Abstract
Streptomycetes are very important industrial bacteria, which produce two thirds of all clinically relevant secondary metabolites. Furthermore, they produce large numbers of eukaryotic cell differentiation and apoptosis inducers. Streptomyces is a mycelial soil bacterium characterized by a complex developmental cycle that includes programmed cell death (PCD) phenomena and sporulation in solid cultures. Industrial fermentations are usually performed in liquid cultures, conditions in which Streptomyces strains generally do not sporulate, and it was traditionally assumed that there was no differentiation. Recently, novel aspects concerning differentiation during the presporulation phases were described in solid and liquid cultures, as well as in natural soils. In this review, we analyze the status of knowledge regarding the above-named aspects of Streptomyces differentiation and their relationships with secondary metabolite production.
Keywords: Streptomyces, differentiation, programmed cell death, sporulation, secondary metabolism, antibiotic
1. Introduction
Actinomycetales are gram-positive, mycelium-forming, soil bacteria that play an important role in mineralization processes in nature. They are characterized by a complex development cycle that includes differentiation processes and programmed cell death phenomena (PCD) [1-7]. Streptomyces PCD is a genetically programmed lytic process with some intriguing homologies with eukaryotic apoptosis. Some authors consider that bacteria having complex life cycles (streptomycetes, cyanobacteria, etc.) are the evolutionary origin of some of the protein domains involved in PCD processes, including eukaryotic apoptosis: AP-ATPases (apoptotic ATPases), kinases, caspases, nucleases, etc. As such, these bacteria would constitute a simple model for studying this important phenomenon [8-13]. From a biotechnological point of view, Streptomyces is extremely important, given that two thirds of all industrially manufactured antibiotics are synthesized by members of this genus [14]. Thus, streptomycin is produced by Streptomyces griseus (Reviewed in [15]); kanamycin by S. kanamyceticus [16]; neomycin and phosphomycin by S. fradiae [17], and thienamycin by S. catleya [18], to name just a few examples. In addition, Streptomycetes produce large numbers of eukaryotic cell differentiation inducers, apoptosis inhibitors and inducers [19], and protein C kinase inhibitors with antitumoral activity (staurosporine, etc.) [20, 21].
In summary, the Streptomyces genus is of great socio-economic relevance for two reasons: a) from the point of view of basic research, it has a complex developmental cycle, making it a “multicellular” prokaryotic model that includes PCD phenomena and that might be the evolutionary origin of some of the genes that participate in eukaryotic apoptosis; b) in terms of applications, it produces an ample variety of secondary metabolites of medical and agricultural interest, different antitumoral agents, as well as many cell differentiation effectors of higher cells, such as apoptosis inducers and inhibitors.
2. Streptomyces developmental cycle and secondary metabolite production
2.1. Streptomyces classical developmental model
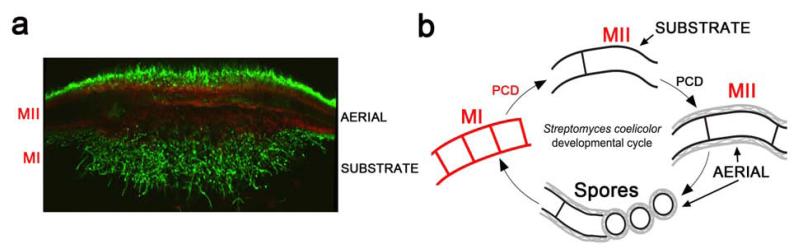
The traditional developmental cycle for Streptomyces confluent cultures on agar surface describes the development of branched filaments consisting of a network of multinucleoid hyphae known as the “substrate mycelium”. After several days, growth of the substrate mycelium produces specialized aerial hyphae, known collectively as the “aerial mycelium”, which extend away from the substrate mycelium into the air. Next, the aerial hyphae undergo massive septation to create a series of uninucleoid compartments. Finally, these compartments differentiate to create spore chains [22]. Both substrate and aerial mycelia are multinucleated (Fig. 1).
Fig. 1. Streptomyces developmental cycle in solid cultures.
(a) Cross section of a 36-hour colony (0.5 cm diameter) of Streptomyces antibioticus ATCC11891 stained with SYTO 9 and propidium iodide and observed under the confocal microscope. (b) Cell-cycle features of Streptomyces development. In red, newly described structures and the proposed nomenclature: MI, first compartmentalized mycelium; MII, second multinucleated mycelium. Classical nomenclature (substrate and aerial mycelium) and hydrophobic layers (in grey) are also indicated. PCD, programmed cell death.
Streptomyces differentiation in liquid cultures has scantly been studied, mainly due to the fact that most Streptomyces strains do not sporulate under these conditions. Despite this, most industrial processes for secondary metabolite production are performed in liquid cultures and it was considered that antibiotics were produced by substrate mycelium at the end of the proliferation phase (Fig. 2). Work carried out in the area of Streptomyces differentiation in submerged cultures has largely focused on the analysis of mycelial morphology, media composition, and bioreactor design [23-25]. Four morphological classes have been distinguished in Streptomyces submerged cultures: “pellets” (compact masses of 950 μm in diameter), “clumps” (less compact masses of 600 μm in diameter), branched hyphae, and non-branched hyphae [26, 27]. Streptomyces pellets have also been proven to develop in the form of a biofilm consisting of sticky extracellular polymers and insoluble substrates [28]. It has been almost unanimously accepted that mycelial morphology is correlated with the production of secondary metabolites, albeit the cause-effect relationship is controversial: some authors hold that cellular aggregation, and hence pellet and clump formation, is fundamental for obtaining good production of secondary metabolites; for instance, retamycin in the case of S. olindensis [29, 30] and nikkomycin from S. tendae [31]. On the other hand, other authors maintain that there is no relationship between morphology and secondary metabolite production, as in the case of virginiamycin production by S. virginiae [32]. In conclusion, there is no general consensus that correlates morphology with production. Therefore, the lack of a reliable developmental model in streptomycetes has hindered the precise identification of reliable phenotypes for use in the analysis and optimization of industrial fermentations.
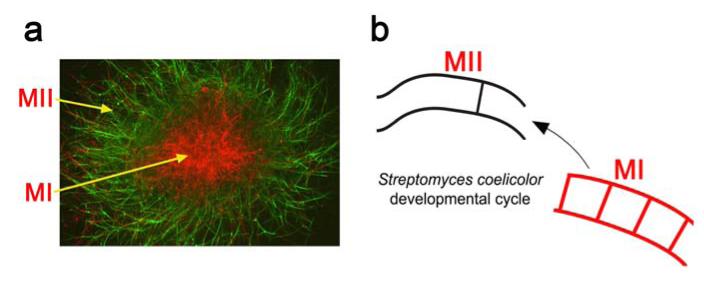
Fig. 2. Streptomyces developmental cycle in liquid cultures.
(a) Mycelial pellet of Streptomyces antibioticus ATCC11891 stained with SYTO 9/propidium iodide and observed under the confocal microscope. (b) Cell-cycle features (see Fig. 1): MI, first compartmentalized mycelium (newly described structure); MII, second multinucleated mycelium (formerly, substrate mycelium).
2.2. A new Streptomyces developmental model
In the recent years, the classical developmental cycle of Streptomyces was refined by describing novel aspects during the presporulation phases in liquid and solid cultures [2, 3, 33, 34]. The existence of a previously unidentified compartmentalized mycelium (MI) that initiates the developmental cycle after spore germination was characterized [6] (Figs. 1 and 2). MI undergoes a highly ordered programmed cell death (PCD) process and the remaining viable segments of this compartmentalized mycelium begin to enlarge in the form of a multinucleated mycelium (MII). In solid cultures, two types of MII have been defined based on the absence (in early development) or presence (in late development) of the hydrophobic layers characteristic of aerial hyphae [6]. The traditionally denominated substrate mycelium corresponds to MII, lacking hydrophobic layers, and the aerial mycelium, to MII coated with these hydrophobic layers (Fig. 1) [6]. The unique mycelial phases present in liquid cultures were MI and MII without hydrophobic layers [33, 35] (Fig. 2). MI is the Streptomyces vegetative mycelium and MII is the Streptomyces differentiated mycelium producing antibiotics and other secondary metabolites [33-35] (see also section 3.2). The new Streptomyces developmental cycle opens a new scenario for studying differentiation [33-35].
MI and the early PCD are new aspects not contemplated in the classical developmental cycle of Streptomyces and are the only phases present in liquid cultures (Fig. 2). The physical nature of the septa delimitating MI compartments was analyzed in detail by using cell-wall and membrane fluorescent stains, as well as electron microscopy [2]. There are two different kinds of septa: a few thick, sporadic straight transversal septa, which correspond to the previously described vegetative mycelium septa [36], and other more numerous, thinner, less-rigid septa, which delimit irregular, rounded compartments conceivably formed by membranes without an associated thick cell wall [2]. The septation process of the Streptomyces presporulation mycelium is not well understood at present. FtsZ, the bacterial tubulin homologue, is the main player in the formation of cytokinetic rings required for the formation of both the widely spaced hyphal cross walls in substrate mycelium and the specialized septa that turn sporogenic aerial hyphae into spores [37]. FtsZ mutants are considered to be syncytium not containing any septa; however, they tolerate surprisingly strong mechanical breakage after which they are capable of growing in the same way as the wild strain (revised in [38]). This strongly suggests the existence of some kind of septa without an associated thick cell wall. Further work will be necessary to define more precisely the nature of these septa and the biomolecular pathways regulating their formation.
2.3. Streptomyces development in nature
Most Streptomyces development studies have been performed using high cell densities, rich culture media, and high temperatures (28-30°C), far removed from natural conditions in soils [39, 40]. Natural growth conditions imply discontinuous growth and limited colony development [41, 42]. To resemble them, works using poor soils inoculated with diluted spore inocula were performed [40]. In these conditions, spore germination was a very slow, non-synchronous process that commenced at about 7 days and lasted for at least 21 days, peaking at around 14 days. The mycelium did not clump into dense pellets and remained in the MI phase for the time period analyzed (1 month) (Fig. 3). PCD and the MII formation were not observed, even after one month of incubation. It is clear that in nature, cell death and sporulation must take place at the end of the long vegetative phase when the imbalance of nutrients will give rise to bacterial differentiation [39, 43], but we were not able to observe it under our experimental conditions. Overall, MI is the predominant phase in nature and it represents the genuine Streptomyces vegetative growth phase (see also section 3.2).
Fig. 3. Cell-cycle features of Streptomyces growing in natural soils.
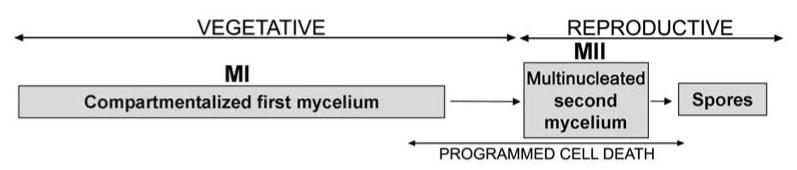
Mycelial structures (MI, first mycelium; MII, second mycelium), vegetative and reproductive phases and PCD are indicated.
3. Biochemical pathways regulating Streptomyces differentiation
3.1. Streptomyces hydrophobic covers formation, septation and sporulation
Regulation of Streptomyces differentiation was classically studied by analyzing mutant strains defective in morphological development. “Bald”, or bld, mutants are blocked earlier in development: they grow as substrate mycelium on rich media and most fail to produce antibiotics [44, 45]. Mutants blocked in the later stages of development are known as “white,” or whi. They erect aerial hyphae that fail to complete metamorphosis into spore chains and hence, do not exhibit the grey colour characteristic of mature spores [46]. Recent findings indicate the existence of an additional regulatory pathway that operates after aerial hyphae begin to grow into the air and is known as the sky pathway. This pathway controls the expression of chaplin and rodlin genes, which encode the proteins that assemble into a rodlet layer that provides surface hydrophobicity to aerial hyphae and spores (Reviewed in [47]) (Figs. 1 and 2). Another well characterized differentiation process is hypha septation during sporulation. In contrast to vegetative crosswalls, multiple sporulation septa are produced simultaneously and in a highly coordinated way within the sporogenic aerial hypha (Reviewed in [48]). A first step in sporulation-specific cell division is the localization of FtsZ at multiple, regularly spaced sites along the aerial hyphal wall [37]. Septa are then simultaneously synthesized in a process in which proteins required for the formation of bacterial divisomes are presumably sequentially directed to the Z ring [49, 50]. The segregation of DNA into the prespore compartments is partially dependent on ParA and ParB [51, 52] and appears to be completed by a process involving septum-located FtsK [53]. After septum closure, the wall of the spore compartments thickens and becomes pigmented, and mature spores are separated by means of a poorly understood process of autolytic cleavage.
3.2. Streptomyces presporulation developmental phases
As described above, the classical Streptomyces developmental cycle focused on sporulation. New issues about presporulation phases of Streptomyces differentiation (spore germination and MI/MII transition) have emerged in recent years, although much remains to be learned. For instance, a new protein (NepA) has been described as a structural cell wall protein involved in the maintenance of spore dormancy in Streptomyces coelicolor [7]. Noens et al., [54] have described a new protein (SsgA) which marks the cell-wall sites where germination takes place. Manteca et al., [34] have performed a quantitative proteomic analysis of Streptomyces coelicolor development demonstrating that MI is enriched in proteins that participate in primary metabolism and therefore, this mycelium is the genuine vegetative phase in Streptomyces. Moreover, they have identified a vast number of regulatory proteins (transcriptional regulators, kinases, etc.) differentially expressed in MI and MII, which conceivably play a role in regulating differentiation. These and other system biology works [55-58] will be the basis upon which to design future experiments that will seek to contribute to elucidating the biochemical pathways regulating the transition from MI (vegetative mycelium) to MII (reproductive mycelium). Research into Streptomyces presporulation developmental phases is a hot topic subject of intense work and will likely provide new and critical knowledge regarding the biology of this important bacterium.
4. Streptomyces programmed cell death
Genetically regulated cell death was described in bacteria from very different taxa, such as Bacillus [16], Anabaena [17], Caulobacter [18], Escherichia coli [16], Streptococcus [19], Staphylococcus [20], or Myxobacteria, [21]. Consideration of bacterial development-associated death as programmed is controversial and depends on how we define programmed cell death (PCD). The best known PCD is that occurring in eukaryotic cells (apoptosis), whose main characteristics are the activation of several regulatory proteins and effectors inside dying cells, and the maintenance of membrane integrity [16]. The eukaryotic alternative to apoptosis would be necrosis, a passive cell death in which cells loss membrane permeability and die [16]. Recent reviews defined bacterial PCD as “any form of cell death mediated by an intracellular death programme”, statement which also includes the eukaryotic process [16]. This definition was created on the basis of the best known bacterial cell death mechanisms: the toxin — antitoxin modules, and cell death occurring during the sporulation of Bacillus. Both processes were autolytic (Reviewed in [16]), and this definition did not include another type of bacterial cell death, allolysis, in which part of the bacterial population induces the lysis in trans of the rest, as it happens for instance in Streptococcus pneumoniae [22]. Allolysis is also a kind of PCD, since it is genetically regulated and it is crucial for Streptococcus development and differentiation [22]. We propose here to define PCD as any type of genetically controlled cell dismantling involving the activation of specific cell death transducers, regulators, and effectors. This definition would include all types of bacterial genetically regulated cell death (autolysis and allolysis), eukaryotic apoptosis and of course, Streptomyces PCD. With few exceptions, the biochemical pathways controlling bacterial PCD and the specific biological role of this process in bacteria are poorly understood (Reviewed in [16]).
Miguelez et al., [59] and Manteca et al., [5] demonstrated that Streptomyces death phenomena occurring during development (see also section 2) present the characteristics of programmed cell death: it is a highly regulated cell suicide in which several degradative enzymes are involved in cellular dismantling (cell-wall, nucleic acids, and protein degradation) [5]. A proteomic analysis revealed that PCD in S. coelicolor was accompanied by the appearance of enzymes involved in the degradation of cellular macromolecules, regulatory proteins, and stress-induced proteins [4]. The increased amounts of several antioxidant proteins suggested oxidative stress as either the cause or consequence of the cell death. Interestingly, regulatory proteins were detected in dying cells and not in the live cells: AAA+ ATPase (SCO1648), whose eukaryotic homologue is one of the enzymes that comprise the eukaryotic proteasome; ClpC1 (SCO3373), the ATP binding subunit of the clp-type proteolytic complex, which is widely distributed in living organisms and plays a key role in cellular homeostasis, particularly under conditions of stress; an inositol-1-phosphate synthase (SCO3899) which participates in the formation of inositol-1-phosphate, an important second messenger in bacteria and eukaryotic cells; several AMPc binding proteins with unknown functions (SCO2368, SCO4277); several transcriptional regulators (SCO5405, SCO1490), etc. [4]. Further work will be needed to characterize the biochemical pathways regulating this interesting process, and, more importantly, to determine their role in Streptomyces biology.
5. Screening for new secondary metabolites in nature
New drugs, especially antibiotics, are urgently required in clinic, and the most propitious natural source remains in environmental bacteria, especially Streptomycetes. The screening for new secondary metabolites was traditionally performed by means of strain isolations, cultures and bioassays [60], a process tremendously productive during the “Golden Age” of antibiotics (1940s-1960s), but which was becoming more and more difficult once the most common antibiotics were already discovered. The main handicap of this screening approach was the existence of several false negatives: i.e., strains that did not reach the production stage (MII) in the lab. At present, most research groups working in the search for new bioactive compounds are making it by means of metagenomics: large genome sequencing and identification of biosynthetic pathways by sequence homology. However, this genetic approach is totally dependent on a previous knowledge in order to find homologies, and it is not suitable to discover totally new bioactive compounds. Finding new secondary metabolites is becoming critical in biomedicine, and the only way will be to resume screening from natural strains. The latest discoveries about Streptomyces differentiation will be crucial to design new experimental approaches to optimize the selection process, and reduce the incidence of false negatives.
6. Conclusions and future perspectives regarding Streptomyces developmental cycle
Streptomyces growth in nature differs substantially from that observed in ordinary laboratory cultures, a fact that must be born in mind when development is analyzed. As stated above, the biological role of MI and the early PCD was obscured by its short lifespan in typical laboratory culture conditions [2, 3]. However, if we consider that MI is the vegetative structure in nature and that the early PCD is an artefact of the stressful conditions found in the laboratory, a new Streptomyces developmental model emerges: after spore germination there is a vegetative phase in the form of a fully compartmentalized mycelium (MI) (Fig. 3); under stress conditions (nutrient/oxygen limitation, etc.) there is activation of a PCD which switches on the differentiation of a multinucleated mycelium (MII) that forms spore chains at the end of the cycle. The MI phase would be the predominant phase in nature and the multinucleated MII would facilitate rapid growth and nucleoid division, crucial for the processes of spore formation. Moreover, as previously mentioned, MII produces antibiotics [33], which would be decisive in helping the bacterium to compete with other microorganims.
Aerial mycelium and sporulation in Streptomyces have been assumed to arise as a result of a kind of cannibalism in which the purpose of PCD is to release nutrients for the aerial mycelium and spore formation [59, 61]. In fact, some cellular components released from dying cells are recycled during aerial mycelium formation and sporulation [61], although there is an excess of nutrients and most of the precursors originated by the lysis and degradation of MI cellular components (amino acids, nucleotides, phospholipids, etc.) are not recycled in the spores [5]. On the other hand, several authors have hypothesized the existence of horizontal gene transmission phenomena in Streptomyces [62-69], albeit a DNA competence status in this bacterium has not yet been described. An interesting alternative function of Streptomyces PCD would be their involvement in a DNA competence status similar to those of other gram positive bacteria such as Streptococcus pneumonia or Bacillus [70, 71]: appropriate DNA fragments would be produced by nucleolytic activities [72] and the lysis of MI mycelium [5] and incorporated by the multinucleated MII followed by recombination and the formation of a huge battery of variable spores. Streptomyces chromosome has been described to be extremely unstable [73], which means that even a single clonal colony has enough genetic variability to generate tremendously diverse spores by means of DNA recombination. The evolutionary advantages of this process in nature are clear and would constitute a process for genetic variability generation in bacteria analogous to sexual reproduction in eukaryotes. Further work will be necessary to support or discard the existence of these processes in Streptomyces.
Streptomyces research has classically focused on the sporulation phases taking place in solid cultures. By contrast, presporulation, and differentiation in liquid cultures have been largely ignored. The new Streptomyces developmental model combined with the use of system biology methodologies (proteomics, transcriptomics, etc.) is generating important data about proteins and genes regulating differentiation. All this information will facilitate future experiments aimed at characterizing the biochemical pathways regulating Streptomyces differentiation and will represent an important step forward in the knowledge about Streptomyces biology from the perspective of basic and applied research. All this information will be the key to analyze and optimize hyphae differentiation in industrial fermentations [35], including the search for new secondary metabolites reassuming the screening processes from natural Streptomyces strains.
ACKNOWLEDGEMENTS
Authors wish to thank the European Research Council for economic support (ERC Starting Grant; Strp-differentiation 280304) and Subdirección General de Proyectos de Investigación (DGI MICINN, Spain, grant BIO2010-16303).
REFERENCES
- 1.Chater KF, Losick R. The mycelial lyfe-style of Streptomyces coerlicolor A(3)2 and its relatives. Oxford University Press; New York: 1997. [Google Scholar]
- 2.Manteca A, Fernandez M, Sanchez J. Microbiology. 2005;151:3689. doi: 10.1099/mic.0.28045-0. [DOI] [PubMed] [Google Scholar]
- 3.Manteca A, Fernandez M, Sanchez J. BMC Microbiol. 2005;5:51. doi: 10.1186/1471-2180-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manteca A, Mader U, Connolly BA, Sanchez J. Proteomics. 2006;6:6008. doi: 10.1002/pmic.200600147. [DOI] [PubMed] [Google Scholar]
- 5.Manteca A, Fernandez M, Sanchez J. Res. Microbiol. 2006;157:143. doi: 10.1016/j.resmic.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Manteca A, Claessen D, Lopez-Iglesias C, Sanchez J. FEMS Microbiol. letters. 2007;274:118. doi: 10.1111/j.1574-6968.2007.00825.x. [DOI] [PubMed] [Google Scholar]
- 7.de Jong W, Manteca A, Sanchez J, Bucca G, Smith CP, Dijkhuizen L, Claessen D, Wösten HA. Mol. Microbiol. 2009;71:1591. doi: 10.1111/j.1365-2958.2009.06633.x. [DOI] [PubMed] [Google Scholar]
- 8.Cal S, Aparicio JF, De los Reyes-Gavilan CG, Nicieza RG, Sanchez J. Biochem. J. 1995;306:93. doi: 10.1042/bj3060093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang CC. Mol. Microbiol. 1996;20:9. doi: 10.1111/j.1365-2958.1996.tb02483.x. [DOI] [PubMed] [Google Scholar]
- 10.Aravind L, Dixit VM, Koonin EV. Trends Biochem. Sci. 1999;24:47. doi: 10.1016/s0968-0004(98)01341-3. [DOI] [PubMed] [Google Scholar]
- 11.Koonin EV, Aravind L. Cell Death Differ. 2002;9:394. doi: 10.1038/sj.cdd.4400991. [DOI] [PubMed] [Google Scholar]
- 12.Petrickova K, Petricek M. Microbiology. 2003;149:1609. doi: 10.1099/mic.0.26275-0. [DOI] [PubMed] [Google Scholar]
- 13.Manteca A, Pelaez AI, Zardoya R, Sanchez J. J. Mol. Evol. 2006;63:719. doi: 10.1007/s00239-005-0130-3. [DOI] [PubMed] [Google Scholar]
- 14.Champness WC. Actinomycete development, antibiotic production and phylogeny: questions and challenges. American Society for Microbiology; Washington, DC: 2000. [Google Scholar]
- 15.Horinouchi S, Beppu T. Antonie Van Leeuwenhoek. 1993;64:177. doi: 10.1007/BF00873026. [DOI] [PubMed] [Google Scholar]
- 16.Takeuchi T, Hikiji T, Nitta K, Yamazaki S, Abe S, Takayama H, Umezawa H. J. Antibiot. 1957;3:107. [PubMed] [Google Scholar]
- 17.Perlman D, Cowan SK. J. Antibiot. 1974;27:637. doi: 10.7164/antibiotics.27.637. [DOI] [PubMed] [Google Scholar]
- 18.Kahan JS, Kahan FM, Goegelman R, Currie SA, Jackson M, Stapley EO, Miller TW, Miller AK, Hendlin D, Mochales S, Hernandez S, Woodruff HB, Birnbaum J. J. Antibiot. 1979;3:1. doi: 10.7164/antibiotics.32.1. [DOI] [PubMed] [Google Scholar]
- 19.Umezawa K. Adv. Enzyme Regul. 1997;37:393. doi: 10.1016/s0065-2571(96)00022-2. [DOI] [PubMed] [Google Scholar]
- 20.Tamaoki T, Nakano H. Biotechnology. 1990;8:732. doi: 10.1038/nbt0890-732. [DOI] [PubMed] [Google Scholar]
- 21.Omura S. Gene. 1992;115:141. doi: 10.1016/0378-1119(92)90552-z. [DOI] [PubMed] [Google Scholar]
- 22.Chater KF. Ann. Rev. Microbiol. 1993;47:685. doi: 10.1146/annurev.mi.47.100193.003345. [DOI] [PubMed] [Google Scholar]
- 23.Mayer AF, Deckwer WD. Appl. Microbiol. Biotechnol. 1996;45:41. doi: 10.1007/s002530050646. [DOI] [PubMed] [Google Scholar]
- 24.Theobald U, Schimana J, Fiedler HP. Antonie Van Leeuwenhoek. 2000;78:307. doi: 10.1023/a:1010282818272. [DOI] [PubMed] [Google Scholar]
- 25.Torres-Bacete J, Arroyo M, Torres-Guzmán R, De La Mata I, Acebal C, Castillon MP. Appl. Biochem. Biotechnol. 2005;126:119. doi: 10.1385/abab:126:2:119. [DOI] [PubMed] [Google Scholar]
- 26.Stocks SM, Thomas CR. Biotechnol. Bioeng. 2001;75:702. doi: 10.1002/bit.10017. [DOI] [PubMed] [Google Scholar]
- 27.Pamboukian CRD, Guimaraes LM, Candida M. Braz. J. Microbiol. 2002;33:17. [Google Scholar]
- 28.Kim YM, Kim JH. J. Microbiol. 2004;42:64. [PubMed] [Google Scholar]
- 29.Hobbs G, Frazer CM, Gardner DCJ, Cullum JA, Oliver SG. App. Microbiol. Biotechnol. 1989;31:272. [Google Scholar]
- 30.Pamboukian CRD, Candida M. Process. Biochemistry. 2004;39:2249. [Google Scholar]
- 31.Vecht-Lifshitz SE, Sasson Y, Braun S. J. Appl. Bacteriol. 1992;72:195. doi: 10.1111/j.1365-2672.1992.tb01823.x. [DOI] [PubMed] [Google Scholar]
- 32.Yang YK, Morikawa M, Shimizu H, Shioya S, Suga KI, Nihira T, Yamada Y. J. Ferm. Bioengineering. 1996;81:7. doi: 10.1002/(SICI)1097-0290(19960220)49:4<437::AID-BIT11>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 33.Manteca A, Alvarez R, Salazar N, Yagüe P, Sanchez J. Appl. Environ. Microbiol. 2008;74:3877. doi: 10.1128/AEM.02715-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manteca A, Sanchez J, Jung HR, Schwämmle V, Jensen ON. Mol. Cell. Proteomics. 2010;9:1423. doi: 10.1074/mcp.M900449-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yagüe P, Manteca A, Simon A, Diaz-Garcia ME, Sanchez J. Appl. Environ. Microbiol. 2010;76:3401. doi: 10.1128/AEM.00120-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeJong PJ, McCoy E. Can. J. Microbiol. 1966;12:985. doi: 10.1139/m66-133. [DOI] [PubMed] [Google Scholar]
- 37.Grantcharova N, Lustig U, Flärdh K. J. Bacteriol. 2005;187:3227. doi: 10.1128/JB.187.9.3227-3237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCormick JR. Curr. Opin. Microbiol. 2009;12:689. doi: 10.1016/j.mib.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Wellington EM, Cresswell N, Saunders VA. Appl. Environ. Microbiol. 1990;56:1413. doi: 10.1128/aem.56.5.1413-1419.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manteca A, Sanchez J. Appl. Environ. Microbiol. 2009;75:2920. doi: 10.1128/AEM.02288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams ST. Streptomycetes in the soil ecosystem. Fischer Verlag; New York: 1978. [Google Scholar]
- 42.Williams ST. Oligotrophy in soil: fact or fiction? Academic Press, Inc; 1985. [Google Scholar]
- 43.Anukool U, Gaze WH, Wellington EM. Appl. Environ. Microbiol. 2004;70:5222. doi: 10.1128/AEM.70.9.5222-5228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merrick MJ. Gen. Microbiol. 1976;96:299. doi: 10.1099/00221287-96-2-299. [DOI] [PubMed] [Google Scholar]
- 45.Champness WC. J. Bacteriol. 1988;170:1168. doi: 10.1128/jb.170.3.1168-1174.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chater KF. J. Gen. Microbiol. 1972;72:9. doi: 10.1099/00221287-72-1-9. [DOI] [PubMed] [Google Scholar]
- 47.Claessen D, de Jong W, Dijkhuizen L, Wösten HA. Trends Microbiol. 2006;14:313. doi: 10.1016/j.tim.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 48.Flardh K, Buttner MJ. Nat. Rev. Microbiol. 2009;7:36. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 49.Bramhill D. Annu. Rev. Cell. Dev. Biol. 1997;13:395. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- 50.Errington J, Daniel RA, Scheffers DJ. Microbiol. Mol. Biol. Rev. 2003;67:52. doi: 10.1128/MMBR.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jakimowicz D, Chater K, Zakrzewska-Czerwinska J. Mol. Microbiol. 2002;45:1365. doi: 10.1046/j.1365-2958.2002.03102.x. [DOI] [PubMed] [Google Scholar]
- 52.Jakimowicz D, Gust B, Zakrzewska-Czerwinska J, Chater KF. J. Bacteriol. 2005;187:3572. doi: 10.1128/JB.187.10.3572-3580.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L, Yu Y, He X, Zhou X, Deng Z, Chater KF, Tao M. J. Bacteriol. 2007;189:2310. doi: 10.1128/JB.01660-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noens EE, Mersinias V, Willemse J, Traag BA, Laing E, Chater KF, Smith CP, Koerten HK, van Wezel GP. Mol. Microbiol. 2007;64:1244. doi: 10.1111/j.1365-2958.2007.05732.x. [DOI] [PubMed] [Google Scholar]
- 55.Manteca A, Ye J, Sanchez J, Jensen ON. J. Proteome Res. 2011;10:5481. doi: 10.1021/pr200762y. [DOI] [PubMed] [Google Scholar]
- 56.Nieselt K, Battke F, Herbig A, Bruheim P, Wentzel A, Jakobsen ØM, Sletta H, Alam MT, Merlo ME, Moore J, Omara WA, Morrissey ER, Juarez-Hermosillo MA, Rodríguez-García A, Nentwich M, Thomas L, Iqbal M, Legaie R, Gaze WH, Challis GL, Jansen RC, Dijkhuizen L, Rand DA, Wild DL, Bonin M, Reuther J, Wohlleben W, Smith MC, Burroughs NJ, Martín JF, Hodgson DA, Takano E, Breitling R, Ellingsen TE, Wellington EM. BMC Genomics. 2010;6:11. [Google Scholar]
- 57.Jayapal KP, Philp RJ, Kok YJ, Yap MG, Sherman DH, Griffin TJ, Hu WS. PLoS One. 2008;3:2097. doi: 10.1371/journal.pone.0002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gubbens J, Janus M, Florea BI, Overkleeft HS, van Wezel GP. Mol. Microbiol. 2012;86:1490. doi: 10.1111/mmi.12072. [DOI] [PubMed] [Google Scholar]
- 59.Miguelez EM, Hardisson C, Manzanal MB. J. Cell. Biol. 1999;145:515. doi: 10.1083/jcb.145.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Donadio S, Maffioli S, Monciardini P, Sosio M, Jabes D. J. Antibiot. 2010;63:423. doi: 10.1038/ja.2010.62. [DOI] [PubMed] [Google Scholar]
- 61.Mendez C, Braña AF, Manzanal MB, Hardisson C. Can. J. Microbiol. 1985;31:446. doi: 10.1139/m85-083. [DOI] [PubMed] [Google Scholar]
- 62.Wiener P, Egan S, Huddleston AS, Wellington EM. Mol. Ecol. 1998;7:1205. doi: 10.1046/j.1365-294x.1998.00450.x. [DOI] [PubMed] [Google Scholar]
- 63.Ueda K, Seki T, Kudo T, Yoshida T, Kataoka M. J. Bacteriol. 1999;181:78. doi: 10.1128/jb.181.1.78-82.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Egan S, Wiener P, Kallifidas D, Wellington EM. Antonie Van Leeuwenhoek. 2001;79:127. doi: 10.1023/a:1010296220929. [DOI] [PubMed] [Google Scholar]
- 65.Metsä-Ketelä M, Halo L, Munukka E, Hakala J, Mäntsälä P, Ylihonko K. Appl. Environ. Microbiol. 2002;68:4472. doi: 10.1128/AEM.68.9.4472-4479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garcia-Vallve S, Guzman E, Montero MA, Romeu A. Nucleic Acids Res. 2003;31:187. doi: 10.1093/nar/gkg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nishio Y, Nakamura Y, Usuda Y, Sugimoto S, Matsui K, Kawarabayasi Y, Kikuchi H, Gojobori T, Ikeo K. Mol. Biol. Evol. 2004;21:1683. doi: 10.1093/molbev/msh175. [DOI] [PubMed] [Google Scholar]
- 68.Kawase T, Saito A, Sato T, Kanai R, Fujii T, Nikaidou N, Miyashita K, Watanabe T. Appl. Environ. Microbiol. 2004;70:1135. doi: 10.1128/AEM.70.2.1135-1144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Manteca A, Kamphausen T, Fanghanel J, Fischer G, Sanchez J. FEBS Lett. 2004;572:19. doi: 10.1016/j.febslet.2004.06.091. [DOI] [PubMed] [Google Scholar]
- 70.Guiral S, Mitchell TJ, Martin B, Claverys JP. Proc. Natl. Acad. Sci. USA. 2005;102:8710. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. PLoS Genet. 2006;2:135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nicieza GR, Huergo J, Connolly BA, Sanchez J. J. Biol. Chem. 1999;274:20366. doi: 10.1074/jbc.274.29.20366. [DOI] [PubMed] [Google Scholar]
- 73.Dary A, Martin P, Wenner T, Leblond P, Decaris B. Res. Microbiol. 1999;150:439. doi: 10.1016/s0923-2508(99)00113-8. [DOI] [PubMed] [Google Scholar]