Abstract
Bone metastasis is a frequent complication of breast cancer that is often accelerated by TGF-β signaling; however, little is known about how the TGF-β pathway is regulated during bone metastasis. Here we report that deleted in liver cancer 1 (DLC1) is an important regulator of TGF-β responses and osteolytic metastasis of breast cancer cells. In murine models, breast cancer cells lacking DLC1 expression exhibited enhanced capabilities of bone metastasis. Knockdown of DLC1 in cancer cells promoted bone metastasis, leading to manifested osteolysis and accelerated death in mice, while DLC1 overexpression suppressed bone metastasis. Activation of Rho-ROCK signaling in the absence of DLC1 mediated SMAD3 linker region phosphorylation and TGF-β–induced expression of parathyroid hormone–like hormone (PTHLH), leading to osteoclast maturation for osteolytic colonization. Furthermore, pharmacological inhibition of Rho-ROCK effectively reduced PTHLH production and breast cancer bone metastasis in vitro and in vivo. Evaluation of clinical breast tumor samples revealed that reduced DLC1 expression was linked to elevated PTHLH expression and organ-specific metastasis to bone. Overall, our findings define a stroma-dependent paradigm of Rho signaling in cancer and implicate Rho–TGF-β crosstalk in osteolytic bone metastasis.
Introduction
Breast cancer is one of the major causes of cancer-related death worldwide, mainly due to outgrowth of cancer cells in vital organs, including bone, lungs, liver, and brain (1). The majority of patients with advanced breast cancer will develop bone metastases and suffer from severe pain and eventually death (2). Current treatments for bone metastasis have limited efficacy; therefore, there is an urgent need to identify functional molecules in cancer cell bone colonization as new therapeutic targets.
TGF-β signaling is a critical regulator of breast cancer metastasis to the bone, which is a rich reservoir of various growth factors, such as TGF-β, IGF, and EGF (2, 3). TGF-β binds to and activates a pair of cell surface receptors, which in turn phosphorylate SMAD2 and SMAD3. These receptor-regulated SMAD (R-SMAD) proteins then bind to SMAD4 and translocate into the nucleus for transcriptional regulation. The TGF-β–activated transcriptional program is involved in various steps of cancer metastasis, including angiogenesis, extracellular matrix remodeling, chemoattraction of protumor stroma, metastatic homing, cancer cell survival, and colonization (4–6). In particular, TGF-β in the bone milieu enhances the expression of soluble factors or cell surface proteins such as parathyroid hormone–like hormone (PTHLH; also called PTHrP), Jagged 1 (JAG1), and matrix metalloproteinase 1 (MMP1) by tumor cells, which in turn tip the balance of bone remodeling in favor of osteolysis by promoting osteoclast maturation (7–9). Bone destruction leads to release of additional TGF-β embedded in the bone matrix and further cancer cell stimulation, the vicious cycle of osteolytic bone metastasis. Although numerous studies have firmly established the central role of TGF-β signaling in bone metastasis, how this molecular pathway is regulated during the process is largely unknown.
Human deleted in liver cancer 1 (DLC1) is located at chromosome 8q22, a region frequently lost in cancer cells, and encodes a RhoGTPase-activating protein (RhoGAP), which inhibits RHOA, RHOB, RHOC, and CDC42 by stimulating hydrolysis of GTPase-bound GTP. These GTPases are genuine regulators of cytoskeleton organization (10). Once activated by GTP binding, they initiate a signaling cascade of their downstream effectors, including protein kinases and actin-binding proteins. These effectors act on the dynamic assembly and disassembly of F-actin, leading to cell morphological responses that are crucial in several cancer-related biological processes, such as cell motility and cytokinesis (11–14). Rho GTPases and DLC1 are dysregulated in a wide array of cancers and have been found to play opposite roles in tumorigenesis as well as in migration and invasion of tumor cells (13, 15). Thus far, the roles of these molecules in cancer have been mainly attributed to their regulation of the tumor cell cytoskeleton, and it is not clear whether and how the Rho pathway plays a cytoskeleton-independent role in cancer progression and metastasis. Indeed, it is difficult to extrapolate the cancer cell–autonomous role of Rho signaling to tumor microenvironment remodeling, a key aspect of cancer metastasis. In the present study, we found that the DLC1-Rho pathway regulated the metastatic colonization of circulating breast cancer cells in bone, but not lungs. Such an organ-specific phenotype could not be explained by the roles of DLC1 and Rho in cytoskeleton rearrangement, but rather was attributed to their ability to regulate the response of cancer cells to TGF-β stimulation from bone and the remodeling of the osteolytic microenvironment for metastatic colonization.
Results
DLC1 suppresses bone metastasis of breast cancer cells.
MDA-MB-231 (MDA231) derivative cell lines, obtained by in vivo selection and single-cell subcloning from the parental breast cancer cell line, are useful models for studying tissue tropism of breast cancer metastasis. Upon direct inoculation into the circulation of immunodeficient mice, these syngeneic cells display distinct capabilities to colonize to the bone and lungs (16, 17). In these cells, DLC1 expression was negatively correlated with bone metastasis at both mRNA and protein levels (Figure 1A). Actually, DLC1 was among the bone metastasis signature genes (16) that could segregate cancer cells with different bone metastasis traits via unsupervised clustering of gene expression profiles (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI71812DS1). However, there was no obvious difference in DLC1 expression among cells with different lung metastasis proclivities (Supplemental Figure 1B).
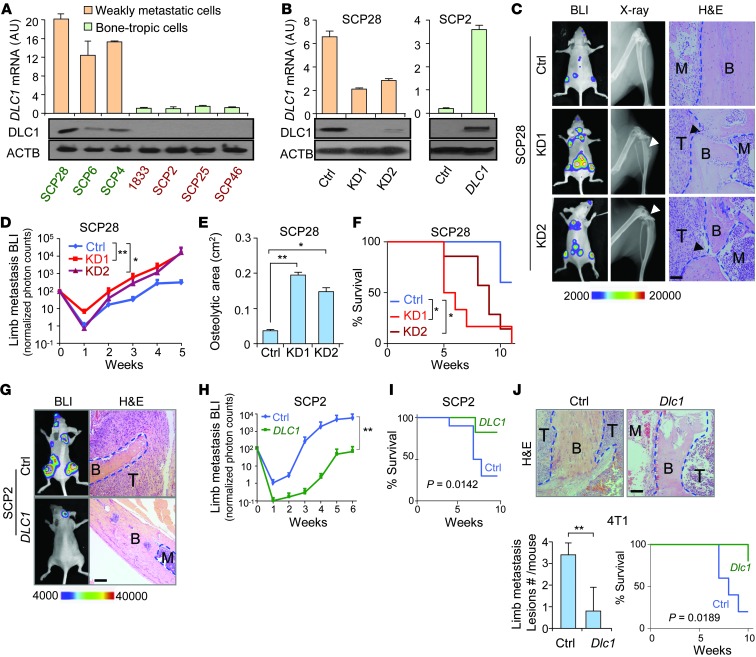
Figure 1. DLC1 suppresses breast cancer osteolytic metastasis.
(A) DLC1 expression in MDA231 derivative cell lines (n = 3). Green and red text denotes cell lines with high and low DLC1 expression, respectively. (B) DLC1 KD and OE in SCP28 and SCP2 cells (n = 3). (C) Representative BLI, X-ray, and H&E images of bone metastases by SCP28 cells. Arrowheads denote areas of overt osteolysis. (D) BLI quantitation of limb metastasis by SCP28 cells (n = 10 per group). (E) Osteolytic area sizes caused by SCP28 cells. (F) Survival of mice injected with SCP28 cells. (G) Representative BLI and H&E images of animals injected with SCP2 cells. (H) BLI limb metastasis burden by SCP2 cells (n = 10 per group). (I) Survival of mice injected with SCP2 cells. (J) In vivo bone metastasis analysis of 4T1 cells with Dlc1 OE in Balb/c mice (n = 10 per group). Shown are H&E images, quantitation of metastasis lesions, and animal survival. Scale bars: 100 μm. B, bone; T, tumor; M, bone marrow or marrow with scattered cancer cell. *P < 0.05, **P < 0.01.
We then analyzed the role of DLC1 in breast cancer organ-specific metastasis by knockdown (KD) and overexpression (OE) approaches. We first used 2 different shRNA constructs to stably silence DLC1 in SCP28 cells, a line with abundant DLC1 expression (Figure 1B), and then intracardially injected the cells into nude mice. Both shRNA constructs significantly enhanced bone metastasis, as revealed by bioluminescent imaging (BLI), X-ray analysis, and histology examination (Figure 1C). The metastasis burden became more than 10-fold higher in KD cells at the fifth week after transplantation (Figure 1D). DLC1 KD also manifestly aggravated bone damage and accelerated death (Figure 1, E and F). Notably, the first KD construct caused more pronounced changes in metastasis than the second, which correlated to their respective efficiencies in silencing DLC1 (Figure 1B). We then analyzed DLC1 function by inducing DLC1 OE in bone-tropic SCP2 cells (Figure 1B). Concordantly, we observed a stark decrease of cancer cell colonization to the skeleton and significantly longer life span after DLC1 OE (Figure 1, G–I).
The function of DLC1 in bone metastasis was further studied in immunocompetent mice by OE of murine Dlc1 in 4T1 mouse cells (Supplemental Figure 1C). Inoculation of 4T1 control and Dlc1 OE cells into Balb/c mice revealed that DLC1 greatly diminished metastatic lesions in the limbs and improved animal survival as well (Figure 1J).
We also analyzed the role of DLC1 in breast cancer lung colonization via i.v. transplantation of SCP28 and 4T1 cells (Supplemental Figure 1, D–I). Concordant to the organ-specific correlation of its expression with metastasis, DLC1 did not obviously alter the lung colonization of these cancer cells in nude or immunocompetent mice. Although the BLI intensity of one of the KD cell lines was slightly higher than that of control SCP28 cells (Supplemental Figure 1D), we did not observe evident differences in the number of lung metastasis foci, nor in animal survival time, by DLC1 KD or OE (Supplemental Figure 1, E–I). Together, these data demonstrated that DLC1 specifically inhibits bone colonization of circulating breast cancer cells.
Rho-inhibiting activity is critical for DLC1 in suppressing bone metastasis.
DLC1 is known to inhibit the small GTPases RHOA/B/C and CDC42. In addition to the RhoGAP domain, which is immediately involved in GTPase inhibition, DLC1 carries a N-terminal sterile α motif (SAM), a focal adhesion targeting (FAT) domain, and a C-terminal steroidogenic acute regulatory protein-related lipid transfer (START) domain, all of which can be involved in cancer-related signaling (18–20). Therefore, we investigated the role of Rho in inhibiting DLC1’s function in bone metastasis. In SCP28 cells, DLC1 KD activated GTPases (RHOA/B/C and CDC42), but had no effects on total protein levels (Figure 2A). GTPase activation by DLC1 KD also affected downstream signaling events, including phosphorylation of vimentin, a substrate of the Rho downstream kinase ROCK (21), and p21-activated protein kinase 4 (PAK4), the crucial effector of active CDC42 (22). Reciprocally, DLC1 OE in SCP2 cells led to GTPase inactivation and inhibition of downstream effectors (Figure 2A). We further tested the in situ regulation of GTPase activities by analyzing stress fiber formation in cancer cells. DLC1 affected cytoskeleton rearrangement and prevented stress fiber formation in these cells (Figure 2, B and C).
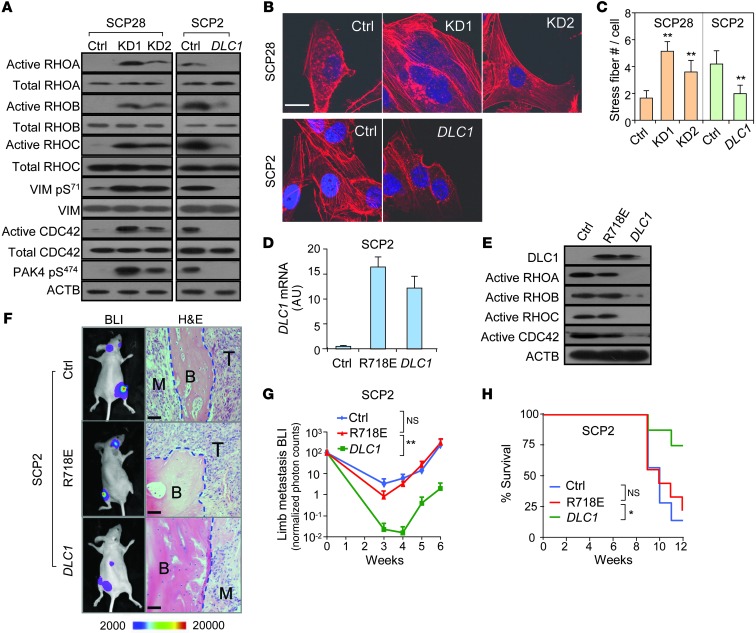
Figure 2. The GTPase pathway mediates DLC1’s role in breast cancer metastasis.
(A) Activation of Rho, CDC42, and their downstream effectors in cells with DLC1 KD and OE. (B) Effect of DLC1 on stress fiber formation. (C) Stress fiber density (n = 6). (D) WT DLC1 and R718E mutant OE in SCP2 cells. (E) The R718E mutant failed to inactivate Rho and CDC42. (F–H) In vivo bone metastasis analysis of the R718E mutant (n = 10 per group). Shown are representative BLI images and H&E staining (F), BLI quantitation (G), and animal survival (H). Scale bars: 10 μm (B); 50 μm (F). *P < 0.05, **P < 0.01.
To determine whether Rho inhibition mediates the suppressive role of DLC1 in bone colonization, we stably overexpressed the R718E DLC1 mutant (point substitution in the RhoGAP domain to abolish its catalytic activity on GTPases; ref. 23) and compared its function with OE of WT DLC1 in SCP2 cells. Both forms of DLC1 were overexpressed to comparable levels in SCP2 cells, while the mutant failed to inactivate GTPase proteins (Figure 2, D and E). Concordantly, the RhoGAP mutation completely abolished the suppressive function of DLC1 in bone metastasis when cancer cells were inoculated into nude mice. Metastasis burden caused by R718E DLC1 OE cancer cells was similar to that caused by control cells, but more than 40-fold higher than that by WT DLC1 OE cells (Figure 2, F and G). Animal survival time was also similar between the mutant and control groups (Figure 2H), which corroborates the notion that RhoGAP activity is critical for DLC1 in regulating breast cancer bone metastasis.
DLC1-Rho signaling regulates both intrinsic malignant traits of cancer cells and osteoclast maturation in the metastatic microenvironment.
Next we sought to understand how DLC1 specifically inhibits bone metastasis of breast cancer. Rho signaling is important for cancer cell cytoskeleton rearrangement, which is crucial in cell proliferation and motility. Concordantly, we found that DLC1 KD in SCP28 cells significantly facilitated anchorage-independent colony formation, wound-healing migration, and transwell invasion, whereas DLC1 OE suppressed such malignant traits of SCP2 cells (Supplemental Figure 2, A–D). Similar results were also observed in 2 other breast cancer cell lines, MCF10CA1h and MCF10CA1a (Supplemental Figure 2, B–D). The effect of DLC1 on cancer cell invasiveness was dependent on regulation of its target GTPases, as treatment of cancer cells with a Rho inhibitor (C3 transferase) or a peptide inhibitor of CDC42 (24) completely abolished the changes conferred by DLC1 KD and OE. R718E DLC1 OE, as well as RhoA(63L), a constitutively active form of RHOA (25), also disabled DLC1 in suppressing cancer cell invasiveness (Supplemental Figure 2, E–G). DLC1 also suppressed tumor growth at the orthotopic site, but did not obviously affect tumor cell proliferation in the bone metastases, as revealed by Ki67 staining (Supplemental Figure 2, H and I).
Although these assays confirmed the roles of DLC1-Rho signaling in regulating cancer cell tumorigenicity, migration, and invasion, we reasoned that these could not explain the organ-specific function of DLC1 in breast cancer metastasis, because such autonomous features of cancer cells would affect metastasis to all distant organs. In addition, the experimental metastasis assay used in our studies bypassed the early steps of cancer cell spreading, which heavily rely on intrinsic features of cancer cells. Therefore, we investigated whether DLC1 specifically regulates the biological events for the circulating cancer cells to colonize in the bone. First, we analyzed the ability of SCP2 and SCP28 cells to interact with bone endothelium that are important during endothelium arrest and extravasation at target organs. DLC1 suppressed cancer cell adherence to and invasion through the monolayer of human bone marrow endothelial cell line HBMEC-60. However, similar phenotypes were also observed with lung endothelial cells (Supplemental Figure 2J). Therefore, the altered tumor-endothelium interaction was also not likely to account for the specific function of DLC1 in bone colonization.
Next, we investigated whether DLC1 regulates the capability of cancer cells to reshape the metastatic microenvironment in the bone. Bone-tropic breast cancer cells are usually characterized by an enhanced ability to drive maturation of osteoclasts, the bone host cells responsible for osteolysis during breast cancer metastasis (2, 26). Therefore, we performed osteoclastogenesis assays with mouse primary bone marrow cultured together with SCP28 cells. Although DLC1 KD in SCP28 cells did not directly promote osteoclastogenesis of cocultured bone marrow, treatment with TGF-β, the cytokine abundantly present in bone, significantly enhanced the ability of DLC1 KD cells, but not of control cells, to drive osteoclast differentiation. DLC1 KD activated the response of SCP28 cells to TGF-β for driving osteoclastogenesis (Figure 3A). The same phenomenon was observed when bone marrow was cultured in conditioned media (CM) from SCP28 cells (Figure 3A), which indicated that DLC1 altered SCP28 secretome to affect osteoclast maturation. In addition, DLC1 OE had the opposite effect in the highly metastatic SCP2 cells (Figure 3B). In a broader range of MDA231 derivative cells, we further observed a reverse correlation of DLC1 expression, along with the ability to induce osteoclast differentiation when cells were treated with TGF-β (Figure 1A and Figure 3C). Expressing the DLC1-resistant RhoA(63L) in SCP2 cells antagonized the effect of DLC1 OE, while treatment of cancer cells with the Rho inhibitor C3 transferase or the ROCK inhibitor Y27632 phenocopied DLC1 expression in both SCP2 and SCP28 cells (Figure 3, A and D), which suggests that DLC1 acts through Rho-ROCK signaling to regulate TGF-β–enhanced osteoclastogenesis. Meanwhile, adding these inhibitors directly to the primary bone marrow culture did not produce any differences in osteoclast maturation (Figure 3D). Therefore, it was Rho-ROCK signaling in cancer cells, not that in bone stromal cells, that mediated the role of DLC1 in osteoclastogenesis.
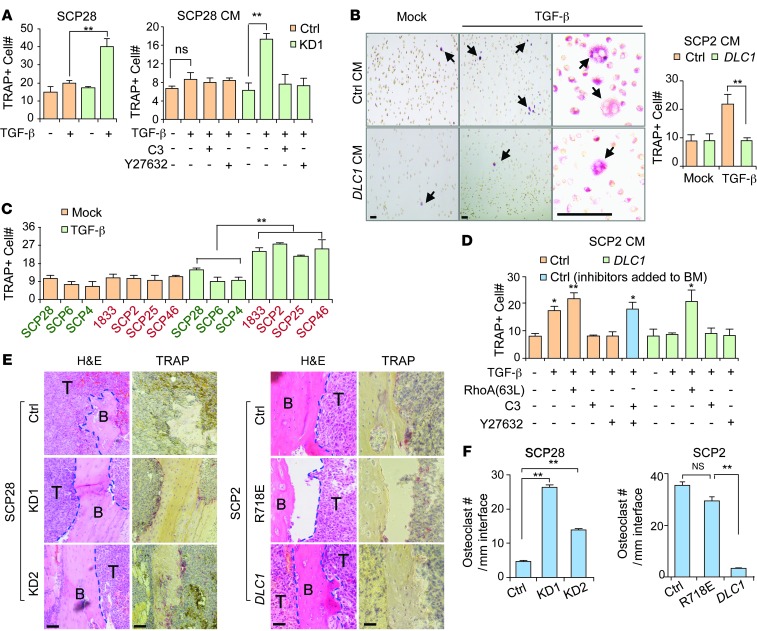
Figure 3. DLC1-Rho signaling regulates osteoclast maturation in the bone microenvironment.
(A) Osteoclastogenesis assays of primary bone marrow cultured together with DLC1 KD or control cells treated with TGF-β, or in CM from SCP28 cells treated with TGF-β, C3, and/or Y27632 (n = 3). (B) Osteoclastogenesis assays of primary bone marrow culture in SCP2 CM (n = 3). Representative TRAP staining images are also shown. Arrows denote TRAP-positive osteoclasts. (C) Osteoclastogenesis assays of CM from additional MDA231 derivative cell lines (n = 3). Green and red text denotes cell lines with high and low DLC1 expression, respectively. (D) Osteoclastogenesis assays of CM from cancer cells treated with various genes or chemical reagents (n = 3). (E) TRAP staining of bone metastasis tumors from animals injected with DLC1 KD or OE cancer cells. (F) Osteoclast cells along the tumor-bone interface (n = 3). Scale bars: 100 μm. *P < 0.05, **P < 0.01.
To further analyze the effect of DLC1 on osteoclastogenesis in vivo, we performed tartrate-resistant acid phosphatase (TRAP) staining of bone metastasis lesions from mice with intracardiac injection of DLC1 KD and OE cancer cells. DLC1 KD in SCP28 cells markedly enhanced mature osteoclast density along the tumor-bone interface, with the more efficient shRNA construct producing a more profound difference. In addition, OE of WT DLC1, but not of R718E DLC1, annihilated the ability of SCP2 cells to induce osteoclast maturation in the bone (Figure 3, E and F). Therefore, our results demonstrated that the DLC1-Rho-ROCK signaling pathway regulates the response of tumor cells to TGF-β for osteolytic microenvironment remodeling.
DLC1-Rho signaling regulates osteoclastogenesis by blocking TGF-β–induced PTHLH secretion.
To identify the mechanism by which DLC1 regulates TGF-β–induced osteoclast maturation, we performed microarray analyses of SCP2 cells to find genes that were responsive to TGF-β, but affected by DLC1. The results showed that DLC1 was not able to interfere with the overall response of cancer cells to TGF-β, but only affected a small subset of TGF-β downstream genes. Gene Set Enrichment Analysis (GSEA) revealed that the previously identified TGF-β signature genes (27) were regulated similarly in both control and DLC1 OE cells (Supplemental Figure 3A). Moreover, among the 285 transcripts (392 probes) that were up- or downregulated by TGF-β in SCP2 cells, only 29 (10.2%) were reversed by DLC1 OE (Figure 4A and Supplemental Figure 3B). These 29 genes (Supplemental Table 1) consisted of those that were upregulated by TGF-β but blocked by DLC1 (group A), and those that were downregulated by TGF-β but reversed by DLC1 (group B). Interestingly, the influence of DLC1 seemed fundamentally different in these 2 groups, as DLC1 altered the expression of group A genes after TGF-β treatment, while its effect on group B was significant only in the absence of TGF-β (Figure 4B).
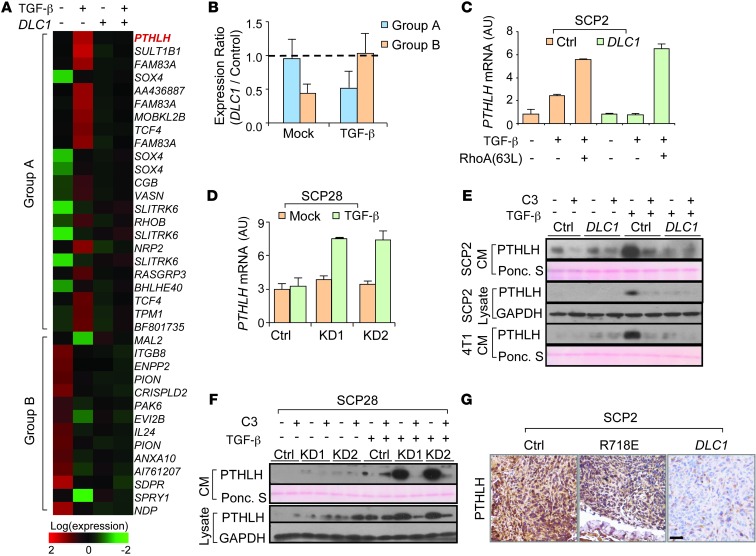
Figure 4. DLC1 blocks TGF-β–induced PTHLH expression and secretion.
(A) Expression heatmap of genes regulated by TGF-β, but reversed by DLC1, in SCP2 cells. (B) Expression ratios of the 2 gene groups in DLC1 OE and control cells when cells were untreated or treated with TGF-β (n = 3). Data represent ratio median ± median absolute deviation (MAD). (C) PTHLH mRNA levels in SCP2 cells with DLC1 OE (n = 3). (D) PTHLH mRNA levels in SCP28 cells with DLC1 KD (n = 3). (E) Intracellular and extracellular PTHLH protein levels in SCP2 and 4T1 cells with DLC1 OE. (F) Intracellular and extracellular PTHLH protein levels in SCP28 cells with DLC1 KD. (G) PTHLH IHC analyses in bone metastases from animals inoculated by control SCP2 cells or the cells with WT and mutant DLC1 OE. Scale bar: 50 μm.
Thus, we primarily focused on the genes in group A, as DLC1 only had a marked effect on osteoclastogenesis with TGF-β treatment. Among these, PTHLH (which encodes a secretory protein) became our candidate for further investigation, because it ranked at the top when these genes were sorted by TGF-β response ratios in DLC1 OE and control cells (Figure 4A and Supplemental Table 1). More importantly, PTHLH has been previously reported to be an important regulator of osteoclastogenesis (9, 28, 29). We first validated that PTHLH expression was promoted by TGF-β in SCP2 cells, and this effect was neutralized by DLC1. Reciprocally, DLC1 KD activated the response of PTHLH to TGF-β in SCP28 cells (Figure 4, C and D). Regulation of PTHLH protein expression and secretion by DLC1 were also observed in multiple cell lines, including SCP2, SCP28, SCP4, MCF10Ca1a, and 4T1 (Figure 4, E and F, and Supplemental Figure 4). Furthermore, constitutive activation of RHOA or treatment with C3 or Y27632 masked the effects of DLC1 on PTHLH production by these cancer cells after TGF-β treatment (Figure 4, C, E, and F, and Supplemental Figure 4). In the bone metastasis tumors of nude mice inoculated with SCP2 cells, OE of WT DLC1, but not that of R718E DLC1, markedly reduced PTHLH protein levels (Figure 4G). These results indicated that DLC1-Rho signaling regulates the response of PTHLH to TGF-β in tumor cells. In contrast, other known TGF-β target genes, including JAG1, IL11, CTGF, and CDKN1A, were not affected by DLC1 (Supplemental Figure 5A). In particular, whereas JAG1 was previously found to mediate the response of stromal Notch pathway to tumor cell TGF-β signaling for osteoclast maturation and bone metastasis (7), we found that DLC1 did not alter basal or TGF-β–induced JAG1 expression, nor could JAG1 OE change DLC1 expression (Supplemental Figure 5, A and B). In addition, JAG1 OE in SCP28 cells promoted bone colonization to the same extent as DLC1 KD, while simultaneous JAG1 OE and DLC1 KD further enhanced the metastatic capability of SCP28 cells (Supplemental Figure 5C). Therefore, the molecular route regulated by DLC1 is likely to be independent of stromal JAG1/Notch signaling.
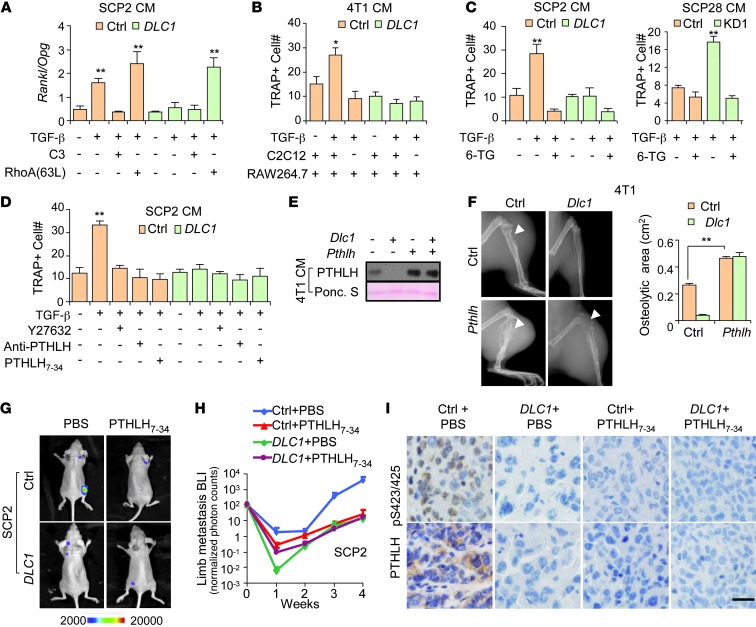
Previous reports showed that PTHLH binds to its receptor on the osteoblast cell surface to enhance RANKL secretion and suppress osteoprotegerin (OPG) secretion by osteoblasts (30). RANKL, which acts upon the RANK receptor on osteoclasts, is the primary cytokine for osteoclast maturation, whereas OPG is a decoy receptor and RANKL inhibitor. We found that when DLC1-deficient cancer cells were treated with TGF-β, their CM significantly enhanced Rankl expression and suppressed Opg expression by murine preosteoblast C2C12 cells. Such effects were augmented by constitutively active RhoA(63L), but blocked by DLC1 OE as well as by C3 treatment (Figure 5A). In addition, when C2C12 was cocultured with the preosteoclast cell line RAW264.7 in 4T1 CM, the results were similar to those of the primary bone marrow osteoclastogenesis assays. However, DLC1 OE caused no difference when C2C12 was absent from the coculture system (Figure 5B). These data suggested that DLC1 signaling of tumor cells regulates osteoclastogenesis through osteoblast-derived cytokines, consistent with the notion that PTHLH regulation mediates DLC1’s role in osteolytic microenvironment remodeling.
Figure 5. DLC1 inhibits bone metastasis by regulating PTHLH expression.
(A) Rankl/Opg expression ratios of C2C12 preosteoblasts cultured in CM of SCP2 cells with DLC1 OE (n = 3). (B) Osteoclastogenesis of RAW264.7 cells in 4T1 CM, with or without C2C12 coculture (n = 3). (C) Osteoclastogenesis of primary bone marrow in CM from SCP2 and SCP28 cells treated with 6-TG (n = 3). (D) Osteoclastogenesis of primary bone marrow in CM from SCP2 cells treated with anti-PTHLH neutralizing antibody, PTHLH7–34, or Y27632. (E) Simultaneous OE of Pthlh and Dlc1 in 4T1 cells. Shown are Western blot results after cells were treated with TGF-β. (F) Osteolysis (arrowheads) caused by 4T1 cells with double OE of Pthlh and Dlc1 (n = 4). (G) Representative BLI images of mice injected with SCP2 cells and treated with PTHLH7–34. (H) BLI quantitation of mice injected with SCP2 cells and treated with PTHLH7–34 (n = 10 per group). (I) IHC analyses of PTHLH and C-tail phosphorylated SMAD3 (Ser423/425) in bone metastases of mice injected with SCP2 cells and/or treated with PTHLH7–34. Scale bar: 25 μm. *P < 0.05, **P < 0.01.
To determine whether tumor-derived PTHLH is critical for osteoclast maturation regulated by DLC1, we treated cancer cells with 3 types of PTHLH inhibitors — the chemical inhibitor 6-thioguanine (6-TG; ref. 31), the peptide antagonist PTHLH7–34 (32, 33), and a neutralizing antibody — followed by osteoclastogenesis analyses. All inhibitors efficiently blunted the osteoclast-promoting effects of CM from cancer cells. Importantly, PTHLH inhibition completely abolished the effects of DLC1 OE and KD on osteoclast differentiation (Figure 5, C and D).
We further investigated the role of PTHLH in DLC1-Rho signaling in vivo. Murine Pthlh was overexpressed together with Dlc1 in 4T1 cells, which were then tested for bone metastasis (Figure 5E). Pthlh OE markedly increased protein secretion and led to overt bone damage, regardless of Dlc1 expression. Most importantly, DLC1 was no longer able to reduce osteolysis when PTHLH was abundantly expressed (Figure 5F).
We also intracardially injected SCP2 cells with DLC1 OE into nude mice and treated them with PTHLH7–34, which starkly suppressed the bone metastasis signals. Furthermore, there was no additional benefit from DLC1 OE when PTHLH was inhibited (Figure 5, G and H), which suggests that PTHLH acts downstream of DLC1. Notably, DLC1 OE, as well as PTHLH7–34 treatment, not only reduced PTHLH levels in bone lesions, but also attenuated TGF-β activity, as assessed by C-tail phosphorylation of SMAD3 in the lesions (Figure 5I). To rule out the possibility of PTHLH-independent side effects of the inhibitor in the bone microenvironment, we repeated the experiment with 6-TG and observed similar trends in bone metastasis intensity and survival time (Supplemental Figure 6, A–C). Taken together, these data suggested that DLC1 inhibits bone destruction caused by tumor-derived PTHLH secretion and also reduces TGF-β activity in bone, which is crucial for the osteolytic vicious cycle in metastasis.
DLC1 suppresses TGF-β–induced PTHLH expression by regulating SMAD3 linker region phosphorylation.
We further investigated how the DLC1-Rho pathway regulates TGF-β signaling to alter PTHLH expression. TGF-β exerts its biological effects mainly through SMAD transcription factors. However, SMAD-independent pathways, including PI3K, p38, JNK, and ERK, have also been reported to mediate TGF-β signaling (34). Therefore, we first tested whether these pathways play a role in DLC1 regulation of PTHLH. Treatment of SCP2 cells with specific inhibitors of these pathways, as well as CDC42 and RAC1 GTPase inhibitors, did not influence the effects of DLC1 and TGF-β on PTHLH expression. In contrast, the ROCK inhibitor Y27632 efficiently suppressed PTHLH expression driven by TGF-β treatment (Figure 6A). Therefore, only Rho proteins, not other small GTPases, mediate the role of DLC1 to regulate PTHLH production, and the downstream signaling of Rho-ROCK was not the reported SMAD-independent pathway.
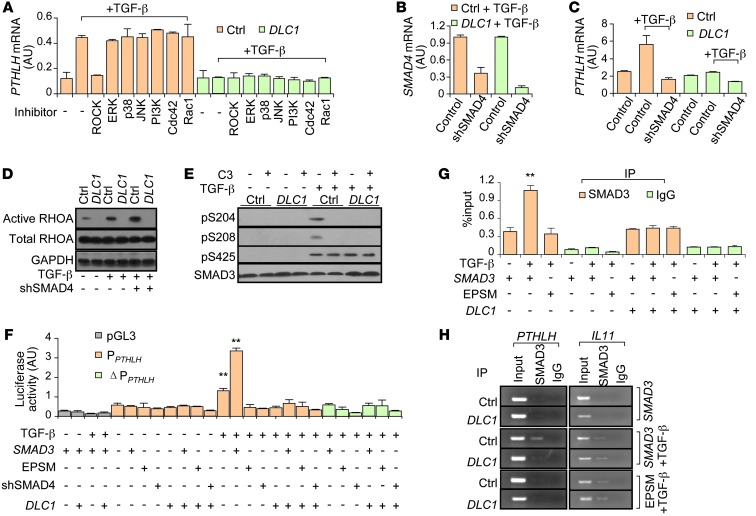
Figure 6. DLC1-Rho signaling regulates SMAD3 linker phosphorylation to affect PTHLH expression.
(A) PTHLH expression in SCP2 cells treated with inhibitors of various pathways (n = 3). (B) shRNA-mediated SMAD4 KD (shSMAD4) in SCP2 cells. (C) PTHLH expression in SCP2 cells with SMAD4 KD (n = 3). (D) TGF-β activation of RHOA in SCP2 cells. (E) SMAD3 linker (Ser204, Ser208) and C-tail (Ser425) phosphorylation in SCP2 cells affected by TGF-β, C3, and DLC1. (F) PTHLH promoter activities in SCP2 cells affected by TGF-β, DLC1 OE, SMAD4 KD, and WT and mutant SMAD3 (n = 4). (G) ChIP-qPCR analyses of SMAD3 binding on PTHLH promoter in SCP2 cells (n = 3). (H) ChIP analyses of SMAD3 binding on PTHLH and IL11 promoters in SCP2 cells. **P < 0.01 vs. TGF-β–untreated control.
We subsequently tested the role of SMADs in regulation of PTHLH expression by the DLC1-Rho-ROCK pathway. We used a shRNA construct to achieve SMAD4 KD in SCP2 cells, which abolished the PTHLH response to TGF-β and DLC1 (Figure 6, B and C). TGF-β treatment noticeably enhanced RHOA activation, which was suppressed by DLC1 OE, but not by SMAD4 shRNA (Figure 6D). These data indicated that Rho signaling acts between TGF-β and SMADs to promote PTHLH expression.
We further investigated how Rho signaling regulates SMAD activities. TGF-β activates the SMAD complex by phosphorylating the C-tail of R-SMADs. However, we found that DLC1, as well as the RHOA inhibitor, did not alter SMAD3 expression or its C-tail phosphorylation (Figure 6E). Previously, Liu and colleagues reported that phosphorylation of Ser204 and Ser208 in the linker region of SMAD3 could also contribute to SMAD activation (35). We found that TGF-β–induced SMAD3 phosphorylation in this region was dependent on Rho-ROCK activity, as DLC1 OE or C3 treatment sufficiently blocked this modification. We also noted that prior to TGF-β activation, Rho was not able to stimulate SMAD3 phosphorylation, even without interruption by DLC1 OE or C3 treatment (Figure 6E), consistent with the notion that SMAD3 C-tail phosphorylation is necessary for the linker modification (35).
To determine whether and how SMAD3 linker phosphorylation regulates PTHLH transcription, we used luciferase reporter assays to test the effects of TGF-β treatment, DLC1 OE, SMAD4 KD, and OE of WT SMAD3 and the SMAD3 mutant EPSM (linker phosphorylation sites mutated; ref. 36) on PTHLH promoter activities. Whereas TGF-β and WT SMAD3 activated the PTHLH promoter in a synergistic manner, DLC1 OE, SMAD4 KD, and EPSM SMAD3 OE inhibited TGF-β–induced promoter activity (Figure 6F). Interestingly, EPSM SMAD3 OE and DLC1 OE could not repress the general SMAD activity induced by TGF-β, as revealed by luciferase reporter analyses of a consensus SMAD binding element (SBE) construct (Supplemental Figure 7A). These results were concordant with our microarray analysis and indicated that although TGF-β–induced SMAD3 C-tail phosphorylation was sufficient to regulate most target genes, activation of the PTHLH promoter required additional linker phosphorylation of SMAD3, which was mediated by Rho-ROCK signaling.
The SMAD complex could activate the PTHLH promoter directly or through other transcription factors. Previous studies reported that GLI2, a SMAD target gene, could enhance PTHLH transcription in some cancer cell lines (37), suggestive of a possible indirect mechanism for SMAD regulation of PTHLH. However, we found that although GLI2 was induced by TGF-β in SCP2 and SCP28 cells, it was not affected by DLC1 (Supplemental Figure 7B). In addition, GLI2 OE could not rescue the suppression of PTHLH by DLC1 (Supplemental Figure 7C), thus excluding the possibility of GLI2 involvement in DLC1-mediated PTHLH regulation. Instead, mutating the SMAD binding site in the PTHLH promoter (Supplemental Figure 7D) totally erased its response to TGF-β treatment, DLC1 OE, SMAD4 KD, and WT and EPSM SMAD3 OE (Figure 6F). Furthermore, ChIP coupled with quantitative PCR (qPCR) analysis revealed direct binding of SMAD3 to the PTHLH promoter. EPSM SMAD3 OE and DLC1 OE expelled SMAD3 from the PTHLH promoter, but not from that of other TGF-β target genes, such as IL11 (Figure 6, G and H).
Inhibition of Rho-ROCK signaling reduces osteolytic metastasis of breast cancer.
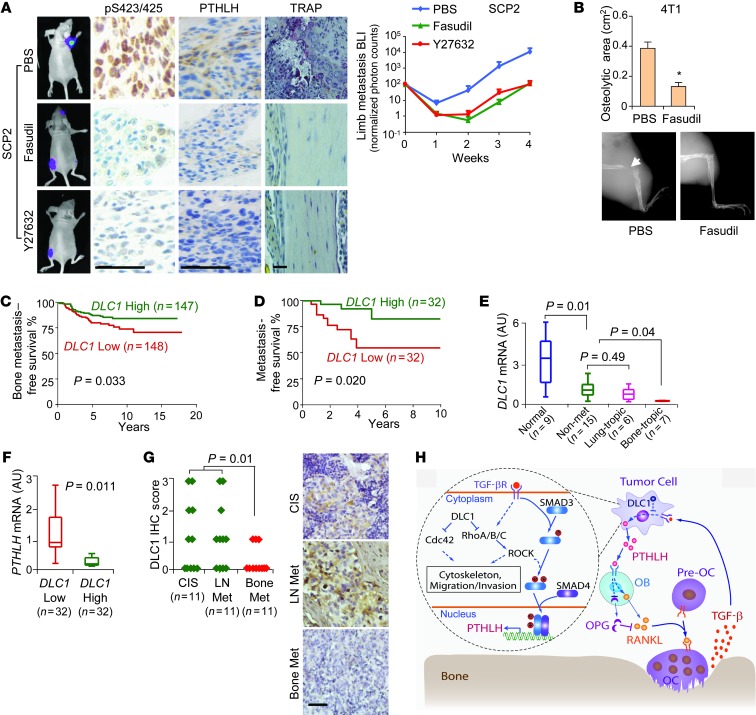
Thus far, our data suggested that Rho-ROCK signaling could be a potential therapeutic target to treat breast cancer bone metastasis that is enhanced by TGF-β–driven osteolysis. To test this, we injected SCP2 cells into nude mice intracardially, followed by treatment with 2 different Rho-ROCK inhibitors, Y27632 and fasudil (38). Both inhibitors effectively prevented the bone metastasis of SCP2 cells. By the fourth week, the bone metastasis burden of drug-treated mice was reduced 100-fold compared with control mice (Figure 7A). The reduced bone metastasis was associated with decreased PTHLH expression and osteoclast activities in the bone (Figure 7A and Supplemental Figure 8). Interestingly, although Rho inhibition did not directly affect SMAD3 C-tail phosphorylation in the in vitro system (Figure 6E), this region was much less phosphorylated in the bone lesions when animals were treated with the inhibitors (Figure 7A). This was consistent with our observations with PTHLH inhibitor treatment (Figure 5I), suggestive of a possible secondary effect: less TGF-β release caused by reduced bone breakdown.
Figure 7. Effect of in vivo targeting Rho-ROCK signaling and clinical correlation of DLC1 with bone metastasis.
(A) The ROCK inhibitors fasudil and Y27632 prevented SCP2 cell bone metastasis in nude mice (n = 10 per group). Shown are BLI images of bone metastases, IHC analyses of SMAD3 C-tail phosphorylation and PTHLH, osteoclast TRAP staining, and BLI quantitation. (B) Effect of fasudil on bone metastasis in Balb/c mice injected with 4T1 cells (n = 4). Arrowhead denotes area of overt osteolysis. *P < 0.05 vs. PBS. (C) Bone metastasis of NKI patients stratified by DLC1 expression. (D) Metastasis-free survival analysis of Qilu patients. (E) DLC1 expression in paracancerous normal tissues and in nonmetastatic, lung-tropic, or bone-tropic fresh-frozen primary tumors. (F) Correlation of DLC1 and PTHLH expression in Qilu tumors. (G) DLC1 IHC analyses in CIS primary tumors, lymph node metastases, and bone metastases. See Methods for IHC scoring. (H) DLC1-Rho-ROCK signaling in breast cancer bone metastasis. Dashed lines denote multistep processes of regulation. OB, osteoblast; Pre-OC, preosteoclast. Scale bars: 50 μm.
We also assessed the efficacy of ROCK inhibition for treating bone metastasis in the immunocompetent setting. 4T1 cells were inoculated into Balb/c mice, followed by daily fasudil treatment. The drug rescued the mice from massive bone damage caused by the cancer cells, as demonstrated by the 3-fold decrease of osteolytic areas in the limbs after treatment (Figure 7B). These results thus confirmed the pivotal role of Rho-ROCK signaling in bone metastasis and supported the application of Rho inhibitors in breast cancer therapy.
DLC1 expression is negatively associated with PTHLH and bone metastasis in clinical samples.
Finally, we assessed the clinical significance of DLC1 in breast cancer. We first analyzed a publicly available NKI microarray dataset (17, 39, 40). In these clinical samples, lower DLC1 expression was linked to obviously higher risk of overall metastasis and bone relapse, but not relapse in lungs (Figure 7C and Supplemental Figure 9, A and B), indicative of an organ-specific correlation of DLC1 with clinical metastases of breast cancer. Notably, DLC1 expression was not associated with molecular subtypes of breast cancer (luminal, basal, HER2+, or normal-like; Supplemental Figure 9C), which was confirmed by additional analyses of breast invasive carcinoma data available in the Cancer Genome Atlas (TCGA; ref. 41) database (Supplemental Figure 9D).
To further analyze the clinical relevance of DLC1 expression, we collected a set of 64 fresh-frozen breast tumors from Qilu Hospital and analyzed the correlation of DLC1 expression with metastasis. In this cohort, DLC1 expression was also a prognostic factor of metastasis independent of ER, PR, HER2, or triple-negative breast tumor status (Figure 7D, Supplemental Figure 9E, and Supplemental Table 2). Closer examination revealed that DLC1 expression was profoundly lower in tumors prone to bone, but not lung, recurrence (Figure 7E). In addition, DLC1 expression was negatively correlated with that of PTHLH in these tumors (r = –0.43; Figure 7F), a phenomenon observed in the TCGA dataset as well (Supplemental Figure 9F). We further analyzed DLC1 protein levels in 33 carcinomas in situ (CIS), LN metastases, and bone metastases of mammary origin and found significantly weaker DLC1 expression in bone metastases than in primary tumors and LN metastases (Figure 7G). Thus, our data confirmed that DLC1 silencing is clinically associated with enhanced PTHLH expression and breast cancer bone metastasis.
Discussion
Here, we reported the organ-specific role of DLC1-Rho signaling in bone colonization of breast cancer. DLC1 displayed lower expression in breast cancer cell lines and tumors with higher affinity for the skeleton. DLC1 silencing enhanced bone colonization of circulating cancer cells, and its restoration significantly diminished the metastatic capability of bone-tropic cells. Apparently, DLC1 plays its role in bone metastasis by inhibiting Rho GTPases, which on the one hand work synergistically with CDC42 to regulate cancer cell cytoskeleton and motility, and on the other hand regulate TGF-β–induced SMAD3 phosphorylation to activate PTHLH transcription. Tumor-derived PTHLH acts on osteoblasts to change the RANKL/OPG ratio in the bone stroma and thus instigate osteoclast maturation, which causes metastatic bone destruction. Interestingly, although DLC1 only regulated the linker phosphorylation of SMAD3 in tumor cells in vitro (Figure 6E), it suppressed SMAD3 C-tail phosphorylation in bone metastases as well (Figure 5I), likely resulting from the altered bioavailability of TGF-β from PTHLH-induced bone demolition. Therefore, DLC1 serves as a “molecular brake” to the positive feedback of TGF-β signaling and osteolysis. Taken together, our findings delineate a new pathway regulating TGF-β–driven breast-to-bone metastasis (Figure 7H).
Distant metastasis of tumor cells is an organ-specific, multistep process consisting of basement membrane infringe, migration through nearby stroma, blood vessel entrance and extravasation, and colonization at foreign sites. The metastasis events prior to circulation entrance, relying on cancer cell migration and invasion, are shared prerequisites for the tumors to spread to multiple distant organs. On the contrary, the late metastasis steps dictate the organotropism of circulating tumor cells and are usually driven by the ability of tumor cells to interact with the residential components of target organs. Our present findings demonstrated that DLC1-Rho signaling not only regulated the intrinsic motility and invasiveness of cancer cells, but also influenced their ability to respond to and affect the metastatic microenvironment. The latter role seems to be specific in the context of bone, as DLC1 only regulated the thriving of circulating tumor cells in bone, not in lungs. In addition, we observed an organ-specific correlation of DLC1 expression with bone metastasis. Interestingly, we also found in the NKI dataset that DLC1 silencing was associated with bone metastasis only for luminal-type breast tumors (Supplemental Figure 9G), which are known to be predisposed to bone recurrence (42, 43), although such findings need to be validated in additional datasets. Previous studies have shown the association of DLC1 silencing with poor prognosis in breast cancer (15). Our data indicated that this association is more likely attributable to an elevated risk for metastasis in bone compared with other organs. Numerous studies have reported that DLC1 inhibited the lung metastasis of MDA-MB-435, a cancer cell line of debatable origin (44, 45), and hepatocellular carcinoma cells (46, 47). These studies seem to be at variance with our work, but likely reflect a role of DLC1-Rho signaling in cell invasiveness that is critical for the dissemination of cancer cells into circulation, as in these studies, the cancer cells were transplanted subcutaneously or into the mammary fat pads of mice. In contrast, we inoculated the cells directly into the circulation to assess the late phase of metastasis. Taken together, these data are indicative of the different roles of DLC1 in early and late stages of cancer spreading. In line with our present findings regarding the organ-specific role of DLC1 is the complicated association of PTHLH with breast cancer bone metastasis. Although it is well established that PTHLH promotes skeleton remodeling for cancer cell bone colonization and is upregulated in bone metastases compared with primary tumors, a large clinical study found that patients with PTHLH-positive primary tumors were instead less likely to develop bone metastasis, suggesting that PTHLH production by cancer cells might link to less malignancy in general invasiveness, but a stronger ability to induce osteolysis (48, 49). Therefore, as a PTHLH upstream regulator, DLC1’s function is supported by PTHLH suppression during metastasis to the bone, but might be compromised by the downstream protein during metastasis to other sites.
DLC1 is a multidomain protein and can interact with small GTPases and other molecules that might be involved in cancer progression (18–20). We found here that the role of DLC1 in bone metastasis was mediated by Rho-ROCK signaling. Rho GTPases are important regulators of oncogenic transformation, migration, and invasion (14). However, most of these functions have been accounted for by their widely accepted role in cytoskeleton conformation. Here, we presented solid evidence for the role of cancer cell Rho signaling in nonautonomous regulation of the tumor microenvironment. Our data showed that Rho was activated by TGF-β and enhanced PTHLH induction to influence the osteoblast/osteoclast balance, resulting in an osteolytic vicinity of tumor cells. Importantly, such function was independent of cytoskeleton reorganization and was specific to Rho proteins, not to other small GTPases (Figure 6A). Therefore, our results provided a new paradigm for the Rho signaling pathway in cancer, in which it not only responds to the environmental signal, but also maneuvers the efficacy of cancer cells to reshape the surrounding stroma to enhance metastasis.
TGF-β signaling is one of the most important molecular pathways in cancer metastasis. It is a potent inducer of epithelial-mesenchymal transition of cancer cells, an event thought to be critical for tumor dissemination, and also mediates communication of cancer cells with other tumor components during metastasis of cancer cells to various organs (4–6, 50). TGF-β is of particular importance in bone metastasis because of its abundance in bone matrix and its pivotal role in the osteolytic vicious cycle (3, 51). The pathway has been reported to regulate other prominent pathways, such as PI3K, MAPK, Rho/RAC1, NF-κB, Notch, and TNF-α (52). However, how TGF-β is regulated by other pathways in distant metastasis largely remains a mystery. Our data showed that Rho-ROCK signaling regulated SMAD3 activity by phosphorylating its linker region. TGF-β stimulation appeared insufficient to spark a full response from cancer cells when Rho activity was repressed. Thus, obstacle clearance for Rho activation by DLC1 silencing may be necessary in order for cancer cells arriving at the bone to adapt to the foreign environment and repopulate to secondary tumors. This would explain why some tumor cells keep indolent in the skeleton for a long time, while others erupt quickly to macrometastases. Interestingly, Rho-mediated SMAD3 linker phosphorylation was required only for a small subset of TGF-β target genes. A question raised by our present findings is what underlies such specificity. One of the conceivable hypotheses would be the sequence context of SMAD binding sites in the target promoters. Nevertheless, these results enrich our understanding of the TGF-β network in tumor metastasis and may lead to new approaches to targeting TGF-β signaling.
Rho and ROCK inhibitors are highly desired for cancer therapeutics (53, 54). Our findings supplied additional grounds by which to target these molecules for breast cancer treatment, as bone metastasis is a frequent symptom affecting the majority of advanced-stage breast cancer patients. Previous reports have showed that ROCK inhibition can suppress TGF-β–induced PTHLH production by cancer cells cultured on the bone-like rigid matrix (55) and hinder bone metastasis in a “human breast cancer metastasis to human bone” mouse model (56). Here, we extended these studies with preclinical experiments using 2 different ROCK inhibitors in both immunodeficient and immunocompetent animals, and also revealed the underlying mechanism of the efficacy of ROCK inhibition in stopping bone metastasis. Therefore, Rho-ROCK inhibitors join TGF-β and PTHLH inhibitors in the drug family targeting tumor-driven osteoclast activities. Currently, effective chemical inhibitors of PTHLH have been lacking, other than 6-TG and the closely related 6-thioguanosine, and these guanine analogs inevitably have cytotoxic side effects (31, 57). Rho-ROCK inhibition might provide a valid alternative, with the additional benefit of restraining cancer cell migration and invasion. Moreover, targeting Rho-ROCK could be more specific than directly inhibiting TGF-β, as it only affects PTHLH expression, without revoking the proliferation suppression mediated by other TGF-β target genes, such as CDKN1A (Supplemental Figure 5A). Overall, our present data support the therapeutic application of Rho-ROCK inhibitors and/or DLC1 surrogates against breast cancer bone metastasis.
Methods
For cell line cultures, primers, colony formation assay, migration and invasion assays, endothelia adhesion assay, trans-endothelial adhesion assay, and luciferase reporter assay, see Supplemental Methods.
Plasmids and reagents.
The plasmids of human DLC1 and the R718E missense mutant within the RhoGAP catalytic domain (23) were gifts from C.J. Der (University of North Carolina, Chapel Hill, North Carolina, USA). The constitutively active RhoA mutant RhoA(63L) was provided by L. Ma (Fudan University, Shanghai, People’s Republic of China). The GLI2 expression plasmid (58) was provided by A.-M. Frischauf (University of Salzburg, Salzburg, Austria). The SBE luciferase reporter plasmid was previously described (59). Mouse Dlc1 was expressed in pMSCV-puro. Human JAG1 and mouse Pthlh were expressed in pMSCV-hygro. The human PTHLH isoform 3 promoter segment, covering –309 to +100 bp, and the mutant were cloned into the pGL3-basic vector. shRNA target sequences were as follows: DLC1 KD1, 5′-CCTTGACTGGAATATGTAA-3′; DLC1 KD2, 5′-CCCGATTGCAAATAGTGAT-3′; SMAD4 KD, 5′-GGATGAATATGTGCATGAC-3′. shRNA sequences were cloned into the pSuper-Retro-Puro retrovirus vector (OligoEngine).
For Western blot and immunohistochemistry (IHC) analyses, the following antibodies were used: mouse anti-human DLC1 (BD); rabbit anti-human RHOA, RHOB, RHOC, phospho-PAK4 (Ser474), and SMAD3 (Cell Signaling Technology); rabbit anti-human PTHLH (Aogma), rabbit anti-human phospho-SMAD3 (Ser423/425) (ab51177, ab9523S; Cell Signaling Technology); Ki67 (ab86373; Abcam); rat anti-human phospho-vimentin (Ser71) (MBL International Corp.); rabbit anti-human SMAD3 pSer204 and pSer208 (gifts from F. Liu, Rutgers University, Piscataway, New Jersey, USA). C3 transferase (1 μg/ml; Cytoskeleton) was used as a Rho inhibitor. A synthetic peptide (10 μg/ml) corresponding to aa 17–32 of CDC42 tagged with a TAT internalization sequence (GRKKRRQRRRPPQC) on the C terminus (24) was used as a CDC42 inhibitor. NSC23766 (50 μM; Santa Cruz) was used as a RAC1 inhibitor. PTHLH inhibitors were PTHLH neutralizing antibody (T-4512; Bachem), 6-TG (Sigma-Aldrich), and PTHLH7–34 (LLHDKGKSIQDLRRRFFLHHLIAEIHTA; GL Biochem). Y27632 and Fasudil (Selleck) were used as ROCK inhibitors. PD98059 (50 μM), P38 MAPK inhibitor (10 μM), JNK inhibitor II (10 μM), and LY294002 (4 μM) were purchased from Calbiochem and used in cell culture to inhibit ERK, P38, JNK, and PI3K, respectively.
Active Rho and Cdc42 assay.
Cancer cells of 80% confluence were gently rinsed once with ice-cold TBS and then lysed. Cell lysis was centrifuged at 16,000 g at 4°C for 15 minutes, and the supernatant was used for active Rho and Cdc42 assays with Active Rho and Cdc42 Detection Kits (nos. 16116 and 16119; Pierce), according to the manufacturer’s protocols.
Stress fiber staining.
When cells reached 40% confluence, they were starved in DMEM containing 0.5% BSA. After 24 hours, cells were fixed with 10% formalin for 15 minutes, permeated with 0.1% Triton X-100 for 10 minutes, and stained with 5 U/ml rhodamine phalloidin (Invitrogen) for 20 minutes. Stained cells were then imaged with a laser confocal microscope. 6 random fields of view per sample were assessed for stress fiber intensity with Image-Pro Plus 5.1 software, as previously described (60, 61).
Osteoclastogenesis assays.
Primary bone marrow osteoclastogenesis analysis was performed with bone marrow from 4- to 6-week-old Balb/c mice essentially as previously described (7), except that 25 ng/ml M-CSF (PeproTech) and no RANKL was used in coculturing media of bone marrow with cancer cells or their CM. Cancer cells were treated with 5 ng/ml TGF-β, 1.0 μg/ml C3, 10 μM Y27632, or 10 μM 6-TG for 72 hours, or infected with RhoA(63L) virus for 48 hours, and the resultant CM were mixed with α-MEM at a 1:9 ratio for bone marrow culturing. 10 μg/ml PTHLH neutralizing antibody (62) and 0.5 μM PTHLH7–34 (63) were added into the bone marrow culture system for PTHLH inhibitor analysis. TRAP staining was performed with a TRAP kit (387A; Sigma-Aldrich). Osteoclasts were defined as TRAP-positive multinucleated cells containing more than 3 nuclei.
For RAW264.7 osteoclastogenesis assay, 105 RAW264.7 cells were added into 48-well plates alone or together with 105 C2C12 cells that were already induced to mature with WNT3a (250 ng/ml; R&D). Osteoclasts were counted on day 6 as described above.
Microarray hybridization and data analysis.
Total RNA from 4 groups of SCP2 cells — with or without DLC1 OE, and treated with 10 ng/ml TGF-β or vehicle for 24 hours — was subjected to hybridization with Affymetrix U133 plus 2.0 arrays, performed by Shanghai Biotechnology Corp. Microarray data were normalized according to the median intensity of each sample.
Genes with untreated/TGF-β–treated cell expression ratios greater than 2 were defined as TGF-β–responsive genes in SCP2 cells. These genes were further defined as those regulated by DLC1 if (a) they were no longer responsive to TGF-β treatment in DLC1 OE cells, and (b) the response difference (expression ratio of OE cells/expression ratio of control cells) exceeded 2-fold. These genes were further divided to groups A and B according to the direction of their response to TGF-β. Microarray data were deposited in GEO (accession no. GSE46214).
ChIP.
ChIP assays were performed using the fast ChIP method (64) with some modifications. Briefly, SCP2 cells with or without DLC1 OE were transfected with WT SMAD3 or EPSM SMAD3 mutant expression plasmids. 1 day later, tumor cells were grown in DMEM with or without TGF-β (10 ng/ml) for 24 hours. Cells were crosslinked with 1% formaldehyde, and 125 mM glycine was used to quench the formaldehyde. Nuclear extracts were sonicated and incubated with control IgG or anti-SMAD3 antibody for immunoprecipitation. Precipitated complexes were eluted and reverse-crosslinked. Captured genomic DNA was purified with the silica membrane purification kit (TIANGEN) and used for PCR analysis. 1% of the total genomic DNA from the nuclear extract was used as input.
Animal studies.
Female Balb/c or Balb/c nude mice aged 4–8 weeks were used in all studies. Orthotopic injection, i.v. injection, and intracardiac injection to study primary tumor growth, lung metastasis, and bone metastasis were performed as previously described (65). Beginning the day after cancer cell injection, mice were treated with i.p. injection of Y27632 (8 mg/kg in 100 μl PBS) every other day, or daily s.c. injection of fasudil (50 mg/kg in 100 μl PBS), 6-TG (1.0 mg/kg in 100 μl PBS), or PTHLH7–34 (200 μg/kg in 100 μl PBS) for 4 weeks in the pharmacological experiments. Control mice received PBS injection. BLI was acquired with a NightOWL II LB 983 Imaging System (Berthold). Bone damages were detected by X-ray radiography with a Faxitron instrument (Faxitron Bioptics) as previously described (65). Osteolytic areas were identified on radiographs as demarcated radiolucent lesions in bone and quantified using ImageJ (NIH).
Histological analysis.
Forelimb and hindlimb long bones of mice were excised, fixed in 10% neutral-buffered formalin for 24 hours, decalcified (10% EDTA, 2 weeks), dehydrated through a graded alcohol series, embedded in paraffin, and stained with H&E.
Clinical analysis.
NKI microarray data and clinical information were obtained from previous publications (17, 39, 40). There were 2 probes for DLC1 in the NKI dataset, and the average intensity of these probes was used for DLC1 expression in each sample. TCGA microarray data and clinical information of breast invasive carcinomas (41) were accessed through bulk download from TCGA ( https://tcga-data.nci.nih.gov/tcga/). As of October 2013, the complete data of 580 samples were obtained. The patients in NKI and TCGA datasets were classified into 2 groups according to median DLC1 level for analyses of Cox survival and PTHLH expression.
Frozen primary breast tumor specimens were obtained from Qilu Hospital of Shandong University, and paraffin-embedded specimens of primary breast tumors and of lymph node and bone metastases were from Shanghai Changzheng Hospital. RNA was extracted from the frozen tumors and corresponding paratumor tissues. See Supplemental Table 2 for detailed clinical information of frozen tumors. DLC1 and PTHLH mRNA levels were measured by qPCR. Patients were classified according to DLC1 median expression level for Cox survival analysis. DLC1 protein levels were analyzed with IHC in the archived tissues, and each sample was scored according to staining intensity, as follows: 0, negative; 1, low; 2, medium; 3, high.
Statistics.
Unless otherwise indicated, data in the figures are presented as mean ± SD. 2-tailed Student’s t test was performed to compare in vitro data. 2-sided Wilcoxon rank test was used to compare BLI data at each time point. Tumor growth and BLI curves were compared by ANOVA. Log-rank test was performed to compare animal and patient survival. A P value less than 0.05 was considered significant.
Study approval.
Human subject studies were approved by the Institutional Review Boards of Qilu Hospital and Changzheng Hospital with informed patient consent. All animal experiments were performed according to the guidelines for the care and use of laboratory animals and were approved by the institutional biomedical research ethics committee of Shanghai Institutes for Biological Sciences.
Supplementary Material
Acknowledgments
We thank F. Liu for SMAD3 plasmids and phosphorylation antibodies; C.J. Der for the R718E mutant DLC1 plasmid; L. Ma for the RhoA(63L) plasmid; A-M. Frischauf for the GLI2 plasmid; C.J. Kirkpatrick for the ST1.6R cell line; and L. Fu, S. Yan, X. Miao, and P. Zhou at the Institute of Health Sciences core facilities for technical support. G. Hu was funded by Chinese National 973 program (2011CB510105, 2013CB910904) and by grants from National Natural Science Foundation of China (81222032, 81071754, 31371409), the Ministry of Science and Technology of China (2012ZX09506-001-005), and Chinese Academy of Sciences (2009OHTP08).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article:J Clin Invest. 2014;124(4):1646–1659. doi:10.1172/JCI71812.
References
- 1.Hess KR, et al. Metastatic patterns in adenocarcinoma. Cancer. 2006;106(7):1624–1633. doi: 10.1002/cncr.21778. [DOI] [PubMed] [Google Scholar]
- 2.Weilbaecher KN, Guise TA, McCauley LK. Cancer to bone: a fatal attraction. Nat Rev Cancer. 2011;11(6):411–425. doi: 10.1038/nrc3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buijs JT, Stayrook KR, Guise TA. TGF-β in the bone microenvironment: role in breast cancer metastases. Cancer Microenviron. 2011;4(3):261–281. doi: 10.1007/s12307-011-0075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bierie B, Moses HL. Tumour microenvironment: TGF-β: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6(7):506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 5.Padua D, Massague J. Roles of TGF-β in metastasis. Cell Res. 2009;19(1):89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 6.Peinado H, Lavotshkin S, Lyden D. The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Semin Cancer Biol. 2011;21(2):139–146. doi: 10.1016/j.semcancer.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Sethi N, Dai X, Winter CG, Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell. 2011;19(2):192–205. doi: 10.1016/j.ccr.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duivenvoorden WC, Hirte HW, Singh G. Transforming growth factor beta1 acts as an inducer of matrix metalloproteinase expression activity in human bone-metastasizing cancer cells. Clin Exp Metastasis. 1999;17(1):27–34. doi: 10.1023/a:1026404227624. [DOI] [PubMed] [Google Scholar]
- 9.Guise TA. Parathyroid hormone-related protein and bone metastases. Cancer. 1997;80(8 suppl):1572–1580. doi: 10.1002/(SICI)1097-0142(19971015)80:8+<1572::AID-CNCR7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 10.Sit ST, Manser E. Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci. 2011;124(pt 5):679–683. doi: 10.1242/jcs.064964. [DOI] [PubMed] [Google Scholar]
- 11.Jeong KJ, et al. The Rho/ROCK pathway for lysophosphatidic acid-induced proteolytic enzyme expression and ovarian cancer cell invasion. Oncogene. 2012;31(39):4279–4289. doi: 10.1038/onc.2011.595. [DOI] [PubMed] [Google Scholar]
- 12.Vasiliev JM, Omelchenko T, Gelfand IM, Feder HH, Bonder EM. Rho overexpression leads to mitosis-associated detachment of cells from epithelial sheets: a link to the mechanism of cancer dissemination. Proc Natl Acad Sci U S A. 2004;101(34):12526–12530. doi: 10.1073/pnas.0404723101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2(2):133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 14.Tang Y, Olufemi L, Wang MT, Nie D. Role of Rho GTPases in breast cancer. Front Biosci. 2008;13:759–776. doi: 10.2741/2718. [DOI] [PubMed] [Google Scholar]
- 15.Liao YC, Lo SH. Deleted in liver cancer-1 (DLC-1): a tumor suppressor not just for liver. Int J Biochem Cell Biol. 2008;40(5):843–847. doi: 10.1016/j.biocel.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang Y, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3(6):537–549. doi: 10.1016/S1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 17.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong D, et al. The SAM domain of the RhoGAP DLC1 binds EF1A1 to regulate cell migration. J Cell Sci. 2009;122(pt 3):414–424. doi: 10.1242/jcs.027482. [DOI] [PubMed] [Google Scholar]
- 19.Li G, Du X, Vass WC, Papageorge AG, Lowy DR, Qian X. Full activity of the deleted in liver cancer 1 (DLC1) tumor suppressor depends on an LD-like motif that binds talin focal adhesion kinase (FAK). Proc Natl Acad Sci U S A. 2011;108(41):17129–17134. doi: 10.1073/pnas.1112122108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75(23):11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goto H, et al. Phosphorylation of vimentin by Rho-associated kinase at a unique amino-terminal site that is specifically phosphorylated during cytokinesis. J Biol Chem. 1998;273(19):11728–11736. doi: 10.1074/jbc.273.19.11728. [DOI] [PubMed] [Google Scholar]
- 22.Callow MG, et al. Requirement for PAK4 in the anchorage-independent growth of human cancer cell lines. J Biol Chem. 2002;277(1):550–558. doi: 10.1074/jbc.M105732200. [DOI] [PubMed] [Google Scholar]
- 23.Healy KD, et al. DLC-1 suppresses non-small cell lung cancer growth invasion by RhoGAP-dependent independent mechanisms. Mol Carcinog. 2008;47(5):326–337. doi: 10.1002/mc.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajnicek AM, Foubister LE, McCaig CD. Temporally and spatially coordinated roles for Rho, Rac, Cdc42 and their effectors in growth cone guidance by a physiological electric field. J Cell Sci. 2006;119(pt 9):1723–1735. doi: 10.1242/jcs.02896. [DOI] [PubMed] [Google Scholar]
- 25.Moorman JP, Luu D, Wickham J, Bobak DA, Hahn CS. A balance of signaling by Rho family small GTPases RhoA, Rac1 and Cdc42 coordinates cytoskeletal morphology but not cell survival. Oncogene. 1999;18(1):47–57. doi: 10.1038/sj.onc.1202262. [DOI] [PubMed] [Google Scholar]
- 26.Roodman GD. Mechanisms of bone metastasis. N Engl J Med. 2004;350(16):1655–1664. doi: 10.1056/NEJMra030831. [DOI] [PubMed] [Google Scholar]
- 27.Padua D, et al. TGFβ primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133(1):66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roodman GD. Genes associate with abnormal bone cell activity in bone metastasis. Cancer Metastasis Rev. 2012;31(3–4):569–578. doi: 10.1007/s10555-012-9372-x. [DOI] [PubMed] [Google Scholar]
- 29.Guise TA, et al. Evidence for a causal role of parathyroid hormone-related protein in the pathogenesis of human breast cancer-mediated osteolysis. J Clin Invest. 1996;98(7):1544–1549. doi: 10.1172/JCI118947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Datta NS, Abou-Samra AB. PTH and PTHrP signaling in osteoblasts. Cell Signal. 2009;21(8):1245–1254. doi: 10.1016/j.cellsig.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallwitz WE, Guise TA, Mundy GR. Guanosine nucleotides inhibit different syndromes of PTHrP excess caused by human cancers in vivo. J Clin Invest. 2002;110(10):1559–1572. doi: 10.1172/JCI0211936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nissenson RA, Diep D, Strewler GJ. Synthetic peptides comprising the amino-terminal sequence of a parathyroid hormone-like protein from human malignancies. Binding to parathyroid hormone receptors and activation of adenylate cyclase in bone cells and kidney. J Biol Chem. 1988;263(26):12866–12871. [PubMed] [Google Scholar]
- 33.Thota CS, Reed LC, Yallampalli C. Effects of parathyroid hormone like hormone (PTHLH) antagonist, PTHLH(7-34), on fetoplacental development growth during midgestation in rats. Biol Reprod. 2005;73(6):1191–1198. doi: 10.1095/biolreprod.105.044628. [DOI] [PubMed] [Google Scholar]
- 34.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature. 2003;425(6958):577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 35.Wang G, Matsuura I, He D, Liu F. Transforming growth factor-β-inducible phosphorylation of Smad3. J Biol Chem. 2009;284(15):9663–9673. doi: 10.1074/jbc.M809281200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bae E, et al. Smad3 linker phosphorylation attenuates Smad3 transcriptional activity TGF-β1/Smad3-induced epithelial-mesenchymal transition in renal epithelial cells. Biochem Biophys Res Commun. 2012;427(3):593–599. doi: 10.1016/j.bbrc.2012.09.103. [DOI] [PubMed] [Google Scholar]
- 37.Sterling JA, et al. The hedgehog signaling molecule Gli2 induces parathyroid hormone-related peptide expression osteolysis in metastatic human breast cancer cells. Cancer Res. 2006;66(15):7548–7553. doi: 10.1158/0008-5472.CAN-06-0452. [DOI] [PubMed] [Google Scholar]
- 38.Ying H, et al. The Rho kinase inhibitor fasudil inhibits tumor progression in human and rat tumor models. Mol Cancer Ther. 2006;5(9):2158–2164. doi: 10.1158/1535-7163.MCT-05-0440. [DOI] [PubMed] [Google Scholar]
- 39.van ‘t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 40.Bos PD, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sihto H, et al. Breast cancer biological subtypes and protein expression predict for the preferential distant metastasis sites: a nationwide cohort study. Breast Cancer Res. 2011;13(5):R87. doi: 10.1186/bcr2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong MH, Pavlakis N. Optimal management of bone metastases in breast cancer patients. Breast Cancer (Dove Med Press). 2011;3:35–60. doi: 10.2147/BCTT.S6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross DT, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24(3):227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 45.Chambers AF. MDA-MB-435 and M14 cell lines: identical but not M14 melanoma? Cancer Res. 2009;69(13):5292–5293. doi: 10.1158/0008-5472.CAN-09-1528. [DOI] [PubMed] [Google Scholar]
- 46.Goodison S, et al. The RhoGAP protein DLC-1 functions as a metastasis suppressor in breast cancer cells. Cancer Res. 2005;65(14):6042–6053. doi: 10.1158/0008-5472.CAN-04-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou X, et al. DLC1 suppresses distant dissemination of human hepatocellular carcinoma cells in nude mice through reduction of RhoA GTPase activity, actin cytoskeletal disruption and down-regulation of genes involved in metastasis. Int J Oncol. 2008;32(6):1285–1291. doi: 10.3892/ijo_32_6_1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henderson MA, et al. Parathyroid hormone-related protein localization in breast cancers predict improved prognosis. Cancer Res. 2006;66(4):2250–2256. doi: 10.1158/0008-5472.CAN-05-2814. [DOI] [PubMed] [Google Scholar]
- 49.Henderson M, et al. Parathyroid hormone-related protein production by breast cancers, improved survival, and reduced bone metastases. J Natl Cancer Inst. 2001;93(3):234–237. doi: 10.1093/jnci/93.3.234. [DOI] [PubMed] [Google Scholar]
- 50.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 51.Kaplan RN, Psaila B, Lyden D. Bone marrow cells in the ‘pre-metastatic niche’: within bone and beyond. Cancer Metastasis Rev. 2006;25(4):521–529. doi: 10.1007/s10555-006-9036-9. [DOI] [PubMed] [Google Scholar]
- 52.Massague J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mardilovich K, Olson MF, Baugh M. Targeting Rho GTPase signaling for cancer therapy. Future Oncol. 2012;8(2):165–177. doi: 10.2217/fon.11.143. [DOI] [PubMed] [Google Scholar]
- 54.Micuda S, Rosel D, Ryska A, Brabek J. ROCK inhibitors as emerging therapeutic candidates for sarcomas. Curr Cancer Drug Targets. 2010;10(2):127–134. doi: 10.2174/156800910791054202. [DOI] [PubMed] [Google Scholar]
- 55.Ruppender NS, et al. Matrix rigidity induces osteolytic gene expression of metastatic breast cancer cells. PLoS One. 2010;5(11):e15451. doi: 10.1371/journal.pone.0015451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu S, Goldstein RH, Scepansky EM, Rosenblatt M. Inhibition of rho-associated kinase signaling prevents breast cancer metastasis to human bone. Cancer Res. 2009;69(22):8742–8751. doi: 10.1158/0008-5472.CAN-09-1541. [DOI] [PubMed] [Google Scholar]
- 57.Martin TJ. Manipulating the environment of cancer cells in bone: a novel therapeutic approach. J Clin Invest. 2002;110(10):1399–1401. doi: 10.1172/JCI0217124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winklmayr M, et al. Non-consensus GLI binding sites in Hedgehog target gene regulation. BMC Mol Biol. 2010;11:2. doi: 10.1186/1471-2199-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korpal M, et al. Imaging transforming growth factor-beta signaling dynamics and therapeutic response in breast cancer bone metastasis. Nat Med. 2009;15(8):960–966. doi: 10.1038/nm.1943. [DOI] [PubMed] [Google Scholar]
- 60.Acharya PS, et al. Fibroblast migration is mediated by CD44-dependent TGF beta activation. J Cell Sci. 2008;121(pt 9):1393–1402. doi: 10.1242/jcs.021683. [DOI] [PubMed] [Google Scholar]
- 61.Lei R, et al. Oncogene. Suppression of MIM by microRNA-182 activates RhoA promotes breast cancer metastasis [published online ahead of print March 11, 2013]. doi: 10.1038/onc.2013.65 . [DOI] [PubMed] [Google Scholar]
- 62.Mak IW, et al. PTHrP induces autocrine/paracrine proliferation of bone tumor cells through inhibition of apoptosis. PLoS One. 2011;6(5):e19975. doi: 10.1371/journal.pone.0019975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sherafat-Kazemzadeh R, Schroeder JK, Kessler CA, Handwerger S. Parathyroid hormone-like hormone (PTHLH) represses decidualization of human uterine fibroblast cells by an autocrine/paracrine mechanism. J Clin Endocrinol Metab. 2011;96(2):509–514. doi: 10.1210/jc.2010-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1(1):179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 65.Hu G, et al. MTDH activation by 8q22 genomic gain promotes chemoresistance metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15(1):9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.