Abstract
Candida albicans biofilms on most medical devices are exposed to a flow of body fluids that provide water and nutrients to the fungal cells. While C. albicans biofilms grown in vitro under static conditions have been exhaustively studied, the same is not true for biofilms developed under continuous flow of replenishing nutrients. Here, we describe a simple flow biofilm (FB) model that can be built easily with materials commonly available in most microbiological laboratories. We demonstrate that C. albicans biofilms formed using this flow system show increased architectural complexity compared to biofilms grown under static conditions. C. albicans biofilms under continuous medium flow grow rapidly, and by 8 h show characteristics similar to 24 h statically grown biofilms. Biomass measurements and microscopic observations further revealed that after 24 h of incubation, FB were more than 2 fold thicker than biofilms grown under static conditions. Microscopic analyses revealed that the surface of these biofilms was extremely compact and wrinkled, unlike the open hyphal layer typically seen in 24 h static biofilms. Results of antifungal drug susceptibility tests showed that C. albicans cells in FB exhibited increased resistance to most clinically used antifungal agents.
Keywords: Candida albicans, biofilm, flow, shear stress
Introduction
Candida albicans can colonize and develop a biofilm on almost any medical device [1, 2]. The initial step in biofilm formation is adhesion of organisms to the surface. Material surfaces adsorb proteins or other organic materials when exposed to surrounding body fluids such as urine, blood, saliva and synovial fluid thereby promoting C. albicans adhesion [3]. Additionally, adherent C. albicans cells encounter conditions of high blood flow within the circulatory system surrounding venous catheters and heart valves [3]. Biofilms associated with devices such as dentures and/or urinary catheters are intermittently bathed in saliva and urine respectively. The exposure of C. albicans cells to body fluids may provide a continuous source of nutrients and a means for the removal of waste products, thereby promoting robust biofilm formation.
The majority of C. albicans biofilm studies in vitro have utilized static models. These models have proven to be useful due to ease of operation, low costs and the ability for rapid processing of large number of samples. These models typically involve growth of adherent cell populations on the several different types of surfaces such as silicon elastomer, plastic, glass slides, or acrylic strips/disks under static conditions [4-8]. For high throughput processing of multiple samples, our group has previously described a simple and robust 96-well microtiter plate model of C. albicans biofilms [6, 9, 10]. However, regardless of the material used, static biofilms are typically incubated for 24-48 h without replenishing nutrients and most phenotypic and molecular analyses are undertaken following partial or total depletion of these resources. Thus, these static models hardly imitate the conditions of flow, nutrient provision and shear forces that fungal cells normally encounter within the host. Recent studies have tried to overcome this problem by developing C. albicans biofilm models under conditions of flow. In a flow model, C. albicans biofilms can be formed either by rocking in a single compartment such as in a tissue culture flask or Erlenmeyer flask with limited amounts of medium, or through perfusion in more elaborate systems such as CDC reactors, modified Robbins device or microfermentors [11-15]. Characteristics of FB have not been studied in detail, but a few studies have indicated that biofilms grown under flow conditions favor the development of more extensive matrix and enhanced cell proliferation compared to statically grown biofilms [12, 14].
Here we describe a simple FB model that can be easily assembled with materials commonly available in most microbiological laboratories. We show that under conditions of flow, C. albicans biofilms are more robust and show increased architectural complexity compared to those formed under static conditions. A potential advantage of this particular flow model system is also that it speeds up the process of biofilm formation since in less than 8 hours it leads to the formation of a biofilm that closely resembles a 24 h statically grown biofilm.
Material and Methods
Strain and growing conditions
C. albicans SC5314 was used in the course of this study. It was routinely grown on yeast extract-peptone-dextrose (YPD) medium [US Biological, Swampscott, MA]. Briefly, batches of medium (about 20 ml) were inoculated from YPD agar plates containing freshly grown C. albicans, and incubated overnight in an orbital shaker at 30°C. Cells were harvested and washed in sterile phosphate buffered saline (PBS: 10 mM phosphate buffer, 2.7 mM potassium chloride, 137 mM sodium chloride, pH 7.4 [Sigma, St Louis, MO]). For biofilm seeding, cells were resuspended and adjusted to the desired cell density in RPMI-1640 supplemented with L-glutamine and buffered with morpholinepropanesulfonic acid (MOPS, final pH 7) (Angus Buffers and Chemicals, Niagara Falls, NY).
Biofilm growth conditions
Two in vitro models were used to assess biofilm growth - static and flow models. For the static model, biofilms were grown on silicon elastomer (SE) surfaces as described previously [16]. Briefly, strains were grown overnight in YPD medium at 30° C and diluted to an optical density at 600 nm (OD600) of 0.5 in RPMI medium (see above). The suspension (2 ml) was added to the wells of a sterile 24-well plate containing bovine serum (B-9433; Sigma) treated SE pieces (1 cm2, Cardiovascular instrument Corp. silicone sheets, Wakefield, MA). The inoculated plates were incubated at 37°C for 90 min at 150 rpm agitation for the initial adhesion of cells. After the initial attachment phase, the squares were washed with phosphate-buffered saline (PBS), transferred to fresh plates containing fresh RPMI medium, and incubated statically at 37°C to allow biofilm formation.
Biofilms were also developed under continuous flow of fresh medium. We manually assembled the flow system using commonly available laboratory materials. This model involves a controlled flow of fresh medium via Tygon® tubing (Cole-Parmer®, Vernon Hills, IL) into a 15 ml polypropylene conical tube (BD, Franklin, NJ) holding a SE strip. The apparatus was constructed by first, boring a hole, such that it could tightly fit one end of a silicon tubing (2.0 mm internal diameter), in the cap of the 15 ml conical tube and by cutting off approximately the bottom 1 cm of the tube. This part of the apparatus was autoclaved for 20 minutes. Once autoclaved, the other end of the silicon tubing was immersed aseptically into a sterile flask containing RPMI medium. Medium flow was controlled by connecting the silicon tubing to a peristaltic pump (Masterflex L/S® Easy-Load® II, Cole-Parmer®). The whole apparatus was placed inside a 37° C incubator to facilitate biofilm development at a controlled temperature. For biofilm development, SE strips (1 cm × 9 cm) were sterilized by autoclaving and pre-treated for 3 h with bovine serum. C. albicans SC5314 was grown overnight in YPD medium at 30° C, washed and diluted to an optical density at 600 nm (OD600) of 0.5 in RPMI medium. The SE strips were incubated with the diluted C. albicans suspension at 37° C for 90 min at 150 rpm agitation for the initial adhesion of cells for 90 min, to allow attachment of the cells to the SE. Next, the SE strip was inserted into the conical tube and the peristaltic pump was turned on. The strips containing attached yeast cells were first exposed for a few seconds with a liberal flow of medium, mainly intended to uniformly impregnate the whole strip (and not only a part of the strip while trickling down from the sides: initial experiments demonstrated that an even coverage of the whole strip by medium was key for the development of a uniform biofilm). Once this was accomplished, medium flow was set at a constant rate of 0.75 ml/min.
Estimation of biofilm formation
Biofilm formation was assessed by both dry weight measurements and the semi quantitative colorimetric XTT (2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide) reduction assay. Briefly, SE strips from the flow model were cut into 1 cm2 pieces. For XTT assay, the SE pieces from both static and flow models were treated with 250 μg/ml XTT (Sigma, St. Louis, MO) and 0.1 mM menadione (Sigma), and incubated for 90 m at 37° C in the dark. After incubation, the liquid was removed from each well, clarified by centrifugation, and XTT formazan production was measured by determining the absorbance at 490 nm using a microtiter plate reader (Bechmark Microplate Reader, BioRad, Hercules, CA). For dry weight measurements, the SE pieces were cut and weighed prior to biofilm development. After the cell adherence step, the pieces were stacked one over other in the tube before starting the medium flow for biofilm development. After 48 hr of biofilm formation, the biofilms were washed gently with sterile PBS and air dried at 37° C for 24 hr. The total biomass of each biofilm was calculated by subtracting the weight of the silicone prior to biofilm growth from the weight of the silicone after biofilm growth. Statistically significant differences between SB and FB, using both XTT-readings and dry weight measurements) were analyzed using ANOVA test, with p ≤ 0.05 considered statistically significant.
Scanning electron microscopy (SEM)
Biofilms were placed in fixative (4% formaldehyde v/v, 1% glutaraldehyde v/v in phosphate buffered saline) overnight. The samples were rinsed in 0.1 M phosphate buffer (2 ×3 min) and placed in 1% Zetterquist's osmium for 30 min. The samples were subsequently dehydrated in a series of ethanol washes (70% for 10 min, 95% for 10 min, 100% for 20 min), treated (2 × 5 min) with hexamethyldisilizane (HMDS: Polysciences Inc., Warrington, PA), and finally air dried in desiccators. The samples were coated with gold/palladium (40%/60%) and observed in a scanning electron microscope (Leo 435 VP) in high vacuum mode at 15kV. The images were processed for display using Photoshop software (Adobe, Mountain View, Calif.).
Confocal Scanning Laser Microscopy
Biofilms were stained with 25 μg/ml Concanavalin A-Alexa Fluor 594 conjugate (C-11253; Molecular Probes, Eugene, Oregon, United States) for 1 hr in the dark at 37°C. Confocal scanning laser microscopy (CSLM) was performed with a Zeiss LSM 510 upright confocal microscope using a Zeiss Achroplan 40×/0.8W objective. Concanavalin A conjugate staining was observed using a HeNe1 laser with an excitation wavelength of 543 nm. Images were assembled into side and depth views using the Zeiss LSM Image Browser v4.2 software.
Antifungal drug susceptibility of cells in biofilms
For biofilms grown under static conditions drug susceptibility was performed as reported earlier [9]. Briefly, mature biofilms were washed gently with sterile PBS and transferred to fresh 24 well plates containing RPMI medium supplemented with appropriate dilutions of fluconazole (0.25-1024 μg/ml), amphotericin B (0.03-32 μg/ml) and caspofungin (0.007 – 4 μg/ml). Four replicate biofilms were used for each condition. The biofilms were further incubated for 24 h and metabolic activities of cells within the biofilms were estimated by the XTT-reduction assay. For biofilms grown under flow conditions FB were first cut into pieces before being placed into individual wells of 24 well plates with different dilutions of antifungal drugs (as above). After 24h of incubation in the presence of drug, the metabolic activities of cells within the biofilms were estimated by the XTT-reduction assay. Sessile minimum inhibitory concentrations were determined at 50% inhibition (SMIC50) and at 80% inhibition (SMIC80) compared to drug-free control wells using the XTT-reduction assay described above.
RNA extraction and Real time reverse transcriptase PCR (RT-RTPCR)
Cells were collected by vigorous vortexing of biofilm-containing SE pieces in ice cold PBS and pelleted by centrifugation. The cells were stored at -80° C before RNA extraction. Total RNA was isolated using the standard hot acid phenol method following grinding frozen cells using a mortar and pestle in liquid nitrogen [17]. The RNA preparation was DNAse treated and the absence of DNA contamination was confirmed with the housekeeping gene EFB1. RNA quality and quantity were determined as described previously [17]. cDNA was synthesized from known amounts total RNA, and equal amounts of cDNA were used as starting templates for RT-RT–PCR reactions. Analysis of transcript was carried out using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) in an ABI Prism 7300 Sequence Detection System (Applied Biosystems). Each reaction was set up in triplicate in 25.0 μl volume with 1.0 μl cDNA for 40 cycles (thermal cycling conditions: initial steps of 50 °C for 2 min and 95 °C for 10 min; then 40 cycles of 95 °C for 15 s; 60 °C for 1 min). The primers are given in Table 1. The target genes were normalized to C. albicans 18S rRNA. Relative gene expression was quantified by the 2 –ΔΔCT method and analyzed using the inbuilt ABI analysis software.
Table 1.
Primers for analysis of selected genes by RT-RT–PCR. The direction of primers is indicated as forward (F) or reverse (R).
| Gene | Primer | Sequence |
|---|---|---|
|
|
||
| HWP1 | HWP1F | TCAGTTCCACTCATGCAACCA |
| HWP1R | AGCACCGAAAGTCAATCTCATGT | |
| ALS3 | ALS3F | GTGATGCTGGATCTAACGGTATTG |
| ALS3R | GTCTTAGTTTTGTCGCGGTTAGG | |
| TUP1 | TUP1F | GCTTCAGGTAACCCATTGTTGAT |
| TUP1R | CTTCGGTTCCCTTTGAGTTTAGG | |
| NRG1 | NRG1F | CACCTCACTTGCAACCCC |
| NRG1R | GCCCTGGAGATGGTCTGA | |
| SAM2 | SAM2F | GGTTCCTTGCCATGGTTGAG |
| SAM2R | TTGTGTCGACTCTTTTTGGGATAA | |
| CYS3 | CYC3F | GTGGTATCGAGTCGTTGATCGA |
| CYS3R | ACCATTGGCTTCTCTTTCTTCCT | |
| 18S | 18SF | CACGACGGAGTTTCACAAGA |
| 18SR | CGATGGAAGTTTGAGGCAAT | |
| EFB1V (to test for genomic DNA contamination) | EFB1VF | ACGAATTCTTGGCTGACAAATCA |
| EFB1VR | TCATCTTCTTCAACAGCAGCTTGT |
Results and Discussion
While in vitro flow models are more realistic representation of biofilm development in vivo, the majority of studies have preferred the use of static models for C. albicans biofilm research. This is because most current FB systems are tedious to set up and normally require the use of highly specialized equipment not generally available in regular laboratories. Thus, the objective of our study was twofold: first, to design and develop a simple FB model; and second, to study the resulting characteristics of these biofilms formed under conditions of flow.
Development of a simple C. albicans biofilm model under conditions of flow
To allow for the formation of biofilms under conditions of flow we assembled an apparatus with materials commonly available in most microbiological laboratories (Figure 1). These include 15 ml disposable conical tubes, Tygon® tubing, a peristaltic pump and an Erlenmeyer flask as a reservoir of fresh medium. The whole apparatus was placed inside an incubator for maintenance of a controlled temperature (in our case 37 °C). For biofilm formation, C. albicans cells were allowed to adhere to SE strips (precoated with serum) for 90 min at 37 °C outside the apparatus, and after the initial attachment phase these SE strips containing adherent populations of C. albicans were placed inside the 15 ml conical tubes (one strip per tube) and bathed with continuous flow of liquid medium over predetermined periods of time. After 24 h, the SE strips were found to be completely covered with a thick layer of biofilm. We found this model to be highly reproducible based on the uniform coverage of SE strips, microscopic analysis, XTT and dry weight measurements (discussed below). Depending upon the number of ports in the peristaltic pump (two to eight roller pump), this system allows formation of multiple independent biofilms under flow conditions.
Figure 1.
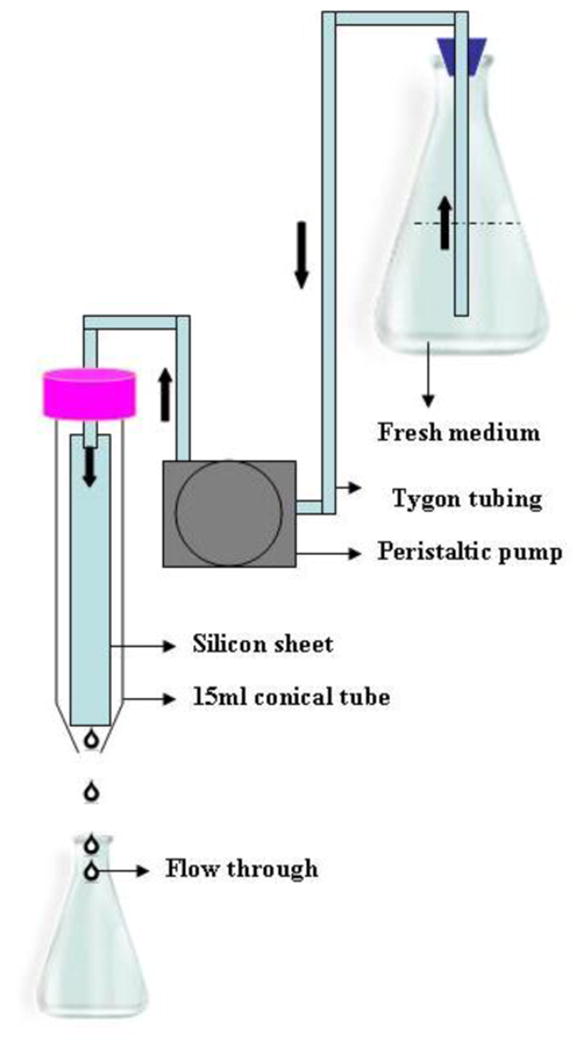
Schematic representation of a simple flow biofilm model. The model was assembled using commonly available laboratory materials. The model involves a controlled flow of fresh medium via Tygon® tubing into a 15 ml conical tube holding a silicon elastomer strip adhered with C. albicans cells.
A major advantage of this model is that it is user friendly and can be assembled using commonly available laboratory materials. Many of the previously described flow models are either too complicated to construct, or do not fully emulate in vivo biofilm growth conditions. For example, models such as the CDC reactor or modified Robbins device need specialized equipment not generally available in regular laboratories [11, 12, 14, 15]. Biofilms have also been developed in tissue culture or Erlenmeyer flasks under slow shaking conditions but these models do not mimic in vivo flow conditions nor permit removal of dispersed cells from the biofilms that eventually contribute to further depletion of nutrients. As with all of the flow models, a clear limitation of this model is that it is not as amenable to high throughput screening as the 96-well microtiter plate model previously described by our group [6, 10].
Kinetics of biofilm formation and architectural characteristics of C. albicans biofilms under flow
We used RPMI medium with a flow rate of 0.75 ml/min to promote biofilms for 24 h at 37° C. Growth kinetics of the biofilms under medium flow were compared to biofilms grown under static conditions. The extent of biofilm formation was estimated by both the XTT assay and by dry weight measurements, and results were analyzed using ANOVA. As seen in Figure 2, results revealed that at a given time-point, the FB were at least 1.8 fold more metabolically active and greater in biomass than the corresponding SB (p ≤0.05, considered significant). In fact, the enhanced robustness of the FB was apparent as early as 8 h of incubation when both the biomass and metabolic activity of the 8 hour FB was comparable to those of a 24 h SB (p > 0.3). We speculate that continuous flow of fresh liquid medium provides adhered C. albicans a condition of growth uninterrupted by factors such as nutrient starvation and toxic metabolite accumulation, thereby promoting rapid biofilm development.
Figure 2.
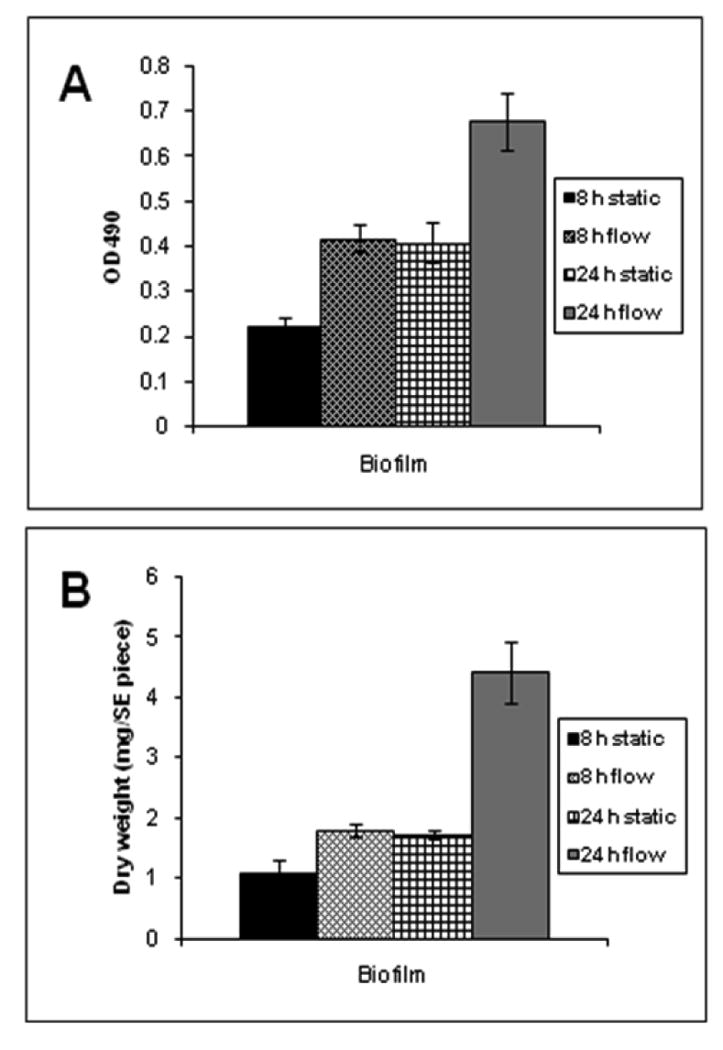
Measurements of metabolic activity and biomass of biofilms (4 replicates) grown in static or flow conditions. Metabolic activity of the C. albicans cells, and biomass of the biofilms formed in the flow model and static model were compared using XTT assay (panel A) and dry weight measurements (panel B), respectively. Values shown are mean and standard deviations for four replicate biofilms under each condition.
Macroscopically we found that the 8 h FB were similar in appearance to 24 h SB. However, as the FB continued to develop, a wrinkly appearance was noticeable, and the frequency of the wrinkles increased and the folds between the wrinkles grew deeper and wider with time leading to an overall undulating topography by 12 and 24 h. The FB also seemed to produce considerable amounts of extracellular matrix (ECM); as indicated by the mucous macroscopic appearance (Figure 3A). Interestingly we found that, the mature biofilms had only a few points of adhesive contact with the silicon elastomer, with presence of empty space in between.
Figure 3.
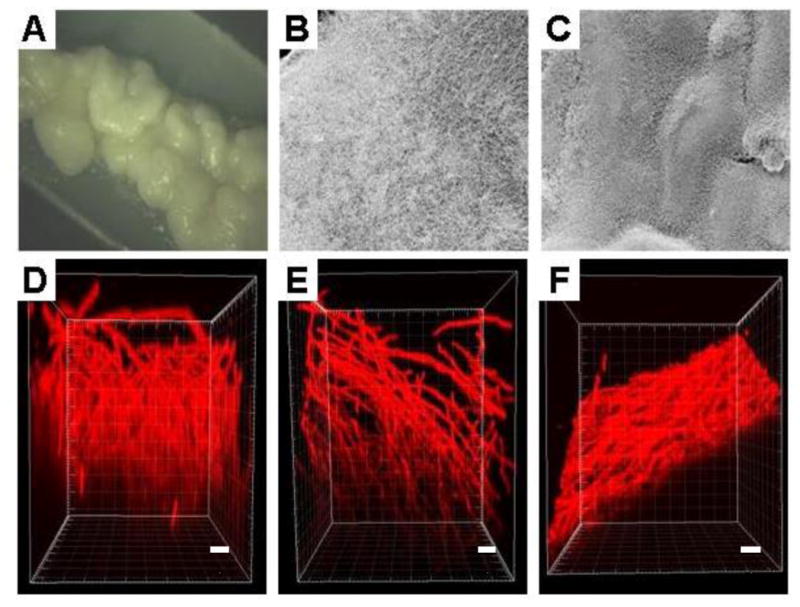
Macroscopic and microscopic observations of biofilms grown in the static and flow model. Macroscopically, biofilms grown in the flow biofilm model appeared mucoid and highly wrinkled in appearance (panel A). Scanning electron microscopy (SEM) observations indicated that, compared to the statically grown biofilms (panel B), biofilms forms under conditions of flow demonstrated an undulating topography (panel C) (250× magnification for both panels). Biofilms grown in the static model (24 h), flow model (8 h and 24 h) were stained with Con A and visualized by Confocal scanning laser microscope (CSLM). Both 24 h static biofilm and the 8 h flow biofilm comprised of an open hyphal layer with a depth of 326 μm and 342 μm respectively (panels D and E). The 24 h flow biofilm was largely impermeable to Con A, and the top 302 μm imaged displayed a layer of tightly compacted interwoven hyphae (panel F). Scale bars correspond to 20 μm.
Next, biofilms were observed microscopically by light microscopy, CSLM and SEM. Light microscopy at various time points of biofilm development revealed that the FB, like the SB grew in three distinct phases (bottom layer of yeasts and germ tubes, middle layer of yeast, pseudohyphae and hyphae and the top layer comprising predominantly of hyphae; data not shown). However, as mentioned earlier, kinetics of biofilm formation were approximately 3 times more rapid in FB than the SB. CSLM revealed that the biofilm depth and architecture of 8 h FB was comparable to 24 h SB (depth of 326 μm and 342 μm respectively), with the topmost part of both the biofilms comprising entirely of an open hyphal layer (Figure 3D and 3E). Unfortunately, it was difficult to scan the entire depth of 24 h FB because high density of the intertwined hyphae at this time point, prevented penetration of Con A into deeper sections of the biofilms as reported previously by other groups [18]. The top 302 μm that could be imaged, revealed an interwoven mesh of hyphae (Figure 3F). In fact due to the extremely tight and wrinkled nature of the biofilm, the image on scanning appeared slanted (indicating a groove of the wrinkled biofilm surface). This semblance was in stark contrast to the open hyphal layer observed in 8 h FB or 24 h SB. The wrinkled nature of the 24 h biofilm was also clearly seen on SEM (Figure 3C).
In general our observations seem to be in agreement with those displayed by C. albicans biofilms formed under conditions of flow reported by a few other studies. Under these conditions, C. albicans are known to produce high amounts of extracellular matrix and have a high cell density compared to those grown under static conditions [12, 14, 15].
Levels of expression of key biofilm-associated genes in C. albicans biofilms formed under flow compared to statically grown biofilms
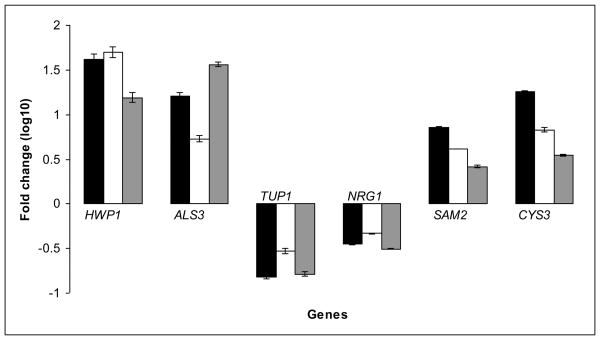
We used RT-RTPCR to examine the expression levels of key genes associated with biofilm development in FB and SB. Besides including genes encoding for hyphal and adhesion molecules such as HWP1 and ALS3, which play a pivotal role as complementary adhesins on the surface of filamentous cells within the biofilm (19], we chose two genes belonging to the sulfur amino acid biosynthesis process; which have previously been reported to be exclusively up-regulated at high levels under biofilm conditions [13]. We also quantified gene expression for two negative regulators of filamentation TUP1 and NRG1. RNA was extracted from 24 h SB, 8 h FB and 24 h FB, reverse transcribed and cDNA quantified by RT-RTPCR. Results revealed a certain degree of variability in the quantitative transcript levels of the biofilm associated genes between the three biofilm growth conditions (Fig 4). However, the overall patterns of up and/or down-regulation for each gene transcript were similar for all three conditions. Thus, it would seem that, as early as 8 h, biofilms formed under flow conditions exhibit a characteristic gene expression signature similar to that of a mature biofilm. This, together with the architectural characteristics described above, opens up possibilities for the development of robust, physiologically relevant biofilms in less than one third of the time taken to grow conventional SB.
Figure 4.
Expression levels of biofilm-associated genes in biofilm cells grown under static or flow conditions. Sessile cells were recovered from 24 h static biofilms (black bars), 8 h flow biofilms (white bars) and 24 h flow biofilms (grey bars). Gene expression profiles of biofilm associated genes (HWP1, ALS3, TUP1, NRG1, SAM2, and CYS3) were quantified by real time RTPCR. Values shown are mean and standard deviations for triplicate biofilms.
Drug susceptibility of biofilms grown under static or flow conditions
Biofilms of C. albicans grown under static or flow conditions were exposed to different concentrations of amphotericin B, fluconazole and caspofungin at 37°C for 24 h. After incubation, the metabolic activity of the biofilms was compared with control biofilms grown in the absence of the drugs. C. albicans FB were completely resistant to fluconazole with SMIC50> 1024 μg/ml (Table 2). FB were also significantly resistant to amphotericin B with SMIC50 values up to 3 dilutions higher than the SB. When treated with caspofungin, the FB was only slightly more resistant, with SMIC50 values of one dilution higher than SB (0.06 μg/ml vs 0.03 μg/ml) (Table 2). It was also found that the SMIC80 of caspofungin was two dilutions higher than SB. Increased resistance of biofilms under flow conditions to amphotericin and fluconazole has been reported earlier [12, 14]. In agreement with previous reports [7, 20, 21], our results using FB corroborated the efficacy of caspofungin against C. albicans biofilms, although it would seem that caspofungin SMIC values for FB biofilms are slightly elevated compared to those obtained for SB. We posit that the decreased susceptibility to amphotericin B and caspofungin of FB could be directly attributed to the increase in the biomass and matrix production in biofilms formed under conditions of flow, since both increased cellular density and matrix production are contributors to drug resistance in biofilms [12, 14, 22].
Table 2.
Results of antifungal susceptibility testing of biofilms formed under conditions of flow (FB) compared to statically grown biofilms (SB).
| Drug | Model | SMIC50 (μg/ml) | SMIC80 (μg/ml) |
|---|---|---|---|
| Fluconazole | FB | > 1024 | > 1024 |
| SB | > 1024 | > 1024 | |
| Amphotericin B | FB | 4 | >32 |
| SB | 1 | 16 | |
| Caspofungin | FB | 0.06 | 0.12 |
| SB | 0.03 | 0.03 |
In summary, we have introduced a simple model for developing C. albicans biofilms under conditions of flowing medium. Our model can be easily assembled using materials commonly available in most laboratories. Biofilms in our model develop three times faster than static biofilms and show distinct phenotypic properties. Flow biofilms also showed decreased susceptibility to clinically used antifungal drugs, although they remained susceptible to caspofungin, a drug known to be highly effective against biofilms grown under static conditions.
Work in the laboratory is supported by Public Health Service grants numbered RO1 AI 064562 from the National Institute of Allergy and Infectious Diseases and R21 DE 017294 from the National Institute of Dental and Craniofacial Research to J.L.L.-R. We would like to thank the Research Center for Minority Institutions (RCMI) Advance Imaging Center, supported by Grant numbered 5G12 RR01 3646-10, for use of the confocal microscope, and Colleen Witt for assistance with confocal microscopy. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, the National Institute of Dental and Craniofacial Research or the National Institutes of Health.
References
- 1.Ramage G, Martinez JP, Lopez-Ribot JL. Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res. 2006;6:979–86. doi: 10.1111/j.1567-1364.2006.00117.x. [DOI] [PubMed] [Google Scholar]
- 2.Ramage G, Saville SP, Thomas DP, Lopez-Ribot JL. Candida biofilms: an update. Eukaryot Cell. 2005;4:633–8. doi: 10.1128/EC.4.4.633-638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nett J, Andes D. Candida albicans biofilm development, modeling a host-pathogen interaction. Curr Opin Microbiol. 2006;9:340–5. doi: 10.1016/j.mib.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001;183:5385–94. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhn DM, Chandra J, Mukherjee PK, Ghannoum MA. Comparison of biofilms formed by Candida albicans and Candida parapsilosis on bioprosthetic surfaces. Infect Immun. 2002;70:878–88. doi: 10.1128/iai.70.2.878-888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pierce CG, Uppuluri P, Tristan AR, Wormley FL, Jr, Mowat E, Ramage G, et al. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat Protoc. 2008;3:1494–500. doi: 10.1038/nport.2008.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachmann SP, VandeWalle K, Ramage G, Patterson TF, Wickes BL, Graybill JR, et al. In vitro activity of caspofungin against Candida albicans biofilms. Antimicrob Agents Chemother. 2002;46:3591–6. doi: 10.1128/AAC.46.11.3591-3596.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uppuluri P, Sarmah B, Chaffin WL. Candida albicans SNO1 and SNZ1 expressed in stationary-phase planktonic yeast cells and base of biofilm. Microbiology. 2006;152:2031–8. doi: 10.1099/mic.0.28745-0. [DOI] [PubMed] [Google Scholar]
- 9.Ramage G, Lopez-Ribot JL. Techniques for antifungal susceptibility testing of Candida albicans biofilms. Methods Mol Med. 2005;118:71–9. doi: 10.1385/1-59259-943-5:071. [DOI] [PubMed] [Google Scholar]
- 10.Ramage G, Vande Walle K, Wickes BL, Lopez-Ribot JL. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob Agents Chemother. 2001;45:2475–9. doi: 10.1128/AAC.45.9.2475-2479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honraet K, Goetghebeur E, Nelis HJ. Comparison of three assays for the quantification of Candida biomass in suspension and CDC reactor grown biofilms. J Microbiol Methods. 2005;63:287–95. doi: 10.1016/j.mimet.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Hawser SP, Baillie GS, Douglas LJ. Production of extracellular matrix by Candida albicans biofilms. J Med Microbiol. 1998;47:253–6. doi: 10.1099/00222615-47-3-253. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Sanchez S, Aubert S, Iraqui I, Janbon G, Ghigo JM, d'Enfert C. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot Cell. 2004;3:536–45. doi: 10.1128/EC.3.2.536-545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Fattani MA, Douglas LJ. Biofilm matrix of Candida albicans and Candida tropicalis: chemical composition and role in drug resistance. J Med Microbiol. 2006;55:999–1008. doi: 10.1099/jmm.0.46569-0. [DOI] [PubMed] [Google Scholar]
- 15.Ramage G, Wickes BL, Lopez-Ribot JL. A seed and feed model for the formation of Candida albicans biofilms under flow conditions using an improved modified Robbins device. Rev Iberoam Micol. 2008;25:37–40. doi: 10.1016/s1130-1406(08)70009-3. [DOI] [PubMed] [Google Scholar]
- 16.Uppuluri P, Nett J, Heitman J, Andes D. Synergistic effect of calcineurin inhibitors and fluconazole against Candida albicans biofilms. Antimicrob Agents Chemother. 2008;52:1127–32. doi: 10.1128/AAC.01397-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uppuluri P, Perumal P, Chaffin WL. Analysis of RNA species of various sizes from stationary-phase planktonic yeast cells of Candida albicans. FEMS Yeast Res. 2007;7:110–7. doi: 10.1111/j.1567-1364.2006.00143.x. [DOI] [PubMed] [Google Scholar]
- 18.Richard ML, Nobile CJ, Bruno VM, Mitchell AP. Candida albicans biofilm-defective mutants. Eukaryot Cell. 2005;4:1493–502. doi: 10.1128/EC.4.8.1493-1502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, et al. Complementary adhesin function in C albicans biofilm formation. Curr Biol. 2008;18:1017–24. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cateau E, Rodier MH, Imbert C. In vitro efficacies of caspofungin or micafungin catheter lock solutions on Candida albicans biofilm growth. J Antimicrob Chemother. 2008;62:153–5. doi: 10.1093/jac/dkn160. [DOI] [PubMed] [Google Scholar]
- 21.Ramage G, VandeWalle K, Bachmann SP, Wickes BL, Lopez-Ribot JL. In vitro pharmacodynamic properties of three antifungal agents against preformed Candida albicans biofilms determined by time-kill studies. Antimicrob Agents Chemother. 2002;46:3634–6. doi: 10.1128/AAC.46.11.3634-3636.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perumal P, Mekala S, Chaffin WL. Role for cell density in antifungal drug resistance in Candida albicans biofilms. Antimicrob Agents Chemother. 2007;51:2454–63. doi: 10.1128/AAC.01237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]