Abstract
The development of new technologies to isolate and identify microbial genomes has markedly increased our understanding of the role of microbiomes in health and disease. The idea, first proposed as part of the hygiene hypothesis, that environmental microbes influence the developmental trajectories of the immune system in early life, has now been considerably extended and refined. The abundant microbiota present in mucosal surfaces, especially the gut, is actively selected by the host through complex receptor systems that respond differentially depending on the molecular patterns presented to mucosal cells. Germ-free mice are more likely to develop allergic airway inflammation and show alterations in normal motor control and anxiety. These effects can be reversed by neonatal microbial recolonization but remain unchanged if recolonization occurs in adults. What emerges from these recent studies is the discovery of a complex, major early environmental determinant of lifetime human phenotypes. To change the natural course of asthma, obesity, and other chronic inflammatory conditions, active manipulation of the extensive bacterial, phage, and fungal metagenomes present in mucosal surfaces may be required, specifically during the developing years. Domesticating the human microbiome and adapting it to our health needs may be a challenge akin to, but far more complex than, the one faced by humanity when a few dozen species of plants and animals were domesticated during the transition between hunter-gatherer and sedentary societies after the end of the Pleistocene era.
In 1903, in a far-reaching book entitled The Nature of Man, Élie Metchnikoff, one of Pasteur’s most distinguished disciples, defined what would be the slant of most scientists and of the lay public toward our guest microbes for the next 9 decades (1). He noted that the “human intestine contains an enormous quantity of bacteria, which…increase at the rate of 128,000,000,000,000 each day.” This flora, he rightfully asserted, “is very varied, and contains an immense number of different species, among which are bacilli, cocci, and many kinds of other bacteria, about which little is known.” But he also argued that “it contributes nothing to the well-being of man…The useless bacterial flora may give rise to serious or fatal maladies. Wounds of the abdomen are really serious only when they penetrate the large intestines and so allow the entrance of bacteria from that region to the peritoneal cavity. In such an event, the microbes rapidly multiply in the organism and produce a grave and frequently mortal illness.” He reasoned that the large intestine in mammals had developed because, “by storing the products of digestion, it allowed them to run long distances without stopping, and so was an advantage in the struggle for existence,” an unnecessary function for modern humans. Best would be to get rid of the large intestine and its woeful inhabitants, which were to be blamed even for the aging process, but he lamented that “in spite of the progress of surgery, I do not expect to find in our time that the large intestine will be removed by operation.”
This utterly antagonistic approach toward microbes dominated human medical sciences during the most of the twentieth century, and it was not wholly unjustified. Metchnikoff and his mentor and colleagues were pioneers in the discovery that pathogenic bacteria and viruses caused the great majority of premature deaths, including those of more than 10% of infants in the first year of life. This discovery was a major accomplishment in human history, and the beneficial role of antibiotics and hygiene in reversing this tragic heritage cannot be overstated. It is thus not surprising that most human microbiology since Pasteur and Koch has been the study of disease-causing bacteria and other microorganisms. Metchnikoff himself believed that bacteria contained in kefir could have positive health effects, but only because they could inhibit the growth of “bacteria of putrefaction.” He also knew of the prescient studies performed by several of his most accomplished colleagues showing that animals raised in aseptic conditions from birth failed to thrive and rapidly died, but he overlooked their importance and dismissed them as methodologically flawed.
As often occurs in the history of science, a major change in the attitude of mainstream life scientists toward the role of the microbes in human biology only occurred when a series of seminal and rather unexpected observations regarding the epidemiology of asthma and allergies was explained by way of a radically new paradigm, the so-called hygiene hypothesis (2, 3). It is my contention that, because of its role in modifying the conceptual framework on which most microbiology was built, the hygiene hypothesis opened the way to a new understanding of the role of microbes in health and disease.
The Hygiene Hypothesis: Environmental Microbes as Protectors
In 1989, Strachan (4) noticed an inverse relation between hay fever and number of older siblings in the household and proposed that allergies could be prevented by early life infections. Subsequent studies of the relation of asthma and allergies and early life infection provided contradictory results, and this seemed to disprove the hypothesis (5). However, several new lines of evidence emerged suggesting that, more than any single infection or infections, what could be more important in determining protection against asthma and allergies was the global “microbial burden” to which the child was exposed in early life (6). Studies in rural communities in German-speaking Europe consistently showed lower prevalence of these conditions among children living on farms, and protection was strongly associated with increased diversity of microbial communities present in dust obtained from the homes of these children as compared with those of children of nonfarmers (7). Similarly, exposure to pets and enrollment in day care in early life both appeared to have a protective effect against asthma and allergies (8, 9). As was true for farming/nonfarming comparisons, significantly different microbial communities were identified in dust obtained from homes with pets or with young children enrolled in day care as compared with those without pets or day care enrollees, respectively (10).
These studies suggested for the first time that different sets of microbial communities present in the environments of infants and young children could influence the developmental trajectories of the immune system and perhaps the lungs and airways and, by this mechanism, could influence the risk for chronic diseases such as asthma and allergies. In support of this contention, genetic variations in innate immunity genes such as CD14 and toll-like receptor 2 (TLR2), which are known to encode for pattern recognition receptors directly involved in responses to microbial products, were found to modulate the protective effects of both farming environments (11) and exposure to day care (12). Moreover, animal models confirmed that exposure to bacterial products and bacteria isolated from farms could indeed inhibit allergic airway inflammation (13).
Although these studies indicated a potential role of environmental microbes in modulating immune responses associated with asthma and allergies, they did not establish a connection between these exposures and the microbial communities in the gut and respiratory tract.
Mucosal Microbiome in Health and Disease
More recently, marked differences were observed in the microbial communities present in samples obtained from airway brushings and bronchoalveolar lavage (14, 15) or sputum (16) of subjects with asthma as compared with those of subjects without asthma. Other reports suggested that pathogenic bacteria were more likely to be present in infants who would subsequently develop asthma symptoms as toddlers than in those who would not (17). These findings emerged together with those of several other studies showing alterations in gut microbial communities in subjects affected by other chronic conditions, such as eczema, diabetes, and obesity, as compared with unaffected individuals (18–20). What remained unclear were the causal pathways and connections involved in these associations and, specifically, the role of the human microbiome in mediating the effects of environmental microbial communities on human health. In one scenario, environmental microbes (i.e., those present in the external surroundings) could directly influence developmental immune pathways, which in turn determine disease risk or protection and, concomitantly, make mucosal surfaces more or less permissive for the subsistence of certain microbial communities. In this scenario, it is still possible for these communities to play a role in determining severity of the underlying disease, but the commensal microbiome is more a passive spectator or secondary byproduct than a causal factor. In a second scenario, colonization of mucosal surfaces by certain microbial communities present in the environment in susceptible individuals is the primal event. These local communities, in turn, establish a long-term, active interaction with the mucosal immune system.
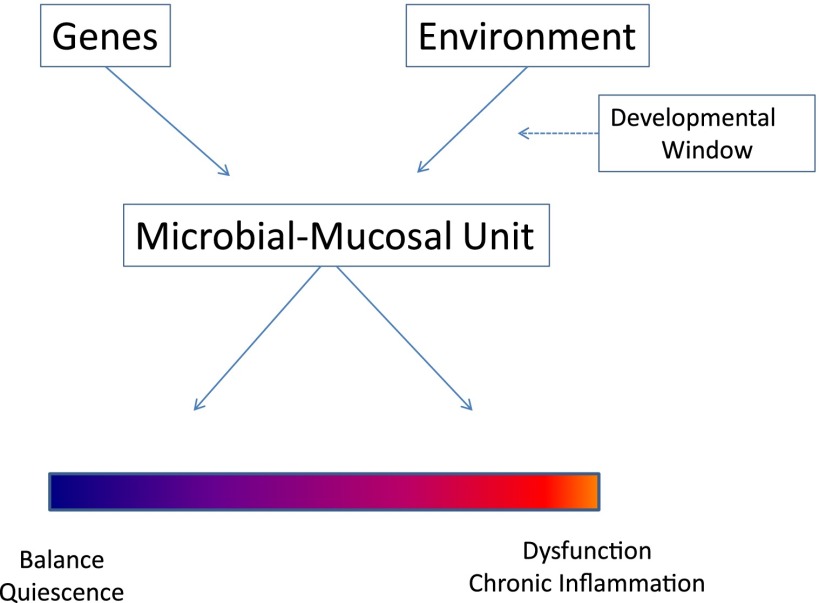
Recent studies have convincingly shown that what is really at play is the development, probably early in life, of a complex, functional interaction between microbes and mucosa and that at least some components of the microbiome are not passive inhabitants of mucosal surfaces but active participants in host biology. Round and colleagues (21) reported that Bacteroides fragilis, a regular component of the gut microbiome, has a symbiosis factor, polysaccharide A (PSA), which signals on FoxP3+ T-regulatory cells through TLR2 to induce immunologic tolerance. T-regulatory cells suppress IL-17A responses in the mucosa, thus allowing for B. fragilis to occupy specific niches within the intestinal mucosal crypts, an actively permissive process that gives rise to a microbial population that is in intimate contact with the mucosal immune system. Animals colonized with B. fragilis lacking PSA displayed profoundly reduced numbers of tissue-associated (but not lumen-associated) bacteria when compared with animals colonized with wild-type B. fragilis. These studies thus suggested that the mucosal immune system of the gut recognizes and tolerates commensal bacteria using the same TLR2 receptor system with which it can also activate inflammatory responses against pathogens. This new concept blurs the distinction between commensalism and parasitism as an intrinsic characteristic of any single microbe; commensals can be treated as pathogens if they do not carry what has been dubbed commensal-associated molecular patterns (CAMPs), such as PSA, as opposed to pathogen associated molecular patterns (PAMPs), which do activate inflammatory responses (22). It is thus the CAMP–PAMP–mucosal receptor system interplay, modulated by the genetic background of the individual and the microbiota, which in final analysis determines if certain bacteria are tolerated or repelled. The final result of this process (Figure 1) is the establishment of a microbial-mucosal unit, which could give rise, at one end of the continuum, to a balanced interface and immunological quiescence and, at the other end, to an interface permissive of aberrant microbiota, which would allow the activation of the heterogeneous inflammatory responses that are characteristic of asthma and other chronic mucosal diseases.
Figure 1.
Schematic of the interaction between genetic background and exposure to environmental microbiota as determinant of the mucosal responses–resident microbiome status. The final result is the establishment of a microbial-mucosal unit, whose “set-point” may range from a balanced, quiescent state to predominance of inflammatory signals and an aberrant local microbiome.
Critical Role of Early Life Exposure to Commensal Microbiome
Timing of exposure to environmental microbiota is likely to play a critical role in determining both lung and also more global health outcomes. Olszak and colleagues (23) showed increased airway resistance and increased inflammatory responses in germ-free (GF) mice sensitized and challenged with ovalbumin, as compared with specific pathogen-free (SPF) mice. When GF mice were recolonized with a conventional microbiota starting at birth, responses to ovalbumin were very similar to those observed in SPF mice. However, airway responses in mice that were recolonized in adult life were not different from those observed in GF mice. These same GF mice also showed greater intestinal inflammation and lethality than SPF mice in a model of ulcerative colitis (23); once again, enhanced responses could be reversed when recolonization occurred in the neonatal period but not in adult life. A limitation of these studies is the highly artificial nature of experiments performed in GF mice, which are not representative of the microbiome, dysfunctional as it may be, that is still present in asthma and ulcerative colitis. These limitations notwithstanding, the results support the contention, first proposed in the framework of the hygiene hypothesis, that microbial exposure in early life can modify immune developmental trajectories, which in turn can influence the risk for asthma and other chronic inflammatory conditions.
Interestingly, experiments performed in GF mice have also suggested that early microbial exposures can affect not only the mucosal response systems that the microbiota come directly in contact with but also distant organs with no mucosal surfaces. Diaz Heijtz and colleagues (24) showed that GF mice exhibited increased motor activity and reduced anxiety when compared with SPF mice. They reported that GF mice showed altered expression profiles of four canonical signaling pathways, neurotransmitter turnover, and synaptic-related proteins. When GF mice were exposed to normal gut microbiota early in life, both behavior and gene expression profiles reverted to those of SPF mice, whereas colonization in adult life yielded similar results to those observed for SPF mice. These results corroborate the conclusion proposed earlier that the microbiome may have profound effects on developmental pathways of both local and distant organs (25, 26). The mechanisms through which the microbiota can determine these long-range effects remain speculative.
Not All Microbiota Are Protective
In most of the previous descriptions, the accent has been placed on the potential favorable role of the microbiota in determining health outcomes. Evidence is emerging, however, suggesting the microbiome may also play a role in predisposing for human disease. Urinary and circulating levels of a proatherosclerotic metabolite of dietary phosphatidylcholine and l-carnitine, trimethylamine-N-oxide (TMAO), are strongly associated with the risk for subsequent major adverse cardiovascular events in humans (27). Koeth and colleagues (28) showed that, in mice, chronic dietary supplementation with l-carnitine altered cecal microbial composition, markedly enhanced synthesis of TMAO, and increased atherosclerosis, but this did not occur if the intestinal microbiota was concurrently suppressed. Similarly in humans, Tang and colleagues (27) reported that an oral phosphatidylcholine challenge significantly increased plasma TMAO levels, but these effects were blocked by the previous administration of an antibiotic, which presumably suppressed gut microbiota. Thus, it appears that the gut microbiota plays a critical role in the synthesis of TMAO, which, in turn, increases the risk of atherosclerosis and adverse cardiovascular outcomes.
The Microbiome: Potential Therapeutic Approaches
The evidence outlined in the succinct discussion above clearly suggests that a new level of intricacy needs to be added to the study of the pathogenesis of complex diseases. The environmental microbiota that humans are exposed to, especially during early life, may exert strong influence on long-term health and disease outcomes of many organs and systems. It seems clear that this influence does not depend on a single set or even a few microbial communities but is the result of interactions between the genetic background of the host, the timing of the initial contact, and the distribution of the different components of the microbiota to which the host is exposed. It also seems plausible to surmise that extended and successful strategies to increase hygiene and treat serious acute bacterial infections may have drastically decreased human morbidity and mortality, especially in infancy, but may also have radically changed the microbiota to which humans are exposed and the guest microbiomes of our mucosal surfaces. It is impossible to envision a situation in which these putative changes could be reversed and humans return to the spontaneous ecological balances of centuries past, which were the result of long-term evolutionary coadaptations between microbiota and hosts. It could certainly be possible to encourage parents to subject their children to more social interactions during the early years, such as those occurring in day care centers, or to foster increased pet ownership in households with young children, but these types of strategies, even if eventually proven to be efficacious, face serious feasibility obstacles. They may not be beneficial or may even be harmful for subgroups of young children who may be susceptible to infections or who may be genetically unresponsive to an increase microbial burden. The opposite strategy, that is, to further decrease exposure to unfavorable elements in the microbiome, as is implicit in the antibiotic trial attempted by Tang and colleagues (27) to block TMAO synthesis, could decrease cardiovascular risk but could also, and even more radically, alter the favorable commensal microbiome. In other words: crude, blind attempts to reestablish the lost balance are unlikely to be successful and could result in more harm than benefit.
Two alternative strategies that could attempt to reproduce therapeutically the overall effects of the microbiome on human health seem theoretically possible. First, it is conceivable that a better understanding of the specific microbial components associated with positive or negative health outcomes may allow us to synthesize pharmaceutical products that reproduce or block the effects of the microbiota, much like streptomycin was derived from the actinobacterium Streptomyces griseus. Interestingly, extracts from different combinations of killed pathogenic bacteria have been used as oral naturopathic products to prevent infections in Europe and elsewhere (29). Recently, these products have been tested in animal models of airway inflammation with encouraging results (30, 31). Their mechanism of action appears to be loosely analogous to that described for B. fragilis PSA: these products interact with dendritic cells in the gut mucosa, which in turn induced an increase in the frequency and number of FoxP3+ T-regulatory cells in the intestinal lamina propria, the airway mucosa, and to a smaller extent in the vagina, but not in secondary lymphoid organs (30). Whether these same products can prevent the development of asthma in young children is still unknown (32, 33). A different approach could be administration of live microorganisms, as has been attempted with probiotic and synbiotic preparations containing a small number of allegedly beneficial bacteria. Although these preparations are currently widely used, their benefit for allergic diseases (34) or for infant health in general (35) has not been clearly established. Moreover, a recent, large, placebo-controlled, randomized trial of the use of lactobacilli and bifidobacteria in the prevention of antibiotic-associated diarrhea due to Clostridium difficile showed no benefit (36). It is thus unlikely that empirically manufactured products derived from a few putatively beneficial microorganisms could act as surrogates for the multiplicity of favorable responses (or could reverse the negative outcomes) that appear to be associated with the human microbiome. Moreover, stimulation of certain immune pathways that have favorable effects on certain outcomes may have unfavorable pleiotropic effects on others.
Taming the Microbiome: The Second Big Domestication
A second approach to the potential use of the microbiota for therapeutic purposes could entail the development of a comprehensive understanding of the different components, favorable or unfavorable, of the human microbiome, with the final goal of exposing very young children to cocktails of microbiota that foster developmental pathways associated with health and block those associated with disease. This process of controlling the microbiota already started with unspecific strategies, such as the availability of clean water supplies, refrigeration, and cooking, but would now require actively raising and administering the appropriate microbial mixture during the correct developmental window. It is intriguing to acknowledge how similar, but at the same time how much more daunting, this challenge would be to the one humanity faced in the transition between hunter-gatherer and agricultural societies (reviewed by Diamond [37]). The transition from controlling predators and passively collecting food (which, in this scenario, I see analogous to providing clean water and refrigerating food) to domestication of animals and plants occurred during a relatively short period of time starting around 8500 B.C. Why it happened then, and not before or after, is the matter of speculation, but available data suggest that domestication was humanity’s active response to a dramatic change in its circumstances. Confronted with marked population growth, climate changes, dwindling wild species, and unpredictable variations in food supplies, humans had no other alternative to survive but to tame and actively control a new source of food. Similarly, we have lived as the behaviorally modern Homo sapiens for the last 50,000 to 80,000 years, but the need to not only control microorganisms but also actively control our microbial environment has only emerged in the last 100 years. The initial adversarial approach toward microbes is now being slowly replaced by an understanding that the symbiotic relation that humans evolved to develop with our microbiota has been disrupted by our own successes in controlling pathogens. Instead of providing ourselves with new sources of food, humans need now to artificially colonize our mucosa with beneficial microbiota. However, there is a major difference between the challenges we face today and those faced by humans after the end of the Pleistocene era, especially in Eurasia. Domestication of only a few hundred species of plants and no more than a handful of animals was enough to provide sufficient food to support the transition to sedentary societies. The study of specific cases of domestication provides important clues as to what the process, undertaken by Holocene humans without any knowledge of the mechanisms involved, entailed. Diamond (37) points out that oak trees, for example, are the most important wild food plant in Eurasia and North America, but were never domesticated. As oaks, almonds, which are a major source of food today, carry bitter poisons in the wild state, but determination of bitterness is polygenic in oak and monogenic dominant in almonds. As expected, controlling a polygenic trait has proven vastly more difficult than simply amplifying carriers of a dominant mutation; oak trees remain undomesticated to this day. In this framework, domesticating the microbiome may mean taming a polygenic community that is several dimensions larger than that of any single animal or plant species. If we add to that the polygenic nature of human responses to environmental stimuli and the potential role of the phageome as a genetic reservoir for bacterial adaptation (38), the challenge to adapt cocktails of human microbiota to the competing needs of different biological response systems appears daunting to say the least.
Conclusions
The whole range of interactions that microbiomes develop with their human hosts is in no way exclusive to or most advanced in our species compared with other animals and plants. The fact that essential metabolic and developmental human functions and disorders, including alterations in mood and other superior brain functions, may be potentially influenced by the guests we carry in our mucosae underlies the importance of better understanding of our interactions with our microbiota. Eventually, manipulation of the microbiota and its products may offer a challenging but fruitful new avenue for the prevention and treatment of human disease.
Footnotes
Funded by National Heart, Lung, and Blood Institute grant HL-56177.
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Metchnikoff É. New York: Putman; 1903. The nature of man, studies in optimistic philosophy; p. 251. [Google Scholar]
- 2.Martinez FD. The coming-of-age of the hygiene hypothesis. Respir Res. 2001;2:129–132. doi: 10.1186/rr48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strachan DP. Family size, infection and atopy: the first decade of the “hygiene hypothesis”. Thorax. 2000;55:S2–S10. doi: 10.1136/thorax.55.suppl_1.s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douwes J, Pearce N. Commentary: the end of the hygiene hypothesis? Int J Epidemiol. 2008;37:570–572. doi: 10.1093/ije/dyn077. [DOI] [PubMed] [Google Scholar]
- 6.Martinez FD, Holt PG. Role of microbial burden in aetiology of allergy and asthma. Lancet. 1999;354:SII12–SII15. doi: 10.1016/s0140-6736(99)90437-3. [DOI] [PubMed] [Google Scholar]
- 7.Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrländer C, Heederik D, Piarroux R, von Mutius E GABRIELA Transregio 22 Study Group. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364:701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 8.Ownby DR, Johnson CC, Peterson EL. Exposure to dogs and cats in the first year of life and risk of allergic sensitization at 6 to 7 years of age. JAMA. 2002;288:963–972. doi: 10.1001/jama.288.8.963. [DOI] [PubMed] [Google Scholar]
- 9.Remes ST, Castro-Rodriguez JA, Holberg CJ, Martinez FD, Wright AL. Dog exposure in infancy decreases the subsequent risk of frequent wheeze but not of atopy. J Allergy Clin Immunol. 2001;108:509–515. doi: 10.1067/mai.2001.117797. [DOI] [PubMed] [Google Scholar]
- 10.Maier RM, Palmer MW, Andersen GL, Halonen MJ, Josephson KC, Maier RS, Martinez FD, Neilson JW, Stern DA, Vercelli D, et al. Environmental determinants of and impact on childhood asthma by the bacterial community in household dust. Appl Environ Microbiol. 2010;76:2663–2667. doi: 10.1128/AEM.01665-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eder W, Klimecki W, Yu L, von Mutius E, Riedler J, Braun-Fahrländer C, Nowak D, Martinez FD ALEX Study Team. Toll-like receptor 2 as a major gene for asthma in children of European farmers. J Allergy Clin Immunol. 2004;113:482–488. doi: 10.1016/j.jaci.2003.12.374. [DOI] [PubMed] [Google Scholar]
- 12.Custovic A, Rothers J, Stern D, Simpson A, Woodcock A, Wright AL, Nicolaou NC, Hankinson J, Halonen M, Martinez FD.Effect of day care attendance on sensitization and atopic wheezing differs by Toll-like receptor 2 genotype in 2 population-based birth cohort studies J Allergy Clin Immunol 2011127390–397.e1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Debarry J, Garn H, Hanuszkiewicz A, Dickgreber N, Blümer N, von Mutius E, Bufe A, Gatermann S, Renz H, Holst O, et al. Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. J Allergy Clin Immunol. 2007;119:1514–1521. doi: 10.1016/j.jaci.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 14.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, et al. Disordered microbial communities in asthmatic airways. PLoS ONE. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, Woyke T, Allgaier M, Bristow J, Wiener-Kronish JP, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma J Allergy Clin Immunol 2011127372–381.e1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD.Asthma-associated differences in microbial composition of induced sputum J Allergy Clin Immunol 2013131346–352.e1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bønnelykke K, Brasholt M, Heltberg A, Vissing NH, Thorsen SV, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. 2007;357:1487–1495. doi: 10.1056/NEJMoa052632. [DOI] [PubMed] [Google Scholar]
- 18.Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 19.Thompson AL. Developmental origins of obesity: early feeding environments, infant growth, and the intestinal microbiome. Am J Hum Biol. 2012;24:350–360. doi: 10.1002/ajhb.22254. [DOI] [PubMed] [Google Scholar]
- 20.Wang M, Karlsson C, Olsson C, Adlerberth I, Wold AE, Strachan DP, Martricardi PM, Aberg N, Perkin MR, Tripodi S, et al. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol. 2008;121:129–134. doi: 10.1016/j.jaci.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirsch AM. Plant-microbe symbioses: a continuum from commensalism to parasitism. Symbiosis. 2004;37:345–363. [Google Scholar]
- 23.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10:735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- 26.Sommer F, Bäckhed F. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 27.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Benedetto F, Sevieri G. Prevention of respiratory tract infections with bacterial lysate OM-85 bronchomunal in children and adults: a state of the art. Multidiscip Respir Med. 2013;8:33. doi: 10.1186/2049-6958-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navarro S, Cossalter G, Chiavaroli C, Kanda A, Fleury S, Lazzari A, Cazareth J, Sparwasser T, Dombrowicz D, Glaichenhaus N, et al. The oral administration of bacterial extracts prevents asthma via the recruitment of regulatory T cells to the airways. Mucosal Immunol. 2011;4:53–65. doi: 10.1038/mi.2010.51. [DOI] [PubMed] [Google Scholar]
- 31.Strickland DH, Judd S, Thomas JA, Larcombe AN, Sly PD, Holt PG. Boosting airway T-regulatory cells by gastrointestinal stimulation as a strategy for asthma control. Mucosal Immunol. 2011;4:43–52. doi: 10.1038/mi.2010.43. [DOI] [PubMed] [Google Scholar]
- 32.Razi CH, Harmancı K, Abacı A, Özdemir O, Hızlı S, Renda R, Keskin F. The immunostimulant OM-85 BV prevents wheezing attacks in preschool children. J Allergy Clin Immunol. 2010;126:763–769. doi: 10.1016/j.jaci.2010.07.038. [DOI] [PubMed] [Google Scholar]
- 33.Martinez FD. New insights into the natural history of asthma: primary prevention on the horizon. J Allergy Clin Immunol. 2011;128:939–945. doi: 10.1016/j.jaci.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osborn DA, Sinn JK. Probiotics in infants for prevention of allergic disease and food hypersensitivity. Cochrane Database Syst Rev. 2007;(4):CD006475. doi: 10.1002/14651858.CD006475.pub2. [DOI] [PubMed] [Google Scholar]
- 35.Mugambi MN, Musekiwa A, Lombard M, Young T, Blaauw R. Synbiotics, probiotics or prebiotics in infant formula for full term infants: a systematic review. Nutr J. 2012;11:81. doi: 10.1186/1475-2891-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen SJ, Wareham K, Wang D, Bradley C, Hutchings H, Harris W, Dhar A, Brown H, Foden A, Gravenor MB, et al. Lactobacilli and bifidobacteria in the prevention of antibiotic-associated diarrhoea and Clostridium difficile diarrhoea in older inpatients (PLACIDE): a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2013;382:1249–1257. doi: 10.1016/S0140-6736(13)61218-0. [DOI] [PubMed] [Google Scholar]
- 37.Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–707. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- 38.Modi SR, Lee HH, Spina CS, Collins JJ. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature. 2013;499:219–222. doi: 10.1038/nature12212. [DOI] [PMC free article] [PubMed] [Google Scholar]