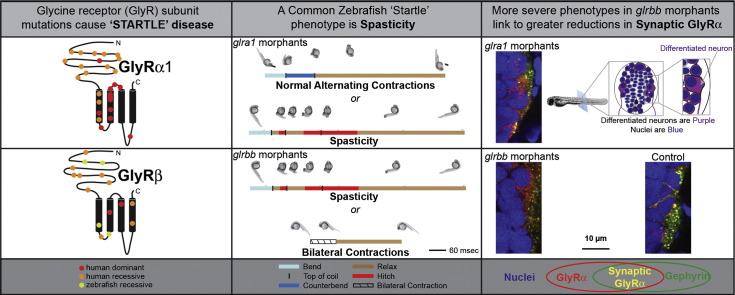
Abstract
Startle disease is an inherited neurological disorder that causes affected individuals to suffer noise- or touch-induced non-epileptic seizures, excessive muscle stiffness and neonatal apnea episodes. Mutations known to cause startle disease have been identified in glycine receptor subunit (GLRA1 and GLRB) and glycine transporter (SLC6A5) genes, which serve essential functions at glycinergic synapses. Despite the significant successes in identifying startle disease mutations, many idiopathic cases remain unresolved. Exome sequencing in these individuals will identify new candidate genes. To validate these candidate disease genes, zebrafish is an ideal choice due to rapid knockdown strategies, accessible embryonic stages, and stereotyped behaviors. The only existing zebrafish model of startle disease, bandoneon (beo), harbors point mutations in glrbb (one of two zebrafish orthologs of human GLRB) that cause compromised glycinergic transmission and touch-induced bilateral muscle contractions. In order to further develop zebrafish as a model for startle disease, we sought to identify common phenotypic outcomes of knocking down zebrafish orthologs of two known startle disease genes, GLRA1 and GLRB, using splice site-targeted morpholinos. Although both morphants were expected to result in phenotypes similar to the zebrafish beo mutant, our direct comparison demonstrated that while both glra1 and glrbb morphants exhibited embryonic spasticity, only glrbb morphants exhibited bilateral contractions characteristic of beo mutants. Likewise, zebrafish over-expressing a dominant startle disease mutation (GlyR α1R271Q) exhibited spasticity but not bilateral contractions. Since GlyR βb can interact with GlyR α subunits 2–4 in addition to GlyR α1, loss of the GlyR βb subunit may produce more severe phenotypes by affecting multiple GlyR subtypes. Indeed, immunohistochemistry of glra1 morphants suggests that in zebrafish, alternate GlyR α subunits can compensate for the loss of the GlyR α1 subunit. To address the potential for interplay among GlyR subunits during development, we quantified the expression time-course for genes known to be critical to glycinergic synapse function. We found that GlyR α2, α3 and α4a are expressed in the correct temporal pattern and could compensate for the loss of the GlyR α1 subunit. Based on our findings, future studies that aim to model candidate startle disease genes in zebrafish should include measures of spasticity and synaptic development.
Keywords: glra1, glrb, Glycine receptor, Startle disease, Hyperekplexia, Zebrafish
Graphical abstract
Highlights
-
•
Morpholinos and dominant-negative mutants disrupted GlyR α1 subunit function.
-
•
Spasticity and delayed onset of rhythmic motor behaviors were observed.
-
•
GlyR βb subunit morphants exhibit both spasticity and bilateral muscle contractions.
-
•
Bilateral contractions likely result from disrupting multiple GlyR subtypes.
-
•
Spasticity may provide a common zebrafish ‘startle’ phenotype.
Introduction
Rhythmic motor behaviors require a balance between nervous system excitation and inhibition (E/I balance). The importance of E/I balance is illustrated by genetic mutations that selectively disrupt either excitation or inhibition and result in nervous system dysfunction (Ganser and Dallman, 2009; Gatto and Broadie, 2010). For example, in humans, startle disease/hyperekplexia results from excessive excitation due to damaging mutations in genes encoding key components of the inhibitory glycinergic synapse (Harvey et al., 2008). As the major inhibitory neurotransmitter in vertebrate hindbrain and spinal cord, glycine plays a critical role in the control of motor behaviors and reflexes. When glycinergic signaling is disrupted in newborn children, the result is exaggerated startle reflexes and hypertonia in response to unexpected auditory, tactile or visual stimuli. This abnormal startle response may also be accompanied by apnea episodes, i.e. the suspension of breathing (Thomas et al., 2010).
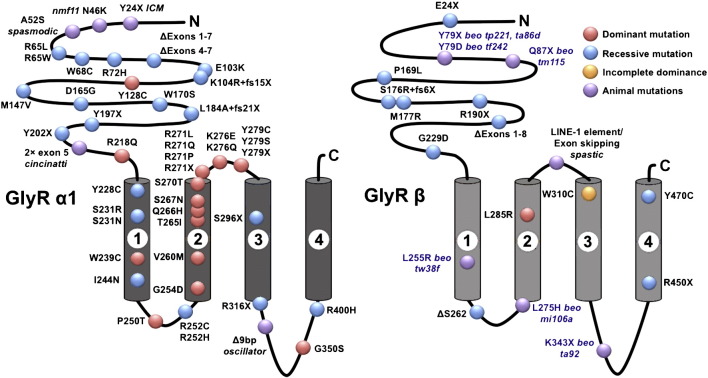
The majority of human startle disease cases are caused by dominant and recessive mutations in GLRA1, encoding the α1 subunit of the glycine receptor, GlyR α1 (Chung et al., 2010; Shiang et al., 1993; Fig. 1A). Mutations in this gene cause similar disorders in mice (Buckwalter et al., 1994; Holland et al., 2006; Ryan et al., 1994; Traka et al., 2006) and Poll Hereford cattle (Pierce et al., 2001). Dominant and recessive mutations in SLC6A5, encoding the presynaptic glycine transporter GlyT2, are now emerging as a second major cause of startle disease (Carta et al., 2012; Giménez et al., 2012; Rees et al., 2006), and also occur in Belgian Blue cattle (Charlier et al., 2008) and Irish wolfhounds (Gill et al., 2011). Mutations in GLRB, encoding the GlyR β subunit were thought to be a rare cause of human startle disease (Rees et al., 2002), although mutations in this gene were also reported in the mouse mutant spastic (Kingsmore et al., 1994; Mülhardt et al., 1994) and the zebrafish mutant bandoneon (beo; Granato et al., 1996; Hirata et al., 2005). However, several recent reports have identified novel dominant and recessive mutations in GLRB (Al-Owain et al., 2012; Chung et al., 2013; James et al., 2013; Lee et al., 2013; Fig. 1B), often associated with additional phenotypic consequences, including gaze disorders, apnea episodes, learning difficulties and developmental delay. Since GlyR α1 and β subunits occur in the same pentameric GlyR complex, it is currently unclear why mutations in GLRB can give rise to a more severe clinical phenotype than mutations in GLRA1.
Fig. 1.
Pathogenic mutations in the postsynaptic GlyR α1 and β subunits in startle disease. The predicted four-membrane spanning domain (M1–M4) topology of GlyR α1 and GlyR β subunits is depicted. Numbered columns indicate four predicted membrane-spanning domains 1–4 in each subunit. Red (dominantly inherited) and blue circles (recessively inherited) indicate the relative positions of amino acid alterations known to cause human startle disease. Purple circles indicate the relative positions of amino acid alterations found in mouse, cattle and zebrafish glycinergic disorders. For primary references to previously reported specific mutations, see Harvey et al. (2008), Chung et al. (2010, 2013) and James et al. (2013).
To model human startle disease, zebrafish are an attractive model because genes can be ‘inactivated’ using antisense morpholino knockdown (Eisen and Smith, 2008). Moreover, the resulting swimming phenotypes are readily quantified (Burgess and Granato, 2007). In contrast to mammalian genomes with five known GlyR subunit genes (GLRA1, GLRA2, GLRA3, GLRA4 and GLRB), the zebrafish genome encodes seven GlyR subunit genes (glra1, glra2, glra3, glra4a, glra4b, glrba, and glrbb; Hirata et al., 2010). The two duplicate genes, glra4b and glrbb resulted from whole genome duplication early in the evolution of teleosts (Hurley et al., 2007a). This phenomenon can be advantageous, since individual paired genes often differ in terms of expression patterns and functional roles (Hurley et al., 2007b; Ogino et al., 2011). For example, the zebrafish mutant bandoneon (beo) harbors mutations in one of the paired GlyR β subunit genes (glrbb), resulting in touch-induced simultaneous bilateral contractions of the axial muscles due to the loss of reciprocal glycinergic inhibition of motor circuits (Hirata et al., 2005, 2010). Despite the duplication of the GlyR β subunit genes in zebrafish, glrba is unable to compensate for the loss of glrbb function due to a different expression pattern, suggesting that it forms part of a distinct GlyR with a unique function (Hirata et al., 2005). Since mutations in human GLRA1, GLRB and SLC6A5 all result in startle disease (Harvey et al., 2008), it is unusual that to date no mutations in glra1 or slc6a5 have been discovered that produce bilateral contractions in larval zebrafish. We therefore sequenced the remaining beo alleles and have confirmed that they all harbor damaging mutations affecting glrbb.
To directly compare zebrafish glra1 and glrbb startle disease models, we designed splice-site morpholinos to knockdown the expression of these genes (Draper et al., 2001). As expected, injecting glrbb morpholinos produced a phenotype similar to beo mutants, characterized by simultaneous bilateral contractions strong enough to shorten the body. In addition, both glrbb and glra1 morphants produced spastic and erratic behaviors at early stages, but in contrast to glrbb morphants, glra1 morphants only rarely produced bilateral contractions and by 48 hpf glra1 morphants produced normal behaviors. Immunohistochemistry of glrbb and glra1 morphants also demonstrated distinct GlyR immunostaining patterns on early-differentiating spinal neurons. While glra1 morphants exhibited reduced but still synaptic GlyR staining, glrbb morphants exhibited GlyR α subunit trapping in intracellular, non-synaptic compartments, suggesting that the more severe beo phenotype results from a loss of multiple GlyR subtypes. Our quantitative analysis of mRNA expression for glycinergic genes in early development identified glra2 and glra3 as having high expression levels early in development that could ameliorate the glra1 morphant phenotype. In summary, we identify a phenotypic range in zebrafish startle disease models that should be taken into account when using zebrafish to study novel candidate startle disease genes.
Materials and methods
Fish care and embryo rearing
Experiments were carried out using Danio rerio wild type strains AB, Tubingen, and BWT (a fish store strain from Long Island). Adults were kept on a 14 hour light/10 hour dark circadian cycle at 28.5 °C. Embryos were collected from natural crossings shortly after removing a divider at first light. Embryos were then reared in glass Petri dishes containing system water (water that houses the adult fish) in a 28.5 °C incubator with the same 14 hour light/10 hour dark cycle. All experiments were conducted in accordance with the University of Miami Institutional Animal Care and Use Committee guidelines.
Detection of beo mutations
DNA was extracted from fin clips of beo mutation carriers using a QIAamp DNA mini kit (Qiagen, Manchester, UK). The nine coding exons and flanking sequences of glrbb were amplified from purified genomic DNA. Forward and reverse primers each of 20–30 bp in length were designed to cover the start and end of the regions to be amplified and ordered from Eurofins MWG Operon (Ebersberg, Germany). DNA polymerase was ordered from Clontech, Saint-Germain-en-Laye, France. PCR products were gel purified and cloned into pCR4-TOPO (Invitrogen, Paisley, UK). Sanger sequencing was performed by the DNA Sequencing Service (University of Dundee, UK). DNA sequences were analyzed using Sequencher 5.1 software (GeneCodes Corporation, Ann Arbor, USA) by alignment with reference sequences downloaded from NCBI or UCSC databases. For mutation identification, single nucleotide variants (SNVs) were noted and the effect of these changes on the encoded protein were examined using bioinformatics software including SIFT (Kumar et al., 2009) and Polyphen-2 (Adzhubei et al., 2010).
Morpholino design and injection
Splice site-targeted morpholinos (MOs) were designed against glra1 and glrbb. Intron/exon junctions were selected for MO design based both upon the appropriateness of the sequence for effective MO knockdown (i.e. 50% GC content and multiple mismatches to related genes). Two distinct MOs against glra1 were tested to control for possible off-target effects: glra1MOex4 5′-GAATTGTCCTCTCACCTTATACTGT-3′ and glra1MOex7 5′-CTTCCCTGAAACACAGAGAGTATGT-3′. Off-target effects were not a concern in the case of the glrbb splice morpholino glrbbMOex5 5′-GAGAGCATTAAAGTTCACCTCATGC-3′ because of the previously described beo mutant phenotype (Hirata et al., 2005). For control-injected embryos, we used the standard control MO provided by Gene Tools (Gene Tools, LLC; Philomath OR). Lyophilized MOs were resuspended in water as 1 mM stocks and stored at room temperature. Prior to use, the MO stock solution was heated for 5 min to 65 °C. For injection, stock solutions were diluted in a filtered solution of 1% (w/v) fast-green dye. MOs were injected using filament-lined Kwik-Fil borosilicate capillary glass (World Precision Instruments, Sarasota FL) pulled on a P-97 micropipette puller (Sutter Instruments, Novato CA) to a long taper with a fine tip that was broken back to ~ 1 μm using forceps. A stage micrometer was used to calibrate the injection rig to produce a 100 μm diameter bolus. MOs were injected into wild type embryos at the 1–2 cell stage. Embryos were sorted 6–8 h after injection so that only morphants in which the MO bolus had evenly dissipated were later analyzed for behavioral phenotypes.
RT-PCR
Unless otherwise stated, all reagents were from Life Technologies, Grand Island NY. Once behavioral analyses were completed, embryos were ground in TRIzol for RNA extraction. RNA preparations were then DNase-treated using DNA-free. The reverse transcriptase SuperScript III with either an anchored oligo(dT) primer or a gene specific primer was used to synthesize cDNA for subsequent PCR analysis. Diagnostic PCR primers were designed to detect mis-splicing events induced by MO masking of intron/exon junctions for glra1 (Fex3 5′-TTGGATCCATTGCTGAAACA-3′; Rex8 5′-ATACTCCAGCAGGGCAGAGA-3′) and glrbb (Fex5 5′-CTGAGACAACGCTGGAATGA-3′; Rex8 5′-CTCCACGCACGTGTAGTAGC-3′) (All primers were synthesized by Integrated DNA Technologies, Coralville, IA). Primers targeting slc6a5 (F: 5′-AGGAGTCACTCTGCCTGGAG-3′; R: 5′-CAAATGCAATTCCTGGACCT-3′) were utilized as internal controls. Shifted RT-PCR products caused by MO-induced alterations in pre-mRNA splicing were gel purified using Wizard Gel and PCR Clean-up System (Promega, Madison, WI). Purified bands were then re-amplified and sequenced using Sanger DNA sequencing to determine the predicted impact on targeted proteins.
Behavioral analyses
A high-speed camera (1024 Photron FASTCAM, San Diego, CA) was used to record spontaneous and touch-evoked behaviors of 17 to 50 hpf control, glra1, and glrbb morphants. Embryos were manually dechorionated. Spastic behaviors were characteristic of all groups just after dechorionation. Therefore, behavioral assays were conducted at least 1 h after dechorionation. Videos were scored by hand to generate ethograms that highlight rhythmic and spastic aspects of behaviors.
Immunohistochemistry
Cryosectioning and antibody staining were performed as previously described (Ogino et al., 2011). Briefly, anesthetized fish embryos were embedded in O.C.T. compound (Tissue-Tek, Torrance, CA) and gradually frozen in liquid nitrogen. Samples were then sectioned on a cryostat (CM-1850, Leica) and mounted on poly-l-lysine coated slides (Newcomer Supply, Middleton, WI) prior to a 10 min fixation in 4% (w/v) formaldehyde (diluted from 16%, Pierce Biotechnology, Rockford, IL). Anti-gephyrin (clone mAb7a, mouse IgG1, 1:500, Synaptic Systems, Göttingen, Germany), and a pan anti-GlyR α subunit (clone mAb4a, mouse IgG1, 1:100, Synaptic Systems) were used as primary antibodies. Alexa 488- and Alexa 568-conjugated donkey anti-mouse IgGs were used as secondary antibodies (1:2000, Life Technologies, Carlsbad, CA). Double staining with anti-gephyrin and anti-GlyR α subunit antibodies was performed sequentially. Stained sections were mounted in Vectashield/DAPI (Vector Laboratories, Burlingame, CA) and images were captured on a confocal microscope using a 1.4 NA 63 × oil objective (SP5, Leica Wetzlar Germany).
Quantification of synaptic staining
Images were processed using ImageJ (NIH). The brightness and contrast were adjusted to maximize dynamic range and reduce background noise before analyzing the density of GlyR α subunit and gephyrin puncta. For puncta density analysis, to avoid the impact of variation along the anterior–posterior axis, three images were picked in each stack: one each near top, middle, and the bottom of the stack. The area with puncta expression in each image was calculated in ImageJ. GlyR α subunit and gephyrin puncta in each image were identified and counted by custom MatLab (MathWorks, Natick, MA) programs (Morgan et al., 2008; Soto et al., 2011). For GlyR α subunit and gephyrin puncta co-localization analysis, puncta in the three-dimensional stack were first found by custom MatLab programs (Morgan et al., 2008; Soto et al., 2011). Then the three-dimensional location of each voxel in all GlyR α subunit puncta and gephyrin puncta were compared in MatLab to identify the colocalized puncta.
Quantitative PCR
Primers were designed against genes associated with the glycinergic synapse (Table 1). Primers were designed using Primer3 (Rozen and Skaletsky, 2000) to span introns with the goal of excluding the possibility that residual genomic DNA contributed to estimates of transcript abundance. To generate a developmental time series, RNA from 24, 32, 48 and 72 hpf wild type embryos was harvested. RNA extraction method followed the manufacturer's protocol (TRIzol, Life Technologies, Carlsbad, CA). Extracted RNAs were then DNase-treated (DNA-Free, Life Technologies), quantified on a NanoDrop 2000 (ThermoScientific, West Palm Beach, FL) and electrophoresed on a 1% (w/v) agarose gel to assess quality.
Table 1.
Oligonucleotide primers used for RT-PCR analysis. Key for selected gene names: slc6a5 encodes GlyT2, slc6a9 encodes GlyT1, slc32a1 encodes VGAT/VIAAT, and eef1a1l1 encodes the housekeeping protein EF1.
| Gene | Oligonucleotides | Tm | Size (bp) | Ensembl protein ID |
|---|---|---|---|---|
| glra1 | F: GGATCTCGAGCCTCTCTTCC | 57 | 67 | ENSDARP00000136733 |
| R: CACACTGCCATCCAAATGTC | 54 | |||
| glra2 | F: CGTGGTCAAAAACACACAGG | 55 | 115 | ENSDART00000066192 |
| R: GCGTGATATGGTGTCGATTC | 54 | |||
| glra3 | F: CCTGATCTTTGAGTGGCAAG | 54 | 116 | ENSDARP00000009777 |
| R: TGTTGTAGTGCTTGGTGCAG | 56 | |||
| glra4a | F: GCAGCACAAGGAGTTCATCA | 56 | 90 | ENSDARP00000146445 |
| R: TCCGTACCCTCGGAAGTAAA | 55 | |||
| glrba | F: CCGGAAAACCACAGAACAAG | 54 | 112 | ENSDARP00000074685 |
| R: AACGTGAAAGGGAACAGAGC | 55 | |||
| glrbb | F: TGCTCATCATCTGTCTGTGG | 55 | 73 | ENSDARP00000127692 |
| R: CCTTTCTTCTTTCCCTTTTTAGC | 52 | |||
| gphna | F: GCCCACCATCATCAAAGC | 54 | 65 | ENSDARP0000087952 |
| R: GATGGTACTCTGGACGTGGA | 56 | |||
| gphnb | F: TCAGCAGCAGACTGATGAGC | 57 | 85 | ENSDARP00000005053 |
| R: TTGTGCAGCTCCACGTATTG | 56 | |||
| slc6a5 | F: GGATTCCCATCATGTTTGCT | 53 | 87 | ENSDARP00000071929 |
| R: CGGGTTGAGGAGAACATACC | 55 | |||
| slc6a9 | F: AAGATTGCCATGTCTGAAGGA | 54 | 65 | ENSDARP00000088755 |
| R: GGTCGGGTTTCACAGAGTTC | 55 | |||
| slc32a1 | F: ACAAGCCCAGAATCACTGCT | 57 | 95 | ENSDARP00000083453 |
| R: GTGGAGAATGGCGTAGGGTA | 56 | |||
| eef1a1l1 | F: CTGGAGGCCAGCTCAAACAT | 58 | 82 | ENSDARG00000020850 |
| R: ATCAAGAAGAGTAGTACCGCTAGCA | 58 |
The SuperScript III First-Strand Synthesis System (Life Technologies, Carlsbad CA) was used for cDNA synthesis. However, gene specific primers (GSPs) were used in place of oligo(dT) to better detect rare transcripts encoding synaptic proteins. Each synthesis reaction contained 1 μg of RNA template. To generate standard curves, serial dilutions of gel-purified PCR products were used as template. Products were purified using the Wizard SV Gel and PCR purification kit (Promega, Madison WI). Stocks were made from a 1:1000 dilution of purified PCR product in DNase/RNase free H2O. Serial dilutions were adjusted to encompass the range of expression levels for each gene. qPCR reactions contained 2 μL of RNase/DNase free H2O, 14.5 μL Master mix, 2.5 μL of 10 mM primer and 1 μL of cDNA template at a final volume of 20 μL. The PCR protocol was as follows: 1 step of 95 °C for 5 min; 30 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s; and 1 step of 72 °C for 1 min. For qPCR, we used an Eppendorf Mastercycler ep realplex4 with the realplex 1.5 software using a Twin Tec Semi-Skirted 96 well plate. Each well contained 5 μL of PerfeCTa SYBR Green FastMix (Quanta BioSciences, Gaithersberg MD), 1.5 μL 10 mM combined forward and reverse primer, 2.5 μL of H2O and 1 μL of cDNA template to a final volume of 10 μL. The PCR protocol was as follows: 1 step of 95 °C for 1 min; 40 cycles of 95 °C for 15 s, 55 °C for 20 s, 68 °C for 30 s; and a melting curve and a final step of 95 °C. Three samples of each time point were run in duplicate on the same plate with a standard curve consisting of four 1:10 serial dilutions of purified PCR product as template. Relative mRNA copy numbers were calculated based on the expression levels of the housekeeping gene encoding EF1 (Tang et al., 2007).
Results
Sequencing of additional beo mutants confirms that mutations are found exclusively in glrbb
Mutations in glrbb were previously identified as the basis for the beo mutant phenotype (Hirata et al., 2005). In the original study, the underlying mutations for some, but not all beo alleles were identified (Hirata et al., 2005; Table 2). These resulted in either missense (tw38f, p.L255R in TM1, mi106a, p.L275H in TM2) or nonsense (tp221, p.Y79X) mutations. Since glra1 and glrbb genes are both located on zebrafish chromosome 14 and participate in the same GlyR complex, we speculated that some of the remaining beo alleles might actually be mutations in glra1 that could fail to complement mutations in glrbb due to linkage. To determine the types of mutations in remaining beo alleles ta86d, ta92, tm115 and tf242, we obtained these lines from the zebrafish Stock Center in Tübingen, Germany. We first amplified the nine coding exons of glrbb, which revealed unique missense mutations (tf242, c.T301G, p.Y79D) or nonsense mutations resulting in premature termination of the GlyR βb polypeptide (ta86d, c.T303A, p.Y79X; tm115, c.C325T, p.Q87X and ta92, c.A1093T, p.K343X respectively; Fig. 1, Table 2). It is also noteworthy that both tp221 and ta86d alleles harbor the same mutation — p.Y79X. Since the remaining beo allele, tu230, was lost (Granato et al., 1996), we conclude that all available beo alleles harbor mutations in glrbb.
Table 2.
Mutations identified in beo mutant alleles. SNVs that result in nonsense mutations (p.Y79X, p.Q87X and p.K343X) will lead to a truncated and incomplete proteins, leading to a deleterious phenotype. While ta86d, tp221, and tm115 mutations will truncate the protein close to the N-terminus (p.Y79X or p.Q79X) creating a small non-functional GlyR βb subunit, the viable ta92 allele (p.K343X) will truncate the GlyR βb subunit within the long intracellular loop between transmembrane helices M3 and M4. Notably, the M3–M4 loop is critical for binding gephyrin, which clusters the GlyRs at synapses. Since the gephyrin-binding site on the GlyR βb subunit is predicted to reside between amino acids 396 and 413 (Hirata et al., 2005), truncation of GlyR βb subunit at residue 343 would result in a truncated protein lacking the gephyrin binding site and the fourth membrane-spanning domain (M4). However, the N-terminal extracellular domain and M1–M3 would be intact. This perhaps explains why ta92 is the only viable beo allele. Missense mutations in alleles tf242, tw38f and mi106a, resulting in p.Y79D, p.L255R and p.R275H substitutions were analyzed using the bioinformatics packages SIFT and Polyphen-2, which predicted these amino acid substitutions to be deleterious or probably damaging, respectively.
| beo allele | cDNA | Substitutiond | Consequence | SIFT | Polyphen-2 | Reference |
|---|---|---|---|---|---|---|
| tu230a | Allele lost | – | – | – | – | Hirata et al. (2010) |
| tp221c | c.T303A | p.Y79X | Truncation | – | – | Hirata et al. (2005) |
| tm115 | c.C325T | p.Q87X | Truncation | – | – | This study |
| tf242 | c.T301G | p.Y79D | Missense | Deleterious (< 0.05) | Probably damaging (1.00) | This study |
| ta92b | c.A1093T | p.K343X | Truncation | – | – | This study |
| tw38f | c.T830G | p.L255R | Missense | Deleterious (< 0.05) | Probably damaging (1.00) | Hirata et al. (2005) |
| ta86d | c.T303A | p.Y79X | Truncation | – | – | This study |
| mi106a | c.G990A | p.R275H | Missense | Deleterious (< 0.05) | Probably damaging (1.00) | Hirata et al. (2005) |
Mutant lost.
Viable allele.
Strongest allele.
Amino acid co-ordinates correspond to the mature GlyR βb subunit polypeptide after signal peptide cleavage.
Splice site-targeted morpholino knockdown of glra1 and glrbb genes
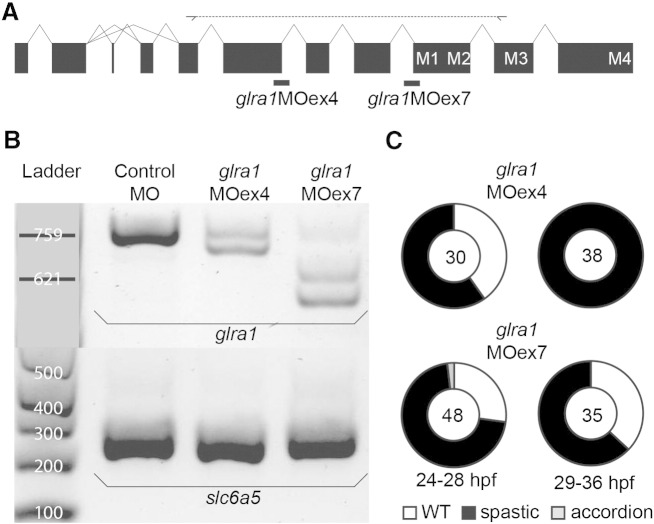
Given that all available beo alleles can be explained by mutations in glrbb, we investigated the phenotype resulting from glra1 inactivation in zebrafish. Two independent, splice site-targeted morpholinos (MOs), glra1MOex4 and glra1MOex7, were designed against intron/exon boundaries shared among all glra1 transcripts (Fig. 2A). The glra1MOex4 targets the 3′ acceptor site of exon 4, an exon that encodes determinants of glycine binding (Lynch, 2004; as well as the epitope recognized by the anti-GlyR antibody mAb4a) while glra1MOex7 targets the 5′ donor site of exon 7 that encodes the first two membrane-spanning domains (M1 and M2) of the GlyR α1 subunit. Both MOs would be expected to disrupt GlyR function by causing exon-skipping or intron-retention, introducing a frameshift, and truncating translated proteins. RT-PCR analysis of RNA harvested from 28 hpf glra1 morphants revealed the nature of mis-splicing events induced by MO injections. Diagnostic primers were designed to detect both exon-skipping and intron-retention events and morphant-specific bands were subsequently gel purified and sequenced to determine the exact nature of the MO-induced disruption (Eisen and Smith, 2008). For glra1MOex4, we identified a single morphant band caused by a 61 bp deletion due to a cryptic-splice-donor site towards the 3′ end of exon 4 (Fig. 2B; middle lane). For glra1MOex7, we identified two morphant-specific PCR products, both smaller than the wild type product (Fig. 2B; right-most lane). Sequencing of glra1MOex7 morphant-specific bands demonstrated that the lower of the PCR products is a result of skipping exon 7 entirely, while the middle band is the result of a cryptic splice-site in the middle of exon 7. The cryptic splice-site results in an in-frame transcript. Nonetheless, the resulting protein would still be expected to lack function because of deletion of the transmembrane domain M1.
Fig. 2.
Efficacy and target specificity of splice-site-blocking glra1ex4 and glra1ex7 morpholinos. A. Schematic representation of the zebrafish GlyR α1 subunit gene (glra1). Exons, shown as black boxes, are connected by introns, shown as lines. Two of the exons, 2a and 2b, are alternatively spliced as indicated by multiple possible intron lines. Exons that encode membrane-spanning domains M1–M4 are indicated. Two distinct splice-site-blocking MOs (thick black bars) were designed to knock down glra1 expression by masking exon/intron junctions. Exon4/intron4 is masked by glra1MOex4, and intron6/exon7 by glra1MOex7. Diagnostic primers were designed to amplify exons 3 to 8 to detect MO-induced mis-splicing events. B. RT-PCR results demonstrate specificity of altered glra1 pre-mRNA splicing caused by MO injection. The leftmost lane contains two size standards that replace the ladder and are based on direct Sanger sequencing of the bands. The following two lanes contain RT-PCR of RNA samples from 28 hpf embryos injected with 2 nL 0.5 mM control MO, 2 nL 0.25 mM glra1MOex4, and 2 nL 0.5 mM glra1MOex7 respectively. In the upper gel, glra1 diagnostic primers are used for PCR from cDNA synthesized using a gene-specific primer for glra1. In contrast to control morphants that show a single strong PCR product at 759 bp, glra1MOex4 morphants exhibit reduced levels of the wild type PCR product and a new product of 698 bp, indicating exon skipping. Likewise, glra1MOex7 morphants exhibit reduced levels of the wild type PCR product and two PCR products of 621 and 544 bp in size. In the lower gel, PCR for a GlyT2 cDNA (slc6a5) is used as a loading control, and is amplified from cDNA synthesized using anchored oligo-dT primers. C. Percentages of glra1MOex4 and glra1MOex7 morphants exhibiting wild type (wt), spastic or accordion phenotypes, plotted at 24–28 hpf and 29–36 hpf. The number of embryos analyzed is indicated at the center of each plot.
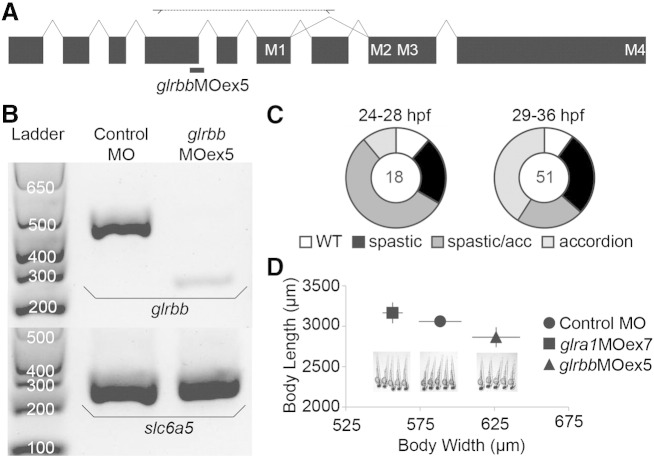
Although a translation-blocking MO had previously been designed to knock down glrbb (Hirata et al., 2005), this MO did not work in our hands, possibly due to sequence variability surrounding the start methionine in different wild-type strains. In addition, we cannot monitor the efficacy of this translation-blocking MO, due to the lack of a specific GlyR βb antibody. Therefore, we designed a new splice-site morpholino against 3′ acceptor site of glrbb exon 5, one of the largest exons upstream of the transmembrane domains (Fig. 3A). Using glrbbMOex5, we were able to faithfully recapitulate the beo mutant accordion phenotype resulting from tonic bilateral muscle contractions (Fig. 3D). RT-PCR analysis of RNA harvested from glrbbMOex5-injected embryos at 28 hpf demonstrates that glrbbMOex5 caused a 156 bp deletion and sequencing this PCR product demonstrated that a cryptic splice-donor site in exon 5 accounted for morpholino-induced mis-splicing (Fig. 3B; right lane).
Fig. 3.
Efficacy and target specificity of splice-site-blocking glrbb exon 5 morpholinos. A. Schematic representation of the zebrafish GlyR βb subunit gene (glrbb). Exons, shown as black boxes, are connected by introns, shown as lines. Exons that encode membrane-spanning domains M1–M4 are indicated. The glrbbMOex5 splice-site-blocking MO (thick black bar) was designed to knockdown glrbb expression by masking exon/intron junction intron4/exon5. Diagnostic primers designed to amplify exons 5 to 8 were used to detect MO-induced mis-splicing events. B. RT-PCR results demonstrate specificity of altered glrbb pre-mRNA splicing caused by MO injection. The leftmost lane contains a size standard ladder followed by two lanes of RT-PCR from RNA samples of 28 hpf embryos injected with 2 nL 0.5 mM control MO and 2 nL 0.5 mM glrbbMOex5 respectively. In the upper gel, glrbb diagnostic primers are used for PCR from cDNA synthesized using a gene-specific primer for glrbb. In contrast to control morphants, where a single strong PCR product of 430 bp is detected, glrbbMOex5 morphants exhibit reduced levels of the wild type PCR product and a new product of 274 bp, indicating exon skipping. In the lower gel, PCR for a GlyT2 cDNA (slc6a5), amplified from cDNA synthesized using anchored oligo-dT primers, is used as a loading control. C. Percentages of glrbbMOex5 morphants exhibiting wild type (wt), spastic or accordion phenotypes are plotted at 24–28 hpf and 29–36 hpf. The number of embryos analyzed is indicated at the center of each plot. D. Pictures of 5 representative 48 hpf larvae demonstrate shortening of the body axis produced by tonic bilateral contraction in glrbb but not glra1 morphants. Length and width (yolk to back) measurements (average n = 5, error bars indicate standard deviation). A Student's t-test indicates that glrbb morphants are significantly shorter (p < 0.05) and wider (p < 0.005) than their control and glra1 morphant counterparts.
Quantification of the onset of swimming behavior
From 24 to 36 hpf, zebrafish larvae transition from coiling behaviors (not propulsive) to rhythmic alternating motor behaviors that can propel the larva away from a threat (Saint-Amant and Drapeau, 1998). Therefore analysis of morphant behaviors at these stages occurs against a rapidly changing but highly stereotyped control baseline. To quantify behaviors, we determined the duration of discrete components of two, sequential touch-induced behaviors in each larva (Fig. 4). For example, in response to touch, a control morphant coils in one direction (light blue bar) until tail touches head (black column), coils in the opposite direction (dark blue bar) until tail touches head (black column) and slowly returns to rest (tan bar). While the exact timing of the two independent behaviors varies, the sequence of components is the same. To capture the developmental progression of control morphant behaviors from 24 to 36 hpf, we plotted single behaviors for 50 individual larva that span the behavioral transition from coiling to propulsive swimming (Fig. 5; left column). We split the 24–36 hpf time period into two qualitatively distinct early, 24–28 hpf, and late, 29–36 hpf bins. Staging is based on a two criteria: time in hpf and morphology (Kimmel et al., 1995). Single coils first transitioned to alternating coils, 24–28 hpf, and then, 29–36 hpf, to swimming characterized by shallower and more rapid alternating bends (Fig. 5; left column).
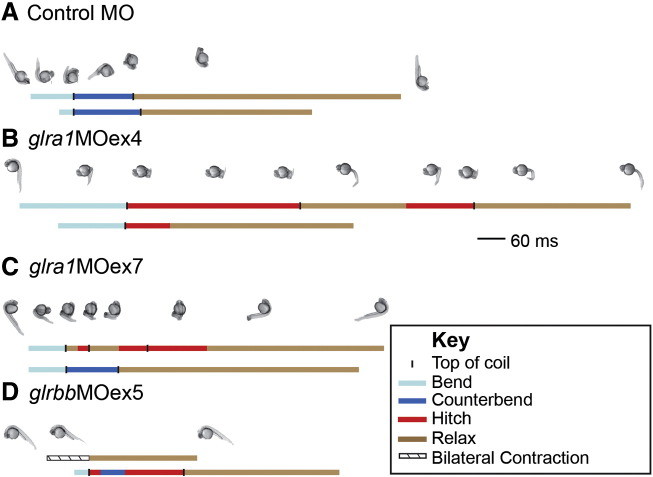
Fig. 4.
Repeated stimulation evokes erratic, sometimes spastic behaviors in glra1 and glrbb morphants. Two sequential, touch-evoked behaviors are plotted for four, representative morphant larvae: control, A; glra1MOex4, B; glra1MOex7, C; and glrbbMOex5, D. Behaviors were recorded using a high-speed camera and plotted as lines with the different behavioral components indicated by line color and each component's duration indicated by line length (scale bar = 60 ms; light blue = initial bend; dark blue = counter-bend; black column = top of coil, when the tail comes closest to touching the head; tan = return to rest; red = stuck or bend to same side; boxed diagonal lines = bilateral contraction). Still frames are included above the first of two plots with the number of pictures/length reflecting relative rates of change in the movement. A. The control morphant exhibited behavioral stereotypy with repeat stimulation: both behaviors consist of two coils in alternating directions followed by relaxation. B. The glra1MOex4 morphant exhibited erratic, spastic behaviors. In the first behavior, the glra1MOex4 morphant produced spastic behavior, getting stuck in the first coil and producing a second coil to the same side as the first. The second behavior was different from the first-the glra1MOex4 larva produced only one coil, and still spastic-the larva got stuck in the coil. C. The glra1MOex7 morphant also exhibited erratic, spastic behaviors. In the first behavior, the glra1MOex7 larva produced three coils, all to the same side. The second behavior was different from the first—this behavior was not spastic, consisting of two smooth alternating coils that return to rest. D. The glrbbMOex5 morphant larvae exhibited erratic, bilateral contraction and spastic behaviors. In the first behavior, the glrbbMOex5 morphant produced bilateral contraction that actually shortened the body axis–the classic accordion phenotype. The second behavior was different from the first—this time the glrbbMOex5 morphant produced two coils, getting stuck in each of these coils.
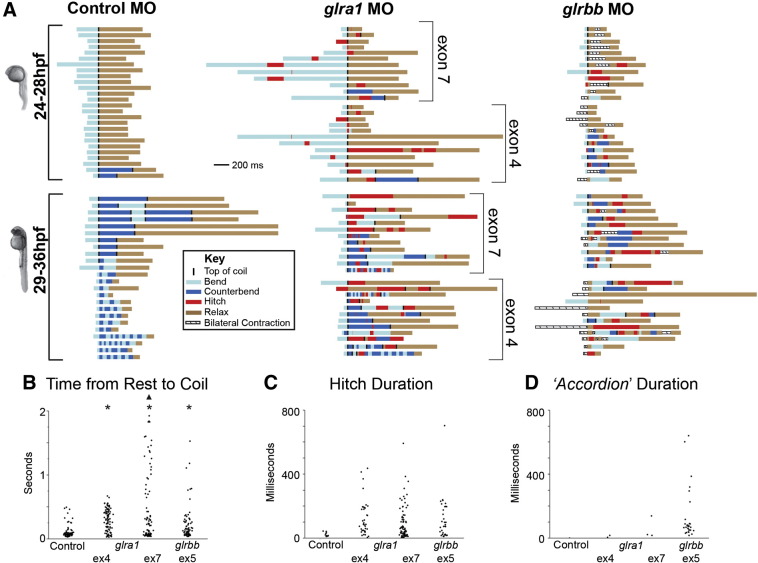
Fig. 5.
Spasticity is characteristic of both glra1 and glrbb morphants but bilateral contraction mainly occurs in glrbb morphants. A. One behavior per larva is plotted for representative populations of 50 control (left column), 48 glra1 (middle column), and 47 glrbb (right column) morphants that span the 24–36 hpf time period. All larvae were stimulated twice but only one of each pair of behaviors was plotted here. (Scale bar = 200 ms; light blue = initial bend direction; dark blue = counterbend; black column = top of coil, when the tail comes closest to touching the head; tan = return to rest; red = stuck or bend to same side; boxed diagonal lines = bilateral contraction). Larval staging was based on using the angle of the head to the long tail axis as diagnostic of stage (Kimmel et al., 1995). B. Time from rest to first coil is plotted for all recorded control (n = 83), glra1ex4 (n = 92), glra1ex7 (n = 92), and glrbbex5 (n = 68) morpholino-injected larvae. ANOVA indicated a significant effect of morpholino injected on time to first coil [F(3,334) = 15.8225, p = 0.0001]. Post hoc Bonferroni-corrected Student's t-tests compared all means and showed significant differences at p < 0.05 (*) that both glra1 morphants and glrbb morphants are significantly different from control morphants. In addition, glra1ex7 morphant times to first coil were significantly longer (p < 0.05, black triangle) than glra1ex4 and glrbbex5 morphants. C. Hitch durations in milliseconds are plotted for any of the control, glra1, or glrbb larva that exhibited this behavior. ANOVA indicated no significant effect of morpholino injected on hitch time [F(3,155) = 2.41, p = 0.07], thus there is no subsequent post hoc analysis. D. ‘Accordion’ durations in milliseconds are plotted for any of the control, glra1, or glrbb larva that exhibited this behavior. The ‘accordion’ behavior is clearly more frequent in glrbbex5 morphants, occurring 27 times compared to never in controls and 5 times in all glra1 morphant larvae.
glra1 and glrbb morphants exhibit spasticity
Both glra1 and glrbb morphants exhibit spastic and erratic behaviors during 24–36 hpf (Figs. 2C & 3C). Spastic behaviors (indicated by red bars) were characterized by hitches. We defined hitches as pauses that disrupted the smooth progression of a movement and/or repeated bends to one side (Figs. 4B, C & D; Fig. 5, middle & right columns). Behaviors were also erratic, lacking stereotypy when elicited multiple times (Supplemental movies; Figs. 4B, C & D). In addition, the duration of the time to first coil was significantly longer in glra1 morphants (glra1MOex4: 296 ± 18 ms, n = 93; glra1MOex7: 444 ± 56 ms, n = 93) and glrbb morphants (glrbbMOex5: 241 ± 33 ms, n = 69) compared to control morphants (Control MO: 109 ± 11 ms, n = 83, means ± standard deviation for all treatments; Fig. 5B). Spasticity in both glra1 and glrbb morphants (Fig. 5C) was associated with a delay in the onset of rhythmic behaviors seen in control morphants at 30–36 hpf. In addition to spasticity, glrbb morphants exhibited simultaneous bilateral contractions strong enough to shorten the body axis–the diagnostic accordion phenotype (Granato et al., 1996; Figs. 3D, 4D & 5D; Supplemental movies). Moreover, in contrast to glra1 morphants in which behavioral disruptions were transient, glrbb morphant phenotypes persisted, resulting in larvae with a shortened axis by 48 hpf (Fig. 3D). Spastic phenotypes were recapitulated by injecting mRNA encoding a dominant-negative GlyR α1 subunit mutant R270Q (Figs. 6B & C). Curiously, by contrast to the dominant-negative glra1 R270Q RNA that lacked a morphological phenotype, over-expressing the wild type GlyR α1 subunit mRNA produced shortened-axis and Cyclops phenotypes (Figs. 6A & C). Cyclopic phenotypes are commonly associated with disrupted sonic hedgehog (shh) signaling in early embryos and likely reflect the disruptive effect of mis-expressing functional glycine receptors.
Fig. 6.
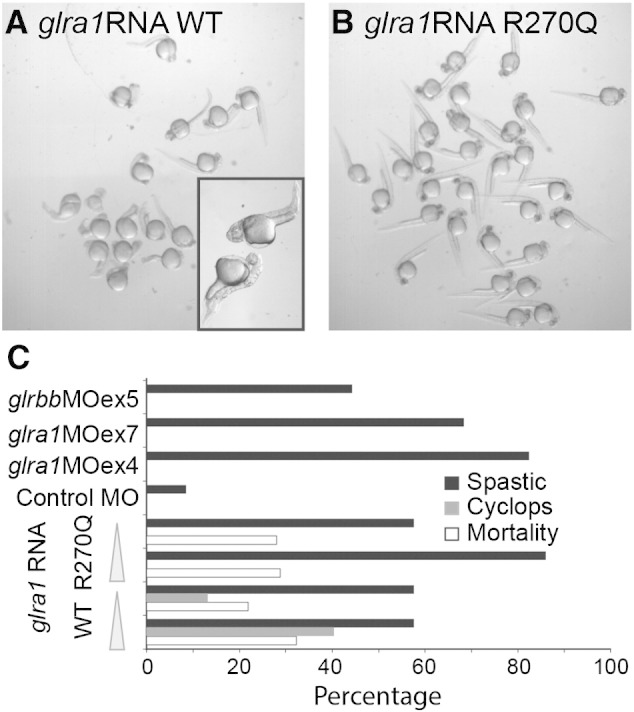
Injection of dominant-negative GlyR α1 subunit R270Q mutant RNA recapitulates glra1 morphant spasticity. A, B. Populations of 30 hpf larvae were injected as zygotes with ~ 2 ng of either wild type, A, or R270Q dominant-negative glra1 RNA, B. While R270Q glra1 RNA-injected larvae were morphologically normal, surprisingly, 60% of wild type glra1 RNA-injected larvae exhibited shortened axis and cyclopic phenotypes. These phenotypes are more commonly associated with defects in early signaling through the sonic hedgehog pathway and are likely a consequence of expressing functional glycine receptors at a time when they are not normally expressed. In A, boxed inset, it is possible to see the medially fused eyes characteristic of the Cyclops phenotype. C. Percentages of larvae from different treatment groups that exhibited spastic or Cyclops phenotypes or mortality are plotted. RNAs encoding wild type and GlyR α1 subunit R270Q mutant were each injected at two concentrations indicated by the wedge (wt high ~ 2 ng, n = 25; low ~ 400 pg, n = 23; R270Q high ~ 2 ng, n = 42; low ~ 400 pg, n = 17).
glrbb is epistatic to glra1 at the level of behavior
To formally test for epistatic relationships between glra1 and glrbb, we co-injected glrbbMOex5 with wither glra1MOex4, glra1MOex7, or control morpholinos and analyzed the resulting phenotypes (Table 3). Embryos co-injected with glrbb and glra1 morpholinos exhibited the classic accordion phenotype of bilateral muscle contractions similar to glrbb morphants co-injected with control morpholino showing that at the level of behavior glrbb is epistatic to glra1.
Table 3.
Epistasis analysis between glra1 and glrbb phenotypes. Batches of embryos were co-injected with either 0.5 mM glrbbMOex5 and 0.5 mM control morpholino, 0.5 mM glrbbMOex5 and 0.25 mM glra1MOex4, or 0.5 mM glrbbMOex5 and 0.5 mM glra1MOex7 and assayed for morphological and behavioral phenotypes between 28 and 36 hpf. Percentages of embryos, with numbers in each treatment in parentheses below, are indicated for each treatment and morphology. For behavior, larvae were tested for whether they ever produced accordion. The ‘yes’ for each treatment indicates that ‘accordion’ was the typical behavioral response.
| 0.5 mM glrbbex5 + 0.5 mM ConMO | 0.5 mM glrbbex5 + 0.25 mM glra1ex4 | 0.5 mM glrbbex5 + 0.5 mM glra1ex7 | |
|---|---|---|---|
| Normal morphology | 87% (n = 118) | 9% (n = 5) | 42% (n = 26) |
| Disrupted morphology | 13% (n = 18) | 91% (n = 52) | 58% (n = 35) |
| Accordion? | Yes | Yes | Yes |
In addition, we also observed a novel morphological disruption in embryos co-injected with glrbbMOex5 and either glra1MOex4, or glra1MOex7 morpholinos but not in embryos co-injected with glrbbMOex5 and control morpholinos suggesting synergistic roles for glra1 and glrbb in development, perhaps relating to their functional roles in spinal cord stem cells.
Glycinergic synaptogenesis in control, glra1, and glrbb morphant zebrafish
To compare the impact of knocking down GlyR α1 and βb subunits on glycinergic synaptogenesis, we carried out immunohistochemistry, double-labeling 24 and 48 hpf fresh-frozen sections with antibodies recognizing GlyR α subunits and the GlyR clustering protein gephyrin (Fig. 7). At 24 and 48 hpf, progenitor cells are still actively dividing in the middle of the spinal cord; surrounding these progenitors are the first neurons to exit the cell cycle and differentiate. Differentiated neurons include primary motor neurons (PMNs) some interneurons, and Rohon Beard sensory neurons. For immunohistochemistry, we focus on the largest neurons, likely PMNs, in the motor neuron/interneuron domain (Fig. 7; white boxes). GlyR α and gephyrin immunohistochemistry were both characterized by bright puncta that likely reflect enrichment of these proteins at postsynaptic densities. Surprisingly, in 24 hpf control morphants, GlyR α subunit and gephyrin puncta did not co-localize (Figs. 7A & 8A). While gephyrin puncta were enriched exclusively in the domain occupied by differentiated neurons, the GlyR α subunit puncta also decorated progenitor cells in the medial cord (Fig. 7A; red arrowheads). Even within single neurons exhibiting both GlyR and gephyrin puncta, there was little overlap (Fig. 7A; red & green arrows).
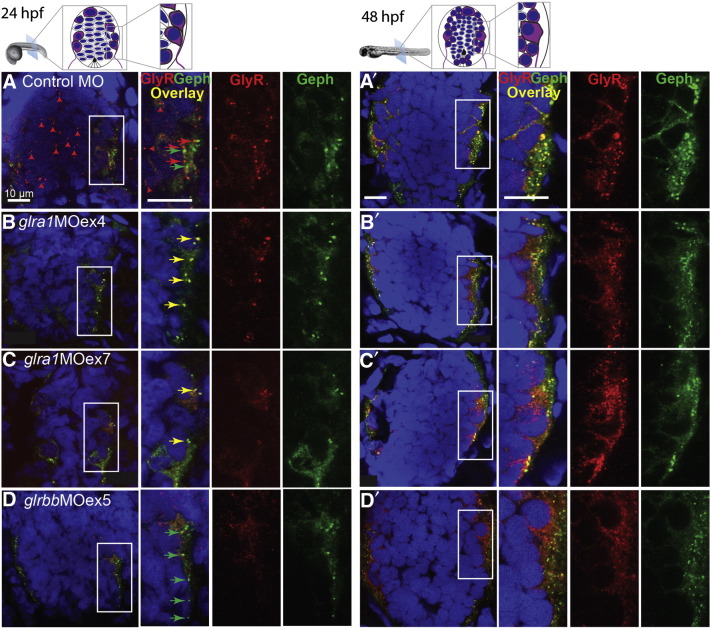
Fig. 7.
GlyR α puncta are reduced in glra1 morphants and absent in glrbb morphants. Transverse sections through fast-frozen 24 hpf (left; A–D) and 48 hpf (right; A′–D′) larvae were stained for GlyR α subunits (red), gephyrin (green), and nuclei (blue). Diagrams above the micrographs show the tight packing of spinal cord cells. In these diagrams, cells with purple cytoplasm represent the differentiated neurons: primary motor neurons (PMNs) that exit the spinal cord to innervate muscle, early differentiating interneurons, and dorsal sensory Rohon Beard neurons. These differentiated neurons form in a rind that surrounds the medial stem cells, outlined in gray. By 48 hpf more neurons have differentiated and become incorporated into the circuit as reflected by an increased synaptic staining on lateral neurons. Representative images for each stage and morpholino treatment are displayed as groupings of four images with the leftmost image giving a low magnification view of the entire spinal cord with the motor/inter-neuron region boxed in white magnified in the subsequent three images. A. In 24 hpf control morphants, GlyR α and gephyrin staining are non-overlapping. GlyR puncta decorate precursor cells near the spinal cord midline (red arrowheads) as well as lateral neurons (red arrows). Gephyrin puncta (green arrows) are solely associated with lateral neurons, but do not co-localize with GlyRs at this developmental time point. B, C. In both glra1 morphants, GlyR α subunit staining is absent in precursors near the midline. Curiously, in glra1 morphants, residual GlyR α subunits did co-localize with gephyrin (yellow arrows) in lateral neurons. D. In 24 hpf glrbbMOex5 morphants, gephyrin puncta (green arrows) occur in the absence of GlyR α subunit puncta. A′. In 48 hpf control morphants, GlyR α subunit and gephyrin puncta commonly co-localized. B′, C′. In 48 hpf glra1 morphants, a subset of GlyR α subunit and gephyrin puncta commonly co-localized but these co-localized puncta are positioned more laterally than in control morphants. D′. 48 hpf glrbbMOex5 morphants lacked clear GlyR α subunit puncta, but exhibited smaller gephyrin puncta that tended towards the lateral neuropil rather than circum-nuclear as in control morphants. Scale bars: 10 μm.
Fig. 8.
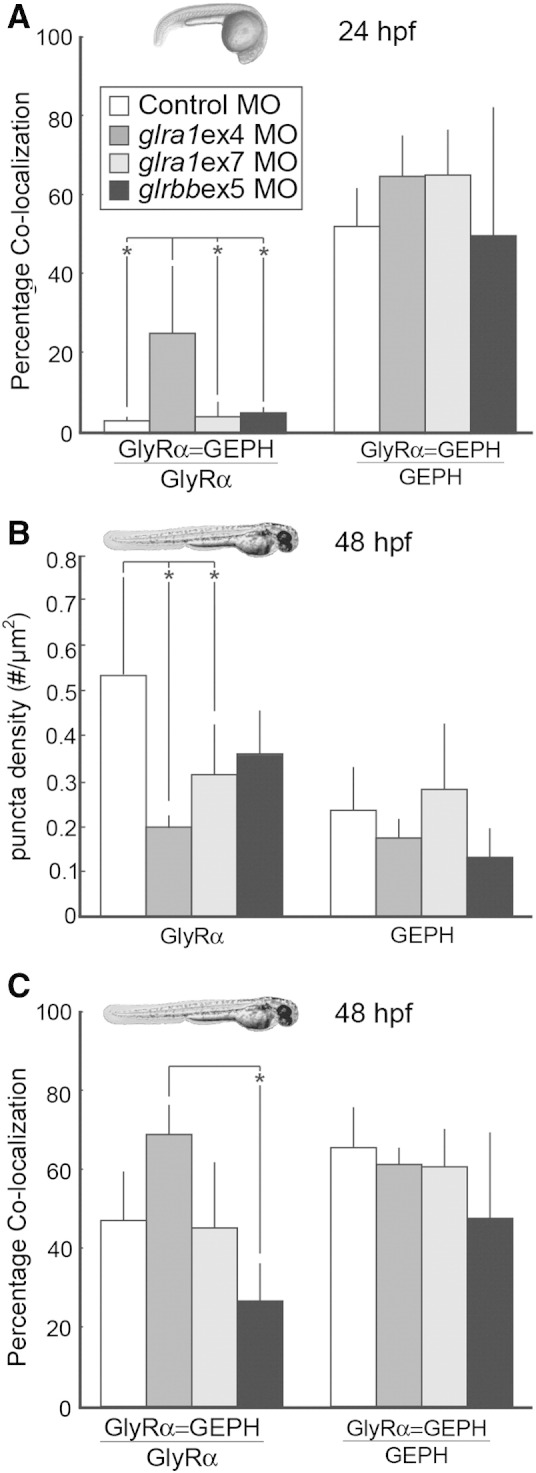
GlyR densities are significantly reduced in glra1 morphants, while the percentage of glycine receptors that co-localize with gephyrin is reduced in glrbb morphants. A. After GlyR α and gephyrin (GEPH) puncta were found using a customized MatLab program, co-localization was calculated in control (n = 7), glra1 (ex4 n = 6; ex7 n = 3), and glrbb (n = 3) 24 hpf morphants in two ways: either as the fraction of GlyRs that co-localized with GEPH (ANOVA indicated a significant effect of morpholino injected on percent of GlyR α puncta that co-localizes with GEPH [F(3,15) = 6.55, p = 0.005]), or as the fraction of GEPH that colocalized with GlyR (ANOVA indicated no significant effect of morpholino injected [F(3,15) = 1.27, p = 0.32]). Bar graphs show means with error bars indicating standard deviation for all groups. Post hoc Bonferroni-corrected Student's t-tests compared all means and demonstrated that at p < 0.05 (* in graph), only glra1MOex4 had significantly more co-localization than control, glra1MOex7, and glrbbMOex5 morpholino treatments. Surprisingly fewer GlyR α puncta co-localize with GEPH at 24 hpf than at 48 hpf, C, while the proportion of GEPH colocalizing with GlyR puncta remains fairly constant. The proportion of GlyR α puncta that co-localize with GEPH is greatest in the glra1ex4 morphants that lack the GlyR α epitope, supporting the conclusion that in glra1 morphants, residual co-localization reflects synaptic staining of GlyR α2, α3, or α4a. B. GlyR and GEPH densities were calculated in control (n = 6), glra1 (ex4 n = 3; ex7 n = 7), and glrbb (n = 5) morphants at 48 hpf. Bar graphs show means with error bars indicating standard deviation. ANOVA indicates a significant effect of the mopholino injected on GlyR puncta density [F(3, 16) = 4.39, p = 0.02] but not on GEPH puncta [F(3,16) = 2.03, p = 0.15]. Post hoc Bonferroni-corrected Student's t-tests compared all means and demonstrated that at p < 0.05 (* in graph), only glra1MOex4 and glra1MOex7 were significantly reduced control morpholino treatments, although glrbbMOex5 treatments also trends in the same direction. C. Co-localization between GlyR and GEPH was quantified in control (n = 5), glra1(ex4 n = 3; ex7 n = 6), and glrbb (n = 4) 48 hpf larva in two ways: either as the fraction of GlyRs that co-localized with GEPH (ANOVA indicated a significant effect of morpholino injected on percent of GlyR α puncta that co-localizes with GEPH [F(3,16) = 5.89, p = 0.007]), or as the fraction of GEPH that colocalized with GlyR (ANOVA indicated no significant effect of morpholino injected [F(3,14) = 1.56, p = 0.24]). Bar graphs show means with error bars indicating standard deviation for all groups. Post hoc Bonferroni-corrected Student's t-tests compared all means and demonstrated that at p < 0.05 (* in graph), only the glrbbMOex5 treatment had significantly less co-localization than the glra1MOex4 treatment. In glra1MOex7 morphants, many peri-nuclear GlyR puncta fail to co-localize with GEPH. These likely represent GlyRs that are trapped intracellularly. Differences between glra1MOex4 and glra1MOex7 are likely explained by fewer intracellular GlyRs being recognized by mAb4a in glra1MOex4, since this antibody targets amino acids 96–105 that are lacking in glra1ex4 morphants. Therefore, residual staining in glra1MOex4 morphants is likely to reflect other GlyR α subunits, e.g. 2, 3, or 4a.
In contrast to wild type embryos, when the GlyR α1 subunit was knocked down either using glra1MOex4 or glra1MOex7, GlyR puncta in the medial spinal cord were dramatically reduced (Figs. 7B & C). Remaining GlyR puncta were enriched in the lateral spinal cord where, in contrast to control morphants, more of the GlyR puncta co-localized with gephyrin (Figs. 7B, C & 8A glra1MOex4). As with GlyR α1 morphants, knocking down the GlyR βb subunit using glrbbMOex5 also reduces GlyR staining in the medial spinal cord. In addition, however, glrbb morphants also exhibited reduced GlyR α puncta in the lateral spinal cord with fewer puncta that co-localize with gephyrin (Fig. 7D). By 48 hpf in control morphants, GlyR and gephyrin puncta colocalized in the lateral spinal cord (Fig. 7A′). Knocking down the GlyR α1 subunit reduced the density of GlyR puncta (Figs. 7B′, C′ & 8B). Knocking down the GlyR βb subunit, as would be expected, also disrupted GlyR and gephyrin co-localization (Figs. 7D′ & 8B–C).
In summary, punctate GlyR immuno-staining was reduced in both glra1 and glrbb morphants at both 24 and 48 hpf. At 24 hpf, GlyR puncta in glra1 morphants were dramatically reduced in the medial spinal cord, although some remaining puncta – presumably containing other GlyR α subunits – co-localizing with gephyrin could be detected in the lateral spinal cord. This staining pattern contrasts with glrbb morphants, in which loss of both GlyR clusters and co-localization with gephyrin was evident.
Quantification of gene expression for components of the glycinergic synapse in developing zebrafish
To quantify the relative mRNA expression levels of genes known to be critical to glycinergic synapse function, we generated a developmental time-course for eleven zebrafish genes implicated in the function of glycinergic synapses using quantitative PCR (Fig. 9). While quantitative PCR yields no spatial information, it does reveal two distinct groupings of genes that share developmental expression trajectories. Expression of the majority of genes including glra1, glra2, glra4a, glrba, glrbb, slc6a5 (GlyT2), and gphnb exhibited a steady increase with development. By contrast, expression of glra3, slc6a9 (GlyT1) and slc32a1 (VIAAT) increased until 48 hpf and then either leveled off (GlyT1/VIAAT) or decreased (glra3, gphna). These shared developmental expression trajectories could reflect shared transcriptional regulation. In the context of multiple duplicated subunits, qPCR also provides information about the dominant subunits expressed at different developmental time periods at the mRNA level. For example, in the case of the GlyR β subunits, the βb subunit mRNA is ~ 5 fold more abundant at both 48 and 72 hpf than βa. Likewise, the expression of gphna mRNA dominates at 48 hpf being 35 times more abundant than gphnb while at 72 hpf, it is only four times more abundant. The levels of GlyR α subunit transcripts differ with the most abundant being glra2 and glra3. Expression of glra1, glra4a and glrbb, is barely detectable at 24 and 32 hpf, but increases onwards from 48 hpf.
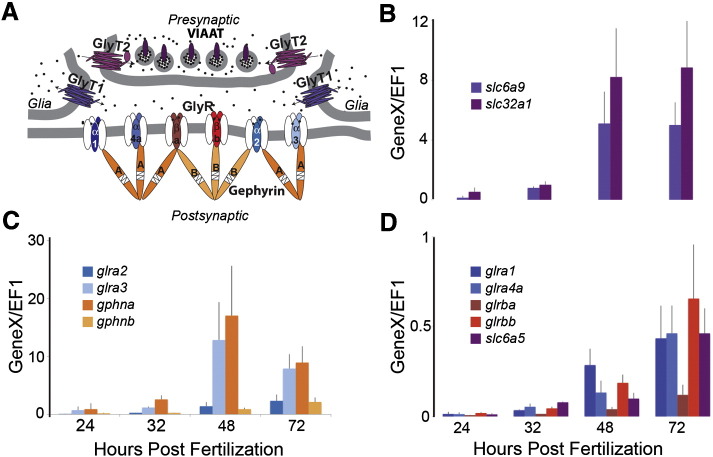
Fig. 9.
Developmental expression profile of glycinergic synapse components. A. A schematic diagram of a glycinergic synapse depicts important proteins found in presynaptic, glial, and postsynaptic compartments. The functional classes of genes are color-coded with glycine transporters (GlyT1, GlyT2, VIAAT) in shades of purple, GlyR α1–4a and GlyR βa–b are red and blue respectively, and gephyrin A and B in shades of orange. GlyT2 transports glycine into the presynaptic terminal, where VIAAT loads glycine into vesicles for release into the synaptic cleft. Glycine then binds to postsynaptic GlyRs. Glycinergic signaling is terminated when glial GlyT1 removes glycine from the synaptic cleft. Six of the seven zebrafish GlyR subunits, α1, α3, α3, α4a, βa and βb, are shown assembled into pentameric, ligand-gated Cl− channels with a 2α:3β subunit stoichiometry. These GlyRs are shown clustered at the synapse via interactions between GlyR β subunit and the scaffolding protein gephyrin. B–D. Average expression levels for all genes and time points (24, 32, 48, 72 hpf) are shown for an average of three independent mRNA preparations ± standard deviation. Genes are grouped into graphs according to their levels of expression and all gene expression is normalized to the housekeeping gene EF1. B. GlyT1 (slc6a9) and VIAAT (slc32a1) expression levels. C. GlyR α1, α4a, βa and βb subunit (glra1, glra4a, glrba, glrbb) and GlyT2 (slc6a9) expression levels. D. GlyR α2 subunit, α3 subunit, gephyrin-a and gephyrin-b (glra2, glra3, gphna, gphnb) expression levels.
Discussion
Our study demonstrated that all available alleles of the GlyR mutant bandoneon correspond to defects in glrbb, encoding the GlyR βb subunit. Consistent with recent studies on human startle disease mutations in the GlyR β subunit gene (Al-Owain et al., 2012; Chung et al., 2013; James et al., 2013; Lee et al., 2013), mutations resulting in protein truncation are the predominant mechanism disrupting GlyR βb subunit function in the beo allele series (4/7 alleles), followed by missense mutations affecting residues in the membrane spanning domains M1/M2 (2/7 alleles) or large extracellular domain (1/7 alleles). Y79D introduces a negative charge and a hydrophilic residue into a packed area of hydrophobic side-chains, which is likely to disrupt the local β-sheet fold on which it resides in the extracellular domain, possibly indirectly affecting glycine binding. The substitution L255R may disrupt packing of hydrophobic and aromatic residues between TM helices due to the large, positively charged side-chain of arginine, thus affecting the correct fold of the subunit or insertion into the membrane. R275 is equivalent to R252 in the human GlyR α1 subunit (James et al., 2013). A spontaneous mutation causing the same substitution in this residue (R252H) is known to cause startle disease when inherited in compound heterozygote manner (Vergouwe et al., 1999). Substitutions at R252 in the GlyR α1 subunit have been shown to severely disrupt the correct function of the protein, specifically affecting GlyR membrane trafficking (Rea et al., 2002). Thus, R275H in the zebrafish GlyR βb subunit is predicted to operate by a similar pathogenic mechanism.
Since mutations in GlyR α1, β subunit or GlyT2 genes all cause startle disease in dogs, cattle, mice and humans, one mystery is why no mutations in glra1 or slc6a5 have been discovered to date in phenotypic screens of zebrafish mutants generated by ENU mutagenesis (Granato et al., 1996). We now have a potential explanation for this finding, since knocking down glra1 (encoding the GlyR α1 subunit) produced a relatively mild phenotype compared to beo, characterized by spasticity that delayed the onset of rhythmic motor behaviors. This phenotype is likely to be genuine, since it was reproduced using two different MOs, and using a dominant-negative zebrafish GlyR α1 subunit mutant, R270Q — equivalent to the most common dominant startle disease mutation found in humans (R271Q; Chung et al., 2010; Shiang et al., 1993).
Notably, this phenotype is distinct from the ‘accordion’ behavior produced by inactivation of glrbb, encoding the GlyR βb subunit (Hirata et al., 2005). While our study clearly implicates GlyRs containing the α1 subunit in the smooth transition from spontaneous coiling behaviors to propulsive locomotion, the finding that glra1 loss-of-function causes this transient phenotype in zebrafish was unexpected. Several potential compensatory mechanisms have been observed in animals with loss-of-function mutations in the GlyR α1 subunit gene, including increased presynaptic glycine uptake, increased inhibitory GABAergic transmission, or up-regulation of additional GlyR α subunit genes (Graham et al., 2003; Gundlach et al., 1988; Kling et al., 1997; Lummis et al., 1990; Wässle et al., 1998). However, a strong visible phenotype is always observed in these animal models and in recessive human startle disease caused by GLRA1 mutations (Chung et al., 2010). So what could be different in the zebrafish? Since the beo phenotype can also be reproduced by applying the GlyR antagonist strychnine (Granato et al., 1996; Hirata et al., 2005), our interpretation is that the beo phenotype results from the loss-of-function of multiple GlyR subtypes. This supported by epistasis experiments that demonstrate glrbb is epistatic to glra1 at the level of behavior. Consistent with epistasis, immunohistochemistry showed that glra1 morphants exhibited residual GlyR α subunit puncta, while in glrbb morphants, GlyR α subunits were trapped in intracellular aggregates. Moreover, qPCR results also identified glra2, glra3, and glra4a in developing embryos, which could participate in α2βb, α3βb or α4βb GlyRs. Certainly, compared with mammals, the zebrafish has additional opportunities for compensating for the loss of glra1, due to the duplication of the α4 subunit genes (glra4a, glra4b; Hirata et al., 2010). In our view, the zebrafish GlyR gene expansion (Hirata et al., 2010) reflects strong selective pressure for effective escape responses in this organism that develops externally with minimal parental care. Taken together, our data suggest that knocking down zebrafish orthologs of human hyperekplexia genes will likely produce a phenotypic range from more mild spasticity (glra1 and glrbb) to bilateral contractions (glrbb).
The behavioral transition from slow coiling to fast rhythmic flexions (Saint-Amant and Drapeau, 1998; Warp et al., 2012), which is delayed in glra1 morphants, reflects changes taking place in the connectivity of neural circuits. While embryonic slow coiling behaviors rely upon gap-junctional coupling among neurons (Brustein et al., 2003; Ganser and Dallman, 2009; Saint-Amant and Drapeau, 2001), larval rhythmic flexions rely upon synaptic inputs (Brustein et al., 2003; Kinkhabwala et al., 2011; Koyama et al., 2011; McLean and Fetcho, 2009; McLean et al., 2008). Our data show that zebrafish express α1βb GlyRs throughout this period of behavioral transition. These findings contrast with rodent spinal cord (Agaki et al., 1991; Becker et al., 1988; Harvey et al., 2004; Singer et al., 1998) and embryonic spinal cultures (Hoch et al., 1989) in which the GlyR α2 subunit predominates at early stages with the GlyR α1 and α3 subunits appearing postnatally. By contrast, in zebrafish, we show that GlyR α subunit immunostaining on neuronal precursors and differentiating neurons was disrupted by knocking down either glra1 or glrbb, suggesting that the majority of early GlyR puncta in 24 hpf zebrafish spinal cord are composed of α1 and βb subunits.
In addition to glycine (Flint et al., 1998), non-synaptic release of both glutamate and GABA are known to cause Ca2 + transients in neural precursors that promote growth and maturation (Akerman and Cline, 2006; Ben-Ari and Spitzer, 2010; Haydar et al., 2000; Komuro and Rakic, 1996; Owens and Kriegstein, 2002). Both immature neurons and muscle express a variety of neurotransmitter receptors prior to innervation (Borodinsky et al., 2004; Moody and Bosma, 2005) at which point neurotransmitters influence a range of processes including proliferation, migration, and differentiation (Ben-Ari et al., 2007; LoTurco et al., 1995; Moody and Bosma, 2005; Owens and Kriegstein, 2002; Spitzer, 2006). For both glycine and GABA, the developmental transition from depolarizing to hyperpolarizing involves a shift in the chloride concentration gradient that is brought about by the up-regulation of the Cl− transporter KCC2 (Ben-Ari et al., 2007). In zebrafish, prematurely expressing KCC2 to block the depolarizing influence of glycine and GABA reduces motor and interneuron differentiation (Reynolds et al., 2008). KCC2 expression is actually triggered by GABA-induced Ca2 + transients indicating a mechanism of developmental negative feedback (Fiumelli et al., 2005; Ganguly et al., 2001). Such feedback is likely a common feature of the developing nervous system (Dallman et al., 1998; Moody and Bosma, 2005) creating sequential check-points so that development only progresses when coordinate regulation of channels, receptors, and transporters successfully produce signals that feedback to promote maturation and qualitatively change excitability of the circuit (Ben-Ari and Spitzer, 2010). One possibility is that by knocking down glra1 we delay the onset of rhythmic behaviors by disrupting signaling associated with depolarizing glycine (Ben-Ari and Spitzer, 2010).
Conclusions
Recent proteomic analyses of glutamatergic synapses suggest that up to a thousand proteins participate in this neuronal specialization (Grant, 2012). Even if glycinergic synapse function requires a fraction of this number of specialized proteins, the door is certainly open to complex forms of inheritance for startle disease. Despite the significant successes in identifying genetic defects that cause startle disease, many idiopathic cases remain in which the genetic cause is unresolved (Carta et al., 2012; Chung et al., 2010, 2013; James et al., 2013). Exome sequencing in these idiopathic cases will identify new candidate startle disease genes in the near future, necessitating animal models in which to test the validity of new candidate genes (Chung et al., 2013; James et al., 2013). Our study improves the utility of zebrafish as a startle disease model by identifying early phenotypes produced by two known startle disease genes in zebrafish that can be used to screen newly identified candidate genes via morpholino knockdown and in overexpression models using proteins harboring potential disease-causing mutations.
Acknowledgments
We would like to thank UM Biology Molecular Core manager Dr. Hurt and Imaging Core manager Dr. Baker for their help with sequencing and confocal imaging respectively. We thank Drs. Baker and Wikramanayake for their feedback on writing, Anthony Sanchez for his assistance in analyzing embryonic movements, and Ricardo Cepeda (UM), Prof. Steve Wilson and Carole Wilson (UCL) for fish care. We would like to acknowledge Dr. Hiromi Hirata for his assistance in generating fast-frozen tissue sections and members of Dr. R. O. Wong's lab, a Travel Grant from the Company of Biologists, and Dr. Jiang for teaching Qing Yan to program MatLab to quantify immunohistochemistry. We also thank Lingyu Wang for assisting in the development of an image analysis work station. Grant/Other Support: Medical Research Council (G0601585 to RJH), University of Miami Provost Award and National Institutes of Health NINDS (K01NS048200 to JED).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.nbd.2013.09.002.
Appendix A. Supplementary data
Supplementary video 1.
Supplementary video 2.
Supplementary video 3.
Supplementary video 4.
Supplementary video 5.
Supplementary video 6.
Supplementary video 7.
Supplementary video 8.
Supplementary video 9.
Supplementary video 10.
References
- Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agaki H., Hirai K., Hishinumu F. Cloning of a glycine receptor subtype expressed in rat brain and spinal cord during a specific period of neuronal development. FEBS Lett. 1991;281:160–166. doi: 10.1016/0014-5793(91)80383-e. [DOI] [PubMed] [Google Scholar]
- Akerman C.J., Cline H.T. Depolarizing GABAergic conductances regulate the balance of excitation to inhibition in the developing retinotectal circuit in vivo. J. Neurosci. 2006;26:5117–5130. doi: 10.1523/JNEUROSCI.0319-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Owain M., Colak D., Al-Bakheet A., Al-Hashmi N., Shuaib T., Al-Hemidan A., Aldhalaan H., Rahbeeni Z., Al-Sayed M., Al-Younes B., Ozand P.T., Kaya N. Novel mutation in GLRB in a large family with hereditary hyperekplexia. Clin. Genet. 2012;81:479–484. doi: 10.1111/j.1399-0004.2011.01661.x. [DOI] [PubMed] [Google Scholar]
- Becker C.M., Hoch W., Betz H. Glycine receptor heterogeneity in rat spinal cord during postnatal development. EMBO J. 1988;7:3717–3726. doi: 10.1002/j.1460-2075.1988.tb03255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Y., Spitzer N.C. Phenotypic checkpoints regulate neuronal development. Trends Neurosci. 2010;33:485–492. doi: 10.1016/j.tins.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Y., Gaiarsa J.L., Tyzio R., Khazipov R. GABA: a pioneer transmitter that excites immature neurons and generates primitive oscillations. Physiol. Rev. 2007;87:1215–1284. doi: 10.1152/physrev.00017.2006. [DOI] [PubMed] [Google Scholar]
- Borodinsky L.N., Root C.M., Cronin J., Sann S.B., Gu X., Spitzer N.C. Activity-dependent homeostatic specification of transmitter expression in embryonic neurons. Nature. 2004;429:523–530. doi: 10.1038/nature02518. [DOI] [PubMed] [Google Scholar]
- Brustein E., Saint-Amant L., Buss R.R., Chong M., McDearmid J.R., Drapeau P. Step during the development of the zebrafish locomotor network. J. Physiol. Paris. 2003;97:77–86. doi: 10.1016/j.jphysparis.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Buckwalter M.S., Cook S.A., Davisson M.T., White W.F., Camper S.A. A frameshift mutation in the mouse α1 glycine receptor gene (Glra1) results in progressive neurological symptoms and juvenile death. Hum. Mol. Genet. 1994;3:2025–2030. doi: 10.1093/hmg/3.11.2025. [DOI] [PubMed] [Google Scholar]
- Burgess H.A., Granato M. Sensorimotor gating in larval zebrafish. J. Neurosci. 2007;27:4984–4994. doi: 10.1523/JNEUROSCI.0615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carta E., Chung S.-K., James V.M., Robinson A., Gill J.L., Remy N., Vanbellinghen J.F., Drew C.J.G., Cagdas S., Cameron D., Cowan F.M., Del Toro M.D., Graham G.E., Manzur A.Y., Masri A., Rivera S., Scalais E., Shiang R., Sinclair K., Stuart C.A., Tijssen M.A.J., Wise G., Zuberi S.M., Harvey K., Pearce B.R., Topf M., Thomas R.H., Supplisson S., Rees M.I., Harvey R.J. Mutations in the GlyT2 gene (SLC6A5) are a second major cause of hyperekplexia. J. Biol. Chem. 2012;287:28975–28985. doi: 10.1074/jbc.M112.372094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier C., Coppieters W., Rollin F., Desmecht D., Agerholm J.S., Cambisano N., Carta E., Dardano S., Dive M., Fasquelle C., Frennet J.C., Hanset R., Hubin X., Jorgensen C., Karim L., Kent M., Harvey K., Pearce B.R., Simon P., Tama N., Nie H., Vandeputte S., Lien S., Longeri M., Fredholm M., Harvey R.J., Georges M. Highly effective SNP-based association mapping and management of recessive defects in livestock. Nat. Genet. 2008;40:449–454. doi: 10.1038/ng.96. [DOI] [PubMed] [Google Scholar]
- Chung S.K., Vanbellinghen J.F., Mullins J.G., Robinson A., Hantke J., Hammond C.L., Gilbert D.F., Freilinger M., Ryan M., Kruer M.C., Masri A., Gurses C., Ferrie C., Harvey K., Shiang R., Christodoulou J., Andermann F., Andermann E., Thomas R.H., Harvey R.J., Lynch J.W., Rees M.I. Pathophysiological mechanisms of dominant and recessive GLRA1 mutations in hyperekplexia. J. Neurosci. 2010;30:9612–9620. doi: 10.1523/JNEUROSCI.1763-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S.K., Bode A., Cushion T.D., Thomas R.H., Hunt C., Wood S.E., Pickrell W.O., Drew C.J., Yamashita S., Shiang R., Leiz S., Longhardt A.C., Raile V., Weschke B., Puri R.D., Verma I.C., Harvey R.J., Ratnasinghe D.D., Parker M., Rittey C., Masri A., Lingappa L., Howell O.W., Vanbellinghen J.F., Mullins J.G., Lynch J.W., Rees M.I. GLRB is the third major gene of effect in hyperekplexia. Hum. Mol. Genet. 2013;22:927–940. doi: 10.1093/hmg/dds498. [DOI] [PubMed] [Google Scholar]
- Dallman J.E., Davis A.K., Moody W.J. Spontaneous activity regulates calcium-dependent K+ current expression in developing ascidian muscle. J. Physiol. 1998;511:683–693. doi: 10.1111/j.1469-7793.1998.683bg.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper B.W., Morcos P.A., Kimmel C.B. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- Eisen J.S., Smith J.C. Controlling morpholino experiments: don't stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- Fiumelli H., Cancedda L., Poo M.M. Modulation of GABAergic transmission by activity via postsynaptic Ca2 +-dependent regulation of KCC2 function. Neuron. 2005;48:773–786. doi: 10.1016/j.neuron.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Flint A.C., Liu X., Kriegstein A.R. Nonsynaptic glycine receptor activation during early neocortical development. Neuron. 1998;20:43–53. doi: 10.1016/s0896-6273(00)80433-x. [DOI] [PubMed] [Google Scholar]
- Ganguly K., Schinder A.F., Wong S.T., Poo M.M. GABA itself promotes the developmental switch of neuronal GABAergic responses from excitation to inhibition. Neuron. 2001;105:521–532. doi: 10.1016/s0092-8674(01)00341-5. [DOI] [PubMed] [Google Scholar]
- Ganser L.R., Dallman J.E. Glycinergic synapse development, plasticity, and homeostasis in zebrafish. Front. Mol. Neurosci. 2009;2:30. doi: 10.3389/neuro.02.030.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto C.L., Broadie K. Genetic controls balancing excitatory and inhibitory synaptogenesis in neurodevelopmental disorder models. Front. Synaptic Neurosci. 2010;2:4. doi: 10.3389/fnsyn.2010.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill J.L., Capper D., Vanbellinghen J.F., Chung S.K., Higgins R.J., Rees M.I., Shelton G.D., Harvey R.J. Startle disease in Irish wolfhounds associated with a microdeletion in the glycine transporter GlyT2 gene. Neurobiol. Dis. 2011;43:184–189. doi: 10.1016/j.nbd.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez C., Pérez-Siles G., Martínez-Villarreal J., Arribas-González E., Jiménez E., Núñez E., de Juan-Sanz J., Fernández-Sánchez E., García-Tardón N., Ibáñez I., Romanelli V., Nevado J., James V.M., Topf M., Chung S.K., Thomas R.H., Desviat L.R., Aragón C., Zafra F., Rees M.I., Lapunzina P., Harvey R.J., López-Corcuera B. A novel dominant hyperekplexia mutation Y705C alters trafficking and biochemical properties of the presynaptic glycine transporter GlyT2. J. Biol. Chem. 2012;287:28986–29002. doi: 10.1074/jbc.M111.319244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham B.A., Schofield P.R., Sah P., Callister R.J. Altered inhibitory synaptic transmission in superficial dorsal horn neurones in spastic and oscillator mice. J. Physiol. 2003;551:905–916. doi: 10.1113/jphysiol.2003.049064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato M., van Eeden F.J., Schach U., Trowe T., Brand M., Furutani-Seiki M., Haffter P., Hammerschmidt M., Heisenberg C.P., Jiang Y.J., Kane D.A., Kelsh R.N., Mullins M.C., Odenthal J., Nüsslein-Volhard C. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development. 1996;123:399–413. doi: 10.1242/dev.123.1.399. [DOI] [PubMed] [Google Scholar]
- Grant S.G.N. Synaptopathies: diseases of the synaptome. Curr. Opin. Neurobiol. 2012;22:522–529. doi: 10.1016/j.conb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Gundlach A.L., Dodd P.R., Grabara C.S., Watson W.E., Johnston G.A., Harper P.A., Dennis J.A., Healy P.J. Deficit of spinal cord glycine/strychnine receptors in inherited myoclonus of Poll Hereford calves. Science. 1988;241:1807–1810. doi: 10.1126/science.2845573. [DOI] [PubMed] [Google Scholar]
- Harvey R.J., Depner U.B., Wässle H., Ahmadi S., Heindl C., Reinold H., Smart T.G., Harvey K., Schütz B., Abo-Salem O.M., Zimmer A., Poisbeau P., Welzl H., Wolfer D.P., Betz H., Zeilhofer H.U., Müller U. GlyR α3: an essential target for spinal PGE2-mediated inflammatory pain sensitization. Science. 2004;304:884–887. doi: 10.1126/science.1094925. [DOI] [PubMed] [Google Scholar]
- Harvey R.J., Topf M., Harvey K., Rees M.I. The genetics of hyperekplexia: more than startle! Trends Genet. 2008;24:439–447. doi: 10.1016/j.tig.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Haydar T.F., Wang F., Schwartz M.L., Rakic P. Differential modulation of proliferation in the neocortical ventricular and subventricular zones. J. Neurosci. 2000;20:5764–5774. doi: 10.1523/JNEUROSCI.20-15-05764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H., Saint-Amant L., Downes G.B., Cui W.W., Zhou W., Granato M., Kuwada J.Y. Zebrafish bandoneon mutants display behavioral defects due to a mutation in the glycine receptor β-subunit. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8345–8350. doi: 10.1073/pnas.0500862102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H., Carta E., Yamanaka I., Harvey R.J., Kuwada J.Y. Defective glycinergic synaptic transmission in zebrafish motility mutants. Front. Mol. Neurosci. 2010;2:26. doi: 10.3389/neuro.02.026.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch W., Betz H., Becker C.M. Primary cultures of mouse spinal cord express the neonatal isoform of the inhibitory glycine receptor. Neuron. 1989;3:339–348. doi: 10.1016/0896-6273(89)90258-4. [DOI] [PubMed] [Google Scholar]
- Holland K.D., Fleming M.T., Cheek S., Moran J.L., Beier D.R., Meisler M.H. De novo exon duplication in a new allele of mouse Glra1 (spasmodic) Genetics. 2006;174:2245–2247. doi: 10.1534/genetics.106.065532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley I.A., Mueller R.L., Dunn K.A., Schmidt E.J., Friedman M., Ho R.K., Prince V.E., Yang Z., Thomas M.G., Coates M.I. A new time-scale for ray-finned fish evolution. Proc. Biol. Sci. 2007;274:489–498. doi: 10.1098/rspb.2006.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley I.A., Scemama J.L., Prince V.E. Consequences of hoxb1 duplication in teleost fish. Evol. Dev. 2007;9:540–554. doi: 10.1111/j.1525-142X.2007.00195.x. [DOI] [PubMed] [Google Scholar]
- James V.M., Bode A., Chung S.K., Gill J.L., Nielsen M., Cowan F.M., Vujic M., Thomas R.H., Rees M.I., Harvey K., Keramidas A., Topf M., Ginjaar I., Lynch J.W., Harvey R.J. Novel missense mutations in the glycine receptor β subunit gene (GLRB) in startle disease. Neurobiol. Dis. 2013;52:137–149. doi: 10.1016/j.nbd.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kingsmore S.F., Giros B., Suh D., Bieniarz M., Caron M.G., Seldin M.F. Glycine receptor β-subunit gene mutation in spastic mouse associated with LINE-1 element insertion. Nat. Genet. 1994;7:136–141. doi: 10.1038/ng0694-136. [DOI] [PubMed] [Google Scholar]
- Kinkhabwala A., Riley M., Koyama M., Monen J., Satou C., Kimura Y., Higashijima S., Fetcho J. A structural and functional ground plan for neurons in the hindbrain of zebrafish. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1164–1169. doi: 10.1073/pnas.1012185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kling C., Koch M., Saul B., Becker C.M. The frameshift mutation oscillator (Glra1spd-ot) produces a complete loss of glycine receptor α1-polypeptide in mouse central nervous system. Neuroscience. 1997;78:411–417. doi: 10.1016/s0306-4522(96)00567-2. [DOI] [PubMed] [Google Scholar]
- Komuro H., Rakic P. Intracellular Ca2 + fluctuations modulate the rate of neuronal migration. Neuron. 1996;17:275–285. doi: 10.1016/s0896-6273(00)80159-2. [DOI] [PubMed] [Google Scholar]
- Koyama M., Kinkhabwala A., Satou C., Higashijima S., Fetcho J. Mapping a sensory–motor network onto a structural and functional ground plan in the hindbrain. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1170–1175. doi: 10.1073/pnas.1012189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lee C.G., Kwon M.J., Yu H.J., Nam S.H., Lee J., Ki C.S., Lee M. Clinical features and genetic analysis of children with hyperekplexia in Korea. J. Child Neurol. 2013;28:90–94. doi: 10.1177/0883073812441058. [DOI] [PubMed] [Google Scholar]
- LoTurco J.J., Owens D.F., Heath M.J.S., Davis M.B.E., Kriegstein A.R. GABA and glutamate depolarize cortical progenitor cells and inhibit DNA synthesis. Neuron. 1995;15:1287–1298. doi: 10.1016/0896-6273(95)90008-x. [DOI] [PubMed] [Google Scholar]
- Lummis S.C., Gundlach A.L., Johnston G.A., Harper P.A., Dodd P.R. Increased γ-aminobutyric acid receptor function in the cerebral cortex of myoclonic calves with an hereditary deficit in glycine/strychnine receptors. J. Neurochem. 1990;55:421–426. doi: 10.1111/j.1471-4159.1990.tb04153.x. [DOI] [PubMed] [Google Scholar]
- Lynch J.W. Molecular structure and function of the glycine receptor chloride channel. Physiol. Rev. 2004;84:1051–1095. doi: 10.1152/physrev.00042.2003. [DOI] [PubMed] [Google Scholar]
- McLean D.L., Fetcho J.R. Spinal interneurons differentiate sequentially from those driving the fastest swimming movements in larval zebrafish to those driving the slowest ones. J. Neurosci. 2009;29:13566–13577. doi: 10.1523/JNEUROSCI.3277-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean D.L., Masino M.A., Koh I.Y., Lindquist W.B., Fetcho J.R. Continuous shifts in the active set of spinal interneurons during changes in locomotor speed. Nat. Neurosci. 2008;11:1419–1429. doi: 10.1038/nn.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody W.J., Bosma M.M. Ion channel development, spontaneous activity, and activity-dependent development in nerve and muscle cells. Physiol. Rev. 2005;85:883–941. doi: 10.1152/physrev.00017.2004. [DOI] [PubMed] [Google Scholar]
- Morgan J.L., Schubert T., Wong R.O. Developmental patterning of glutamatergic synapses onto retinal ganglion cells. Neural Dev. 2008;3:8. doi: 10.1186/1749-8104-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mülhardt C., Fischer M., Gass P., Simon-Chazottes D., Guénet J.L., Kuhse J., Betz H., Becker C.M. The spastic mouse: aberrant splicing of glycine receptor β subunit mRNA caused by intronic insertion of L1 element. Neuron. 1994;13:1003–1015. doi: 10.1016/0896-6273(94)90265-8. [DOI] [PubMed] [Google Scholar]
- Ogino K., Ramsden S.L., Keib N., Schwarz G., Harvey R.J., Hirata H. Duplicated gephyrin genes showing distinct tissue distribution and alternative splicing patterns mediate molybdenum cofactor biosynthesis, glycine receptor clustering, and escape behavior in zebrafish. J. Biol. Chem. 2011;286:806–817. doi: 10.1074/jbc.M110.125500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens D.F., Kriegstein A.R. Developmental neurotransmitters? Neuron. 2002;36:989–991. doi: 10.1016/s0896-6273(02)01136-4. [DOI] [PubMed] [Google Scholar]
- Pierce K.D., Handford C.A., Morris R., Vafa B., Dennis J.A., Healy P.J., Schofield P.R. A nonsense mutation in the α1 subunit of the inhibitory glycine receptor associated with bovine myoclonus. Mol. Cell. Neurosci. 2001;17:354–363. doi: 10.1006/mcne.2000.0934. [DOI] [PubMed] [Google Scholar]
- Rea R., Tijssen M.A., Herd C., Frants R.R., Kullmann D.M. Functional characterisation of compound heterozygosity for GlyR α1 mutations in the startle disease hyperekplexia. Eur. J. Neurosci. 2002;16:186–196. doi: 10.1046/j.1460-9568.2002.02054.x. [DOI] [PubMed] [Google Scholar]
- Rees M.I., Lewis T.M., Kwok J.B., Mortier G.R., Govaert P., Snell R.G., Schofield P.R., Owen M.J. Hyperekplexia associated with compound heterozygote mutations in the β-subunit of the human inhibitory glycine receptor (GLRB) Hum. Mol. Genet. 2002;11:853–860. doi: 10.1093/hmg/11.7.853. [DOI] [PubMed] [Google Scholar]
- Rees M.I., Harvey K., Pearce B.R., Chung S.K., Duguid I.C., Thomas P., Beatty S., Graham G.E., Armstrong L., Shiang R., Abbott K.J., Zuberi S.M., Stephenson J.B., Owen M.J., Tijssen M.A., van den Maagdenberg A.M., Smart T.G., Supplisson S., Harvey R.J. Mutations in the gene encoding GlyT2 (SLC6A5) define a presynaptic component of human startle disease. Nat. Genet. 2006;38:801–806. doi: 10.1038/ng1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A., Brustein E., Liao M., Mercado A., Babilonia E., Mount D.B., Drapeau P. Neurogenic role of the depolarizing chloride gradient revealed by global overexpression of KCC2 from the onset of development. J. Neurosci. 2008;28:1588–1597. doi: 10.1523/JNEUROSCI.3791-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H.J. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S., Misener S., editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press; Totowa, NJ: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Ryan S.G., Buckwalter M.S., Lynch J.W., Handford C.A., Segura L., Shiang R., Wasmuth J.J., Camper S.A., Schofield P., O'Connell P. A missense mutation in the gene encoding the α1 subunit of the inhibitory glycine receptor in the spasmodic mouse. Nat. Genet. 1994;7:131–135. doi: 10.1038/ng0694-131. [DOI] [PubMed] [Google Scholar]
- Saint-Amant L., Drapeau P. Time course of the development of motor behaviors in the zebrafish embryo. J. Neurobiol. 1998;37:622–632. doi: 10.1002/(sici)1097-4695(199812)37:4<622::aid-neu10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Saint-Amant L., Drapeau P. Synchronization of an embryonic network of identified spinal interneurons solely by electrical coupling. Neuron. 2001;31:1035–1046. doi: 10.1016/s0896-6273(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Shiang R., Ryan S.G., Zhu Y.Z., Hahn A.F., O'Connell P., Wasmuth J.J. Mutations in the α1 subunit of the inhibitory glycine receptor cause the dominant neurologic disorder, hyperekplexia. Nat. Genet. 1993;5:351–358. doi: 10.1038/ng1293-351. [DOI] [PubMed] [Google Scholar]
- Singer J.H., Talley E.M., Bayliss D.A., Berger A.J. Development of glycinergic synaptic transmission to rat brain stem motoneurons. J. Neurophysiol. 1998;80:2608–2620. doi: 10.1152/jn.1998.80.5.2608. [DOI] [PubMed] [Google Scholar]
- Soto F., Bleckert A., Lewis R., Kang Y., Kerschensteiner D., Craig A.M., Wong R.O. Coordinated increase in inhibitory and excitatory synapses onto retinal ganglion cells during development. Neural Dev. 2011;6:31. doi: 10.1186/1749-8104-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer N.C. Electrical activity in early neuronal development. Nature. 2006;444:707–712. doi: 10.1038/nature05300. [DOI] [PubMed] [Google Scholar]
- Tang R., Dodd A., Lai D., McNabb W.C., Love D.R. Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim. Biophys. Sin. 2007;3:384–390. doi: 10.1111/j.1745-7270.2007.00283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R.H., Stephenson J.B.P., Harvey R.J., Rees M.I. Hyperekplexia: stiffness, startle and syncope. J. Pediatr. Neurol. 2010;8:11–14. [Google Scholar]
- Traka M., Seburn K.L., Popko B. Nmf11 is a novel ENU-induced mutation in the mouse glycine receptor α1 subunit. Mamm. Genome. 2006;17:950–955. doi: 10.1007/s00335-006-0020-z. [DOI] [PubMed] [Google Scholar]
- Vergouwe M.N., Tijssen M.A., Peters A.C., Wielaard R., Frants R.R. Hyperekplexia phenotype due to compound heterozygosity for GLRA1 gene mutations. Ann. Neurol. 1999;46:634–638. doi: 10.1002/1531-8249(199910)46:4<634::aid-ana12>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Warp E., Agarwal G., Wyart C., Friedmann D., Oldfield C.S., Conner A., Del Bene F., Arrenberg A.B., Baier H., Isacoff E.Y. Emergence of patterned activity in the developing zebrafish spinal cord. Curr. Biol. 2012;22:93–102. doi: 10.1016/j.cub.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wässle H., Koulen P., Brandstätter J.H., Fletcher E.L., Becker C.M. Glycine and GABA receptors in the mammalian retina. Vision Res. 1998;38:1411-1130. doi: 10.1016/s0042-6989(97)00300-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary video 1.
Supplementary video 2.
Supplementary video 3.
Supplementary video 4.
Supplementary video 5.
Supplementary video 6.
Supplementary video 7.
Supplementary video 8.
Supplementary video 9.
Supplementary video 10.