Abstract
MicroRNAs (miRNAs) are a class of small ~22 nt noncoding (nc) RNAs that regulate gene expression post-transcriptionally by direct binding to target sites on mRNAs. They comprise more than 1,000 novel species in mammalian cells and exert their function by modulating gene expression through several different mechanisms, including translational inhibition, and/or degradation of target mRNAs. Mitochondria maintain and express their own genome, which is distinct from the nuclear transcriptional and translational apparatus. Thus, they provide a potential site for miRNA mediated post-transcriptional regulation. To determine whether they maintain a unique miRNA population, we examined the miRNA profile from highly purified and RNase treated mitochondria from adult rat liver. Fifteen miRNAs were identified by microarray analysis of which, five were confirmed by TaqMan® 5′nuclease assays using rat specific probes. Functional analysis of the miRNAs indicated that they were not targeted to the mitochondrial genome nor were they complementary to nuclear RNAs encoding mitochondrial proteins. Rather, the mitochondria-associated miRNAs appear to be involved in the expression of genes associated with apoptosis, cell proliferation, and differentiation. Given the central role that mitochondria play in apoptosis, the results suggest that they might serve as reservoirs of select miRNAs that may modulate these processes in a coordinate fashion.
Keywords: miRNA, mitochondria, apoptosis, liver, cell division
Introduction
Recently, the role of RNA in the cell was changed by the discovery of a diverse array of non-coding RNA (ncRNA) genes that do not encode proteins but function primarily at the level of RNA. They can regulate DNA replication and chromosome maintenance, transcriptional activity, RNA processing, translation and stability, as well as translocation of proteins.1,2 MicroRNAs (miRNAs) are a class of small ncRNAs that play a critical role in modulating gene expression at sites in the nucleus and cytosol.3 They were originally identified in plants, nematodes and then humans from non-coding, imperfect complementary, stem-loop RNA precursors.4 microRNA genes are transcribed primarily from intronic regions by RNA polymerase II and are capped and polyadenylated similarly to normal RNA transcription.5 Mature miRNAs 18- to 24-nt long are produced from sequential processing of their primary transcripts by several key players, including RNases Drosha, located in the nucleus and Dicer in the cytosol. Mature miRNAs are paired with RISC (RNA Induced Silencing Complex), which aids in binding miRNA to the target mRNA, thereby acting as an active repressor of expression. These mature miRNA:RISC complexes reduce protein expression of specific ‘target’ mRNAs by either translational inhibition or mRNA degradation.6
The miRNAs have been shown to exert their influence predominately at the posttranscriptional level in cytosol,6 in part, through modulation of translational activity following miRNA:RISC binding to the target mRNA.7 The fate of target mRNAs is primarily dependent on the complementarity of the miRNA with the target site, in which perfect or near-perfect pairing with the target will typically result in mRNA decay. However if multiple, partially complementary target sites are present, the miRNAs will inhibit translation without significantly affecting mRNA levels.8 The exact mechanism of miRNA triggered mRNA degradation has not yet been elucidated, but appears to be mediated in part by “Slicer” activity of the Argonaute (Ago) proteins. Cleavage by Ago-Slicer activity is believed to mark the target mRNA for further degradation via deadenylation, decapping and 5′–3′ exonuclease degradation in the cytosol. In contrast, miRNA translational repressed mRNAs are translocated to specialized processing (P) bodies located in the cell cytoplasm.9 It should be noted, however, that miRNA-mediated translational repression occurs independently of the movement of miRISC:mRNA to P-bodies.10 However, once released from the polysome, the miRNA-targeted transcripts are transferred to cytoplasmic processing (P)-bodies for either degradation and/or storage, and can return to the actively translated mRNA pool as required.11 However, the cellular conditions that regulate the precise destiny of the transcripts are not entirely understood.7,12,13 In fact, it appears that mRNA degradation occurs significantly less frequently than translational repression from miRNA. Moreover, a single miRNA can target many different mRNAs,14 thus providing a posttranscriptional mechanism to coordinately regulate numerous genes as well as sequester mRNAs for either degradation13,15 or re-expression without de novo transcription.12
Mitochondria are subcellular organelles that contain their own genetic system and include proteins translated from mRNA derived from either the nuclear or mitochondrial genome. Mitochondrial transcripts reflecting the extreme economy of their compact gene organization differ from nuclear transcripts in that there are no intronic sequences in mtDNA, and the 3′ UTR regions are shorter due to the reduced intergenic regions.16,17 Nuclear derived mRNA transcripts are transported to both the endoplasmic reticulum and mitochondria by an elaborate mechanism involving translational stalling of ribosomes and direct physical transport to their respective membranes. In addition to using nuclear derived transcripts, mitochondria express their own genome and regulate cellular events such as induction of programmed cell death without nuclear involvement.18 The central role that mitochondria has in governing the overall fate of a cell involves an orchestrated progression through complex pathways. The myriad discrete pathways used for mitochondrial driven apoptosis in the absence of de novo transcription19-21 suggested that one heretofore unrecognized mechanism could involve a unique role of mitochondrial miRNA in the regulation of gene expression.
Interestingly, ncRNAs have been identified in chloroplasts and mitochondria by size selection and cloning.22 In mammalian cells, these included four from the D-loop region involved in mitochondrial genome replication, and two antisense transcripts of known mitochondrial genes. In addition, a novel ncRNA class was found in mitochondria of the African trypanosome brucei23 whose mitochondrial mRNAs are subject to an editing reaction that involves small non-coding, 3′-oligouridylated (oU) guide RNAs (gRNAs). A multi-enzyme complex the editisome, is responsible for the insertion/ deletion of U residues in the mRNAs. In the U deletion/insertion reaction, gRNAs are the transacting templates, and by employing different gRNAs, alternative-editing events can increase the diversity of mitochondrial transcripts. Thus, in both lower eukaryotes and mammals, small ncRNAs that modulate other RNA class functions have been identified in mitochondria. Although miRNAs have not been reported in mitochondria, their presence would not be surprising in light of the dynamic trafficking of RNAs across the mitochondrial membrane.24,25
Results
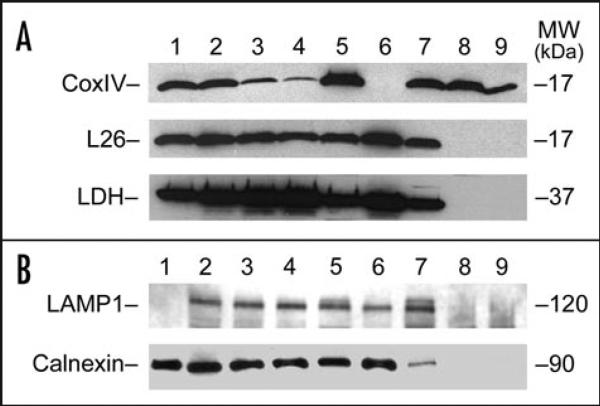
To explore the possibility that mitochondria might contain miRNAs, a highly purified population of mitochondria was isolated from normal adult rat liver, following homogenization in isotonic buffer and Percoll® gradient centrifugation as described in Materials and Methods. The purity of the mitochondrial fraction was established by western blot analysis using the mitochondrial marker protein CoxIV, which was significantly enriched (Fig. 1, lanes 7–9, upper) in the purified fractions. Cytosolic protein and polysome content of the mitochondrial fractions were determined using lactate dehydrogenase (LDH) and the ribosomal L26 protein, respectively. These analyses indicated that while LDH and L26 protein were present in the crude mitochondrial pellet (Fig. 1A, lane 7, middle and lower panels); neither protein was detectable in the mitochondrial fractions following Percoll® gradient centrifugation (Fig. 1A, lanes 8 and 9). We also investigated if other subcellular organelles might be present in the purified mitochondrial fraction by western blot using LAMP1 as a lysosomal marker and calnexin for endoplasmic reticulum (ER). The results indicated that while both LAMP1 and calnexin were present in all the liver fractions up to and including the mitochondrial pellet prior to Percoll® gradient centrifugation (Fig. 1B, lanes 2–7), neither protein was present in the final gradient-derived mitochondria (Fig. 1B, lanes 8 and 9).
Figure 1.
Western blot analysis of Percoll gradient isolated mitochondria. (A) Total proteins (40 μg) isolated from the various fractions during the mitochondria purification process were separated by SDS-PAGE. Following electrophoretic transfer onto nitrocellulose membrane, the immunoblots were processed for western blot analysis to assess purity using primary antibodies against lactate dehydrogenase (LDH, bottom) as a cytosolic marker; cytoplasmic large ribosomal protein L26 for polysomal contamination (middle); and protein CoxIV as a mitochondrial marker (top). Each of the proteins was detected by ECL. The protein is indicated at left, and the molecular weight (MW) in kDa at right. Lane 1, isolated rat liver polysomes; lane 2, whole liver homogenate; lane 3, supernatant post 600 ×g centrifugation; lane 4, supernatant post 1,100 ×g centrifugation; lane 5, pellet from 1,100 ×g centrifugation; lane 6, supernatant post 7,600 ×g centrifugation; lane 7, crude mitochondrial pellet from 7,600 ×g centrifugation; lane 8, mitochondrial fraction from Percoll® gradient; and lane 9, mitochondrial fraction from Percoll® gradient following RNase treatment. (B) Total proteins (25 μg) isolated from the various fractions during the mitochondria purification were separated by SDS-PAGE and processed for immunoblot analysis using primary antibodies anti-LAMP1 (upper) and anti-calnexin (lower) as markers for lysosomal and endoplasmic reticulum, respectively. The proteins were detected using ECL and their identity is shown at left and their molecular weight (MW) in kDa at right. Lane 1, MCF-7 whole cell lysate; lane 2, whole liver homogenate; lane 3, supernatant post 600 ×g centrifugation; lane 4, supernatant post 1,100 ×g centrifugation; lane 5, pellet from 1,100 ×g centrifugation; lane 6, supernatant post 7,600 ×g centrifugation; lane 7, crude mitochondrial pellet from 7,600 ×g centrifugation; lane 8, mitochondrial fraction from Percoll® gradient; and lane 9, mitochondrial fraction from Percoll® gradient following RNase treatment.
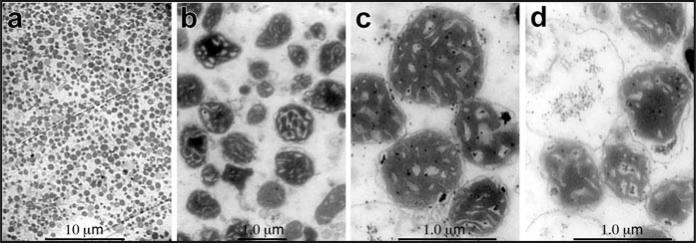
We then characterized the integrity of the isolated mitochondria by transmission electron microscopy. The initial characterization (animals 4–6) indicated that some potentially non-mitochondrial membranous structures were present in the final mitochondrial pellets used for the RNA extractions (data not shown). However, the uppermost portion of the band from the Percoll® gradient contained a highly purified mitochondrial fraction (Fig. 2a–c) that was almost entirely free from contaminating membranes and exhibited little alteration of normal mitochondrial morphology. Purified mitochondria were treated with RNase to ensure that the miRNAs did not result from contaminating sources as previously described for other RNAs.24 Following RNase treatment, some swelling and disruption of the intact mitochondria was observed (Fig. 2d) relative to no treatment (Fig. 2c). Taken together, the data suggested that the isolated mitochondria were highly purified and structurally intact for identifcation of miRNAs within their RNA pool.
Figure 2.
Transmission electron micrographs of purified liver mitochndria. Following the Percoll® gradient isolation and washing, a portion of the mitochondrial pellet was fixed overnight at 4°C in cacodylate buffered 6% glutaraldehyde, then processed and embedded for electron microscopy. Sections 70–100 nm in thickness were cut and stained with uranyl acetate and lead citrate. The morphology and purity of the isolated mitochondria were examined using a JEOL-100 CX electron microscope. The representative digital micrographs indicated that the mitochondria fields did not contain significant amounts of non-mitochondrial material (a), and exhibited classical mitochondrial morphology (b and c). (d) The same Percoll® gradient isolated mitochondria post-treatment with RNase showing minimal loss of mitochondrial structural integrity. The length in μm of the scale bars shown is indicated above the bar.
We used a custom miRNA microarray system to survey miRNA expression at a genome-wide scale in the isolated mitochondria. The array had probes derived from rat (rno), mouse (mmu), or human (hsa) miRNAs. Using this array, 15 miRNA probes were identified that yielded signals significantly above background in each of the mitochondrial fractions and were also present following RNase treatment of the purified structures (Table 1). In contrast, highly abundant miRNAs in liver, such as let-7a and miR-21 were absent, suggesting a unique population of miRNAs in mitochondria independent of total cellular abundance. In addition, the results suggested very selective miRNA retainments in the mitochondrial fractions. Of the miRNA species identified, miR-130a and b, miR-140*, miR-320 and miR-671 have been experimentally verified by northern blot analysis or cloning in rat.26-28 While the other species have not been experimentally verified in rat, all were identified in screens of neuronal or embryonic tissues.29-31
Table 1.
miRNAs identified in RNA isolated from purified rat liver mitochondria
| Species and miRNAa | Detected by microarrayb | TaqMan® miRNA assays | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 4–6d | 7–9d | ||||||||
| 1–3d | 4–6 | 7–9 | Totale | CrudeF | Percollg | Percoll + RNaseh | Percoll | Percol + RNase | |
| Rno miR-21 | no | no | no | 22.19 ± 0.20 | 36.96 ± 2.16 | Udi | Ud | Ud | Ud |
| Rno miR-130a | yes | yes | yes | 27.65 ± 0.59 | 28.29 ± 0.81 | 29.78 ± 0.38 | 31.17 ± 0.85 | 36.79 ± 1.29 | 39.24 ± 1.28 |
| Rno miR-130b | yes | yes | yes | 30.08 ± 0.27 | 30.77 ± 0.31 | 32.72 ± 0.56 | 34.20 ± 1.02 | 39.86 ± 1.42 | 43.11 ± 3.42 |
| Rno miR-140* | yes | no | yes | 28.49 ± 0.55 | 28.70 ± 0.62 | 29.48 ± 0.68 | 30.89 ± 0.32 | 36.21 ± 1.13 | 42.03 ± 4.74 |
| Rno miR-290 | no | no | no | Ud | Ud | Ud | Ud | Ud | Ud |
| Rno miR-320 | yes | yes | yes | 27.73 ± 0.03 | 28.62 ± 1.0 | 29.87 ± 0.56 | 31.84 ± 0.30 | 35.48 ± 2.21 | 37.43 ± 1.64 |
| Rno miR-494 | yes | yes | yes | 37.39 ± 0.31 | 38.55 ± 0.34 | 37.80 ± 1.07 | 34.24 ± 1.3 | 40.89 ± 1.03 | 46.33 ± 2.42 |
| Rno miR-671 | yes | yes | yes | NDj | ND | ND | ND | ND | ND |
| Mmu miR-202 | yes | yes | yes | ND | ND | ND | ND | ND | ND |
| Mmu miR-705 | no | yes | yes | ND | ND | ND | ND | ND | ND |
| Mmu miR-709 | no | no | yes | ND | ND | ND | ND | ND | ND |
| Mmu miR-721 | no | yes | yes | ND | ND | ND | ND | ND | ND |
| Mmu miR-761 | no | no | yes | ND | ND | ND | ND | ND | ND |
| Mmu miR-763 | yes | yes | yes | ND | ND | ND | ND | ND | ND |
| Hsa miR-198 | yes | yes | yes | ND | ND | ND | ND | ND | ND |
| Hsa miR-765 | yes | yes | yes | ND | ND | ND | ND | ND | ND |
Species identity and miRNA from rat (Rno), mouse (Mmu) and human (Hsa). Mmu and Hsa miRNAs are identified in the microarrays, but have no Rno homologs in the Invitrogen array probe set.
Identified by microarray hybridization with the total RNA isolated from the purified mitochondria.
cmiRNAs detected using TaqMan® miRNA assays.
Animal number used for RNA isolated for microarray hybridization and TaqMan® miRNA assays.
Total RNA isolated from whole liver homogenates.
Total RNA isolated from the crude mitochondrial pellets prior to Percoll® gradient centrifugation.
Total RNA isolated from mitochondria purified by Percoll® gradient centrifugation.
Total RNA isolated from mitochondria following Percoll® gradient centrifugation and RNase treatment.
Mean Ct ± 1 STD from the three animals for TaqMan® miRNA assays.
Ud, undetectable by TaqMan® miRNA assays through 50 cycles.
kND, Rno specific TaqMan® miRNA assays not available.
It was important to confirm that the population identified by microarray was, in fact, mature miRNAs. Thus, we performed northern blot analysis of RNA isolated from the mitochondrial pellet prior to gradient fractionation, and the Percoll® gradient fractions both before and after RNase treatment. Northern analysis using probes specific for miR-130, indicated that the mature species was present in the mitochondrial RNA prior to gradient fractionation, but not in the fractionated material (data not shown). Because of relatively low yield from the purified fractions, we used the more sensitive stem-loop qRT-PCR method for detection of other mature miRNAs. Of the 15 miRNAs identified, miR-130a and b, miR-140*, miR-320 and miR-494 rat specific TaqMan® qRT-PCR 5′nuclease assays were available, as well as those for miR-21 and miR-290, which were used as controls. In the first group of animals (Table 1, 4–6) we determined the levels of specific miRNAs in whole liver, mitochondrial fractions before and after Percoll® gradient purification and treatment with RNase (Table 1). The control miR-21 was found in total, significantly diminished in the mitochondria pellet prior to purification, and was undetectable in the gradient isolated material, again suggesting an enriched population of mitochondrial miRNAs. In addition, the negative control miR-290 although identified to be present in rat by homology predictions,32 was not found by microarray or by qRT-PCR in whole liver or any of the fractions assayed. For the 5 miRNAs confirmed by qRT-PCR, the change in Ct cycle number between the total and final mitochondrial fractions treated with RNase was <5 cycles. That was significantly less than the 14 cycles observed for miR-21 between whole liver and the mitochondrial pellet. Interestingly, miR-494 appears to be significantly enriched in mitochondria as its abundance in total RNA increased with purification as measured by qRT-PCR. The more rigorously purified mitochondria (Table 1, animals 7–9) gave similar results, albeit lower abundance of the miRNAs, that was perhaps due to some disruption of mitochondrial integrity (Fig. 2c and d). However, the population of miRNAs is protected from RNase treatment in agreement with other known mitochondrial genome expressed RNAs and the nuclear encoded mitochondrial 5S rRNA.24
Finally, it should be noted that although the TaqMan RT-PCR is designed to detect only the mature miRNA species, it is possible for the pre-miRNA to act as a template albeit at a very low frequency. Given the results of the miRNA detection by RT-PCR in RNAs isolated from whole liver homogenate, the mitochondria pre- and post-gradient purification suggests that it is the mature miRNA species, which is detected in the mitochondria.
To further define whether the miRNAs were present within the mitochondria, we used deoxycholic acid (DCA) to induce opening of the mitochondrial megapore channel33 and induction of the mitochondria permeability transition (MPT) in the isolated mitochondria. This disruption of mitochondrial integrity resulted in a significant loss of miRNAs by RT-PCR in the DCA treated mitochondria relative to the control vehicle group. The release of miR-130a and b, miR-320 and miR-494 was ~50% and that of miR-140 almost 80%.
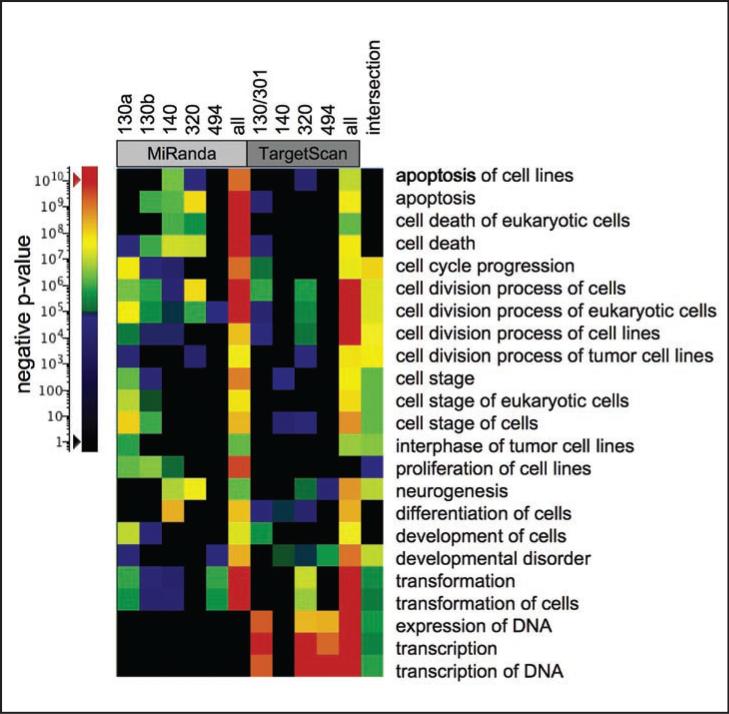
To explore the potential biological relevance of miRNAs in mitochondria, predicted mRNA targets were examined by functional enrichment analyses to identify biological functions or annotations that are overrepresented in a list of genes. The set of targets for each of the individual miRNAs, the union of each set as predicted by the Miranda34 or the TargetScan35 algorithm, multiple miRNAs that can act together to regulate target mRNAs,36 and the intersection between the unions of each set were individually analyzed by IPA functional analyses. The results of these analyses were combined, filtered and visualized by heat map. Functions with a p-values of <10E-6 in 3 or more separate analyses are shown in Figure 3. Apoptosis, cell death, cell cycle/cell division, development and neurogenesis were the most significant processes found in both the target sets calculated by Miranda and TargetScan. Proliferation and cell movement annotations were enriched in the Miranda set, while transcription related functions were identified only in the TargetScan set. The intersection between the two prediction methods showed functions primarily involved in cell cycle/cell division. Interestingly, two genes ZNF148 and CNOT6L were predicted to have conserved binding sites for all the miRNAs found in the mitochondria.
Figure 3.
Functional analyses of mitochondrial-associated miRNA. MiRanda and TargetScan algorithm miRNA target predictions for each miRNA were submitted for functional enrichment analyses to Ingenuity Pathways Analyses individually and as a group to identify over-representation of functional groups in the predicted gene set. The heat map shows annotations with p-values less then 10E-6 in 3 or more separate analyses. The miRNA species are indicated at top, representing the results of miRNA set analyses from each predictive algorithm. The results of the analysis of intersection between the two mitochondrial sets is labeled (top) and the functional groups listed immediately left of the map. The color scale for p-values is shown at left.
Potential binding targets of miRNAs not found in the mitochondria also showed significant signal using enrichment analyses. This was not unexpected, in part due to the critical roles that miRNAs appear play in cell function. Although sequence- and conservation-based methods led to similar functions, there were notable differences between the methods that may be related to the evolutionary constraints imposed when using conservation-based methods. For example the functional enrichment of transcription related genes in the TargetScan set might be related to the evolutionary rigidity in these functions. Both of these target prediction algorithms have predicted valid interactions not found in the other set.36
Discussion
Our results indicate that 15 nuclear encoded miRNAs are uniquely and reproducibly identified in mitochondria isolated from at least 9 different livers from adult rat. Two possibilities to explain the observation include trafficking of nuclear encoded mitochondrial protein mRNAs to the mitochondria for translation,37,38 as well as the association of ribosomes with the mitochondria via protease sensitive proteins.39 However, these were unlikely based on the absence of cytosolic ribosome protein L26 in the purified mitochondria and functional analyses of the miRNAs showing no association with nuclear-encoded mitochondrial protein transcripts. Mitochondria import many small nuclear encoded RNAs including the 5S ribosomal RNA24 and tRNAs.25 Moreover, there is evidence to suggest that the pathways of small RNA uptake into mitochondria are dynamically modulated, with structural and nucleic acid sequence acting as import signals,25,40 suggesting possible mechanisms for miRNA transport.41
The potential role of miRNA in mitochondria is intriguing. One possibility is that they allow additional nuclear control of mitochondrial genomic function/activity.42 If nuclear derived miRNA were controlling translation/degradation of mitochondrial derived mRNA the expectation would be that binding targets would exist in the 3′ region of these transcripts. Examination of the 3′ UTR of mitochondrial derived mRNA as predicted by miRbase version 5 (http://microrna.sanger.ac.uk/index.shtml), which uses a combination of the miRanda algorithm and homology alignment, revealed a single potential interaction between miR-130a and mitochondrial encoded cytochrome c oxidase III (COX3). This was derived from 198 predicted interactions between nuclear miRNA localized to the mitochondria and mRNA transcripts within the mitochondrial genome. A second possibility is that the miRNAs regulate nuclear derived transcripts destined to become mitochondrial proteins. Functional analyses results indicate that the miRNAs were not predicted to bind nuclear-derived mRNAs whose protein products are found in mitochondria. Other possibilities are that they may be preventing inappropriate or detrimental translation from occurring in mitochondria or modulate mitochondrial transcript stability.43
It is conceivable that miRNAs are sequestered in mitochondria to provide an alternate mechanism for control of cellular function. In fact, the predicted gene targets are consistent with those that would be involved in mitochondrial signaling at the onset of cell death. Multicellular organisms that have made the decision to carry out programmed cell death no longer require cell division and development, a tightly coupled mechanism. This is further supported by the observation that one of the two genes which showed conserved binding sites for each of the miRNA species found in mitochondria is involved in inhibition of cell division (CNOT6L/CCR4b). Expression of CCR4b was seen in a wide range of human tissues including liver.44 Depletion of mammalian CCR4b deadenylase triggers elevation of the p27Kip1 mRNA level and impairs cell growth.43 Moreover, other mitochondrial-associated miRNAs identified by the microarray analysis were discovered in screens of embryonic tissue in which both cell division and apoptosis are known to exert key roles in the developmental remodeling.29,30,32
Mitochondria represent a central checkpoint of apoptosis control and can activate apoptosis via the disruption of membrane potential resulting in increased permeability to small molecules, including miRNAs.18 This increase in permeability may represent a mechanism by which miRNAs sequestered in the mitochondria are released into the cell. In fact, when the mitochondrial megapore channel was opened in isolated mitochondria by incubating with the apoptosis inducing agent deoxycholic acid,33 significant loss of miRNAs by RT-PCR was detected. Although the loss was incomplete, in addition to the megapore opening, further alternate channel formation occurs via translocation of the pro-apoptotic protein BAX from the cytosol to mitochondria in rat liver.45 This suggests that additional release of the miRNA in mitochondria in intact cells may proceed via BAX mitochondrial channel formation. This model is further supported by the fact that mitochondrial disruption has been reported to result in translational inhibition,46 one of the known mechanisms by which miRNAs modulate gene expression posttranscriptionally.
The intracellular trafficking and localization of RNA is an important biologic process.47 Our results extend the relevance of RNA localization to miRNA species and indicate that the sequestering and release of miRNA from mitochondria may represent a fundamental mechanism for cell signaling and activation.
Materials and Methods
Isolation of mitochondria and RNA
Liver mitochondria were isolated from adult male 175–200 g Sprague-Dawley rats (Harlan Sprague-Dawley, Indianapolis, In) as previously described48 with the following modifications. Animals were killed by exsanguination under ether anesthesia, and the livers were removed and rinsed in homogenate buffer containing 70 mM sucrose, 220 mM mannitol, 1 mM EGTA, 10 mM EDTA and 10 mM HEPES, pH 7.4. Approximately 10 g of minced liver was prepared as a 10% (wt/vol) homogenate in an ice-cold solution of homogenate buffer using 3 complete strokes of a Tri-R Model K41 skill drill (Tri-R Instruments) and Teflon pestle at 800 rpm in ice-cold homogenate buffer containing EDTA-free Complete®-Mini protease inhibitor cocktail (Roche Diagnostic Corp.,). The homogenate was centrifuged at 600 ×g for 10 min at 4°C in an SS-34 rotor and RC5C centrifuge (Sorvall Instruments); and the post-nuclear supernatant was centrifuged at 1,100 ×g for 10 min at 4°C. The supernantant was centrifuged for 10 min at 7,600 ×g and the crude mitochondrial pellet further purified by sucrose:Percoll® gradient centrifugation.49 The resulting pellet was resuspended in 4 ml of homogenate buffer, and 2 ml of the resuspended pellet was layered unto 24 ml (pre-spun for 30 min) Percoll® gradient containing a mixture of 40% Percoll® (Sigma Chemical Co.,) and 60% ultra-spin buffer of 0.25 M sucrose and 1 mM EGTA (pH 7.4), and centrifuged at 43,000 ×g for 1 hr at 4°C using a Beckman 50.2Ti rotor and Beckman ultra-centrifuge model L5-65. The lower yellowish-brown mitochondrial band was carefully collected and washed once in 30 ml ice-cold wash buffer containing 0.1 M KCl, 5 mM 3-(N-morpholino)-propanesulfonic acid (MOPS) pH 7.4, 1 mM EGTA, and 10 mM EDTA by resuspension followed by centrifugation at 7,700 ×g for 10 min at 4°C. The mitochondrial pellet was washed an additional 2 times under the same conditions except that the EGTA and EDTA were removed from the wash buffer. Aliquots were taken and saved at different steps to monitor enrichment. The resulting purified and washed mitochondrial pellet was then resuspended in ice-cold homogenate buffer. One-half of the purified mitochondria was subjected to RNase A treatment (2 mg RNase A/ml) on ice 30 min while the remaining portion was left untreated.
The mitochondrial permeability transition (MPT) via the mega-pore channel opening observed in hepatic mitochondria undergoing apoptosis was assessed using a spectrophotometric assay measuring high amplitude rapid changes in mitochondrial volume as described previously.33,45 Briefly, the post Percoll® mitochondria (1 mg protein determined using the Bio-Rad protein assay kit according to the manufacturer's specifications) were incubated in 1 ml of chelex-100-treated respiration buffer (0.1 M NaCl, 10 mM MOPS, pH 7.4) for 10 min at 25°C and swelling was monitored at 540 nm in a Beckman DU64 spectrophotometer. Malate and glutamate (1 mM) were added to initiate respiration, and 3 min later 5 μM rotenone, an inhibitor of complex I of the respiratory chain, was also added to the suspension. Basal values of mitochondrial absorbance were measured for 5 min, and the optical density was monitored for an additional 5 min after addition of either 200 μM DCA (Sigma Chemical Co.,) or an equal volume of the vehicle, PBS pH 7.4. Following the MPT assays, mitochondria were centrifuged at 12,000 ×g for 3 min at 4°C. The pellets were washed ×2 by resuspension in chelex-100-treated respiration buffer prior to isolation of total RNA by trizol.
Total RNA was isolated from the various fractions and purified mitochondria using Trizol® LS reagent (Invitrogen, Corp.,) as per the manufacture's instructions and quantitated using the Christian-Warburg program on a Beckman DU64 spectrophotometer.
Western blotting
Total protein was prepared from retained portions of liver homogenates during isolation of mitochondria using buffer50 containing EDTA-free Complete®-Mini protease inhibitor cocktail. The homogenates were incubated on ice for 30 min, sonicated and centrifuged at 2,000 ×g for 10 min at 4°C. The super-natant protein concentrations were determined using the Bio-Rad protein assay kit according to the manufacturer's specifications; and the proteins aliquoted, flash-frozen in liquid nitrogen, and stored at -80°C. The relevant proteins were detected as described previously52 with the following modifications. For western blot analysis, 40 μg of mitochondrial and cytosolic proteins were separated on a 12% sodium dodecylsulfate (SDS)-polyacrylamide gel for detection of lactate dehydrogenase (LDH), COX IV or the cytosolic ribosome protein L26. For LAMP1 and calnexin, 25 μg of liver and mitochondrial extracts were separated on 10% SDS-polyacryl-amide gels. The control MC7F-7 whole cell lysate (ab387, Abcam) was used at 15 μg as per the manufacturer's recommendation. Following electrophoretic transfer onto nitrocellulose membranes, the immunoblots were incubated with 15% H2O2 for 15 min at room temperature. Blots were sequentially incubated with 5% milk blocking solution for 2 hrs at room temperature, followed by primary antibodies overnight at 4°C, and finally secondary antibody conjugated with horseradish peroxidase for 2 hrs at room temperature. The membranes were processed for detection of proteins, using the SuperSignal™ Dura ECL system from Pierce Chemical Co. The membranes were incubated with the primary antibodies for LDH (37 kDa) (ab52488, Abcam, Inc.,) at a dilution of 1:5000, cytosolic ribosomal protein L26 (ab59567, Abcam, Inc.,) at 1 μg/ml, and for mitochondrial protein Cox IV (ab14744, Abcam, Inc.,) at 1:5000. Incubations with secondary monoclonal-goat anti-mouse (Cox IV) and polyclonal-goat anti-rabbit (LDH and L26) antibodies conjugated with horseradish peroxidase were set up at a dilution of 1:5000 each. The secondary antibodies and detection system, SuperSignal™ Dura ECL were from Pierce Chemical, Co. For LAMP1 (ab24170, Abcam) and calnexin (ab22595, Abcam) the primary rabbit polyclonals were used at 1 μg/ml and the secondary HRP-conjugated anti-rabbit antibody was used at a 1:2000 dilution using the reliblot system (Bethyl Laboratories, Inc.,) and detected using SuperSignal™ Dura ECL (Pierce Chemical, Co.,). The blots were then exposed to film for 1 min to 1 hr, the film developed and scanned.
Transmission electron microscopy
Following sucrose:Percoll® gradient purification and washing, a small portion of the mitochondria before and after RNase treatment were processed and examined by transmission electron microscopy51 as described using the following modifications. Prior to RNA isolation, a portion of the mitochondrial pellet was fixed overnight at 4°C in 0.1 M cacodylate buffer (pH 7.2) containing 6% glutaraldehyde. The mitochondria were then rinsed with 0.1 M piperazine-1,4-bis(2-ethanesulfonic acid) (PIPES) buffer (pH 7.3), followed by a 20-min post-fixation incubation at room temperature in 2% cacodylate-buffered osmium tetroxide. The mitochondria were then dehydrated with increasing concentrations of ethanol, infiltrated with propylene oxide, and embedded in Epon 812:Araldite 502 resin. Sections 70–100 nm in thickness were cut using glass knives on a Reichert Ultracut S ultramicrotome and collected on 200-mesh copper grids (Electron Microscopy Sciences) and stained with 2% uranyl acetate and lead citrate. The morphology of the isolated mitochondria was studied using a JEOL-100 CX electron microscope (JEOL) at 80 kV and the images captured digitally.
Microarray analysis
An in-house miRNA microarray system was used to quantify the RNA samples isolated from the purified mitochondria from nine different animals before and after RNase treatment. The miRNA microarray system utilized the NCode Multi-Species miRNA Microarray Probe Set V2 from Invitrogen, based on the Sanger miRBase Sequence Database, Release 9.0 (October 2006). It contained 1,140 different 34–44 nt long oligonucleotides, which were complementary to C. elegans, Drosophila, zebrafish, human, mouse and rat miRNAs, and also included a 28S rRNA probe, 10 human snoRNA probes, and 76 mismatch probes. The mismatch probes aided in extracting information on background hybridization. All oligonucleotides were quadruply printed on GAPSII slides (Corning) by the Biomedical Genomics Center Microarray Facility at the University of Minnesota. Microarray analyses were performed on RNA isolated from 9 different animal mitochondrial preparations before and after RNase treatment. RNAs were labeled and hybridized to slides as described52 using ~10 μg of the purified RNAs that were ligated to a synthetic RNA linker, pCU-DY547 (Dharmacon). Following hybridization and washing, slides were scanned on a ScanArray 5000 machine (Perkin Elmer). BlueFuse (BlueGenome), GeneSpring GX 7.3.1 (Agilent), and Excel were used to quantify and process microarray data. Individual spots on microarrays were inspected to exclude abnormal signals from subsequent calculations. For every probe on a slide, mean hybridization intensity was calculated from its quadruplicate spots. Only probes found in human, mouse or rat were analyzed for the data presented in Table 1. Hybridization signals that were at least 100% above background values, which were typically between 150–300, were considered positive signals for miRNAs.
TaqMan® RT-PCR assays
The rat specific probes for the miRNAs quantified by 5′nuclease qRT-PCR were obtained from Applied Biosystems, Foster City, CA. The RT-PCR assays were carried out as per the manufacturer's recommended protocols using an Applied Biosystems 9700 HT machine. The cDNA synthesis reactions were performed using 10 ng of the isolated RNAs with the rno miRNA specific stem loop primers (4373090, 21; 4373145, 130a; 4373144, 130b; 4381110, 140*; 4373320, 290; 4373055, 320; 4381037, 494) from Applied Biosystems, and their TaqMan® MicroRNA Reverse Transcription Kit (4366596) as per the manufacture's recommended protocol. The cDNAs were used for 5′nuclease assays with their respective miRNA specific FAM labeled probes. These were run using the cycling conditions specified by Applied Biosystems for their 9700 HT real time PCR machine for fifty cycles. Negative controls using water or no reverse transcriptase reactions were run simultaneously. The average Ct cycle number from each of the mitochondrial RNA samples prepared from the groups of three animals was used to generate the mean and standard deviations shown in Table 1.
Functional analyses of miRNA observed in mitochondria
Predicted mRNA targets of miRNAs were downloaded from the internet. Predictions based purely on the rat genome sequence as predicted by the Miranda algorithm were obtained from microRNA. org31 (www.microrna.org 2008-3-28 update). Predicted human orthologs of conserved miRNA binding sequences generated by the TargetScan algorithm were obtained from TargetScan32 (www.targetscan.org Release 4.2 April 2008). These databases were parsed to obtain predicted mRNA targets of the miRNAs verified by qRT-PCR for enrichment in the purified mitochondria. For each method, individual miRNA predictions were merged to generate sets inclusive of all the mitochondrial miRNA predictions. Additionally, the intersection between the two-mitochondrial sets was also determined using the corresponding ortholog group. The predicted miRNA targets are found in supplementary data, Table 1. The resulting dataset was visualized by construction of a heat map to show the presence or absence of prediction and is shown in Figure 3.
All sets of mitochondrial miRNA derived mRNA predictions were submitted for functional enrichment analyses using Ingenuity Pathways Analyses (www.ingenuity.com). This software package leverages a biological knowledge base to identify biological functions or annotations that are overrepresented in a list of genes as compared to the functions represented in the set of all relevant genes. The resulting analyses were combined and visualized using a heat map to show the p-values for the significance of the annotation to the set. For functions that were found to be present with a p-value of less then 10E-6 in 3 or more, separate analyses were included in Figure 3. This cutoff was determined for the following reasons. First, a standard significance p-value of 0.05 was adjusted for the multiple testing problem, using a Bonferroni correction for the 50,000 individual annotations tested. This resulted in a p-value of <10E-6 required for a result to be considered significant. Second, the observation was that for 3 random 1,000 gene sets, no function scored lower then 10E-5. The entire dataset is available as Supplementary Table 2.
Supplementary Material
Acknowledgements
The mitochondria were processed for transmission electron microscopy and analyzed by Gib G. Ahlstrand from the College of Biological Sciences Imaging Center, University of Minnesota, St. Paul, MN. We thank Behnan Sahin for early discussions, and Subbaya Subramanian for critical review of the manuscript.
Abbreviations
- hrs
hours
- hsa
human
- LDH
lactate deyhdrogenase
- min
minute
- miRNA
microRNA
- mtDNA
mitochondrial DNA
- mmu
mus, mouse
- nc
non-coding
- rno
rat
- SDS
sodium dodecylsulfate
Footnotes
Supplementary materials can be found at:
www.landesbioscience.com/supplement/KrenRNA6-1-SupTable1.xls
www.landesbioscience.com/supplement/KrenRNA6-1-SupTable2.doc
References
- 1.Moulton V. Tracking down noncoding RNAs. Proc Natl Acad Sci USA. 2005;102:2269–70. doi: 10.1073/pnas.0500129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuschl T. Functional genomics: RNA sets the standard. Nature. 2003;421:220–1. doi: 10.1038/421220a. [DOI] [PubMed] [Google Scholar]
- 3.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.Meyers BC, Souret FF, Lu C, Green PJ. Sweating the small stuff: microRNA discovery in plants. Curr Opin Biotechnol. 2006;17:139–46. doi: 10.1016/j.copbio.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 6.Behm-Ansmant I, Rehwinkel J, Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb Symp Quant Biol. 2006;71:523–30. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- 7.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–46. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 8.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 9.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–81. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan SP, Slack FJ. microRNA-mediated silencing inside P-bodies. RNA Biol. 2006;3:97–100. doi: 10.4161/rna.3.3.3499. [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol. 2006;71:513–21. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- 13.Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guarnieri DJ, DiLeone RJ. MicroRNAs: a new class of gene regulators. Ann Med. 2008;40:197–208. doi: 10.1080/07853890701771823. [DOI] [PubMed] [Google Scholar]
- 15.Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Exp Physiol. 2003;88:41–56. doi: 10.1113/eph8802514. [DOI] [PubMed] [Google Scholar]
- 17.Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1999;1410:103–23. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 18.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 19.Moll UM, Wolff S, Speidel D, Deppert W. Transcription-independent pro-apoptotic functions of p53. Curr Opin Cell Biol. 2005;17:631–6. doi: 10.1016/j.ceb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Moll UM, Marchenko N, Zhang XK. p53 and Nur77/TR3—transcription factors that directly target mitochondria for cell death induction. Oncogene. 2006;25:4725–43. doi: 10.1038/sj.onc.1209601. [DOI] [PubMed] [Google Scholar]
- 21.Marchenko ND, Moll UM. The role of ubiquitination in the direct mitochondrial death program of p53. Cell Cycle. 2007;6:1718–23. doi: 10.4161/cc.6.14.4503. [DOI] [PubMed] [Google Scholar]
- 22.Lung B, Zemann A, Madej MJ, Schuelke M, Techritz S, Ruf S, Bock R, Huttenhofer A. Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res. 2006;34:3842–52. doi: 10.1093/nar/gkl448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madej M, Niemann M, Hüttenhofer A, Göringer H. Identification of novel guide RNAs from the mitochondria of Trypanosoma brucei. RNA Biol. 2008;5:84–91. doi: 10.4161/rna.5.2.6043. [DOI] [PubMed] [Google Scholar]
- 24.Magalhaes PJ, Andreu AL, Schon EA. Evidence for the presence of 5S rRNA in mammalian mitochondria. Mol Biol Cell. 1998;9:2375–82. doi: 10.1091/mbc.9.9.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharyya SN, Adhya S. The complexity of mitochondrial tRNA import. RNA Biol. 2004;1:84–8. doi: 10.4161/rna.1.2.1180. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA. 2004;101:360–5. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berezikov E, van Tetering G, Verheul M, van de Belt J, van Laake L, Vos J, Verloop R, van de Wetering M, Guryev V, Takada S, van Zonneveld AJ, Mano H, Plasterk R, Cuppen E. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006;16:1289–98. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mineno J, Okamoto S, Ando T, Sato M, Chono H, Izu H, Takayama M, Asada K, Mirochnitchenko O, Inouye M, Kato I. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–71. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wheeler G, Ntounia-Fousara S, Granda B, Rathjen T, Dalmay T. Identification of new central nervous system specific mouse microRNAs. FEBS Lett. 2006;580:2195–200. doi: 10.1016/j.febslet.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 32.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 33.Rodrigues CM, Fan G, Wong PY, Kren BT, Steer CJ. Ursodeoxycholic acid may inhibit deoxycholic acid-induced apoptosis by modulating mitochondrial transmembrane potential and reactive oxygen species production. Mol Med. 1998;4:165–78. [PMC free article] [PubMed] [Google Scholar]
- 34.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:149–53. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 36.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38:S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 37.Sylvestre J, Margeot A, Jacq C, Dujardin G, Corral-Debrinski M. The role of the 3′ untranslated region in mRNA sorting to the vicinity of mitochondria is conserved from yeast to human cells. Mol Biol Cell. 2003;14:3848–56. doi: 10.1091/mbc.E03-02-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaltimbacher V, Bonnet C, Lecoeuvre G, Forster V, Sahel JA, Corral-Debrinski M. mRNA localization to the mitochondrial surface allows the efficient translocation inside the organ-elle of a nuclear recoded ATP6 protein. RNA. 2006;12:1408–17. doi: 10.1261/rna.18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacKenzie JA, Payne RM. Ribosomes specifically bind to mammalian mitochondria via protease-sensitive proteins on the outer membrane. J Biol Chem. 2004;279:9803–10. doi: 10.1074/jbc.M307167200. [DOI] [PubMed] [Google Scholar]
- 40.Smirnov A, Tarassov I, Mager-Heckel AM, Letzelter M, Martin RP, Krasheninnikov IA, Entelis N. Two distinct structural elements of 5S rRNA are needed for its import into human mitochondria. RNA. 2008;14:749–59. doi: 10.1261/rna.952208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 42.Scarpulla RC. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene. 2002;286:81–9. doi: 10.1016/s0378-1119(01)00809-5. [DOI] [PubMed] [Google Scholar]
- 43.Mukherjee S, Mahata B, Mahato B, Adhya S. Targeted mRNA degradation by complex-mediated delivery of antisense RNAs to intracellular human mitochondria. Hum Mol Genet. 2008;17:1292–8. doi: 10.1093/hmg/ddn017. [DOI] [PubMed] [Google Scholar]
- 44.Morita M, Suzuki T, Nakamura T, Yokoyama K, Miyasaka T, Yamamoto T. Depletion of mammalian CCR4b deadenylase triggers elevation of the p27Kip1 mRNA level and impairs cell growth. Mol Cell Biol. 2007;27:4980–90. doi: 10.1128/MCB.02304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodrigues CM, Ma X, Linehan-Stieers C, Fan G, Kren BT, Steer CJ. Ursodeoxycholic acid prevents cytochrome c release in apoptosis by inhibiting mitochondrial membrane depolarization and channel formation. Cell Death Differ. 1999;6:842–54. doi: 10.1038/sj.cdd.4400560. [DOI] [PubMed] [Google Scholar]
- 46.Gagliardi D, Stepien PP, Temperley RJ, Lightowlers RN, Chrzanowska-Lightowlers ZM. Messenger RNA stability in mitochondria: different means to an end. Trends Genet. 2004;20:260–7. doi: 10.1016/j.tig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Corral-Debrinski M. mRNA specific subcellular localization represents a crucial step for fine-tuning of gene expression in mammalian cells. Biochim Biophys Acta. 2007;1773:473–5. doi: 10.1016/j.bbamcr.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Botla R, Spivey JR, Aguilar H, Bronk SF, Gores GJ. Ursodeoxycholate (UDCA) inhibits the mitochondrial membrane permeability transition induced by glycochenodeoxycholate: a mechanism of UDCA cytoprotection. J Pharmacol Exp Ther. 1995;272:930–8. [PubMed] [Google Scholar]
- 49.Sokol RJ, Devereaux M, Mierau GW, Hambidge KM, Shikes RH. Oxidant injury to hepatic mitochondrial lipids in rats with dietary copper overload. Modification by vitamin E deficiency. Gastroenterology. 1990;99:1061–71. doi: 10.1016/0016-5085(90)90627-d. [DOI] [PubMed] [Google Scholar]
- 50.Trembley JH, Kren BT, Steer CJ. Posttranscriptional regulation of cyclin B messenger RNA expression in the regenerating rat liver. Cell Growth Differ. 1994;5:99–108. [PubMed] [Google Scholar]
- 51.Rodrigues CM, Stieers CL, Keene CD, Ma X, Kren BT, Low WC, Steer CJ. Tauroursodeoxycholic acid partially prevents apoptosis induced by 3-nitropropionic acid: evidence for a mitochondrial pathway independent of the permeability transition. J Neurochem. 2000;75:2368–79. doi: 10.1046/j.1471-4159.2000.0752368.x. [DOI] [PubMed] [Google Scholar]
- 52.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.