Abstract
We conducted a genome-wide association study meta-analysis of mean arterial pressure and pulse pressure among 26,600 East Asian participants (stage-1) followed by replication study of up to 28,783 participants (stage-2). For novel loci, statistical significance was determined by a P<5.0×10−8 in joint analysis of stage-1 and stage-2 data. For loci reported by the previous mean arterial and pulse pressure genome-wide association study meta-analysis in Europeans, evidence of trans-ethnic replication was determined by consistency in effect direction and a Bonferroni-corrected P<1.4×10−3. No novel loci were identified by the current study. Five independent mean arterial pressure variants demonstrated robust evidence for trans-ethnic replication including rs17249754 at ATP2B1 (P=7.5×10−15), rs2681492 at ATP2B1 (P=3.4×10−7), rs11191593 at NT5C2 (1.1×10−6), rs3824755 at CYP17A1 (P=1.2×10−6), and rs13149993 at FGF5 (P=2.4×10−4). Two additional variants showed suggestive evidence of trans-ethnic replication (consistency in effect direction and P<0.05), including rs319690 at MAP4 (P=0.014) and rs1173771 at NPR3 (P=0.018). For pulse pressure, robust evidence of replication was identified for 2 independent variants, including rs17249754 at ATP2B1 (P=1.2×10−5) and rs11191593 at NT5C2 (P=1.1×10−3), with suggestive evidence of replication among an additional 2 variants including rs3824755 at CYP17A1 (P=6.1×10−3) and rs2681492 at ATP2B1 (P=9.0×10−3). Replicated variants demonstrated consistency in effect sizes between East Asian and European samples, with effect size differences ranging from 0.03 to 0.24 mmHg for mean arterial pressure and from 0.03 to 0.21 mmHg for pulse pressure. In conclusion, we present the first evidence of trans-ethnic replication of several mean arterial and pulse pressure loci in an East Asian population.
Keywords: genetics, polymorphism, single nucleotide, blood pressure, hypertension, genome-wide association study, meta-analysis
INTRODUCTION
Hypertension affects nearly 30% of the world’s adult population and has been identified as the leading risk factor for mortality globally1–3. A common complex trait, high blood pressure (BP) is influenced by genomic and environmental factors, as well as their interactions4–7. Recent genome-wide association study (GWAS) meta-analyses have made important strides in advancing hypertension genomic research through the identification of numerous novel loci for systolic blood pressure (SBP) and diastolic blood pressure (DBP)4–7. Mean arterial pressure (MAP), defined as the average pressure in the arteries, and pulse pressure (PP), a measure of large artery stiffness, represent two additional blood pressure components which also predict cardiovascular disease risk and mortality8–11. Despite their public health relevance and established heritability12–14, only one previous GWAS meta-analysis has reported genomic loci influencing these traits15. Wain and colleagues described several novel MAP and PP loci which they identified exclusively in populations of European descent15. GWAS meta-analyses in distinct ethnic groups could enable the discovery of additional novel loci for MAP and PP and help to determine whether the previously reported variants are relevant to populations of non-European ancestry.
We carried out the first ever GWAS meta-analysis of MAP and PP in East Asian participants to: 1) identify novel loci influencing MAP and PP; and 2) determine whether loci previously identified in populations of European ancestry could be replicated among a distinct ethnic group. Here we report the results of our two-stage study that included a meta-analysis of MAP and PP GWAS in 26,600 participants and replication study in up to 28,783 participants.
METHODS
Stage-1 GWAS meta-analysis
The Asian Genetic Epidemiology Network (AGEN)-MAP/PP work-group consists of 9 GWAS conducted in East Asian populations. Each AGEN-MAP/PP study collected at least 2 measurements of systolic blood pressure (SBP) and diastolic blood pressure (DBP) in a clinical setting using methods described previously 6. If participants were taking antihypertension medications, 10 mmHg and 5 mmHg were added to measured SBP and DBP, respectively. Mean MAP and PP were calculated for each participant from SBP and DBP values. Prior to GWAS, each study imputed the HapMap set of approximately 2.4 million single nucleotide polymorphisms (SNPs)16–18. GWAS of MAP and PP in each study were carried out using linear regression models to adjust for age, age2, gender, body mass index, and enrollment site (for multi-site studies). Detailed study-specific information can be found in Table 1, the Supplementary Note, and Table S1.
Table 1.
Characteristics of AGEN-MAP/PP studies.
| Study | N | Ancestry | Blood pressure measurement (device, number of measures) |
Age (SD)* | Women† | BMI (SD)‡ | SBP (SD)§ | DBP (SD)§ | MAP (SD)§,‖ | PP (SD)§,¶ | Hypertension†,# | Anti- hypertension medication† |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stage-1: AGEN-MAP/PP GWAS meta-analysis (n=26,600) | ||||||||||||
| CAGE | 1,547 | Japanese | Standard mercury sphygmomanometer, 2–3/digital, 2–3 | 66.1 (8.0) | 42.8 | 23.5 (3.3) | 134.1 (20.3) | 76.8 (11.9) | 98.4 (14.5) | 59.2 (15.6) | 56.1 | 37.9 |
| CLHNS | 1,787 | Filipino | Standard mercury sphygmomanometer, 3 | 48.4 (6.1) | 100 | 24.3 (4.4) | 120.0 (20.5) | 79.8 (12.7) | 93.2 (14.5) | 40.2 (12.8) | 27.3 | 3.8 |
| GenSalt | 1,881 | Chinese | Random zero sphygmomanometer, 9 | 38.7 (9.5) | 47.2 | 23.3 (3.2) | 116.9 (14.2) | 73.7 (10.3) | 88.2 (10.9) | 43.2 (9.5) | 9.8 | 0.37 |
| KARE | 8,842 | Korean | Standard mercury sphygmomanometer, 3 | 52.2 (8.9) | 52.7 | 24.6 (3.1) | 118.7 (19.4) | 75.6 (12.0) | 90.0 (13.8) | 43.1 (11.8) | 22.3 | 10.9 |
| NHAPC | 2,817 | Chinese | Omron HEM-705 CP Blood Pressure Monitor, 3 | 58.6 (6.0) | 56.9 | 24.5 (3.6) | 143.0 (24.8) | 81.6 (11.7) | 102.0 (14.9) | 61.5 (18.2) | 55.9 | 28.7 |
| SiMES | 2,538 | Malay | Random zero sphygmomanometer, 2–3 | 59.1 (11.1) | 50.5 | 26.4 (5.1) | 150.2 (24.8) | 81.2 (11.4) | 104.2 (10.9) | 69.0 (19.1) | 69.9 | 23.0 |
| SP2 | 2,434 | Chinese | Random zero sphygmomanometer, 2–3 | 48.1 (11.2) | 53.5 | 22.9 (3.7) | 130.8 (21.3) | 77.6 (11.2) | 95.3 (13.6) | 53.2 (14.9) | 18.0 | 14.3 |
| Taiwan Type 2 Diabetes Study | 1,000 | Chinese | Random zero sphygmomanometer, 3 | 51.2 (17.8) | 49.8 | 23.8 (3.5) | 122.6 (19.4) | 76.5 (11.0) | 91.9 (12.7) | 46.1 (14.4) | 8.9 | 6.8 |
| Vanderbilt | 3,754 | Chinese | Sphygmomanometer, 2–3 | 57.1 (8.4) | 76.0 | 24.8 (3.5) | 128.7 (19.4) | 80.5 (10.5) | 96.5 (12.6) | 48.2 (14.4) | 49.0 | 22.7 |
| Stage-2: Replication study (n=28,783) | ||||||||||||
| In-silico genotyping studies (n=5,584) | ||||||||||||
| HEXA | 3,703 | Korean | Standard mercury sphygmomanometer, 2 | 53.2 (8.3) | 55.4 | 24.0 (2.9) | 121.7 (14.4) | 77.1 (9.9) | 91.9 (10.7) | 44.7 (9.2) | 18.7 | 0 |
| SCES | 1,881 | Chinese | Random zero sphygmomanometer, 3 | 58.4 (9.5) | 48.7 | 23.7 (3.5) | 140.8 (20.5) | 80.6 (9.9) | 100.7 (12.1) | 60.2 (16.3) | 56.1 | 51.8 |
| De-novo genotyping studies (n=23,199) | ||||||||||||
| CAGE-Amagasaki | 5,331 | Japanese | Digital, 2–3 | 47.8 (12.3) | 39.8 | 23.0 (3.2) | 124.3 (17.3) | 75.9 (11.0) | 92.1 (12.9) | 48.4 (8.5) | 57.9 | 23.9 |
| JMGP | 11,570 | Japanese | Digital cuff-oscillometric device, 2 | 56.1 (14.0) | 50.0 | 23.0 (3.1) | 131.3 (19.6) | 78.4 (11.6) | 96.0 (13.4) | 52.6 (13.1) | 41.6 | 19.2 |
| SMWHS | 3,237 | Chinese | Sphygmomanometer, 2–3 | 59.3 (8.8) | 56.0 | 25.3 (3.6) | 132.6 (19.6) | 82.6 (10.5) | 100.7 (13.4) | 51.1 (15.0) | 53.0 | 21.3 |
| Suzhou Study | 3,061 | Chinese | Standard mercury sphygmomanometer, 2 | 54.2 (10.5) | 61.7 | 24.8 (3.6) | 134.2 (19.8) | 86.4 (10.2) | 70.5 (9.9) | 47.8 (14.3) | 47.2 | 27.5 |
AGEN=Asian Genetic Epidemiology Network; BP=Blood pressure; CAGE=Cardio-metabolic Genome Epidemiology; CLHNS=Cebu Longitudinal Health and Nutrition Survey; DBP=Diastolic blood pressure; GenSalt=Genetic Epidemiology Network of Salt-Sensitivity; GWAS=Genome-wide association study; HTN=Hypertension; JMPG=Japanese Millenium Genome Project; KARE=Korean Association Resource; MAP=Mean arterial pressure; N=sample size; NHAPC=Nutrition and Health of Aging Population in China; PP=Pulse pressure; SBP=Systolic blood pressure; SCES=Singapore Chinese Eyes Study; SD=Standard deviation; Vanderbilt=Vanderbilt Genome-Wide Association Studies; SiMES=Singapore Malay Eye Study; SMWHS=Shanghai Men’s and Shanghai Women’s Health Studies; SP2=Singapore Prospective Study.
Age in years;
Data are percentages;
Measurement in kilograms per square meters;
Measurement in mmHg;
MAP is calculated as DBP + (SBP–DBP)/3 in each study;
PP is calculated as SBP–DBP in each study
Hypertension is defined as SBP≥140 mmHg and DBP≥90 mmHg or taking antihypertension medication.
Inverse-variance weighted fixed effect meta-analyses of MAP and PP results from the 9 GWAS were carried out using METAL19. SNPs were excluded if they had minor allele frequency (MAF)<0.05, Hardy-Weinberg P<1×10−6, call rate<0.95, imputation quality score<0.5, sample size less than 10,000 or showed evidence of heterogeneity across studies (P for Cochrane’s Q-test<1×10−6). Genomic control was applied to each study (Table S1) and the final meta-analyses (λGC=1.02 for MAP and λGC=1.00 for PP; and Figure S1).
Stage-2 replication studies and joint analyses
Novel SNPs were selected for stage-2 replication genotyping by choosing the most significant SNP from loci which achieved a stage-1 P<1.0×10−5 for MAP or PP. We considered physiological plausibility by also selecting SNPs located within candidate genes20 if they achieved P<1.0×10−4 for either MAP or PP or P<1.0×10−3 for both MAP and PP. For assessment of trans-ethnic replication, previously identified MAP and PP SNPs15 which achieved nominal significance (P<0.05) in stage-1 study were selected for evaluation in the in-silico replication stage.
In-silico or de novo replication genotyping and association analyses were conducted in up to six additional samples of 28,783 participants (Table 1, Supplementary Note). Meta-analysis was again used to combine results across the stage-2 studies and to conduct joint analysis of stage-1 and stage-2 findings. For novel loci, findings were considered significant if they achieved genome-wide significance (P<5.0×10−8) in the joint analysis. For loci previously identified in European populations, a Bonferroni P<1.35×10−3 and consistency in effect direction was considered evidence of replication.
RESULTS
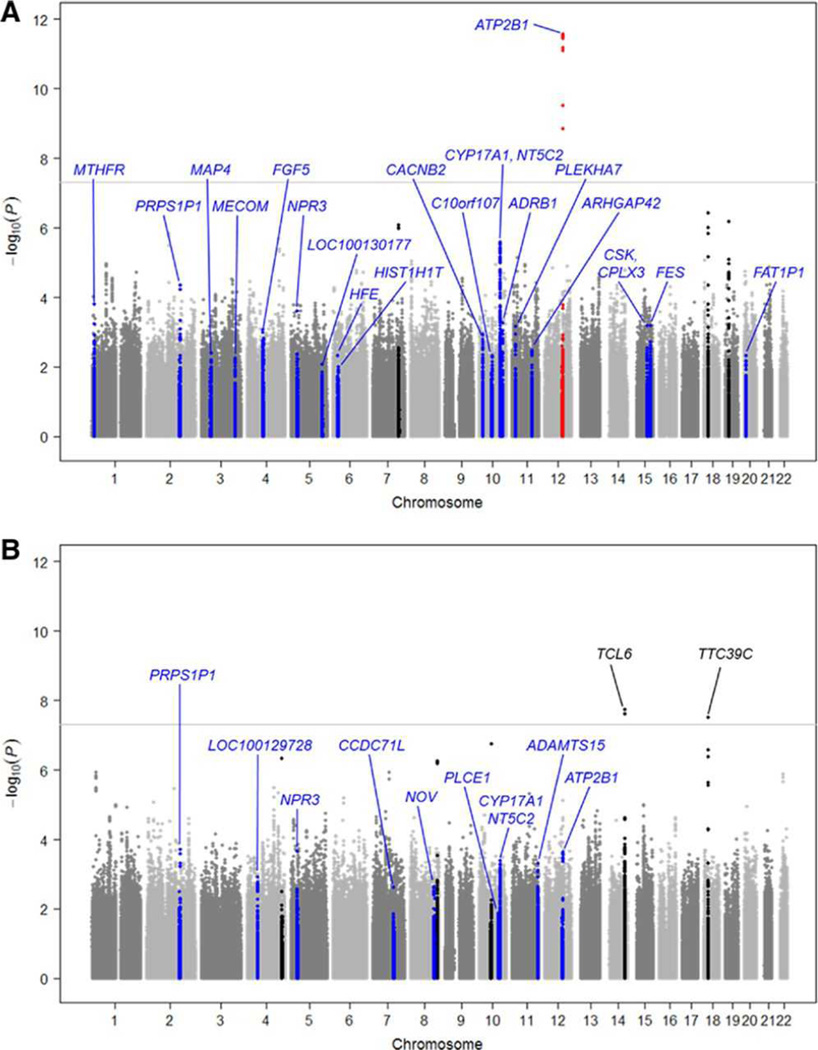
In the stage-1 GWAS meta-analysis of 26,600 participants, genome-wide significance was achieved at 12q21.33 (rs17249754) at the widely reported ATP2B1 locus for the MAP phenotype (P=3.65×10−12; Figure 1 and Table 2). For PP, novel loci TCL6 at 14q32.13 (rs2145975; P=1.90×10−8) and TTC39C at 18q11.2 (rs11874765; P=3.14×10−8) achieved genome-wide significance in stage-1 study (Figure 1 and Table S2). Six additional novel MAP and PP loci achieved borderline significance (P<1.0×10−6; Figure 1 and Table S2). A full list of stage-1 P-values for the associations between each of the 2.4 million SNPs and the MAP and PP phenotypes are publicly available for download at www.agenconsortium.org/publications.php.
Figure 1.
Genome-wide association study (GWAS) meta-analysis results for MAP (a) and PP (b). Loci highlighted in red indicate the 2 Mb regions of SNPs which achieved genome-wide significance in stage-1 and joint analyses of stage-1 and stage-2 studies. Loci highlighted in black indicate the 2 Mb regions of SNPs which achieved borderline significance (P<1E-6) in the stage-1 GWAS meta-analysis. Loci highlighted in blue indicate the 2 Mb regions of SNPs which achieved genome-wide significance in the GWAS meta-analysis of Europeans (unless achieving P<1E-6 in the current study)15. Loci which achieved genome-wide significance in Europeans15 or East Asians are labeled (blue if originally identified in Europeans; black if originally identified in the current study).
Table 2.
Trans-ethnic replication of MAP and PP loci among East Asian samples of the AGEN consortium.
| Locus | Marker | Chromosome | Position (build 36.3) |
Nearest Gene | CA/OA | CAF | Stages | N | MAP |
PP |
Previously reported phenotype |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta* | (SE) | P | Beta* | (SE) | P | ||||||||||
| 3p21.31 | rs319690 | 3 | 47902488 | MAP4† | T/C | 0.71 | Stage 1 | 26263 | 0.28 | (0.12) | 2.06E-02 | 0.11 | (0.11) | 3.42E-01 | MAP |
| Stage 2 | 5583 | 0.20 | (0.22) | 3.65E-01 | 0.06 | (0.21) | 7.65E-01 | ||||||||
| Stages 1+2 | 31845 | 0.26 | (0.11) | 1.36E-02 | 0.10 | (0.10) | 3.28E-01 | ||||||||
| 4q21.21 | rs13149993 | 4 | 81377319 | FGF5 | A/G | 0.41 | Stage 1 | 25964 | 0.32 | (0.11) | 4.57E-03 | 0.37 | (0.11) | 5.67E-04 | MAP |
| Stage 2 | 5495 | 0.51 | (0.21) | 1.35E-02 | 0.06 | (0.20) | 7.56E-01 | ||||||||
| Stages 1+2 | 31459 | 0.37 | (0.10) | 2.39E-04§ | 0.30 | (0.09) | 1.51E-03‖ | ||||||||
| 5p13.3 | rs1173771‡ | 5 | 32850785 | NPR3 | A/G | 0.37 | Stage 1 | 26399 | −0.28 | (0.11) | 1.30E-02 | −0.25 | (0.11) | 1.84E-02 | MAP, PP |
| Stage 2 | 5477 | −0.08 | (0.21) | 7.05E-01 | 0.11 | (0.19) | 5.62E-01 | ||||||||
| Stages 1+2 | 31876 | −0.24 | (0.10) | 1.84E-02 | −0.17 | (0.09) | 7.39E-02 | ||||||||
| 10q24.32 | rs3824755 | 10 | 104585839 | CYP17A1† | C/G | 0.32 | Stage 1 | 26130 | −0.50 | (0.12) | 2.42E-05 | −0.28 | (0.11) | 1.46E-02 | MAP‖ PP |
| Stage 2 | 5571 | −0.51 | (0.21) | 1.65E-02 | −0.25 | (0.20) | 2.12E-01 | ||||||||
| Stages 1+2 | 31700 | −0.50 | (0.10) | 1.21E-06§ | −0.27 | (0.10) | 6.12E-03 | ||||||||
| 10q24.33 | rs11191593‡ | 10 | 104929205 | NT5C2† | T/C | 0.74 | Stage 1 | 26044 | 0.60 | (0.13) | 2.76E-06 | 0.38 | (0.12) | 1.50E-03 | MAP, PP¶ |
| Stage 2 | 5577 | 0.37 | (0.23) | 1.15E-01 | 0.21 | (0.22) | 3.26E-01 | ||||||||
| Stages 1+2 | 31621 | 0.54 | (0.11) | 1.13E-06§ | 0.34 | (0.11) | 1.14E-03§ | ||||||||
| 12q21.33 | rs2681492 | 12 | 88537220 | ATP2B1† | T/C | 0.67 | Stage 1 | 16915 | 0.55 | (0.15) | 1.65E-04 | 0.19 | (0.15) | 1.94E-01 | MAP, PP |
| Stage 2 | 5577 | 0.73 | (0.21) | 4.47E-04 | 0.51 | (0.19) | 8.81E-03 | ||||||||
| Stages 1+2 | 22492 | 0.61 | (0.12) | 3.39E-07§ | 0.31 | (0.12) | 9.02E-03 | ||||||||
| 12q21.33 | rs17249754‡ | 12 | 88584717 | ATP2B1 | A/G | 0.35 | Stage 1 | 25401 | −0.82 | (0.12) | 3.65E-12 | −0.40 | (0.11) | 2.76E-04 | MAP, PP |
| Stage 2 | 5504 | −0.75 | (0.21) | 4.55E-04 | −0.49 | (0.20) | 1.37E-02 | ||||||||
| Stages 1+2 | 30905 | −0.80 | (0.10) | 7.48E-15§ | −0.42 | (0.10) | 1.21E-05§ | ||||||||
CA=Coded allele; CAF=CA frequency; MAP=Mean arterial pressure; N=Effective sample size; OA=Other allele; Position=Physical Position (in basepairs); PP=Pulse pressure; SE=Standard error.
Beta is the effect size in mmHg per coded allele based on an additive genetic model.
Corresponding marker lays within reported gene.
The variant [or its proxy (r2>0.8)] was previously implicated for SBP or DBP in the genome-wide association study meta-analysis of East Asians conducted by Kato and colleagues.
Significant after Bonferonni correction for 37 statistical tests.
Although there is evidence of significance for PP, this variant was only identified for MAP in the study by Wain and colleagues. Therefore, this does not represent evidence of trans-ethnic replication.frs1004467, a proxy for rs3824755 in East Asian samples (r2=0.95), achieved genome-wide significance for MAP in the study by Wain and colleagues.
rs11191548, a proxy for rs1191593 in East Asian samples (r2=1.00), achieved genome-wide significance for PP in the study by Wain and colleagues.
A total of 35 independent, novel trait-associated SNPs were selected for stage-2 replication study. None achieved genome-wide significance in joint analysis of stage-1 and stage-2 findings (Table S2). Among 48 SNPs which previously achieved genome-wide significance for MAP or PP traits in European populations15, 11 independent loci achieved nominal significance in the stage-1 study and were followed-up in stage-2 study (Table S3).
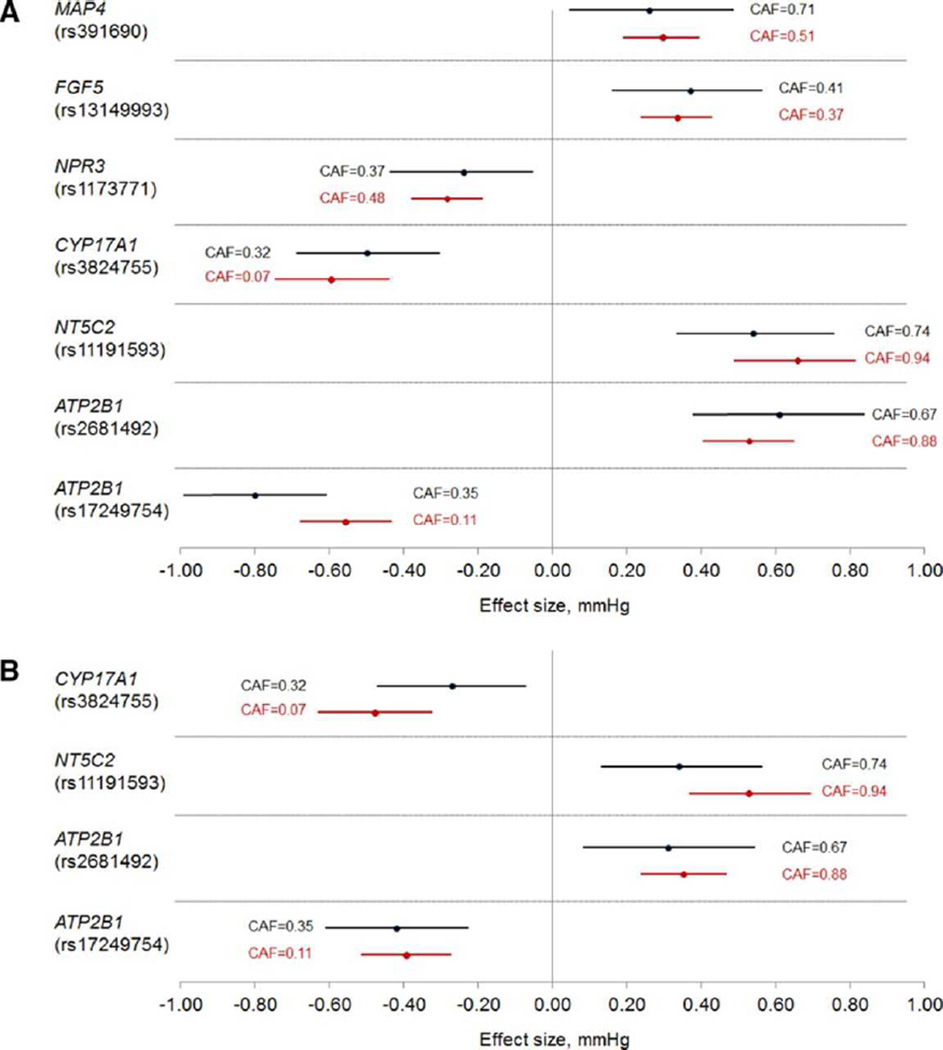
Table 2 provides top trans-ethnic replication results. For the MAP phenotype, rs17249754 at ATP2B1 achieved genome-wide significance in the joint analysis of stage-1 and stage-2 studies (P=7.48×10−15). Four additional SNPs showed robust evidence of replication for MAP (consistency of effect direction and significance after adjustment for multiple testing), including rs13149993 at FGF5 (P=2.39×10−4), rs3824755 at CYP17A1 (P=1.21×10−6), rs11191593 at NT5C2 (1.13×10−6), and rs2681492 at ATP2B1 (P=3.39×10−7). Furthermore, two SNPs showed suggestive replication [consistency of effect direction and nominal significance (P<0.05)], including rs319690 at MAP4 (P=0.01) and rs1173771 at NPR3 (P=0.02). For the PP phenotype, robust evidence of replication was identified for 2 SNPs including rs11191593 at NT5C2 (P=1.14×10−3) and rs17249754 at ATP2B1 (P=1.21×10−5). Suggestive evidence of replication was identified for 2 additional SNPs including rs3824755 at CYP17A1 (P=6.12×10−3) and rs2681492 at ATP2B1 (P=9.02×10−3). Effect sizes of replicated MAP and PP loci were very similar between the previous GWAS meta-analysis of Europeans15 and the current GWAS meta-analysis of East Asians (Figure 2).
Figure 2.
Effect sizes and coded allele frequencies (CAF) for SNPs that showed evidence of trans-ethnic replication for MAP (a) and PP (b) in East Asian participants of the current GWAS meta-analysis. Effect sizes in the current study of East Asians are shown in black while those of the previous GWAS meta-analysis of Europeans15 are shown in red.
DISCUSSION
The current meta-analysis of 26,600 East Asian participants provided robust trans-ethnic replication evidence for 5 independent SNPs at MAP and PP loci previously identified in populations of European ancestry15, including rs13149993 at FGF5, rs3824755 at CYP17A1, rs11191593 at NT5C2, rs2681492 at ATP2B1, and rs17249754 at ATP2B1. In addition, two SNPs, rs319690 at MAP4 and rs1173771 at NPR3, showed suggestive evidence of trans-ethnic replication. Further examination of these 7 variants demonstrated remarkable consistency in per allele effect sizes across populations of East Asian and European ancestry.
Seven SNPs from MAP and PP loci identified in samples of European ancestry showed evidence of trans-ethnic replication in the current study of East Asians. Marker rs17249754 at the ATP2B1 locus (12q21.33) achieved genome-wide significance for MAP and was robustly associated with PP. Furthermore, rs2681492, a moderately correlated intronic ATP2B1 SNP (r2=0.78), also showed evidence of trans-ethnic replication for the MAP and PP phenotypes. ATP2B1 is a widely reported BP-related gene, with marker rs17249754 also identified for the SBP and DBP phenotypes in East Asians4–7, 15. ATP2B1 is thought to exert its influence on BP regulation through alteration of calcium handling and vasoconstriction in vascular smooth muscle cells21. At 3p21.31, the current study provided the first evidence of association for marker rs319690 with a BP-related phenotype among individuals with East Asian ancestry. Marker rs319690 represents an intronic variant of the MAP4 gene, implicated in heart failure through its interference with beta-adrenergic receptor recycling22. At the FGF5 locus (4q21.21), marker rs13149993 was associated with both MAP and PP phenotypes in the current study. The FGF5 rs13149993 variant [or a proxy (r2>0.8)] has been reported previously for its associations with not only MAP but other BP-related phenotypes5,7,15. Furthermore, a variant modestly correlated with rs13149993 at FGF5, rs16998073 (r2=0.49), was previously related to SBP and DBP in East Asians6,23. A fibroblast growth factor gene, FGF5 is expressed in cardiac myocytes and has been shown to promote angiogenesis in the heart24. Near NPR3 (at 5p13.3), rs1173771 was associated with MAP in the current study. Previously reported for its association with MAP in Caucasians and other BP phenotypes in Caucasians and East Asians6,7,15, NPR3 encodes the natriuretic peptide receptor C, a peptide known to regulate BP and fluid homeostasis by modifying glomerular filtration rate and sodium urinary excretion25. Finally, moderately correlated SNPs rs3824755 and rs11191593 (r2=0.67) at 10q24.32-10q24.33 were associated with MAP and PP in the current study. Marker rs3824755 is an intronic variant of CYP17A1, the gene responsible for the monogenic BP disorder congenital adrenal hyperplasia26, while rs11191593 is an intronic variant of NT5C2, a gene involved in DNA synthesis with no known functional role in BP regulation27. Marker rs3824755 (or a proxy) was previously reported to associate with not only MAP and PP15 but also SBP in the manuscript by Levy and colleagues4, while rs11191593 (or a proxy) has been reported previously for numerous BP phenotypes, including SBP and DBP in East Asians5–7, 15.
The current GWAS meta-analysis represents the largest genetic association study of MAP and PP conducted in participants of East Asians ancestry to date. Additional study strengths include the adherence of all studies to a standard analytic protocol and stringent genotyping and imputation quality control procedures at the study and meta-analysis levels. Despite these strengths, the current study failed to identify any novel loci related to MAP and PP traits. Although currently the largest study conducted in East Asians, the stage-1 sample was only one-third the size of that of the prior GWAS meta-analysis of MAP and PP conducted in populations of European ancestry15. Thus, we still may have lacked the statistical power needed to identify novel variants for MAP and PP. Furthermore, we did not replicate 24 of the 31 loci previously identified in Europeans. Lack of replication could be related to differences in linkage disequilibrium (LD) patterns between Europeans and East Asians. To address this concern, we examined inter-population LD variation at these loci28. Our results showed that LD structure was generally similar between populations at all but five regions (see Figure S2). However, examination of variants in LD with the lead SNP in Europeans did not reveal any further significant associations in East Asians, suggesting that differences in LD may not have been a major factor limiting replication in the current study (data not shown). Lack of replication could also be due to limited statistical power. To assess this issue, we compared effect sizes and MAFs of independent SNPs which achieved genome-wide significance in Europeans between the two studies (Table S4). Despite differences in the MAF of many of the variants, very strong correlations in effect sizes between populations were observed (Table S4). Furthermore, power calculations demonstrated that we lacked the statistical power to detect associations for the 24 un-replicated loci (Table S5). These data suggest the existence of additional promising MAP and PP loci in East Asian populations that may be identified by future, larger GWAS meta-analyses.
Supplementary Material
PERSPECTIVES.
The current study of 26,600 East Asian participants from 9 GWAS provides the first evidence of trans-ethnic replication of 7 MAP and PP loci previously identified in populations of European ancestry. In addition, we demonstrate remarkable consistency in allelic effect sizes between populations with vast differences in not only genomic ancestry but also environmental and cultural factors. Our findings add to the accumulating evidence that many genomic associations are reproducible in populations with distinct LD structure, suggesting common genomic mechanisms underlying the development of hypertension and cardiovascular disease across populations.
NOVELTY AND SIGNIFICANCE.
What is New?
The current GWAS meta-analysis of 26,600 East Asians the first evidence of trans-ethnic replication of 7 MAP and PP loci previously identified in populations of European ancestry.
Per allele effect sizes of replicated variants were consistent between Europeans and East Asians.
What is Relevant?
The physiologic effects of many common polymorphisms may be generalizable across populations.
Summary
The current meta-analysis of 26,600 East Asian participants from 9 GWAS provided evidence of trans-ethnic replication for 7 MAP and PP variants previously identified in populations of European ancestry15. These variants demonstrated remarkable consistency in per allele effect sizes across populations of East Asian and European ancestry. We add to the accumulating evidence that many genomic associations are reproducible in populations with distinct linkage disequilibrium structure, suggesting common genomic mechanisms underlying the development of hypertension and cardiovascular disease across populations.
Acknowledgments
SOURCES OF FUNDING
A full list of acknowledgements and funding sources is provided in the Supplementary Note.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
REFERENCES
- 1.Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J. Global burden of hypertension: Analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- 2.Danaei G, Finucane MM, Lin JK, Singh GM, Paciorek CJ, Cowan MJ, Farzadfar F, Stevens GA, Lim SS, Riley LM, Ezzati M Global Burden of Metabolic Risk Factors of Chronic Diseases Collaborating G. National, regional, and global trends in systolic blood pressure since 1980: Systematic analysis of health examination surveys and epidemiological studies with 786 country-years and 5.4 million participants. Lancet. 2011;377:568–577. doi: 10.1016/S0140-6736(10)62036-3. [DOI] [PubMed] [Google Scholar]
- 3.Lim SS, Vos T, Flaxman AD, Danaei G, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990 –2010: A systematic analysis for the global burden of disease study 2010. Lancet. 2013;380:2224–2260. doi: 10.1016/S0140-6736(12)61766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy D, Ehret GB, Rice K, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newton-Cheh C, Johnson T, Gateva V, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato N, Takeuchi F, Tabara Y, et al. Meta-analysis identifies five novel loci associated with blood pressure in east asians. Nat Genet. 2011;43:531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehret GB, Munroe PB, Rice KM, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franklin SS, Khan SA, Wong ND, Larson MG, Levy D. Is pulse pressure useful in predicting risk for coronary heart disease? The framingham heart study. Circulation. 1999;100:354–360. doi: 10.1161/01.cir.100.4.354. [DOI] [PubMed] [Google Scholar]
- 9.Miura K, Soyama Y, Morikawa Y, Nishijo M, Nakanishi Y, Naruse Y, Yoshita K, Kagamimori S, Nakagawa H. Comparison of four blood pressure indexes for the prediction of 10-year stroke risk in middle-aged and older asians. Hypertension. 2004;44:715–720. doi: 10.1161/01.HYP.0000145108.23948.7b. [DOI] [PubMed] [Google Scholar]
- 10.Kengne A-P, Czernichow S, Huxley R, et al. Blood pressure variables and cardiovascular risk: New findings from advance. Hypertension. 2009;54:399–404. doi: 10.1161/HYPERTENSIONAHA.109.133041. [DOI] [PubMed] [Google Scholar]
- 11.Dorjgochoo T, Shu XO, Zhang X, Li H, Yang G, Gao L, Cai H, Gao Y-T, Zheng W. Relation of blood pressure components and categories and all-cause, stroke and coronary heart disease mortality in urban chinese women: A population-based prospective study. J Hypertens. 2009;27:468–475. doi: 10.1097/HJH.0b013e3283220eb9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu D, Rice T, Wang S, Yang W, Gu C, Chen C-S, Hixson JE, Jaquish CE, Yao Z-J, Liu D-P, Rao DC, He J. Heritability of blood pressure responses to dietary sodium and potassium intake in a chinese population. Hypertension. 2007;50:116–122. doi: 10.1161/HYPERTENSIONAHA.107.088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adeyemo AA, Omotade OO, Rotimi CN, Luke AH, Tayo BO, Cooper RS. Heritability of blood pressure in nigerian families. J Hypertens. 2002;20:859–863. doi: 10.1097/00004872-200205000-00019. [DOI] [PubMed] [Google Scholar]
- 14.Pilia G, Chen W-M, Scuteri A, et al. Heritability of cardiovascular and personality traits in 6,148 sardinians. PLoS Genet. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wain LV, Verwoert GC, O'Reilly PF, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011;43:1005–1011. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. American Journal of Human Genetics. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. Mach: Using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willer CJ, Li Y, Abecasis GR. Metal: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat Genet. 2008;40:124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi Y, Hirawa N, Tabara Y, et al. Mice lacking hypertension candidate gene atp2b1 in vascular smooth muscle cells show significant blood pressure elevation. Hypertension. 2012;59:854–860. doi: 10.1161/HYPERTENSIONAHA.110.165068. [DOI] [PubMed] [Google Scholar]
- 22.Cheng G, Qiao F, Gallien TN, Kuppuswamy D, Cooper Gt. Inhibition of beta-adrenergic receptor trafficking in adult cardiocytes by map4 decoration of microtubules. Am J Physiol Heart Circ Physiol. 2005;288:H1193–H1202. doi: 10.1152/ajpheart.00109.2004. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi F, Isono M, Katsuya T, et al. Blood pressure and hypertension are associated with 7 loci in the japanese population. Circulation. 2010;121:2302–2309. doi: 10.1161/CIRCULATIONAHA.109.904664. [DOI] [PubMed] [Google Scholar]
- 24.Vatner SF. Fgf induces hypertrophy and angiogenesis in hibernating myocardium. Circ Res. 2005;96:705–707. doi: 10.1161/01.RES.0000164184.63158.6c. [DOI] [PubMed] [Google Scholar]
- 25.Saulnier P-J, Roussel R, Halimi JM, Lebrec J, Dardari D, Maimaitiming S, Guilloteau G, Prugnard X, Marechaud R, Ragot S, Marre M, Hadjadj S, Surdiagene DN. Impact of natriuretic peptide clearance receptor (npr3) gene variants on blood pressure in type 2 diabetes. Diabetes Care. 2011;34:1199–1204. doi: 10.2337/dc10-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White PC. Disorders of aldosterone biosynthesis and action. N Engl J Med. 1994;331:250–258. doi: 10.1056/NEJM199407283310408. [DOI] [PubMed] [Google Scholar]
- 27.Hotta K, Kitamoto A, Kitamoto T, et al. Genetic variations in the cyp17a1 and nt5c2 genes are associated with a reduction in visceral and subcutaneous fat areas in japanese women. J Hum Genet. 2012;57:46–51. doi: 10.1038/jhg.2011.127. [DOI] [PubMed] [Google Scholar]
- 28.Ong RT, Teo YY. varLD: a program for quantifying variation in linkage disequilibrium patterns between populations. Bioinformatics. 2010;59:1269–1270. doi: 10.1093/bioinformatics/btq125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.