Abstract
Fuelled by ATP hydrolysis, dyneins generate force and movement on microtubules in a wealth of biological processes, including ciliary beating, cell division and intracellular transport. The large mass and complexity of dynein motors have made elucidating their mechanisms a sizable task. Yet, through a combination of approaches, including X-ray crystallography, cryo-electron microscopy, single-molecule assays and biochemical experiments, important progress has been made towards understanding how these giant motor proteins work. From these studies, a model for the mechanochemical cycle of dynein is emerging, in which nucleotide-driven flexing motions within the AAA+ ring of dynein alter the affinity of its microtubule-binding stalk and reshape its mechanical element to generate movement.
Introduction
To move, divide and spatially organize their teeming interiors, eukaryotic cells use ATP-fuelled motor proteins to generate forces and transport cargoes along cytoskeletal tracks. The numerous proteins, organelles and mRNAs that undergo directed transport by motor proteins touch on a wide range of cellular and developmental processes. Underscoring the importance of cytoskeletal motors in biology, it is now clear that serious human and animal diseases arise from motor protein dysfunction1, 2.
Dynein is one of the three families of cytoskeletal motor protein. Originally identified 50 years ago as an ATPase in Tetrahymena pyriformis cilia3, dynein was named by Gibbons and Rowe after the unit of force, the dyne4. A cytoplasmic form of dynein was subsequently isolated from brain tissue5 and shown to drive intracellular transport towards the minus ends of microtubules6, 7, which typically lie in the microtubule-organizing centre near the nucleus in non-dividing cells (Fig. 1). The discovery of dynein thus complemented the finding of kinesins8; microtubule-based motors that typically move towards the plus ends of microtubules and hence the cell periphery.
Figure 1. Sites of dynein action in the cell.
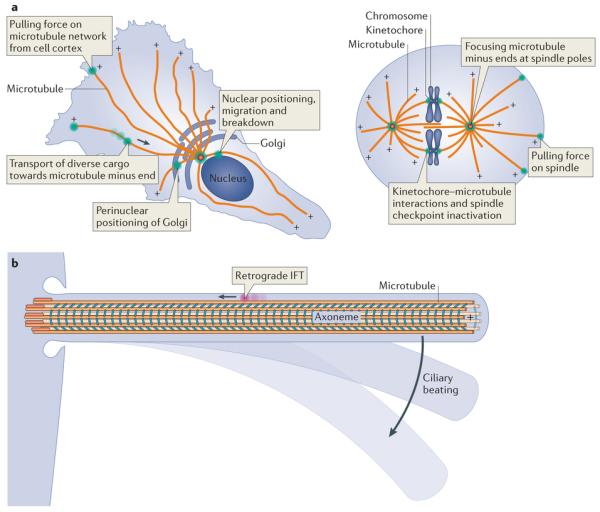
a | Example functions of cytoplasmic dynein (green) are shown in an interphase cell (left) and a dividing cell (right). The polarity of microtubules is indicated by plus signs. The arrow depicts the direction of dynein movement towards the microtubule minus end. Note that in some cell types and regions, such as the dendritic arbors of neurons, the microtubule network can have mixed polarity. b | Dynein functions in cilia. Intraflagellar transport (IFT) dynein (pink) performs retrograde IFT, whereas axonemal dyneins (cyan) power the beating of motile cilia.
Given that dyneins and kinesins both move along microtubules, one might have expected their mechanisms of motility to have more in common with one another than with the actin-based motor, myosin. It was therefore a surprise when crystal structures of myosin and kinesin revealed that — despite moving on different cytoskeletal polymers — these motors share a G protein-related fold and similarities in their core mechanisms9.
Dynein is currently at the frontier of cell motility research at the molecular level, as its mechanism of movement is much less well understood than that of kinesin and myosin. Dubbed the ‘big wheel’ of motor proteins10, dynein belongs to the AAA+ superfamily (ATPases associated with diverse activities)11. Conventional AAA+ ATPases function as hexameric rings that unfold proteins, dismantle DNA and RNA duplexes and pry apart macromolecular complexes and aggregates11. Like conventional AAA+ ATPases, dynein has a ring of six AAA+ modules at its core but, unusually, these are linked together into one large polypeptide, along with several unique appendages that enable motor function. The large size and complexity of dynein have made elucidating its mechanism a formidable task. However, recent studies have risen to this challenge using various approaches. For example, in the past 2 years, long sought-after crystal structures of the motor domain of dynein have been solved, and single-molecule studies, live-cell imaging and electron microscopy have provided key insights into the dynamics of dynein.
In this Review, we focus on advances in two main areas: first, the cellular functions of dyneins, and second, the molecular mechanism of the dynein motor domain. Exciting progress has also been made in understanding how dyneins are regulated and recruited to specific cargoes in the cell, and the reader is referred to recent reviews on these topics12, 13, 14, 15, 16, 17, 18, 19.
Overview of the dynein family
Dyneins operate as protein complexes built around force-generating subunits called heavy chains, so termed because of their large molecular mass (typically ~500 kDa) (Fig. 2). Each heavy chain contains a motor domain that belongs to the AAA+ superfamily11 attached to a divergent amino-terminal tail domain (Fig. 2a). The tail specifies distinct oligomerization properties and serves as a platform for the binding of several types of associated subunit (Fig. 2b), which in turn mediate interactions with cargo either via direct binding or through the recruitment of adaptor proteins.
Figure 2. Overview of dynein composition.
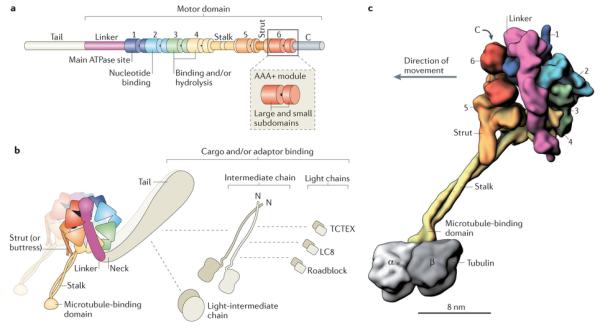
a | Linear representation of domains within the dynein heavy chain. The amino-terminal tail domain is involved in dynein oligomerization, cargo-binding and regulation, but is not part of the minimal motor domain capable of producing movement in vitro. The motor domain comprises the linker domain, six AAA+ modules (1–6), the coiled-coil stalk and strut, and the carboxy-terminal region. The motor domain of Dictyostelium discoideum dynein has a molecular mass of ~380 kDa. In many fungal dyneins, the C-terminal region is shorter and the motor domain is ~330 kDa. Each of the AAA+ modules is composed of a large N-terminal subdomain and a smaller C-terminal subdomain (see inset and Box 2). b | The cytoplasmic dynein complex contains a pair of identical heavy chains. Within each heavy chain, the six AAA+ modules fold into a ring. The stalk protrudes as an extension from the small subdomain of AAA4. The tail is connected to AAA1 by the linker domain, which arches over the AAA+ ring. In Chlamydomonas reinhardtii inner arm dynein-c, a sharp ~90° kink exists between the linker and the tail90, 92, 115. Although not yet visualized in cytoplasmic dynein, a similar kink might exist, as it would prevent a steric clash between the tail and the microtubule. In dynein-c, this neck region of the tail is a natural site of flexibility in the molecule, allowing the angle of the tail to vary with respect to the motor domain90, 92, 115. The cytoplasmic dynein heavy chains assemble with up to five types of associated subunit, which are also dimers148. The associated subunits comprise the intermediate chain, the light-intermediate chain and three classes of light chain: TCTEX, LC8 and Roadblock. Dashed lines indicate reported interactions of the associated subunits with each other and with the tail26. The three-dimensional (3D) arrangement of the associated subunits with respect to the tail is unknown. c | A 3D model of the cytoplasmic dynein motor domain bound to the microtubule (the associated subunits are not shown). As no high-resolution structure currently exists for the entire motor domain bound to a tubulin dimer, this model is based on a 2.8 Å crystal structure of the D. discoideum dynein motor domain lacking the microtubule-binding domain93 (Protein Data Bank ID: 3VKG), joined to a cryo-electron microscopy-derived model of the mouse microtubule-binding domain bound to an α-tubulin–β-tubulin dimer112 (Protein Data Bank ID: 3J1T). Subdomains are shown in surface representation, with the two long α-helices in the stalk rendered separately to emphasize their coiled-coil arrangement. The six AAA+ modules are numerically labelled.
Phylogenetically, there are nine major classes of dynein heavy chain20. The cytoplasmic dynein 1 heavy chain (encoded by DYNC1H1 in humans) is used for nearly all of the minus end-directed transport in the cytoplasm of most eukaryotic cells (Fig. 1a). However, archaeplastidans, which lack dyneins and possess an expanded repertoire of minus end-directed kinesins21, are an exception. Cytoplasmic dynein 2 (encoded by DYNC2H1 in humans) has a specialized role in transporting material along motile and sensory cilia and flagella (Fig. 1b). In this Review, we refer to cytoplasmic dynein 1 as ‘cytoplasmic dynein’ and cytoplasmic dynein 2 as ‘intraflagellar transport (IFT) dynein’ for distinction. The remaining seven dynein classes are built into the axoneme, where they power ciliary beating (Box 1). Some axonemal dynein classes have multiple representatives per genome, so the total number of distinct heavy chain genes in organisms that build a motile axoneme typically exceeds nine. For example, there are 16 dynein heavy chain genes in the human genome22.
Box 1. Functions of dynein in the axoneme.
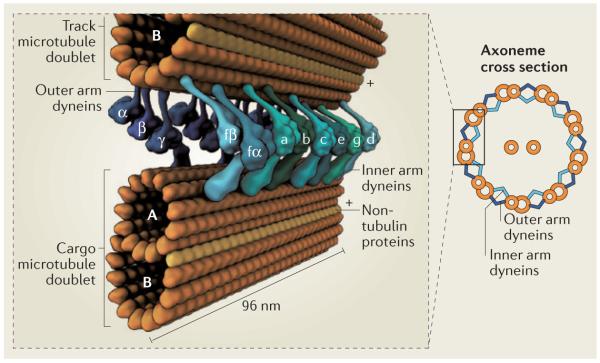
Observed in cross-section, the axoneme has a characteristic ‘9 + 2’ appearance in many eukaryotes, corresponding to nine peripheral doublet microtubules surrounding a central pair of singlet microtubules (see the figure). Each doublet consists of a complete microtubule with 13 protofilaments (termed the A-tubule) and an incomplete microtubule of 10 protofilaments (termed the B-tubule), with non-tubulin proteins at the junction138. Microtubule polarity is indicated by plus signs. Numerous additional proteins, such as radial spokes, nexin–dynein regulatory complexes and microtubule inner proteins (not shown), have crucial roles in axoneme motility, structure and regulation.
Within motile axonemes, dyneins drive the sliding of adjacent microtubule doublets relative to each other139. These sliding motions are resisted by other axonemal components, converting them into bending deformations that propagate along the axoneme. The resulting oscillations can either propel the cell through its fluid environment or create flow over the cell surface. To produce sliding, axonemal dyneins form a stable attachment to a cargo microtubule via their tails and use their motor domains to translocate along an adjacent track microtubule.
Axonemal dyneins subdivide into inner and outer arms, depending on their position3. Their arrangement is currently best characterized in the green algae Chlamydomonas reinhardtii, in which detailed insights have come from molecular genetic and cryo-electron tomography studies140. Each outer arm consists of three different dynein heavy chains (α-, β- and γ-chain; see the figure), which adopt a stacked arrangement and co-purify as a complex with ~15 smaller subunits141. The outer arms repeat at 24 nm intervals along the axoneme and are crucial for generating the proper beat frequency. The inner arms comprise eight different dynein heavy chains (one heterodimer (fα and fβ) and six monomers (a–e and g), each with its own complement of associated subunits), which repeat with a 96-nm periodicity142. Each inner arm dynein has distinct motile properties and roles in shaping the ciliary waveform143.
From this general layout, dynein composition can vary both around and along the axoneme. For example, in C. reinhardtii, three specialized heavy chains replace canonical inner arm dyneins near the base of the axoneme144, and one of the doublets lacks outer arms along its length142. In most metazoans, each outer arm contains two heavy chains, rather than three. The 9 + 2 arrangement also varies in some organisms. For instance, the highly motile sperm axoneme of the eel Anguilla anguilla is a minimal 9 + 0 structure that lacks outer arms, radial spokes and the central pair of microtubules145. These structural variations probably contribute to the distinct waveforms and motile properties displayed by cilia. In all systems, for wave-like bending motions to occur, zones of dynein activity are proposed to switch from one side of the axoneme to the other, and to propagate along its length. Models for the coordination and regulation of dynein activity are discussed in Refs 16,17,18,19. How axonemal dyneins assemble into this configuration and work as an integrated system, with the many regulators and structural proteins in the axoneme, is only beginning to emerge.
Functions of cytoplasmic dynein
Cytoplasmic dynein performs a great variety of cellular functions. The breadth of these activities seems to be greatest in metazoan cells (described below), but cytoplasmic dynein is also used to varying extents in fungi, alveolata, stramenopila and amoebozoa20. For example, in the yeast Saccharomyces cerevisiae, the sole known role of cytoplasmic dynein is positioning the nucleus during cell division23, whereas in filamentous fungi24 and the slime mould Dictyostelium discoideum25 it is also used for vesicle transport.
Cytoplasmic dynein assembles around two identical heavy chains and is thus known as a two-headed motor. A pressing question is how this assembly can be directed to its many sites of action in the cell (Fig. 1a). At least part of the answer lies in the large complement of additional subunits in the complex (Fig. 2b). Metazoan genomes encode five classes of associated subunit, each of which functions as a dimer; these are the intermediate chain, light-intermediate chain and three types of light chain (TCTEX, LC8 and Roadblock). Moreover, in mammals, there are two genes for each class of subunit, some of which have restricted expression patterns in the body26. Diversity in the associated subunits is compounded by differentially spliced and phosphorylated isoforms26. Recent reconstitution of the human cytoplasmic dynein complex has revealed that the associated subunits can serve critical structural roles, as the heavy chain forms aggregates instead of dimers in the absence of the light-intermediate chain27.
In addition to these core components of the complex, cytoplasmic dynein interacts with three ubiquitous regulators: lissencephaly 1 (LIS1; also known as NUDF), nuclear distribution E (NUDE) and the dynactin complex. These important regulators alter the intrinsic mechanical behaviour of cytoplasmic dynein and are involved in most, if not all, of its cellular functions. Therefore, a complete understanding of cytoplasmic dynein motility requires a detailed dissection of how dynactin, NUDE and LIS1 act on dynein, and progress towards this goal has been reviewed recently12, 13, 14, 28.
Cargo transport along cytoplasmic microtubules
Cytoplasmic dynein powers the transport of membrane-bound vesicles and tubules, together with their resident molecules, towards microtubule minus ends (Fig. 1a). Examples of organelles trafficked by cytoplasmic dynein include endosomes29, lysosomes30, phagosomes31, melanosomes32, peroxisomes33, lipid droplets34, mitochondria35 and vesicles from the endoplasmic reticulum (ER) destined for the Golgi36. Such transport can transmit signals between different parts of the cell. For example, at nerve terminals, stimulated tropomyosin-receptor kinase (TRK) receptors are internalized into endosomes and transported by dynein towards the cell body, where they initiate signalling cascades essential for neuron survival37. Additional types of cytoplasmic dynein cargo include components of the centrosome38, transcription factors39, cytoskeletal filaments40 and mRNA-containing ribonucleoprotein complexes41. Cytoplasmic dynein is also involved in clearing material from the periphery of the cell for degradation and recycling42, 43: at the distal tip of neurons, organelles and proteins are engulfed into autophagosomes and transported in a dyneindependent manner towards the cell body for breakdown43. Furthermore, viruses such as HIV, herpesvirus and adenovirus have evolved mechanisms to hijack cytoplasmic dynein to reach the nucleus44. Finally, although cytoplasmic dynein typically moves its cargo through the intracellular space, recent studies raise the idea that it can also translocate membrane-spanning proteins and signalling complexes in the plane of the lipid bilayer, for instance within the nuclear envelope45, 46 and at the immunological synapse47, 48.
Exerting tension on cellular structures
Cytoplasmic dynein executes additional functions by associating with cellular structures and exerting tension on microtubules. For example, dyneins tethered at the cell cortex can apply a pulling force on the microtubule network (Fig. 1a) by either walking towards the minus end of a microtubule or coupling to a disassembling plus end49, 50. In doing so, cytoplasmic dynein is thought to pull the microtubule cytoskeleton towards the leading edge of migrating neurons51 and fibroblasts52 or towards the immunological synapse in T cells53.
The pulling force of dynein is also used during cell division54. By hauling on astral microtubules that emanate from the spindle, cytoplasmic dynein can cause the spindle to oscillate or to localize towards one end of the cell. Remarkably, during spindle oscillation in mammalian cells, cortical dynein dynamically redistributes from one side of the cell to the other, dissociating from its cortical receptors when the spindle pole is within ~2 μm and accumulating with them when the spindle pole moves further away55, 56.
In large cells, such as amphibian embryos, dynein seems to exert pulling forces on microtubule asters even when the microtubules are not long enough to reach the cell cortex57. Recent studies indicate that this can occur as the dynein-driven movement of organelles along microtubules generates sufficient viscous drag to slowly pull the microtubule network in the opposite direction57, 58, 59. An appealing feature of this model is that the pulling force would vary according to microtubule length (as net organelle transport will be greater on longer microtubules). In embryos, length-dependent pulling is proposed to be a key ingredient for aster centring and spacing57, 58.
Further cellular structures that are connected to the microtubule network by cytoplasmic dynein are the Golgi and the nuclear envelope. Indeed, the perinuclear positioning of the Golgi during interphase depends on cytoplasmic dynein60 (Fig. 1a). At the outer nuclear envelope, dynein has been reported to contribute to nuclear rotation61 and positioning51, centrosome separation62 and the breakdown of the nuclear envelope for open mitosis63.
Functions at the spindle and kinetochore
At cell division, cytoplasmic dynein assists in assembling microtubules into the chromosome-segregating device known as the spindle (Fig. 1a). Specifically, studies using frog egg extracts reveal that dynein is required to focus the minus ends of microtubules at the spindle poles64, 65. The mechanism remains to be elucidated, but a leading model is that dynein slides antiparallel microtubules apart so that their plus ends project outward and their minus ends are gathered. The dynein-powered transportation of other spindle components, such as nuclear mitotic apparatus (NUMA), also plays a part in the construction of this large microtubule-based structure65.
Finally, cytoplasmic dynein localizes to the kinetochore — the protein assembly that links centromeric DNA to spindle microtubules. Kinetochore dynein has an important role in the molecular surveillance mechanism that aids faithful chromosome segregation by delaying anaphase until all chromosomes are properly attached to the spindle66. When the plus ends of spindle microtubules have stably engaged with the kinetochores, cytoplasmic dynein is thought to remove spindle-assembly checkpoint proteins by transporting them towards the spindle poles67, 68. In mammalian cells, cytoplasmic dynein is also reported to help maintain initial spindle–kinetochore attachments and drive rapid poleward chromosome movements during their alignment at the metaphase plate69, 70.
IFT dynein
IFT dynein, which is closely related to cytoplasmic dynein, also functions as a cargo transporter but operates within the confines of the cilium (Fig. 1b). Although motile and sensory cilia form stable steady-state structures, their building blocks are continually turning over71. Through the process of IFT, new components are trafficked towards the tip of the cilium, material is moved back to the base, and signalling receptors are distributed in the plane of the membrane. IFT dynein transports cargo towards the base of the cilium72, 73, where the microtubule minus ends reside. Members of the kinesin 2 family drive cargo transport to the tip. IFT dynein functions as a homodimer of heavy chains and associated subunits, although relatively little is known about its motile properties74, 75, 76. Observations in Chlamydomonas reinhardtii indicate that minus end-directed IFT involves the movement of oligomeric particles (also termed ‘trains’) of material ~250 nm long, which form multiple contacts with the outer surface of the axoneme77. Moreover, the forces generated by retrograde IFT trains exceed those of a single motor78, 79, indicating that IFT dynein works in multi-motor collectives in situ. This property is akin to the mode of action of cytoplasmic dynein, which also carries out many of its tasks in teams of motors (reviewed in Ref. 80).
Dynein motor structure and mechanism
At the core of all dynein functions is the action of the motor domain. This carboxy-terminal region of the heavy chain (Fig. 2a) can be recombinantly expressed as a stable monomeric entity81 and contains all of the elements that are needed to convert the energy from ATP hydrolysis into movement27, 82, 83, 84. Evidence suggests that this process occurs through a mechanochemical cycle, in which chemical transitions (ATP binding, ATP hydrolysis and the release of inorganic phosphate (Pi) and ADP) are coupled to structural changes in the motor and, conversely, mechanical events (such as microtubule binding) can influence the rate of chemical transitions. Although certain parameters such as the overall cycle rate vary among dynein classes, probably reflecting their specialization to particular cellular functions, current data suggest that cytoplasmic and axonemal dynein use a conserved basic mechanism.
Mechanochemical cycle
In an overview of the dynein operating cycle, ATP induces dissociation of the motor–microtubule complex85. After detaching from the microtubule, the motor rearranges, becoming primed for a subsequent structural change (termed the powerstroke) that is thought to generate force. Next, following a diffusive search, rebinding of the motor to a new site on the microtubule stimulates the release of ATP hydrolysis products, thus triggering the powerstroke86 (Fig. 3).
Figure 3. Model of the mechanochemical cycle of a cytoplasmic dynein motor domain.
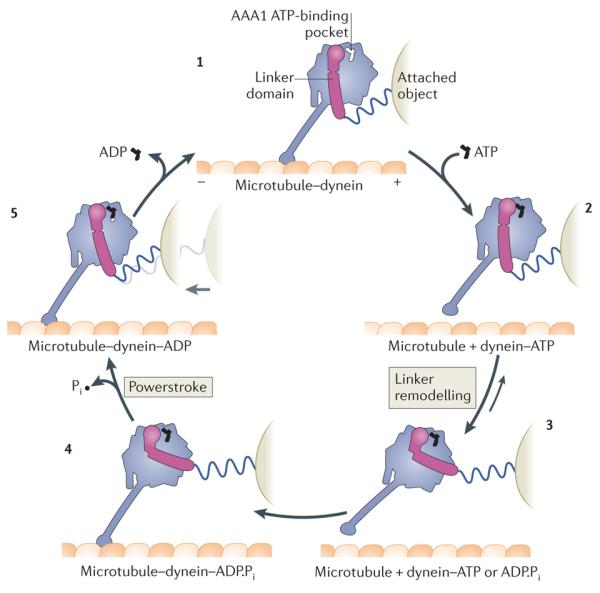
Model for the Dictyostelium discoideum cytoplasmic dynein motor domain85, 86, 91, 93, 111, 118. Plus and minus signs indicate microtubule polarity. The dynein motor domain moves towards the minus end of the microtubule. Force exerted on an attached object is shown schematically by the stretching of a spring, which does not represent a part of the dynein structure. Conceptually, the attached object could represent a partnering motor domain in a cytoplasmic dynein dimer or the cargo microtubule of an axonemal dynein. With no nucleotide bound at the AAA1 ATPase site, the dynein motor domain is tightly bound to the microtubule (1). ATP binding induces rapid dissociation from the microtubule (2) (Fig. 4). The dissociation rate is ~460 s–1 for the D . discoideum dynein motor domain85. A slower ATP-driven change (~200 s–1) is the remodelling of the linker domain85, which is displaced across the AAA+ ring (3) (Fig. 5). Remodelling of the linker extends the search range of the microtubule-binding domain along the microtubule. After hydrolysis of ATP to ADP and inorganic phosphate (Pi), the motor domain is thought to engage a new binding site on the microtubule, initially via a weak interaction (4). Strong binding to the microtubule accelerates the release of Pi from AAA1, inducing the linker to revert to its straight form. This transition is speculated to represent the powerstroke: the main step in which force (indicated by the light grey arrow) is transmitted to the attached object (5). Finally, ADP is released from AAA1 and the cycle restarts. The occupancy of other nucleotide-binding AAA+ modules of dynein, AAA2, AAA3 and AAA4, during the cycle, is unknown. Although the cycle is shown starting with dynein having no nucleotide bound at AAA1, it is not meant to imply that this is the longest-lived state. Indeed, in vivo, where the ATP concentration is in the millimolar range, ATP would rapidly bind at AAA1 once ADP is released, and thus this state would be populated only transiently.
At first glance, the chemical and mechanical events in the dynein cycle bear a strong resemblance to those of the actin-based motor myosin 2. This striking piece of convergent evolution became clear from early studies of outer arm axonemal dyneins87. However, a look ‘under the hood’ of the motor domain of dynein reveals that its ancestry in the AAA+ superfamily makes its mechanism of action quite distinct from the other cytoskeletal motor classes.
Motor domain architecture
Figure 2c depicts the architecture of the dynein motor domain, which has been uncovered through sequence analysis, two-dimensional (2D) electron microscopy and, most recently, X-ray crystallography and three-dimensional (3D) cryo-electron microscopy (cryo-EM)11, 88, 89, 90, 91, 92, 93, 94, 95, 96. The N-terminal part of the motor forms an elongated linker domain that arches over the catalytic region and is thought to deliver the powerstroke. The catalytic core of dynein contains six AAA+ modules, which fold into a ring akin to that of other AAA+ machines. However, unlike canonical AAA+ hexamers, which are oligomeric, the six AAA+ modules of dynein are joined together in the same polypeptide by short connecting sequences and have different properties (Fig. 2a). For example, only the first four modules (AAA1–AAA4) contain functional nucleotide-binding motifs. On the face opposite the linker, the ring is embellished with a C-terminal domain that interacts mainly with AAA5 and AAA6. The microtubule-binding domain lies at the tip of a ~15 nm antiparallel coiled-coil stalk that protrudes as a pair of α-helices from AAA4. Near its base, the stalk interacts with a second coiled-coil strut (also known as the buttress) that emerges from AAA5. This expansive architecture, which places the sites of ATP breakdown and microtubule binding ~25 nm apart, makes long-range structural changes a central part of the dynein mechanism.
The AAA+ ring
The AAA+ ring can be thought of as the engine of dynein as it converts the chemical energy from ATP hydrolysis into motion. Each AAA+ module consists of a large and a small subdomain, which are connected by a flexible joint93, 94, 95, 96 (Box 2). To form a ring, the small subdomain of one AAA+ module packs against the large subdomain of the neighbouring module. This layout produces six tight interfaces (between the AAA+ modules) and six potential hinges (one within each AAA+ module), as seen in prior structures of AAA+ hexamers97, 98. Based on these earlier studies, nucleotide transactions in the AAA+ modules are expected to alter the angles of the hinges and cause flexing of the ring.
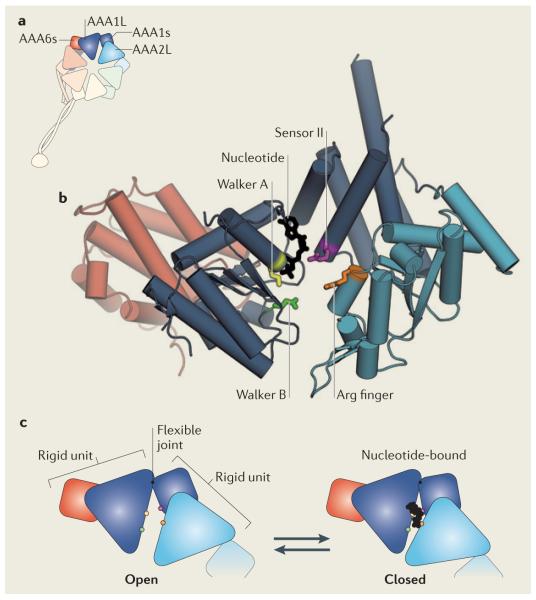
Box 2. The AAA+ engine.
Each AAA+ module in the dynein ring contains a large subdomain (a five-stranded parallel β-sheet flanked by α-helices), which is flexibly joined to a smaller, predominantly α-helical, subdomain93, 95. The small subdomain of AAA1 (AAA1s) packs against the large subdomain of AAA2 (AAA2L), and AAA2s packs against AAA3L, and so on, until the ring is closed by AAA6s packing against AAA1L (see the figure, part a). The highlighted subdomains are shown in detail for Dictyostelium discoideum dynein93 (Protein Data Bank code: 3VKG) (see the figure, part b). AAA1 is the principal ATPase site and contains enzymatic motifs characteristic of the AAA+ superfamily. AAA1L bears Walker A and Walker B motifs, which are crucial for nucleotide binding and hydrolysis, respectively. Within the Walker A consensus sequence (Gly-Lys-Thr), the Lys residue (yellow) interacts with the phosphate groups of the nucleotide. Mutating this residue severely impairs nucleotide binding, thereby eliminating the motility of dynein while rendering it in a state that binds tightly to microtubules85, 100. Within the Walker B sequence (Asp-Glu; at the end of the third β-strand), the Glu residue (green) is thought to be the catalytic base that polarizes H2O for an inline attack on the γ-phosphate of ATP146; mutating this residue to Gln severely impairs ATP hydrolysis and eliminates dynein motility114. However, ATP binding still occurs in the Walker B mutant, trapping dynein in an ATP-bound state with weak microtubule affinity and a primed linker domain85, 118. AAA1L also contains a residue termed sensor I (typically Asp or Thr; not shown) that is implicated in hydrolysis147. AAA1s bears a residue termed sensor II (purple), an Arg residue that is thought to detect the status of the nucleotide pocket and help mediate closure of the AAA+ module in response to nucleotide binding (see the figure, part c). In addition, an Arg finger (orange) from AAA2L contributes to the ATPase site of AAA1. By analogy to other AAA+ proteins, this Arg finger is expected to be crucial for hydrolysis. In contrast to AAA1, the other AAA+ modules in the dynein ring show varying degrees of conservation with the enzymatic motifs of AAA+ superfamily members (Supplementary information S1 (table)). For example, across the dynein classes, AAA2 lacks the catalytic Walker B motif but contains a Walker A sequence, suggesting it can bind but not hydrolyse nucleotide. Consistent with this notion, when the Saccharomyces cerevisiae dynein motor is purified and crystallized in the absence of nucleotide, an ATP molecule seems to remain tightly bound in AAA2, further indicating that dissociation from this site is extremely slow95.
Opening and closing of AAA1
Mutagenesis and biochemical experiments suggest that AAA1 is the main site of ATP hydrolysis in the dynein ring99, 100. Accordingly, it displays the hallmarks of an AAA+ nucleotide-binding pocket, as seen in a 2.8 Å structure of D. discoideum dynein in the presence of ADP93 (Box 2). The nucleotide binds between the large and small subdomains of AAA1, interacting with several signature motifs (detailed in Box 2). Moreover, in an intimate arrangement, the large subdomain of the adjacent module (AAA2) also contributes to the nucleotide-binding pocket of AAA1, providing an Arg finger that is crucial for ATP hydrolysis (Box 2).
Comparison of D. discoideum and S. cerevisiae dynein motor structures in different nucleotide states supports the idea that large and small subdomains of AAA1 move relative to each other during the ATPase cycle. In the D. discoideum structure crystalized in the presence of ADP, AAA1 is partially closed around ADP, placing the backbone atoms of its Walker A and sensor II motifs ~13 Å apart93 (Box 2). In the S. cerevisiae structure crystalized in the absence of nucleotide, AAA1 is more open and the Walker A and sensor II motifs are ~17 Å apart. This open conformation of AAA1 cannot bind nucleotides following crystal soaking95. Thus, the open AAA1 conformer seen in the S. cerevisiae dynein structure seems to represent a state with very low affinity for nucleotide that is fixed by crystallization95. Free from the constraints of a crystal lattice, AAA1 is expected to bind and fully close around ATP, bringing the AAA2 Arg finger close enough to participate in hydrolysis and eliciting changes in the microtubule-binding domain and linker domain of dynein. In D. discoideum dynein, the products of ATP hydrolysis (Pi and ADP) seem to be kinetically trapped in AAA1, based on pre-steady state studies93. The release of Pi from AAA1 is stimulated by microtubules93, suggesting that track binding might allosterically drive AAA1 towards the open conformation. In summary, current studies point to a model in which the opening and closing motions of AAA1 are the principal drivers of the mechanochemical cycle of dynein, coordinating its ATPase activity with microtubule binding and linker movement.
Nucleotide transactions at AAA2, AAA3 and AAA4
In some AAA+ machines, such as the E1 helicase, ATP is thought to bind and hydrolyse in an ordered sequence around the ring101. In dynein, three AAA+ modules adjacent to AAA1 (AAA2, AAA3 and AAA4) can bind nucleotides, and the microtubule-binding stalk protrudes as an outgrowth of two α-helices in the small subdomain of AAA4. Thus, it could be envisioned that AAA1 and the stalk communicate via sequential nucleotide hydrolysis within AAA2–AAA4. However, structural and mutagenesis studies have ruled out this model. First, inspection of AAA2 indicates that it serves as a stable nucleotide-binding pocket rather than as an active site of hydrolysis (detailed in Box 2). Second, eliminating nucleotide binding at AAA2 by mutagenesis slows down, rather than abrogates, dynein motility100. In dynein, long-lived nucleotide binding at AAA2 might keep this module in a comparatively rigid, closed conformation that is optimal for propagating structural changes to and from AAA1.
In most cytoplasmic dynein isoforms, AAA3 and AAA4 contain motifs indicative of active ATPase sites, but this is not the case in all dyneins (Supplementary information S1 (table)). For example, in IFT dynein, the catalytic Walker B motif in AAA3 is not well conserved95. Nevertheless, disabling nucleotide binding or hydrolysis at AAA3 in D. discoideum and S. cerevisiae cytoplasmic dynein severely slows motility; equivalent mutations in AAA4 have a milder effect100, 102. In D. discoideum dynein, pre-steady state kinetic studies hint that AAA3 and AAA4 may spend most of their time in an ADP-bound state. When all other hydrolysis sites are disabled, both AAA3 and AAA4 release Pi rapidly (~60 s−1) on mixing with ATP, then subsequently cycle at a lower rate (<1 s–1), probably owing to the slow release of ADP93. Unlike AAA1, these modules do not autonomously respond to microtubules93. However, it is an open question whether nucleotide turnover in AAA3 and AAA4 can be stimulated by the ATPase cycle of AAA1 (Ref. 86). Alternatively, AAA3 and AAA4 might use hydrolysis principally to enter an ADP-bound state, perhaps giving these modules good geometry or appropriate flexibility to accommodate and propagate the motions of AAA1 in the ring. Interestingly, LIS1, a ubiquitous regulator of cytoplasmic dynein, engages the motor domain between AAA3 and AAA4 (Ref. 103) and biases dynein towards a microtubule-bound state103, 104, 105. Whether LIS1 influences the ATPase activity of AAA3 and AAA4 is unknown.
Functions of AAA5, AAA6 and the C-terminal region
The two non-nucleotide-binding modules within the AAA+ ring, AAA5 and AAA6, share the same large–small subdomain architecture as the other AAA+ modules. However, AAA5 displays two distinct features. Two α-helices in its small subdomain are markedly elongated, forming the coiled-coil strut that interacts with the stalk94, 96 (Fig. 2c). The stalk and strut are extensions of different α-helices in AAA4 and AAA5, respectively, suggesting that they are not directly related94, 96. AAA5 also bears an extension at its C-terminal end, comprising a compact bundle of α-helices, the role of which is not yet clear93. Although AAA5 does not hydrolyse ATP, this does not mean it is a static component in the ring. Indeed, extrinsically driven opening and closing of AAA5, and associated angular shifts between the stalk and the strut, may be fundamental in coupling the ATPase cycle of AAA1 with microtubule binding (see below).
The ring is completed by the small subdomain of AAA6 packing against the large subdomain of AAA1. In addition, a structural element C-terminal to AAA6 is required for motor function. Comprising an α-helix that bridges the small subdomains of AAA5 and AAA6 (Ref. 93,96), this structure would be expected to restrict motion within AAA6 if its position were maintained throughout the mechanochemical cycle106, although this model remains untested. Such a role might be important for transmitting structural changes from AAA1 to AAA5 and the strut93. Most dyneins also possess a further C-terminal domain, which in D. discoideum consists of an incomplete β-barrel and several α-helices that spread out over AAA1, AAA5 and AAA6 (Ref. 93). This C-terminal domain is not strictly required for movement and is absent in most fungal dyneins83, but its truncation reduces the processivity of D. discoideum dynein dimers107.
In summary, the global rearrangements within the dynein AAA+ ring and C-terminal sequence during the mechanochemical cycle are not yet clear. When thinking about how structural changes could propagate through the AAA+ ring, one approach is to mentally fix one segment of the ring and imagine the other segment (or segments) moving relative to this reference93, 94, 96. Alternatively, because the AAA+ ring is a topologically closed structure with six potential hinges, another perspective is that ATP binding and hinge closure in one of the AAA+ modules (for example AAA1) exerts strain on the rest of the ring108, inducing the most compliant hinges (perhaps AAA5) to distort in response.
Cyclic microtubule binding
For microtubule binding to stimulate the release of hydrolysis products from AAA1, a signal must be relayed from the microtubule-binding domain at the tip of the stalk to the AAA+ ring. An inverse pathway also operates, because ATP binding at AAA1 weakens the affinity of the microtubule-binding domain85 (Fig. 3). Accumulating evidence suggests that the basis for transmitting signals to and from the microtubule-binding domain involves sliding between the outward (CC1) and return (CC2) α-helices in the stalk coiled coil, over a distance of 4.5–6 Å (equivalent to one turn along the α-helix). This model began with the deduction that the hydrophobic residues in CC1 and CC2 could potentially pack against each other in two types of registry, termed α- and β-registry, owing to an unusual heptad repeat in CC1 (Ref. 109). Both registries have now been observed structurally: a specific β-registry (called +β-registry) in the context of a short stalk construct from mouse cytoplasmic dynein110 and an α-registry within the entire D. discoideum dynein motor domain93. Moreover, fixing the stalk in different registries by protein engineering enforces distinct mechanochemical states. For example, the +β-registry confers weak microtubule affinity109, 110, 111 and low basal ATPase activity111, whereas the α-registry confers strong microtubule affinity109, 110, 111 and stimulates ATPase activity111.
How might track binding cause the stalk to shift registry? Insight comes from a comparison of the mouse microtubule-binding domain in its weak binding state (visualized by crystallography110) versus its strong binding state on the microtubule (visualized by cryo-EM112). Notably, on forming a strong interface with the microtubule, an α-helix (known as H1) on the periphery of the microtubule-binding domain is displaced inwards (Fig. 4a,b). Because H1 directly attaches to CC1 in the stalk, such a movement could bias the coiled coil from the +β-registry towards the α-registry, as articulated in a pseudo-atomic model from electron microscopy and molecular dynamics analysis112 (Fig. 4b). In the crystal structure of the complete D. discoideum motor, the stalk is in the α-registry, but the distal segment of CC1 is disordered and the microtubule-binding domain adopts the weak binding state93. These results suggest that the strong binding conformation may be critically stabilized by interactions with the microtubule112, analogous to the situation with myosin 2 and actin113.
Figure 4. Cyclic microtubule binding.
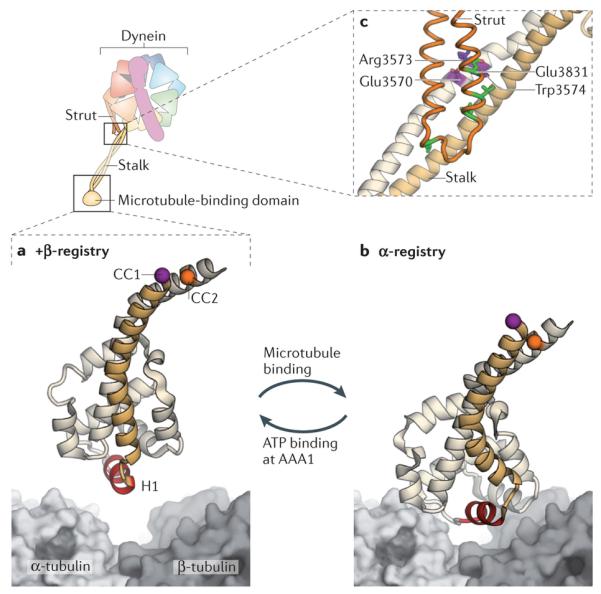
A cartoon of the dynein motor domain is shown, with regions of interest enlarged in parts a–c. a | Crystal structure of the microtubule-binding domain from mouse cytoplasmic dynein in its weak binding state110 (Protein Data Bank (PDB) code: 3ERR), shown above a surface representation of a tubulin dimer. b | Model of the mouse microtubule-binding domain strongly bound to the tubulin dimer, based on a ~10 Å cryo-electron microscopy (cryo-EM) map and steered molecular dynamics112 (PDB code: 3J1T). The binding site of dynein between α-tubulin and β-tubulin overlaps with that of kinesin149. Although the positioning of atoms in the cryo-EM derived model of the microtubule-binding domain is less certain than in the crystal structure, it is clear that helix H1 undergoes a large displacement on forming a strong interface with the microtubule112. Movement of H1 is thought to be associated with changes in the relative alignment or registry of the α-helices (CC1 and CC2) of the stalk, thereby allowing the microtubule-binding domain and the AAA+ ring to communicate through the stalk. This communication is vital for dynein movement, as it allows the microtubule-binding domain to sequentially bind and release its track during the ATPase cycle (see Fig. 3). A marker on CC1 (purple) and CC2 (orange) is coloured in each panel to highlight the registry change between the models. c | Communication between the AAA+ ring and the microtubule-binding domain depends critically93 on interactions between the stalk (an outgrowth of AAA4) and the strut (an outgrowth of AAA5). These stalk–strut interactions are expected to change during the ATPase cycle but, thus far, they have only been visualized in the ADP-bound state of the Dictyostelium discoideum dynein motor, in which the stalk adopts the α-registry93. Four highly conserved residues on the strut (Glu3831, Leu3835, Leu3838 and Leu3846; green) and a highly conserved triad of residues on CC2 of the stalk (Glu3570, Arg3573 and Trp3574; purple) might be important for the structure and dynamics of the stalk–strut interface.
The precise nature of helix sliding in the stalk is unclear. In one scenario, CC1 could move as a unit, undergoing a net translation and small rotation relative to CC2, although side chain rearrangements would be required to accommodate the new registry109. Alternatively, it is possible that CC1 moves by a reptation-like mechanism, in which a bulge propagates along the length of CC1 to generate the register change110. Although reptation would involve distortion along the CC1 backbone, analogous bulge propagation has been reported to occur in canonical coiled coils when strained using optical tweezers114. Furthermore, in the cryo-EM map of the dynein microtubule-binding domain in the α-registry112, the coiled coil seems to open up near the microtubule surface (Fig. 4b), providing a potential site for bulge initiation.
Evidence suggests that the strut may transduce rearrangements in the stalk into enzymatic transitions in the AAA+ ring, and vice versa. When stalk–strut interactions are ablated via truncation of the strut, the ring displays an uncontrolled high ATPase rate in the absence or presence of microtubules93. As revealed by the D. discoideum dynein crystal structure, the strut forms an extensive interaction with the stalk in the α-registry, forming a four-helix bundle93. Specifically, the strut inserts three well-conserved Leu residues between CC1 and CC2 within the stalk (Fig. 4c). There is also a strikingly conserved triad of amino acids on CC2 of the stalk that forms a hydrogen bond interaction with an absolutely conserved Glu on the strut (Fig. 4c). How the stalk–strut interface changes during the mechanochemical cycle is unknown. One idea is that the strut might pivot on CC2 via this conserved triad, and this pivoting would couple with helix sliding or bulge formation in the stalk. In this model, angular shifts between the stalk and strut would be associated with the opening and closing of AAA5, which would feed back to AAA1 via the other AAA+ modules and the C-terminal sequence.
Finally, electron microscopy studies indicate that the stalk is a compliant element, capable of flexing with respect to the rest of the motor domain91, 92, 115. Reinforcing this view of the stalk as a somewhat flexible entity, the two copies of the D. discoideum dynein motor domain in the crystallographic asymmetric unit display different stalk angles93, and the packing between CC1 and CC2 differs between these ‘fleximers’ (Ref. 115). The compliance of the stalk is an intriguing property for a structure that must also transmit subtle conformational changes and sustain force between dynein and the microtubule116. In fact, it is possible that these activities are closely coupled, as strain on the stalk could conceivably influence the registry between CC1 and CC2 and hence alter the affinity of the microtubule-binding domain. In this regard, it is notable that S. cerevisiae cytoplasmic dynein shows an asymmetric response to load, with more force being required to pull it backwards (towards the microtubule plus end) than forwards (towards the minus end)117.
Remodelling of the linker
Arching over the AAA+ ring is the linker domain — the main mechanical element of dynein (Fig. 2c). It is 10 years since the linker was discovered by electron microscopy and proposed to be the principal structure that amplifies conformational changes in the AAA+ modules92. This model has garnered support from 2D and 3D electron microscopy studies of axonemal and cytoplasmic dyneins90, 91, 92, as well as from fluorescence resonance energy transfer (FRET)118 and dynein motility assays83, 119. However, the full extent of the functions of the linker is still emerging. Structurally, the linker comprises five subdomains that form an elongated rod93, 95 (Fig. 5a). The C-terminal end forms an extensive interface with the ring, chiefly involving the large subdomain of AAA1. The N-terminal end of the linker undergoes large-scale movements during the ATPase cycle90, 91, 92, 118 that are crucial for transmitting force to the tail and attached cargo83, 119 (Fig. 3).
Figure 5. Linker domain structure and remodelling.
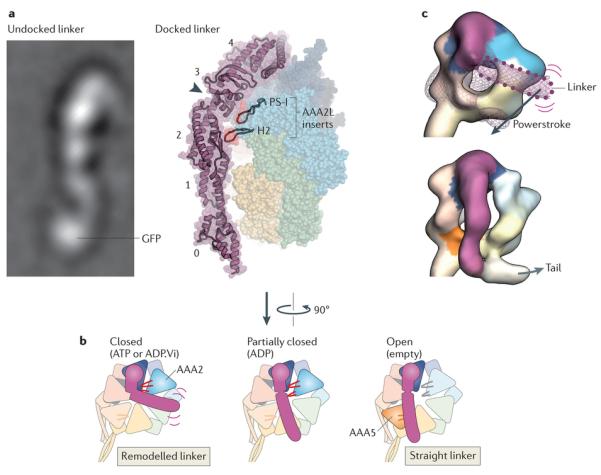
Rearrangements in the linker domain’s structure are crucial for dynein motility. a | Electron microscopy image of the linker domain from Dictyostelium discoideum dynein when it is undocked from the AAA+ ring, showing that the linker is a stable structural entity (left). A GFP tag on the amino-terminal end of the linker is labelled. Linker undocking has, thus far, been observed by electron microscopy only in purified dyneins90, 91, 92, and it is not known whether it can occur under physiological settings. View of the linker arching over the AAA+ ring in the D. discoideum dynein crystal structure in the ADP-bound state93 (right). The five subdomains in the linker are numbered (0–4). The cleft between subdomains 2 and 3, which is spanned by a single α-helix, is indicated by the arrowhead. Two β-hairpin inserts (PS-I and H2) within the large subdomain of AAA2 (AAA2L) that contact the linker near the cleft are highlighted. In addition to the cleft, note that there is a tenuous connection between linker subdomains 0 and 1 that could potentially deform, for example, under strain. b | Model of linker remodelling based on available structural data90, 91, 92, 93, 95, 118. When ATP or ADP.Vi (ADP plus vanadate) is bound in AAA1, the linker seems to be mobile (indicated by purple lines) but bent towards AAA2 in most molecules, according to cryo-EM studies of Chlamydomonas reinhardtii inner arm dynein-c and D. discoideum cytoplasmic dynein90 (left). The PS-I and H2 inserts in AAA2 (shown in red) are strong candidates to mediate this remodelling of the linker93, 95. Following inorganic phosphate (Pi) release, the linker is thought to undergo a powerstroke, in which it straightens to lie over AAA4, based on the D. discoideum dynein crystal structure93 (middle). AAA1 is partially closed around ADP, and the PS-I and H2 inserts form a limited interaction with the linker. Finally, the linker docks at AAA5, as has been observed for Saccharomyces cerevisiae cytoplasmic dynein and C. reinhardtii inner arm dynein-c in the absence of nucleotide (right). This step is proposed to fully open AAA1 and thus eject ADP95. It is unknown if the inserts within the core folds of AAA3 and AAA4 (not shown) can also interact with the linker106. c | Cryo-electron microscopy maps of C. reinhardtii inner arm dynein-c in the ADP.Vi-bound state (upper) and in the absence of nucleotide (lower)90 (Electron Microscopy Data Bank codes: 2156 and 2155, respectively). The maps are coloured as in part b, with AAA+ modules interacting with the linker shown in saturated colours and other modules in pale colours. In the ADP.Vi map, density for the distal linker is missing, owing to variability in its position. The position of the distal linker suggested by a variance map (purple wiremesh) and GFP-based tagging is shown with a dotted outline90. Straightening of the linker is thought to represent the powerstroke of dynein (Fig. 3). Electron microscopy image in part a courtesy of B. Malkova, Biomolecular Research Laboratory, Paul Scherrer Institute, Switzerland.
The question of how the linker moves is therefore key to understanding dynein as a force-generating machine. Several lines of evidence now point towards a model in which the linker is remodelled by interactions with the AAA+ ring. As noted in crystallographic studies, a cleft exists within the linker, suggestive of a hinge93, 95, 96 (Fig. 5a). However, spontaneous sharp bending about this cleft does not seem to occur readily. When the linker is detached or ‘undocked’ from the AAA+ ring (via a C-terminal truncation that destabilizes D. discoideum dynein), the linker retains a rather stable straight form in most molecules, as revealed by electron microscopy and image processing90 (Fig. 5a). Insights into what promotes or stabilizes bending of the linker come from the D. discoideum dynein crystal structure in the ADP-bound state93. Within the large subdomain of AAA2, there are two β-hairpin inserts (the pre-sensor-I (PS-I) and helix 2 (H2) inserts), which contact the linker near the cleft (Fig. 5a). On exchange of ADP for ATP, full closure of AAA1 might bring these inserts into a closer embrace with the linker to elicit bending and/or to stabilize the bent conformation93, 95 (Fig. 5b). EM studies indicate that when ATP or ADP.Vi (ADP plus vanadate, a phosphate analogue) are bound to AAA1, the distal linker is bent towards AAA2 in most molecules (Fig. 5b,c) but remains straight in a subpopulation90, 91. This raises the possibility that linker remodelling is an equilibrium process that is biased by ATP binding, rather than a switch-like transition. It is anticipated that a high-resolution crystal structure of dynein with ATP or an analogue bound to AAA1 will clarify how the linker is remodelled. Moreover, because such a structure may represent a snapshot of a fluctuating state, it will need to be combined with experiments that can reveal the dynamics of the linker to gain a full understanding of the remodelling reaction.
With the linker bent towards AAA2, binding of the motor to the microtubule is thought to stimulate Pi release and trigger the powerstroke, in which the linker straightens to lie over AAA4 (Fig. 5c). This path would take the linker over AAA3; whether there is interplay between the linker and the nucleotide status of AAA3 or AAA4 is unknown. During the powerstroke transition, the end of the linker would move principally along the long axis of the stalk91, and it has been posited that the angle at which the stalk binds the microtubule is an important determinant of the directionality of the powerstroke110. Consistent with this idea, various dyneins have been shown to bind preferentially with the stalk angled from the ring towards the microtubule minus end — the direction of dynein movement110, 112, 120, 121 (Fig. 2c).
The linker can also influence enzymatic transitions within the AAA+ ring. Small N-terminal truncations and mutations within the linker impair the ATPase activity and motility of dynein, although the precise functional boundary may differ between species83, 89, 91, 95. When S. cerevisiae cytoplasmic dynein and C. reinhardtii inner arm dynein-c are visualized in the nucleotide-free state, the N-terminal region of the linker forms a contact with the large subdomain of AAA5 (Refs 90,95) (Fig. 5b). This differs from the ADP-bound D. discoideum dynein structure93, in which the linker lies above AAA4 (Fig. 5b). It has been proposed that, by shifting to interact with AAA5, the linker may function as a spacer that applies tension to the AAA+ ring, prying open AAA1 and promoting ADP release after the powerstroke95. Similarly, it is possible that rearward strain on the linker (imparted by cargo or a partnering motor domain) could favour the closed conformation of AAA1, thus inhibiting ADP release and microtubule detachment. In summary, the linker domain may have functions beyond delivering the powerstroke of dynein, such as sensing strain and regulating progression through the mechanochemical cycle.
Emergent motile properties
According to the model in Fig. 3, a single dynein motor domain would bind to the microtubule, perform a solitary tug and then diffuse away after binding ATP103. However, when multiple dynein motor domains act together, as is typical in living cells, striking additional motile behaviours can emerge. One example is the beating motions of cilia, which are driven by the thousands of dyneins within the axoneme (Box 1). Another is the processive motion produced by cytoplasmic dynein dimers.
When two S. cerevisiae cytoplasmic dynein motor domains are paired (either naturally via the tail or by fusing an exogenous dimerizing moiety to the end of the linker), the resulting dimer can move along the microtubule for ~1,500 nm before detaching, as revealed by in vitro single-molecule studies83, 122. These movements represent dozens of individual steps, with each motor domain typically moving in 8–16 nm increments83, 122, 123, corresponding to a distance of 1–2 tubulin dimers along the long axis of the microtubule. Larger steps, as well as sideways and backwards excursions, are also observed, suggesting there is a diffusive component to each step83, 122, 123. Furthermore, optical trapping has revealed that S. cerevisiae cytoplasmic dynein dimers are high-force motors, capable of moving against resisting loads of up to 7 pN before stalling117.
By labelling each motor domain with a different coloured fluorophore, the relative movements within the S. cerevisiae dimer have recently been unveiled122, 123. Remarkably, there is no strict pattern to the steps: the motor domains do not need to alternately pass each other in space and, in some cases, the rear motor can take multiple steps while the lead motor remains stationary, and vice versa. This contrasts with the methodical ‘hand-over-hand’ movements of kinesin 1 and myosin 5 dimers, which arise from alternating ATPase cycles in their motor domains9. In the case of the less rigid stepping pattern of dynein, what prevents both motor domains detaching from the microtubule simultaneously? One part of the answer may lie in the duty ratio of the S. cerevisiae motor domain122, 123 (that is, the fraction of the mechanochemical cycle spent attached to the track). This parameter has not been measured for S. cerevisiae dynein but, as a theoretical example, if the motor domains acted independently but spent 90% of their cycle attached to the microtubule, the dimer would still be expected to take ~70 steps on average before detaching124. Yet, a purely stochastic model does not capture all of the features of the stepping behaviour in S. cerevisiae dynein. Notably, as the separation between the motor domains along the microtubule increases, the probability of the rear head taking a step becomes greater and the stepping of the lead head is inhibited122, 123. It has been proposed that this reflects a response of the motor domains to internal strain within the dimer122, 123. Whether this strain sensing is mediated via the stalk, the linker or some other unknown mechanism awaits discovery.
Interestingly, in D. discoideum cytoplasmic dynein, the two motor domains may influence one another’s enzymatic cycles more strongly. In this system, the intrinsic duty ratio of each motor domain is low (24%)125. Thus, in the absence of any communication, the dimer would be expected to take only around two steps along the microtubule on average before detaching124 — at odds with the measured run length of ~750 nm107. Moreover, following dimerization, the ATPase rate of the D. discoideum motor domain on the microtubule is slowed down by ~70%, further pointing towards enzymatic regulation within the dimer107. Elucidating the basis for this intra-dimer communication will require structural information on the geometry of D. discoideum dynein dimers on the microtubule, coupled with two-colour tracking of each motor domain. In summary, the mechanism and extent of communication within cytoplasmic dynein dimers from different species is an active area of research.
Conclusions and perspectives
Fifty years since the discovery of its founding member, the dynein family is now known to participate in a broad range of cellular functions involving microtubule-based movement. A model for the basic reaction of dynein with the microtubule is beginning to emerge from mechanistic studies on its motor domain, but crucial features remain untested. Some concepts, such as the motions of AAA1 and the use of inserts in the AAA+ modules to remodel the linker domain, are reminiscent of the actions of other AAA+ mechanoenzymes. However, other aspects of the dynein mechanism, such as the interplay between the stalk, strut and microtubule, are without precedent.
The roles of the subsidiary nucleotide-binding modules of dynein, particularly AAA3 and AAA4, remain mysterious. New tools, perhaps involving module-specific conformation sensors126, are needed to elucidate their function. Simultaneous visualization of stepping and nucleotide binding, as has been achieved for myosin 5 (Ref. 127), would be highly informative. We can also look forward to high-resolution structures of dynein in different nucleotide states, which might clarify how small changes in the active site of AAA1 are propagated through the large dynein motor domain. Simulations and experiments are required to test exactly how the α-helices of the stalk slide relative to one another. And small-molecule inhibitors (such as ciliobrevin128) may be useful in trapping specific conformational states of the motor, as well as in unravelling the cellular functions of dynein.
Our current picture of dynein motility is a synthesis of data from various dynein species and truncated constructs, in part owing to the challenges associated with working with intact dynein holoenzymes. However, it will be important to elucidate how dynein holoenzymes work and how they have diverged from one another to carry out specific biological functions. Ciliary beating, for example, arises from the action of numerous dynein species, each with distinct motile characteristics129. It is known that a subset of inner arm dyneins can rotate microtubules around their long axis as they translocate them in in vitro assays130 and that some have specialized structural features such as curved stalk domains92, but how these properties relate to ciliary motility is unclear at present.
There is also much to learn about variations in the behaviour of cytoplasmic dynein. For instance, one controversial area is the force produced by a single mammalian cytoplasmic dynein complex. Although several studies have measured a stall force of ~1 pN104, 131, 132, 133, reports of higher stall forces of 5–7 pN persist134, 135. Does this discrepancy arise from differences in protein handling? Or are there subpopulations of cytoplasmic dynein in vivo with distinct force-producing capacities that are isolated by different purification methods? Answers to these questions are keenly sought, and they have the potential to provide insights into how dynein is regulated in the cell. In contrast to previous studies with tissue-purified dynein, a recent study using recombinant protein challenges the idea that mammalian cytoplasmic dynein complexes are intrinsically processive, indicating that additional factors could be required for sustained movement27. There are also reports that mammalian cytoplasmic dynein has the ability to move towards the plus end of the microtubule135, 136, although the structural and mechanistic basis for this behaviour remains unknown. Very little is known about the motility of IFT dynein and how it is adapted for IFT.
Looking forward, we anticipate a more complete fusion of cell biological and in vitro analyses of dynein. One challenge will be bringing the high precision of in vitro measurements to single dynein molecules, regulators and cargoes in living cells, but important advances are being made in this area. For example, a recent study was able to track single cytoplasmic dynein molecules within the Schizosaccharomyces pombe cell volume using highly inclined and laminated optical sheet microscopy137, and in-cell optical trapping is revealing the force-producing behaviour of cargo-bound motors in situ80. Fungi have been, and will continue to be, powerful model systems for studying intracellular transport, in part because genes of interest can be readily tagged and modified at their genomic loci. Advances in genome editing technologies should make it easier to perform similar manipulations on the dynein machinery in metazoan cells. And, to complement the insights from living cells, another important challenge will be to reconstitute more complex dynein-powered motile systems in vitro using purified proteins. Analysis of cell extracts, which combine near-cellular complexity with biochemical tractability, offers an interesting way to help bridge the gap between in vivo and in vitro studies.
Finally, structural information will have an important part to play in uncovering how dyneins are harnessed in cells. A high-resolution structure of the dynein tail domain with its associated subunits is expected to shed light on how dyneins oligomerize and engage cargo. Insight into the architecture of the dynein–dynactin complex and other regulatory assemblies will be essential. Moreover, at intermediate resolution, cryo-EM and tomography have the potential to visualize the arrangement of the two motor domains of cytoplasmic dynein on the microtubule, as well as how groups of motors self-organize on macromolecular cargo. Combining such structural data with dynamic information from in vitro and in vivo studies promises to reveal the workings of dyneins in breathtaking detail, sufficient to understand their dysfunction in disease and perhaps design synthetic applications for dynein.
Supplementary Material
Dynein species and isoform are given on the left. Arrows depict the trans action of the Arg finger, which reaches from one AAA+ (ATPases associated with diverse activities) module into the nucleotide-binding pocket of the preceding module. Conserved = black dot; partially conserved = grey dot; not conserved = white dot.
Acknowledgements
The authors apologize to their colleagues whose work could not be cited owing to space limitations. They thank K. Toropova, R. Hernandez-Lopez, J. Huang and S. Reck-Peterson for helpful comments on the manuscript and B. Malkova for providing electron microscopy data for figure 5a. A.J.R. is grateful to J. Iwasa for training on AutoDesk Maya software. The work in the authors laboratory was supported by: a Sir Henry Wellcome Postdoctoral Fellowship (092436/Z/10/Z) to A.J.R.; a Grant-in-Aid for Scientific Research (B) 23370073 from the Japan Society for Promotion of Science (JSPS) and a Japan Science and Technology Agency PRESTO award to T.K.; a Grant-in-Aid for Scientific Research (B) 23370075 from the JSPS to K.S., and grants BB/E00928X/1 and BB/BB/K000705/1 from the BBSRC (UK) and RGP0009/2008-C from the Human Frontiers Science Program to S.A.B.
Glossary
- G protein-related fold
A characteristic arrangement of secondary structure elements and loops (such as switch I and switch II) shared by G proteins, myosins and kinesins, which indicates that these proteins originated from a common ancestor.
- Axoneme
The microtubule-based core of eukaryotic cilia and flagella. The terms cilia and flagella are often used interchangeably, as both describe cellular appendages with an axoneme at their core. In this Review, we use cilia for consistency.
- Autophagosomes
Organelles that enwrap cytoplasmic material in a double-membrane-bound structure and subsequently fuse with lysosomes, leading to degradation of the confined material.
- Immunological synapse
The interface formed between an antigen-presenting cell and a lymphocyte, such as a B cell or a T cell.
- Astral microtubules
Microtubules radiating from the spindle poles that do not contact the kinetochore or overlap with other microtubules in the spindle midzone.
- Crystal soaking
A technique in which a crystallized macromolecule is bathed in a ligand-containing solution. The ligand has the opportunity to bind the macromolecule of interest owing to diffusion through solvent-filled channels in the crystal.
- E1 helicase
A hexameric AAA+ ATPase protein from papillomavirus that encircles and translocates along single-stranded DNA, thereby unwinding DNA duplexes with a 3′ to 5′ directionality.
- Reptation
Snake-like movement of a polymer along a path, originally introduced by De Gennes in the field of polymer theory.
- Highly inclined and laminated optical sheet microscopy
A technique for visualizing fluorescently labelled single molecules in cells. To minimize background signal (which can confound single-molecule detection) the specimen is illuminated with an angled sheet of light.
Footnotes
Competing interests statement: The authors declare no competing financial interests.
References
- 1.Moughamian AJ, Holzbaur E. G protein-related fold. In: King SM, editor. Dyneins: Structure, Biology and Disease. Elsevier Inc.; 2011. pp. 584–601. [Google Scholar]
- 2.Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 3.Gibbons IR. Studies on the protein components of cilia from Tetrahymena pyriformis. Proc. Natl Acad. Sci. USA. 50 doi: 10.1073/pnas.50.5.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibbons I, Rowe A. Dynein: a protein with adenosine triphosphatase activity from cilia. Science. 1965;149:424–426. doi: 10.1126/science.149.3682.424. [References 3 and 4 mark the discovery of dynein motor proteins.] [DOI] [PubMed] [Google Scholar]
- 5.Paschal BM, Shpetner HS, Vallee RB. MAP 1C is a microtubule-activated ATPase which translocates microtubules in vitro and has dynein-like properties. J. Cell Biol. 1987;105:1273–1282. doi: 10.1083/jcb.105.3.1273. [Describes the isolation and characterization of cytoplasmic dynein.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paschal BM, Vallee RB. Retrograde transport by the microtubule-associated protein MAP 1C. Nature. 1987;330:181–183. doi: 10.1038/330181a0. [DOI] [PubMed] [Google Scholar]
- 7.Schroer TA, Steuer ER, Sheetz MP. Cytoplasmic dynein is a minus end-directed motor for membranous organelles. Cell. 1989;56:937–946. doi: 10.1016/0092-8674(89)90627-2. [DOI] [PubMed] [Google Scholar]
- 8.Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vale RD, Milligan RA. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 10.Billington N, Sellers JR. Dynein struts its stuff. Nat. Struct. Mol. Biol. 2011;18:635–636. doi: 10.1038/nsmb.2082. [DOI] [PubMed] [Google Scholar]
- 11.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 12.Vallee RB, McKenney RJ, Ori-McKenney KM. Multiple modes of cytoplasmic dynein regulation. Nat. Cell Biol. 2012;14:224–230. doi: 10.1038/ncb2420. [DOI] [PubMed] [Google Scholar]
- 13.Kardon J, Vale R. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allan VJ. Cytoplasmic dynein. Biochem. Soc. Trans. 2011;39:1169–1178. doi: 10.1042/BST0391169. [DOI] [PubMed] [Google Scholar]
- 15.Akhmanova A, Hammer JA., 3rd. Linking molecular motors to membrane cargo. Curr. Opin. Cell Biol. 2010;22:479–487. doi: 10.1016/j.ceb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riedel-Kruse IH, Hilfinger A, Howard J, Jülicher F. How molecular motors shape the flagellar beat. HFSP J. 2007;1:192–208. doi: 10.2976/1.2773861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brokaw C. Thinking about flagellar oscillation. Cell. Motil. Cytoskeleton. 2009;66:425–436. doi: 10.1002/cm.20313. [DOI] [PubMed] [Google Scholar]
- 18.Lindemann CB, Lesich KA. Flagellar and ciliary beating: the proven and the possible. J. Cell Sci. 2010;123:519–528. doi: 10.1242/jcs.051326. [DOI] [PubMed] [Google Scholar]
- 19.King SM. Integrated control of axonemal dynein AAA+ motors. J. Struct. Biol. 2012;179:222–228. doi: 10.1016/j.jsb.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wickstead B, Gull K. Dyneins across eukaryotes: a comparative genomic analysis. Traffic. 2007;8:1708–1721. doi: 10.1111/j.1600-0854.2007.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 22.Yagi T. Bioinformatic approaches to dynein heavy chain classification. Methods Cell Biol. 2009;92:1–9. doi: 10.1016/S0091-679X(08)92001-X. [DOI] [PubMed] [Google Scholar]
- 23.Moore J, Stuchell-Brereton M, Cooper J. Function of dynein in budding yeast: mitotic spindle positioning in a polarized cell. Cell. Motil. Cytoskeleton. 2009;66:546–555. doi: 10.1002/cm.20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egan MJ, McClintock MA, Reck-Peterson SL. Microtubule-based transport in filamentous fungi. Curr. Opin. Microbiol. 2012;15:637–645. doi: 10.1016/j.mib.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koonce MP. Dictyostelium, a model organism for microtubule-based transport. Protist. 2000;151:17–25. doi: 10.1078/1434-4610-00004. [DOI] [PubMed] [Google Scholar]
- 26.Pfister KK, et al. Genetic analysis of the cytoplasmic dynein subunit families. PLoS Genet. 2006;2:e1. doi: 10.1371/journal.pgen.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trokter M, Mücke N, Surrey T. Reconstitution of the human cytoplasmic dynein complex. Proc. Natl Acad. Sci. USA. 2012;109:20895–20900. doi: 10.1073/pnas.1210573110. [Describes the reconstitution of the human cytoplasmic dynein complex from recombinant subunits. Surprisingly, despite driving robust microtubule gliding, the complexes do not show processive motility, suggesting that additional factors might be required for this behaviour.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroer TA. Dynactin. Annu. Rev. Cell Dev. Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 29.Driskell OJ, Mironov A, Allan VJ, Woodman PG. Dynein is required for receptor sorting and the morphogenesis of early endosomes. Nat. Cell Biol. 2007;9:113–120. doi: 10.1038/ncb1525. [DOI] [PubMed] [Google Scholar]
- 30.Jordens I, et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- 31.Blocker A, et al. Molecular requirements for bi-directional movement of phagosomes along microtubules. J. Cell Biol. 1997;137:113–129. doi: 10.1083/jcb.137.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gross SP, et al. Interactions and regulation of molecular motors in Xenopus melanophores. J. Cell Biol. 2002;156:855–865. doi: 10.1083/jcb.200105055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kural C, et al. Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement? Science. 2005;308:1469–1472. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- 34.Gross SP, Welte MA, Block SM, Wieschaus EF. Dynein-mediated cargo transport in vivo. A switch controls travel distance. J. Cell Biol. 2000;148:945–956. doi: 10.1083/jcb.148.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pilling AD, Horiuchi D, Lively CM, Saxton WM. Kinesin-1 and dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol. Biol. Cell. 2006;17:2057–2068. doi: 10.1091/mbc.E05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Presley JF, et al. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- 37.Heerssen HM, Pazyra MF, Segal RA. Dynein motors transport activated Trks to promote survival of target-dependent neurons. Nature Neurosci. 2004;7:596–604. doi: 10.1038/nn1242. [DOI] [PubMed] [Google Scholar]
- 38.Young A, Dictenberg JB, Purohit A, Tuft R, Doxsey SJ. Cytoplasmic dynein-mediated assembly of pericentrin and gamma tubulin onto centrosomes. Mol. Biol. Cell. 2000;11:2047–2056. doi: 10.1091/mbc.11.6.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrell JM, et al. Evidence for glucocorticoid receptor transport on microtubules by dynein. J. Biol. Chem. 2004;279:54647–54654. doi: 10.1074/jbc.M406863200. [DOI] [PubMed] [Google Scholar]
- 40.Shah JV, Flanagan LA, Janmey PA, Leterrier JF. Bidirectional translocation of neurofilaments along microtubules mediated in part by dynein/dynactin. Mol. Biol. Cell. 2000;11:3495–3508. doi: 10.1091/mbc.11.10.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilkie GS, Davis I. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell. 2001;105:209–219. doi: 10.1016/s0092-8674(01)00312-9. [DOI] [PubMed] [Google Scholar]
- 42.Johnston JA, Illing ME, Kopito RR. Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell. Motil. Cytoskeleton. 2002;53:26–38. doi: 10.1002/cm.10057. [DOI] [PubMed] [Google Scholar]
- 43.Maday S, Wallace KE, Holzbaur ELF. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 2012;196:407–417. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dodding MP, Way M. Coupling viruses to dynein and kinesin-1. EMBO J. 2011;30:3527–3539. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato A, et al. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009;139:907–919. doi: 10.1016/j.cell.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinberg G, et al. Motor-driven motility of fungal nuclear pores organizes chromosomes and fosters nucleocytoplasmic transport. J. Cell Biol. 2012;198:343–355. doi: 10.1083/jcb.201201087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnyder T, et al. B cell receptor-mediated antigen gathering requires ubiquitin ligase Cbl and adaptors Grb2 and Dok-3 to recruit dynein to the signaling microcluster. Immunity. 2011;34:905–918. doi: 10.1016/j.immuni.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 48.Hashimoto-Tane A, et al. Dynein-driven transport of T cell receptor microclusters regulates immune synapse formation and T cell activation. Immunity. 2011;34:919–931. doi: 10.1016/j.immuni.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 49.Laan L, et al. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148:502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hendricks AG, et al. Dynein tethers and stabilizes dynamic microtubule plus ends. Curr. Biol. 2012;22:632–637. doi: 10.1016/j.cub.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai J-W, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nature Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- 52.Dujardin DL, et al. A role for cytoplasmic dynein and LIS1 in directed cell movement. J. Cell Biol. 2003;163:1205–1211. doi: 10.1083/jcb.200310097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Combs J, et al. Recruitment of dynein to the Jurkat immunological synapse. Proc. Natl Acad. Sci. USA. 2006;103:14883–14888. doi: 10.1073/pnas.0600914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McNally FJ. Mechanisms of spindle positioning. J. Cell Biol. 2013;200:131–140. doi: 10.1083/jcb.201210007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 2012;14:311–317. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collins ES, Balchand SK, Faraci JL, Wadsworth P, Lee W-L. Cell cycle-regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol. Biol. Cell. 2012;23:3380–3390. doi: 10.1091/mbc.E12-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitchison T, et al. Growth, interaction, and positioning of microtubule asters in extremely large vertebrate embryo cells. Cytoskeleton. 2012;69:738–750. doi: 10.1002/cm.21050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kimura K, Kimura A. Intracellular organelles mediate cytoplasmic pulling force for centrosome centration in the Caenorhabditis elegans early embryo. Proc. Natl Acad. Sci. USA. 2011;108:137–142. doi: 10.1073/pnas.1013275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Longoria RA, Shubeita GT. Cargo transport by cytoplasmic dynein can center embryonic centrosomes. PLoS ONE. 2013;8:e67710. doi: 10.1371/journal.pone.0067710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corthesy-Theulaz I, Pauloin A, Pfeffer SR. Cytoplasmic dynein participates in the centrosomal localization of the Golgi complex. J. Cell Biol. 1992;118:1333–1345. doi: 10.1083/jcb.118.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levy JR, Holzbaur EL. Dynein drives nuclear rotation during forward progression of motile fibroblasts. J. Cell Sci. 2008:3187–3195. doi: 10.1242/jcs.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raaijmakers JA, et al. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J. 31:4179–4190. doi: 10.1038/emboj.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salina D, et al. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- 64.Heald R, et al. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 65.Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- 66.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2012;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Howell BJ, et al. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wojcik E, et al. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat. Cell Biol. 2001;3:1001–1007. doi: 10.1038/ncb1101-1001. [DOI] [PubMed] [Google Scholar]
- 69.Varma D, Monzo P, Stehman SA, Vallee RB. Direct role of dynein motor in stable kinetochore-microtubule attachment, orientation, and alignment. J. Cell Biol. 2008;182:1045–1054. doi: 10.1083/jcb.200710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Z, Tulu US, Wadsworth P, Rieder CL. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr. Biol. 2007;17:973–980. doi: 10.1016/j.cub.2007.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- 72.Pazour GJ, Dickert BL, Witman GB. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 1999;144:473–481. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Porter ME, Bower R, Knott JA, Byrd P, Dentler W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell. 1999;10:693–712. doi: 10.1091/mbc.10.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mikami A, et al. Molecular structure of cytoplasmic dynein 2 and its distribution in neuronal and ciliated cells. J. Cell Sci. 2002;115:4801–4808. doi: 10.1242/jcs.00168. [DOI] [PubMed] [Google Scholar]
- 75.Perrone CA, et al. A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in Chlamydomonas and mammalian cells. Mol. Biol. Cell. 2003;14:2041–2056. doi: 10.1091/mbc.E02-10-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ichikawa M, Watanabe Y, Murayama T, Toyoshima YY. Recombinant human cytoplasmic dynein heavy chain 1 and 2: observation of dynein-2 motor activity in vitro. FEBS Lett. 2011;585:2419–2423. doi: 10.1016/j.febslet.2011.06.026. [DOI] [PubMed] [Google Scholar]
- 77.Pigino G, et al. Electron-tomographic analysis of intraflagellar transport particle trains in situ. J. Cell Biol. 2009;187:135–148. doi: 10.1083/jcb.200905103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Laib JA, Marin JA, Bloodgood RA, Guilford WH. The reciprocal coordination and mechanics of molecular motors in living cells. Proc. Natl Acad. Sci. USA. 2009;106:3190–3195. doi: 10.1073/pnas.0809849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shih SM, et al. Intraflagellar transport drives flagellar surface motility. Elife. 2013;2:e00744. doi: 10.7554/eLife.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mallik R, Rai AK, Barak P, Rai A, Kunwar A. Teamwork in microtubule motors. Trends Cell Biol. 2013;23:575–582. doi: 10.1016/j.tcb.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 81.Koonce MP, Samsó M. Overexpression of cytoplasmic dynein’s globular head causes a collapse of the interphase microtubule network in Dictyostelium. Mol. Biol. Cell. 1996;7:935–948. doi: 10.1091/mbc.7.6.935. [Details a recombinant expression system for the D. discoideum cytoplasmic dynein motor domain, forming the foundation for numerous structural and functional analyses. Also demonstrates that the motor domain includes the globular head structure seen in earlier electron microscopy images.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nishiura M, et al. A single-headed recombinant fragment of Dictyostelium cytoplasmic dynein can drive the robust sliding of microtubules. J. Biol. Chem. 2004;279:22799–22802. doi: 10.1074/jbc.M313362200. [DOI] [PubMed] [Google Scholar]
- 83.Reck-Peterson SL, et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [Establishes S. cerevisiae as another important cytoplasmic dynein expression system and provides insight into the requirements for processive dynein motion.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Höök P, et al. Long range allosteric control of cytoplasmic dynein ATPase activity by the stalk and C-terminal domains. J. Biol. Chem. 2005;280:33045–33054. doi: 10.1074/jbc.M504693200. [DOI] [PubMed] [Google Scholar]
- 85.Imamula K, Kon T, Ohkura R, Sutoh K. The coordination of cyclic microtubule association/dissociation and tail swing of cytoplasmic dynein. Proc. Natl Acad. Sci. USA. 2007;104:16134–16139. doi: 10.1073/pnas.0702370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mogami T, Kon T, Ito K, Sutoh K. Kinetic characterization of tail swing steps in the ATPase cycle of Dictyostelium cytoplasmic dynein. J. Biol. Chem. 2007;282:21639–21644. doi: 10.1074/jbc.M701914200. [DOI] [PubMed] [Google Scholar]
- 87.Johnson KA. Pathway of the microtubule–dynein ATPase and the structure of dynein: a comparison with actomyosin. Annu. Rev. Biophys. Biophys. Chem. 1985;14:161–188. doi: 10.1146/annurev.bb.14.060185.001113. [DOI] [PubMed] [Google Scholar]
- 88.Samsó M, Radermacher M, Frank J, Koonce MP. Structural characterization of a dynein motor domain. J. Mol. Biol. 1998;276:927–937. doi: 10.1006/jmbi.1997.1584. [DOI] [PubMed] [Google Scholar]
- 89.Gee MA, Heuser JE, Vallee RB. An extended microtubule-binding structure within the dynein motor domain. Nature. 1997;390:636–639. doi: 10.1038/37663. [DOI] [PubMed] [Google Scholar]
- 90.Roberts AJ, et al. ATP-driven remodeling of the linker domain in the dynein motor. Structure. 2012;20:1670–1680. doi: 10.1016/j.str.2012.07.003. [A 2D and 3D electron microscopy study of an axonemal and cytoplasmic dynein, revealing that the linker domain is a stable structural entity that is remodelled by the AAA+ ring.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roberts A, et al. AAA+ ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 93.Kon T, et al. The 2.8 Å crystal structure of the dynein motor domain. Nature. 2012;484:345–350. doi: 10.1038/nature10955. [Reports a high-resolution crystal structure of the D. discoideum dynein motor domain in the ADP-bound state. Structure-guided mutagenesis provides insight into how the linker domain moves and the roles of the AAA+ modules, strut and C-terminal sequence in allosteric communication.] [DOI] [PubMed] [Google Scholar]
- 94.Kon T, Sutoh K, Kurisu G. X-ray structure of a functional full-length dynein motor domain. Nat. Struct. Mol. Biol. 2011;18:638–642. doi: 10.1038/nsmb.2074. [DOI] [PubMed] [Google Scholar]
- 95.Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-Å crystal structure. Nat. Struct. Mol. Biol. 2012;19:492–497. doi: 10.1038/nsmb.2272. [Reports a high-resolution structure of the S. cerevisiae dynein motor domain, crystallized in the absence of nucleotide. Crystal-soaking experiments reveal distinct nucleotide-binding behaviours in AAA1–AAA4, and mutagenesis suggests a functional role for linker docking at AAA5.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang J, et al. Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure. 2001;9:1107–1116. doi: 10.1016/s0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- 99.Gibbons IR, et al. Photosensitized cleavage of dynein heavy chains. Cleavage at the “V1 site” by irradiation at 365 nm in the presence of ATP and vanadate. J. Biol. Chem. 1987;262:2780–2786. [PubMed] [Google Scholar]
- 100.Kon T, Nishiura M, Ohkura R, Toyoshima YY, Sutoh K. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry. 2004;43:11266–11274. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- 101.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 102.Cho C, Reck-Peterson S, Vale R. Cytoplasmic dynein’s regulatory ATPase sites affect processivity and force generation. J. Biol. Chem. 2008;283:25839–25845. doi: 10.1074/jbc.M802951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huang J, Roberts AJ, Leschziner AE, Reck-Peterson SL. Lis1 acts as a “clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell. 2012;150:975–986. doi: 10.1016/j.cell.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yamada M, et al. LIS1 and NDEL1 coordinate the plus-end-directed transport of cytoplasmic dynein. EMBO J. 2008;27:2471–2483. doi: 10.1038/emboj.2008.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cho C, Vale RD. The mechanism of dynein motility: insight from crystal structures of the motor domain. Biochim. Biophys. Acta. 2011;1823:182–191. doi: 10.1016/j.bbamcr.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Numata N, Shima T, Ohkura R, Kon T, Sutoh K. C-Sequence of the Dictyostelium cytoplasmic dynein participates in processivity modulation. FEBS Lett. 2011;585:1185–1190. doi: 10.1016/j.febslet.2011.03.036. [DOI] [PubMed] [Google Scholar]
- 108.Hwang W, Lang MJ. Nucleotide-dependent control of internal strains in ring-shaped AAA+ motors. Cell. Mol. Bioeng. 2013;6:65–73. doi: 10.1007/s12195-012-0264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gibbons IR, et al. The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk. J. Biol. Chem. 2005;280:23960–23965. doi: 10.1074/jbc.M501636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Carter AP, et al. Structure and functional role of dynein’s microtubule-binding domain. Science. 2008;322:1691–1695. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kon T, et al. Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nat. Struct. Mol. Biol. 2009;16:325–333. doi: 10.1038/nsmb.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Redwine WB, et al. Structural basis for microtubule binding and release by dynein. Science. 2012;337:1532–1536. doi: 10.1126/science.1224151. [References 109–112 provide evidence for the helix-sliding hypothesis for allosteric communication through the stalk of dynein, which was put forward in reference 109. Reference 112 also provides insight into the structural changes within the microtubule-binding domain of dynein.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sweeney HL, Houdusse A. The motor mechanism of myosin V: insights for muscle contraction. Philo. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:1829–1841. doi: 10.1098/rstb.2004.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xi Z, Gao Y, Sirinakis G, Guo H, Zhang Y. Single-molecule observation of helix staggering, sliding, and coiled coil misfolding. Proc. Natl Acad. Sci. USA. 2012;109:5711–5716. doi: 10.1073/pnas.1116784109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Burgess SA, Walker ML, Sakakibara H, Oiwa K, Knight PJ. The structure of dynein-c by negative stain electron microscopy. J. Struct. Biol. 2004;146:205–216. doi: 10.1016/j.jsb.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 116.Burgess SA, Knight PJ. Is the dynein motor a winch? Curr. Opin. Struct. Biol. 2004;14:138–146. doi: 10.1016/j.sbi.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 117.Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 2007;131:952–965. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kon T, Mogami T, Ohkura R, Nishiura M, Sutoh K. ATP hydrolysis cycle-dependent tail motions in cytoplasmic dynein. Nat. Struct. Mol. Biol. 2005;12:513–519. doi: 10.1038/nsmb930. [DOI] [PubMed] [Google Scholar]
- 119.Shima T, Kon T, Imamula K, Ohkura R, Sutoh K. Two modes of microtubule sliding driven by cytoplasmic dynein. Proc. Natl Acad. Sci. USA. 2006;103:17736–17740. doi: 10.1073/pnas.0606794103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ueno H, Yasunaga T, Shingyoji C, Hirose K. Dynein pulls microtubules without rotating its stalk. Proc. Natl Acad. Sci. USA. 2008;105:19702–19707. doi: 10.1073/pnas.0808194105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Movassagh T, Bui KH, Sakakibara H, Oiwa K, Ishikawa T. Nucleotide-induced global conformational changes of flagellar dynein arms revealed by in situ analysis. Nat. Struct. Mol. Biol. 2010;17:761–767. doi: 10.1038/nsmb.1832. [DOI] [PubMed] [Google Scholar]
- 122.Qiu W, et al. Dynein achieves processive motion using both stochastic and coordinated stepping. Nat. Struct. Mol. Biol. 2012;19:193–200. doi: 10.1038/nsmb.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dewitt MA, Chang AY, Combs PA, Yildiz A. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science. 2011;335:221–225. doi: 10.1126/science.1215804. [References 122 and 123 use two-colour single-molecule experiments to reveal that S. cerevisiae cytoplasmic dynein dimers can move processively along the microtubule without following a strict stepping pattern. Analysis of the stepping traces provides insight into the spatial configuration of the motor domains and their loose, tension-based coordination.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Veigel C, Wang F, Bartoo ML, Sellers JR, Molloy JE. The gated gait of the processive molecular motor, myosin V. Nat. Cell Biol. 2002;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- 125.Shima T, Imamula K, Kon T, Ohkura R, Sutoh K. Head-head coordination is required for the processive motion of cytoplasmic dynein, an AAA+ molecular motor. J. Struct. Biol. 2006;156:182–189. doi: 10.1016/j.jsb.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 126.Stinson BM, et al. Nucleotide binding and conformational switching in the hexameric ring of a AAA+ machine. Cell. 2013;153:628–639. doi: 10.1016/j.cell.2013.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sakamoto T, Webb MR, Forgacs E, White HD, Sellers JR. Direct observation of the mechanochemical coupling in myosin Va during processive movement. Nature. 2008;455:128–132. doi: 10.1038/nature07188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Firestone AJ, et al. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature. 2012;484:125–129. doi: 10.1038/nature10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kamiya R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int. Rev. Cytol. 2002;219:115–155. doi: 10.1016/s0074-7696(02)19012-7. [DOI] [PubMed] [Google Scholar]
- 130.Kikushima K, Kamiya R. Clockwise translocation of microtubules by flagellar inner-arm dyneins in vitro. Biophys. J. 2008;94:4014–4019. doi: 10.1529/biophysj.107.123083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mallik R, Carter BC, Lex SA, King SJ, Gross SP. Cytoplasmic dynein functions as a gear in response to load. Nature. 2004;427:649–652. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]
- 132.Rai AK, Rai A, Ramaiya AJ, Jha R, Mallik R. Molecular adaptations allow dynein to generate large collective forces inside cells. Cell. 2013;152:172–182. doi: 10.1016/j.cell.2012.11.044. [DOI] [PubMed] [Google Scholar]
- 133.Schroeder HW, Mitchell C, Shuman H, Holzbaur ELF, Goldman YE. Motor number controls cargo switching at actin-microtubule intersections in vitro. Curr. Biol. 2010;20:687–696. doi: 10.1016/j.cub.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Toba S, Watanabe TM, Yamaguchi-Okimoto L, Toyoshima YY, Higuchi H. Overlapping hand-over-hand mechanism of single molecular motility of cytoplasmic dynein. Proc. Natl Acad. Sci. USA. 2006;103:5741–5745. doi: 10.1073/pnas.0508511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Walter WJ, Koonce MP, Brenner B, Steffen W. Two independent switches regulate cytoplasmic dynein’s processivity and directionality. Proc. Natl Acad. Sci. USA. 2012;109:5289–5293. doi: 10.1073/pnas.1116315109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur ELF. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat. Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- 137.Ananthanarayanan V, et al. Dynein motion switches from diffusive to directed upon cortical anchoring. Cell. 2013;153:1526–1536. doi: 10.1016/j.cell.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 138.Nicastro D, et al. Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E845–E853. doi: 10.1073/pnas.1106178108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Summers KE, Gibbons IR. Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc. Natl Acad. Sci. USA. 1971;68:3092–3096. doi: 10.1073/pnas.68.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nicastro D. Cryo-electron microscope tomography to study axonemal organization. Methods Cell Biol. 2009;91:1–39. doi: 10.1016/S0091-679X(08)91001-3. [DOI] [PubMed] [Google Scholar]
- 141.Hom EF, et al. A unified taxonomy for ciliary dyneins. Cytoskeleton. 2011;68:555–565. doi: 10.1002/cm.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bui KH, Yagi T, Yamamoto R, Kamiya R, Ishikawa T. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J. Cell Biol. 2012;198:913–925. doi: 10.1083/jcb.201201120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kagami O, Kamiya R. Translocation and rotation of microtubules caused by multiple species of Chlamydomonas inner-arm dynein. J. Cell Sci. 1992;103:653–664. [Google Scholar]
- 144.Yagi T, Uematsu K, Liu Z, Kamiya R. Identification of dyneins that localize exclusively to the proximal portion of Chlamydomonas flagella. J. Cell Sci. 2009;122:1306–1314. doi: 10.1242/jcs.045096. [DOI] [PubMed] [Google Scholar]
- 145.Woolley DM. Studies on the eel sperm flagellum. I. The structure of the inner dynein arm complex. J. Cell Sci. 1997;110:85–94. doi: 10.1242/jcs.110.1.85. [DOI] [PubMed] [Google Scholar]
- 146.Zhang X, Wigley D. The ‘glutamate switch’ provides a link between ATPase activity and ligand binding in AAA+ proteins. Nat. Struct. Mol. Biol. 2008;15:1223–1227. doi: 10.1038/nsmb.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 148.Pfister KK, et al. Cytoplasmic dynein nomenclature. J. Cell Biol. 2005;171:411–413. doi: 10.1083/jcb.200508078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mizuno N, et al. Dynein and kinesin share an overlapping microtubule-binding site. EMBO J. 2004;23:2459–2467. doi: 10.1038/sj.emboj.7600240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dynein species and isoform are given on the left. Arrows depict the trans action of the Arg finger, which reaches from one AAA+ (ATPases associated with diverse activities) module into the nucleotide-binding pocket of the preceding module. Conserved = black dot; partially conserved = grey dot; not conserved = white dot.