACR, Doody AM, Putnam D. Biophysical and structural characterization of polyethylenimine-mediated siRNA delivery in vitro. Pharmaceut Res. 2006;23:1868–1876. doi: 10.1007/s11095-006-9009-2. [DOI] [PubMed] [Google Scholar]
- 75.Hagerman PJ. Flexibility of RNA. Annu Rev Biophys Biomol Struct. 1997;26:139–156. doi: 10.1146/annurev.biophys.26.1.139. [DOI] [PubMed] [Google Scholar]
- 76.Kebbekus P, Draper DE, Hagerman P. Persistence length of RNA. Biochemistry. 1995;34:4354–4357. doi: 10.1021/bi00013a026. [DOI] [PubMed] [Google Scholar]
- 77.Bolcato-Bellemin AL, Bonnet ME, Creusatt G, Erbacher P, Behr JP. Sticky overhangs enhance siRNA-mediated gene silencing. Proc Natl Acad Sci. 2007;104:16050–16055. doi: 10.1073/pnas.0707831104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li SD, Chen YC, Hackett MJ, Huang L. Tumor-targeted delivery of siRNA by self-assembled nanoparticles. Mol Ther. 2008;16:163–169. doi: 10.1038/sj.mt.6300323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Adair JH, Parette MP, Altinoglu EI, Kester M. Nanoparticulate alternatives for drug delivery. ACS Nano. 2010;4:4967–4970. doi: 10.1021/nn102324e. [DOI] [PubMed] [Google Scholar]
- 80.Forbes DC, Peppas NA. Oral delivery of small RNA and DNA. J Control Release. 2012 doi: 10.1016/j.jconrel.2012.06.037. [DOI] [PubMed] [Google Scholar]
- 81.Dalby B, Cates S, Harris A, Ohki EC, Tilkins ML, Price PJ, Ciccarone VC. Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods. 2004;33:95–103. doi: 10.1016/j.ymeth.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 82.Ma Z, Li J, He FT, Wilson A, Pitt B, Li S. Cationic lipids enhance siRNA-mediated interferon response in mice. Biochem Biophys Res Commun. 2005;330:755–759. doi: 10.1016/j.bbrc.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 83.Palliser D, Chowdhury D, Wang QY, Lee SJ, Bronson RT, Knipe DM, Lieberman J. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature. 2006;439:89–94. doi: 10.1038/nature04263. [DOI] [PubMed] [Google Scholar]
- 84.Chono S, Li SD, Conwell CC, Huang L. An efficient and low immunostimulatory nanoparticle formulation for systemic siRNA delivery to the tumor. J Control Release. 2008;131:64–69. doi: 10.1016/j.jconrel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Judge AD, Bola G, Lee ACH, Maclachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 86.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Akinc A, Goldberg M, Qin J, Dorkin JR, Gamba-Vitalo C, Maier M, Jayaprakash KN, Jayaraman M, Rajeev KG, Manoharan M, et al. Development of Lipidoid-siRNA Formulations for Systemic Delivery to the Liver. Mol Ther. 2009;17:872–879. doi: 10.1038/mt.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lu JJ, Langer R, Chen JZ. A novel mechanism is involved in cationic lipid-mediated functional siRNA delivery. Mol Pharm. 2009;6:763–771. doi: 10.1021/mp900023v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Umeda M, Nojima S, Inoue K. Effect of lipid composition on HVJ-mediated fusion of glycophorin liposomes to erythrocytes. J Biochem. 1985;97:1301–1310. doi: 10.1093/oxfordjournals.jbchem.a135181. [DOI] [PubMed] [Google Scholar]
- 90.Litzinger DC, Huang L. Phosphatidylethanolamine Liposomes - Drug Delivery, Gene-Transfer and Immunodiagnostic Applications. Biochim Biophys Acta. 1992;1113:201–227. doi: 10.1016/0304-4157(92)90039-d. [DOI] [PubMed] [Google Scholar]
- 91.Hafez IM, Cullis PR. Roles of lipid polymorphism in intracellular delivery. Adv Drug Deliv Rev. 2001;47:139–148. doi: 10.1016/s0169-409x(01)00103-x. [DOI] [PubMed] [Google Scholar]
- 92.Heyes J, Palmer L, Bremner K, MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J Control Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 93.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DWY, Stebbing D, Crosley EJ, Hafez IM. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 94.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DW, Stebbing D, Crosley EJ, Yaworski E, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 95.Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, Cantley W, Qin LL, Racie T, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leconet W, Petit P, Peraldi-Roux S, Bresson D. Nonviral Delivery of Small Interfering RNA Into Pancreas-associated Immune Cells Prevents Autoimmune Diabetes. Mol Ther. 2012;20:2315–2325. doi: 10.1038/mt.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. [Accessed 2013];TKM 080301 for Primary or Secondary Liver Cancer. Available at: http://clinicaltrials.gov/ct2/show/NCT01437007.
- 98.Dose Escalation Study to Determine Safety. [Accessed 2013];Pharmacokinetics, and Pharmacodynamics of Intravenous TKM-080301. Available at: http://clinicaltrials.gov/show/NCT01262235.
- 99. [Accessed 2013];Trial to Evaluate Safety and Tolerability of ALN-TTR01 in Transthyretin (TTR) Amyloidosis. Available at: http://clinicaltrials.gov/show/NCT01148953.
- 100. [Accessed 2013];Study With Atu027 in Patients With Advanced Solid Cancer. Available at: http://clinicaltrials.gov/ct2/show/study/NCT00938574.
- 101.Meyer M, Philipp A, Oskuee R, Schmidt C, Wagner E. Breathing life into polycations: Functionalization with pH-responsive endosomolytic peptides and polyethylene glycol enables siRNA delivery. J Am Chem Soc. 2008;130:3272-+. doi: 10.1021/ja710344v. [DOI] [PubMed] [Google Scholar]
- 102.Kakizawa Y, Harada A, Kataoka K. Environment-sensitive stabilization of core-shell structured polyion complex micelle by reversible cross-linking of the core through disulfide bond. J Am Chem Soc. 1999;121:11247–11248. [Google Scholar]
- 103.Kakizawa Y, Harada A, Kataoka K. Glutathione-Sensitive Stabilization of Block Copolymer Micelles Composed of Antisense DNA and Thiolated Poly (ethylene glycol)-b lock-poly (l-lysine): A Potential Carrier for Systemic Delivery of Antisense DNA. Biomacromolecules. 2001;2:491–497. doi: 10.1021/bm000142l. [DOI] [PubMed] [Google Scholar]
- 104.Miyata K, Kakizawa Y, Nishiyama N, Harada A, Yamasaki Y, Koyama H, Kataoka K. Block catiomer polyplexes with regulated densities of charge and disulfide cross-linking directed to enhance gene expression. J Am Chem Soc. 2004;126:2355–2361. doi: 10.1021/ja0379666. [DOI] [PubMed] [Google Scholar]
- 105.Matsumoto S, Christie RJ, Nishiyama N, Miyata K, Ishii A, Oba M, Koyama H, Yamasaki Y, Kataoka K. Environment-Responsive Block Copolymer Micelles with a Disulfide Cross-Linked Core for Enhanced siRNA Delivery. Biomacromolecules. 2009;10:119–127. doi: 10.1021/bm800985e. [DOI] [PubMed] [Google Scholar]
- 106.Christie RJ, Matsumoto Y, Miyata K, Nomoto T, Fukushima S, Osada K, Halnaut J, Pittella F, Kim HJ, Nishiyama N, et al. Targeted Polymeric Micelles for siRNA Treatment of Experimental Cancer by Intravenous Injection. ACS Nano. 2012;6:5174–5189. doi: 10.1021/nn300942b. [DOI] [PubMed] [Google Scholar]
- 107.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. Local and systemic delivery of VEGF siRNA using polyelectrolyte complex micelles for effective treatment of cancer. J Control Release. 2008;129:107–116. doi: 10.1016/j.jconrel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 108.Breunig M, Hozsa C, Lungwitz U, Watanabe K, Umeda I, Kato H, Goepferich A. Mechanistic investigation of poly (ethylene imine)-based siRNA delivery: disulfide bonds boost intracellular release of the cargo. J Control Release. 2008;130:57–63. doi: 10.1016/j.jconrel.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 109.Jeong JH, Christensen LV, Yockman JW, Zhong ZY, Engbersen JFJ, Kim WJ, Feijen J, Kim SW. Reducible poly(amido ethylenimine) directed to enhance RNA interference. Biomaterials. 2007;28:1912–1917. doi: 10.1016/j.biomaterials.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 110.Christensen LV, Chang CW, Kim WJ, Kim SW, Zhong ZY, Lin C, Engbersen JFJ, Feijen J. Reducible poly(amido ethylenimine)s designed for triggered intracellular gene delivery. Bioconjugate Chem. 2006;17:1233–1240. doi: 10.1021/bc0602026. [DOI] [PubMed] [Google Scholar]
- 111.van der Aa L, Vader P, Storm G, Schiffelers R, Engbersen J. Optimization of poly (amido amine)s as vectors for siRNA delivery. J Control Release. 2011;150:177–186. doi: 10.1016/j.jconrel.2010.11.030. [DOI] [PubMed] [Google Scholar]
- 112.van der Aa LJ, Vader P, Storm G, Schiffelers RM, Engbersen JFJ. Optimization of poly(amido amine)s as vectors for siRNA delivery. J Control Release. 2011;150:177–186. doi: 10.1016/j.jconrel.2010.11.030. [DOI] [PubMed] [Google Scholar]
- 113.Vader P, van der Aa LJ, Engbersen JFJ, Storm G, Schiffelers RM. Physicochemical and Biological Evaluation of siRNA Polyplexes Based on PEGylated Poly (amido amine) s. Pharmaceut Res. 2012;29:352–361. doi: 10.1007/s11095-011-0545-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 115.Tzeng SY, Yang PH, Grayson WL, Green JJ. Synthetic poly(ester amine) and poly(amido amine) nanoparticles for efficient DNA and siRNA delivery to human endothelial cells. Int J Nanomedicine. 2012;6:3309–3322. doi: 10.2147/IJN.S27269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vandenbroucke RE, De Geest BG, Bonne S, Vinken M, Van Haecke T, Heimberg H, Wagner E, Rogiers V, De Smedt SC, Demeester J, et al. Prolonged gene silencing in hepatoma cells and primary hepatocytes after small interfering RNA delivery with biodegradable poly(beta-amino esters) J Gene Med. 2008;10:783–794. doi: 10.1002/jgm.1202. [DOI] [PubMed] [Google Scholar]
- 117.Tzeng SY, Hung BP, Grayson WL, Green JJ. Cystamine-terminated poly(beta-amino ester)s for siRNA delivery to human mesenchymal stem cells and enhancement of osteogenic differentiation. Biomaterials. 2012;33:8142–8151. doi: 10.1016/j.biomaterials.2012.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kozielski KL, Tzeng SY, Green JJ. A bioreducible linear poly(beta-amino ester) for siRNA delivery. Chem Commun. 2013 doi: 10.1039/c3cc40718g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mok H, Park TG. Self-crosslinked and reducible fusogenic peptides for intracellular delivery of siRNA. Biopolymers. 2008;89:881–888. doi: 10.1002/bip.21032. [DOI] [PubMed] [Google Scholar]
- 120.Woodrow KA, Cu Y, Booth CJ, Saucier-Sawyer JK, Wood MJ, Saltzman WM. Intravaginal gene silencing using biodegradable polymer nanoparticles densely loaded with small-interfering RNA. Nature Mater. 2009;8:526–533. doi: 10.1038/nmat2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yuan XD, Shah BA, Kotadia NK, Li JA, Gu H, Wu ZQ. The Development and Mechanism Studies of Cationic Chitosan-Modified Biodegradable PLGA Nanoparticles for Efficient siRNA Drug Delivery. Pharmaceut Res. 2010;27:1285–1295. doi: 10.1007/s11095-010-0103-0. [DOI] [PubMed] [Google Scholar]
- 122.Zhou J, Patel TR, Fu M, Bertram JP, Saltzman WM. Octa-functional PLGA nanoparticles for targeted and efficient siRNA delivery to tumors. Biomaterials. 2012;33:583–591. doi: 10.1016/j.biomaterials.2011.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tzeng SY, Lavik EB. Photopolymerizable nanoarray hydrogels deliver CNTF and promote differentiation of neural stem cells. Soft Matter. 2010;6:2208–2215. [Google Scholar]
- 124.Bertram JP, Williams CA, Robinson R, Segal SS, Flynn NT, Lavik EB. Intravenous Hemostat: Nanotechnology to Halt Bleeding. Sci Transl Med. 2009:1. doi: 10.1126/scitranslmed.3000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Davis ME. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol Pharm. 2009;6:659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 126.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jordan M, Schallhorn A, Wurm FM. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 1996;24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tolou H. Administration of oligonucleotides to cultured cells by calcium phosphate precipitation method. Anal Biochem. 1993;215:156–158. doi: 10.1006/abio.1993.1568. [DOI] [PubMed] [Google Scholar]
- 129.Kakizawa Y, Kataoka K. Block copolymer self-assembly into monodispersive nanoparticles with hybrid core of antisense DNA and calcium phosphate. Langmuir. 2002;18:4539–4543. [Google Scholar]
- 130.Kakizawa Y, Furukawa S, Ishii A, Kataoka K. Organic-inorganic hybrid-nanocarrier of siRNA constructing through the self-assembly of calcium phosphate and PEG-based block aniomer. J Control Release. 2006;111:368–370. doi: 10.1016/j.jconrel.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 131.Zhang M, Ishii A, Nishiyama N, Matsumoto S, Ishii T, Yamasaki Y, Kataoka K. PEGylated Calcium Phosphate Nanocomposites as Smart Environment-Sensitive Carriers for siRNA Delivery. Adv Mater. 2009;21:3520–3525. [Google Scholar]
- 132.Connor EE, Mwamuka J, Gole A, Murphy CJ, Wyatt MD. Gold nanoparticles are taken up by human cells but do not cause acute cytotoxicity. Small. 2005;1:325–327. doi: 10.1002/smll.200400093. [DOI] [PubMed] [Google Scholar]
- 133.Daniel MC, Astruc D. Gold nanoparticles: assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology. Chemical Reviews-Columbus. 2004;104:293. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 134.Ghosh PS, Kim CK, Han G, Forbes NS, Rotello VM. Efficient gene delivery vectors by tuning the surface charge density of amino acid-functionalized gold nanoparticles. ACS Nano. 2008 doi: 10.1021/nn800507t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chemical Reviews-Columbus. 2005;105:1103–1170. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 136.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996 doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 137.Decher G, Hong JD. Makromolekulare Chemie. Macromolecular Symposia. Wiley Online Library; 1991. Buildup of ultrathin multilayer films by a self-assembly process, 1 consecutive adsorption of anionic and cationic bipolar amphiphiles on charged surfaces. [Google Scholar]
- 138.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. Gene regulation with polyvalent siRNA-nanoparticle conjugates. J Am Chem Soc. 2009;131:2072–2073. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zheng D, Giljohann DA, Chen DL, Massich MD, Wang XQ, Iordanov H, Mirkin CA, Paller AS. Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proc Natl Acad Sci. 2012;109:11975–11980. doi: 10.1073/pnas.1118425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tanaka T, Mangala LS, Vivas-Mejia PE, Nieves-Alicea R, Mann AP, Mora E, Han HD, Shahzad MM, Liu X, Bhavane R, et al. Sustained small interfering RNA delivery by mesoporous silicon particles. Cancer Res. 2010;70:3687–3696. doi: 10.1158/0008-5472.CAN-09-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Meng H, Liong M, Xia T, Li Z, Ji Z, Zink JI, Nel AE. Engineered design of mesoporous silica nanoparticles to deliver doxorubicin and P-glycoprotein siRNA to overcome drug resistance in a cancer cell line. ACS Nano. 2010;4:4539–4550. doi: 10.1021/nn100690m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 143.Wu X, Liu H, Liu J, Haley KN, Treadway JA, Larson JP, Ge N, Peale F, Bruchez MP. Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat Biotechnol. 2002;21:41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 144.Derfus AM, Chan WCW, Bhatia SN. Intracellular delivery of quantum dots for live cell labeling and organelle tracking. Adv Mater. 2004;16:961–966. [Google Scholar]
- 145.Derfus AM, Chan WCW, Bhatia SN. Probing the cytotoxicity of semiconductor quantum dots. Nano Lett. 2004;4:11–18. doi: 10.1021/nl0347334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mattheakis LC, Dias JM, Choi YJ, Gong J, Bruchez MP, Liu J, Wang E. Optical coding of mammalian cells using semiconductor quantum dots. Anal Biochem. 2004;327:200–208. doi: 10.1016/j.ab.2004.01.031. [DOI] [PubMed] [Google Scholar]
- 147.Chen AA, Derfus AM, Khetani SR, Bhatia SN. Quantum dots to monitor RNAi delivery and improve gene silencing. Nucleic Acids Res. 2005;33:e190–e190. doi: 10.1093/nar/gni188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Derfus AM, Chen AA, Min DH, Ruoslahti E, Bhatia SN. Targeted quantum dot conjugates for siRNA delivery. Bioconjugate Chem. 2007;18:1391–1396. doi: 10.1021/bc060367e. [DOI] [PubMed] [Google Scholar]
- 149.Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 150.Singh N, Agrawal A, Leung AKL, Sharp PA, Bhatia SN. Effect of nanoparticle conjugation on gene silencing by RNA interference. J Am Chem Soc. 2010;132:8241–8243. doi: 10.1021/ja102132e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Medarova Z, Pham W, Farrar C, Petkova V, Moore A. In vivo imaging of siRNA delivery and silencing in tumors. Nat Med. 2007;13:372–377. doi: 10.1038/nm1486. [DOI] [PubMed] [Google Scholar]
- 152.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. PEG conjugated VEGF siRNA for anti-angiogenic gene therapy. J Control Release. 2006;116:123–129. doi: 10.1016/j.jconrel.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 153.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 154.Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 155.Bolcato-Bellemin AL, Bonnet ME, Creusat G, Erbacher P, Behr JP. Sticky overhangs enhance siRNA-mediated gene silencing. Proc Natl Acad Sci. 2007;104:16050–16055. doi: 10.1073/pnas.0707831104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mok H, Lee SH, Park JW, Park TG. Multimeric small interfering ribonucleic acid for highly efficient sequence-specific gene silencing. Nature Mater. 2010;9:272–278. doi: 10.1038/nmat2626. [DOI] [PubMed] [Google Scholar]
- 157.Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nature Mater. 2012 doi: 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Diegelman AM, Kool ET. Generation of circular RNAs and trans-cleaving catalytic RNAs by rolling transcription of circular DNA oligonucleotides encoding hairpin ribozymes. Nucleic Acids Res. 1998;26:3235–3241. doi: 10.1093/nar/26.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cutler JI, Zhang K, Zheng D, Auyeung E, Prigodich AE, Mirkin CA. Polyvalent nucleic acid nanostructures. J Am Chem Soc. 2011;133:9254–9257. doi: 10.1021/ja203375n. [DOI] [PMC free article] [PubMed] [Google Scholar]