Abstract
Variants in SCN10A, which encodes a voltage-gated sodium channel, are associated with alterations of cardiac conduction parameters and the cardiac rhythm disorder Brugada syndrome; however, it is unclear how SCN10A variants promote dysfunctional cardiac conduction. Here we showed by high-resolution 4C-seq analysis of the Scn10a-Scn5a locus in murine heart tissue that a cardiac enhancer located in Scn10a, encompassing SCN10A functional variant rs6801957, interacts with the promoter of Scn5a, a sodium channel–encoding gene that is critical for cardiac conduction. We observed that SCN5A transcript levels were several orders of magnitude higher than SCN10A transcript levels in both adult human and mouse heart tissue. Analysis of BAC transgenic mouse strains harboring an engineered deletion of the enhancer within Scn10a revealed that the enhancer was essential for Scn5a expression in cardiac tissue. Furthermore, the common SCN10A variant rs6801957 modulated Scn5a expression in the heart. In humans, the SCN10A variant rs6801957, which correlated with slowed conduction, was associated with reduced SCN5A expression. These observations establish a genomic mechanism for how a common genetic variation at SCN10A influences cardiac physiology and predisposes to arrhythmia.
Introduction
Genome-wide association studies (GWAS) on ECG measures in diverse populations have consistently associated noncoding genetic variants within introns of the voltage-gated sodium channel gene SCN10A with cardiac conduction system (CCS) function (1–9). Recent work associated noncoding variants at SCN10A with Brugada syndrome, a cardiac rhythm disorder with a high risk of sudden death (10). The implication of SCN10A, which had not been previously studied in the heart, in cardiac rhythm parameters and Brugada syndrome by GWAS (Figure 1A) was unexpected. Given the position of the associated variants within the gene, including one whose minor allele caused a nonsynonymous substitution in SCN10A, it was assumed that the variants were functionally connected with Nav1.8, the protein product of SCN10A. However, functional follow-up analysis of SCN10A provided conflicting evidence about whether Nav1.8 accelerates (5) or slows (2) conduction velocity, and the magnitude of the effects of blocking Scn10a in mice was small (2, 5). Therefore, the role of SCN10A in cardiac physiology remained controversial (11–13).
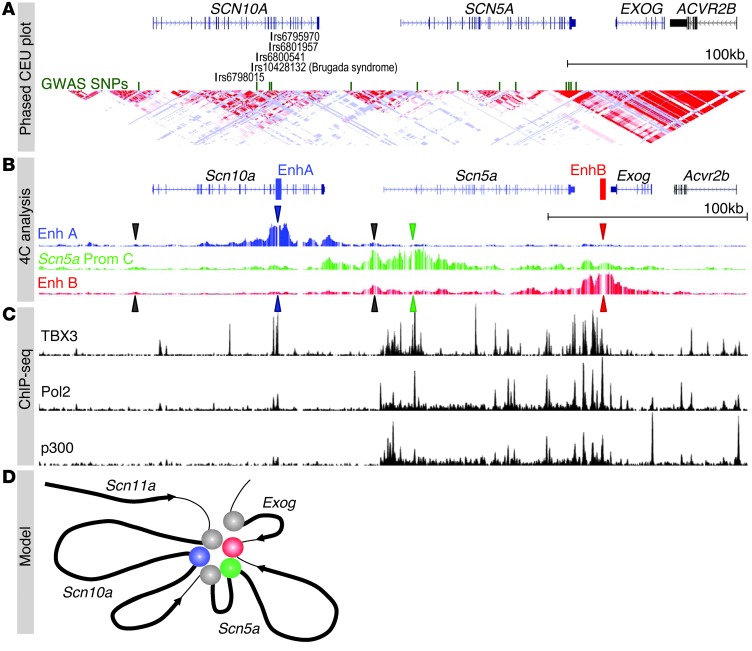
Figure 1. Contact profiles of enhancers and promoters of the Scn10a-Scn5a locus in combination with ChIP-seq data.
(A) UCSC genome browser view of chr3:38,465,426–38,861,154 (hg18), demonstrating distinct LD blocks within the SCN10A-SCN5A locus with SNPs tagged in GWAS studies (green) (1–9). (B) Mouse region (mm9; chr9:119,303,698–119,662,489) depicted with 4C analysis showing contact profiles of EnhA in Scn10a (blue), Scn5a promoter C (green), and EnhB downstream of Scn5a (red). Blue, green, and red arrows correspond to the position of the different viewpoints. Black arrows represent the position of the promoter of Scn10a and alternate promoters of Scn5a (promoters A and B). From the EnhA viewpoint, interactions can be observed between EnhA, EnhB, and the Scn5a promoter regions. From the Scn5a promoter C viewpoint, contacts can be observed with EnhA and EnhB and weakly with the Scn10a promoter region. From EnhB, contacts with the Scn5a promoter region and, weakly, with EnhA can be seen. See Supplemental Figure 1 for quantitative assessment of these interactions. (C) UCSC genome browser views of ChIP-seq data of the TBX3 (21) and of Pol2 and p300 (27–29) binding profiles aligned with the 4C traces, showing the correspondence between contacts and factor occupancy. (D) Model for the interactions of EnhA (blue) and EnhB (red) with the Scn5a and Scn10a promoters (green and gray, respectively). The promoter of flanking gene Exog does not interact.
Interestingly, SCN10A maps immediately adjacent to SCN5A (encoding Nav1.5) on chromosome 3p22.2. In contrast to SCN10A, SCN5A has a well-described role in cardiac physiology and pathophysiology (14–18). CCS function is exquisitely sensitive to Nav1.5 dose, and heterozygous mutations in SCN5A underlie numerous human conduction system diseases, including Brugada syndrome, long QT syndrome, atrial fibrillation, progressive cardiac conduction disease, and sudden cardiac death (14, 19, 20). Furthermore, noncoding variants within introns and downstream of SCN5A have also been associated with ECG parameters in GWAS.
We recently demonstrated the importance of the T-box transcription factors Tbx5 and Tbx3 in the regulation of Scn5a in mice (21, 22). The rapid conducting portions of the CCS require high levels of Nav1.5 for function (23). Strong Scn5a expression is driven in the fast conduction system by TBX5, a transcriptional activator (22). Conversely, repression of Scn5a is a key feature ensuring the slow propagation of the cardiac electrical impulse across the sinus node and atrioventricular node (17, 23). TBX3, a transcriptional repressor, plays a major role in the repression of Scn5a expression in slowly conducting nodal regions of the developing heart (24, 25). We previously investigated the gene regulatory landscape at the Scn10a-Scn5a cluster and identified 2 T-box–regulated enhancers, one positioned downstream of Scn5a and the other in an intron of Scn10a. A SNP, rs6801957, associated with QRS prolongation and in linkage disequilibrium (LD) with variants associated with Brugada syndrome (5, 10) is located in the Scn10a intronic enhancer. This SNP modulates T-box factor binding and activity of the isolated enhancer fragment (21).
We investigated the novel hypothesis that genetic variation at SCN10A associated with cardiac rhythm disease and conduction mechanistically functions by modulating SCN5A expression. We also tested the hypothesis that the regulatory landscape of SCN5A may be regulated by multiple cis-regulatory elements spread over long-range distances, including cis-regulatory elements within introns of SCN10A. In such a scenario, functional SNPs at SCN10A associated with conduction parameters and disease may reflect disruption of these regulatory elements causing altered expression of SCN5A. Using high-resolution chromatin conformation capture (4C), we demonstrated the intricate association of the Scn10a intronic enhancer with the promoters of Scn5a and Scn10a and the downstream Scn5a enhancer. We found that SCN5A was expressed at high levels, whereas SCN10A was expressed at background levels, in the adult human and mouse heart by RNA-seq. We found that the cis-regulatory element located in Scn10a regulated the pattern of Scn5a expression in the heart, and that the rs6801957 SNP affected cardiac expression of Scn5a using a BAC reporter strategy. Finally, we found a direct correlation of SCN5A expression in humans with the presence of the rs6801957 risk-associated SNP in the SCN10A intronic enhancer. Together, our data provided a genomic mechanism explaining how common genetic variants at SCN10A influence cardiac physiology and predispose to arrhythmia.
Results
High-resolution 4C-seq reveals interactions between an enhancer in Scn10a and the Scn5a promoters.
We previously identified 2 regions with conserved enhancer activity able to autonomously drive expression of a reporter construct in the murine CCS (21, 22). The region termed enhancer A (EnhA) spans 2 adjacent introns and 1 exon in Scn10a, and enhancer B (EnhB) is positioned 15 kbp downstream of Scn5a (Figure 1B). EnhA contains a SNP previously found to be associated with QRS prolongation (5). This SNP disrupts a T-box factor binding site that affects EnhA activity in a zebrafish assay (21). Enhancers regulate gene expression by physically interacting with the promoter of a gene. We used the newly developed tool high-resolution 4C-seq (26) to investigate on a genome-wide scale which gene promoters are contacted by EnhA and EnhB, and which genomic regions are contacted by the promoters of Scn5a and Scn10a (Figure 1B). We aligned 4C-seq with cardiac ChIP-seq data for TBX3 (21) as well as for Pol2 and p300 (27–29) to visualize interactions for EnhA and EnhB (Figure 1C). By setting 4C bait regions around 2 known promoter regions for Scn5a, we found that both EnhA and EnhB possessed clear cis interactions with the Scn5a and Scn10a promoters (Figure 1, B and C, and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI73140DS1). Moreover, contacts between Scn5a promoters A and B and the Scn10a promoter were observed. When observed from bait sets in the enhancers themselves, EnhA was found to associate with the Scn5a and Scn10a promoters and the EnhB region, and EnhB was found to associate with the Scn5a promoter region and EnhA. These interaction results revealed that EnhA (in Scn10a) and EnhB (downstream of Scn5a) interacted with both promoters and with each other, which indicates that the enhancers and promoters form a complex (Figure 1D). This topology, in which both enhancers and gene promoters are in close contact, suggests that both enhancers are potential regulators of Scn5a and Scn10a. This is consistent with the observation that both genes show similar spatial patterns of expression in the fetal heart (21). Furthermore, these data indicate that the variants identified in cardiac rhythm and disease GWAS in EnhA may affect the regulation of Scn5a, a dosage-sensitive regulator of cardiac conduction (Figure 1D).
Substantial SCN5A transcripts, but not SCN10A transcripts, in the adult mouse and human heart.
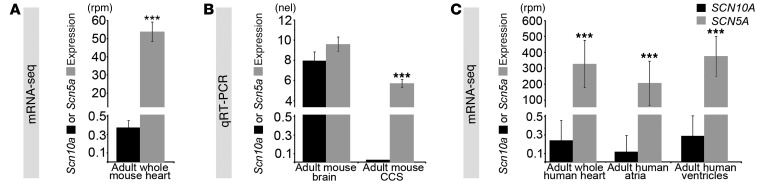
In the fetal heart, expression of cardiac Scn5a is much higher than that of Scn10a (21), which indicates that the contribution of Scn10a to the sodium current before birth is small. To evaluate the expression levels of Scn5a and Scn10a in the adult, we performed RNA-seq on whole mouse hearts (30). We found that Scn5a expression was 145-fold higher than Scn10a expression (53.81 ± 5.39 versus 0.37 ± 0.08 reads per million [rpm]; Figure 2A). Therefore, Scn10a expression in the heart as a whole was considered to be at background levels. Nevertheless, it remained conceivable that Scn10a expression was present in the limited subset of CCS cells based on the pattern of Scn10a expression before birth (21). We therefore evaluated Scn5a and Scn10a expression in the atrioventricular bundle from adult mouse hearts microdissected at 6 weeks of age (Figure 2B). We confirmed the anatomic specificity of isolated tissue, observing robust expression of Gja5 (also known as Connexin40), a gene whose expression in the ventricles is limited to the conduction system (31, 32). In the atrioventricular bundle, expression of Scn5a was high, whereas Scn10a expression was undetectable (Figure 2B).
Figure 2. Scn5a and Scn10a expression analysis.
(A) Expression of Scn5a in the 12-week-old adult mouse heart was 145-fold higher than that of Scn10a (53.81 ± 5.39 versus 0.37 ± 0.08 rpm) by mRNA-seq. (B) CCS expression of Scn5a in the 12-week-old adult mouse heart was significantly higher than that of Scn10a, whose expression was undetectable by qRT-PCR. All samples were analyzed in triplicate. (C) mRNA-seq analysis of adult human heart tissue. SCN5A was expressed 1,357-fold higher than SCN10A in the whole heart (326 ± 149 versus 0.24 ± 0.22 rpm), 1,753-fold higher in the atria (205 ± 138 versus 0.12 ± 0.18 rpm), and 1,299-fold higher in the ventricles (376 ± 126 versus 0.29 ± 0.21 rpm). Data are mean ± SD. ***P < 0.001, Student’s t test.
We next evaluated SCN5A and SCN10A expression from human heart samples by RNA-seq (Figure 2C). SCN5A expression was more than 1,000-fold greater than SCN10A expression in the whole heart, atria, and ventricles (whole heart, 326 ± 149 versus 0.24 ± 0.22 rpm, P = 2.5 × 10–27; atria, 205 ± 138 versus 0.12 ± 0.17 rpm, P = 1.0 × 10–3; ventricles, 376 ± 126 versus 0.29 ± 0.21 rpm, P = 1.1 × 10–26; Figure 2C). The absolute magnitude of SCN10A expression, less than 1 part per million, corresponded to a cellular content of less than 1 SCN10A RNA molecule per 2 cells (33–35) and qualified as not expressed in the heart (30). These data indicated that SCN5A but not SCN10A is expressed at appreciable, physiologically relevant levels in the heart. Therefore, we hypothesized that GWAS SNPs in noncoding regions at the SCN10A locus affecting cardiac conduction (2–5) modulate SCN5A expression.
The enhancer in Scn10a is required for expression of Scn5a in vivo.
We tested the hypothesis that both EnhA and EnhB are necessary for endogenous Scn5a expression by engineering a BAC reporter system for Scn5a in the mouse. Specifically, we used recombineering strategies (36) to replace the endogenous first Scn5a coding exon with the LacZ reporter gene in mouse BAC RP23-198L19. This Scn5a-LacZ BAC included the entire Scn5a coding region, EnhA, and EnhB (Figure 3A). Independent transgenic lines carrying the WT Scn5a-LacZ reporter BAC RP23-198L19 showed a pattern of LacZ expression highly reminiscent of endogenous Scn5a expression (17, 21), including reproducible expression in structures of the ventricular conduction system including the atrioventricular bundle and ventricular myocardium in adult (3 of 3) and embryonic (6 of 6) hearts (Figure 3B, Supplemental Figure 2, A–F, and Supplemental Figure 4A).
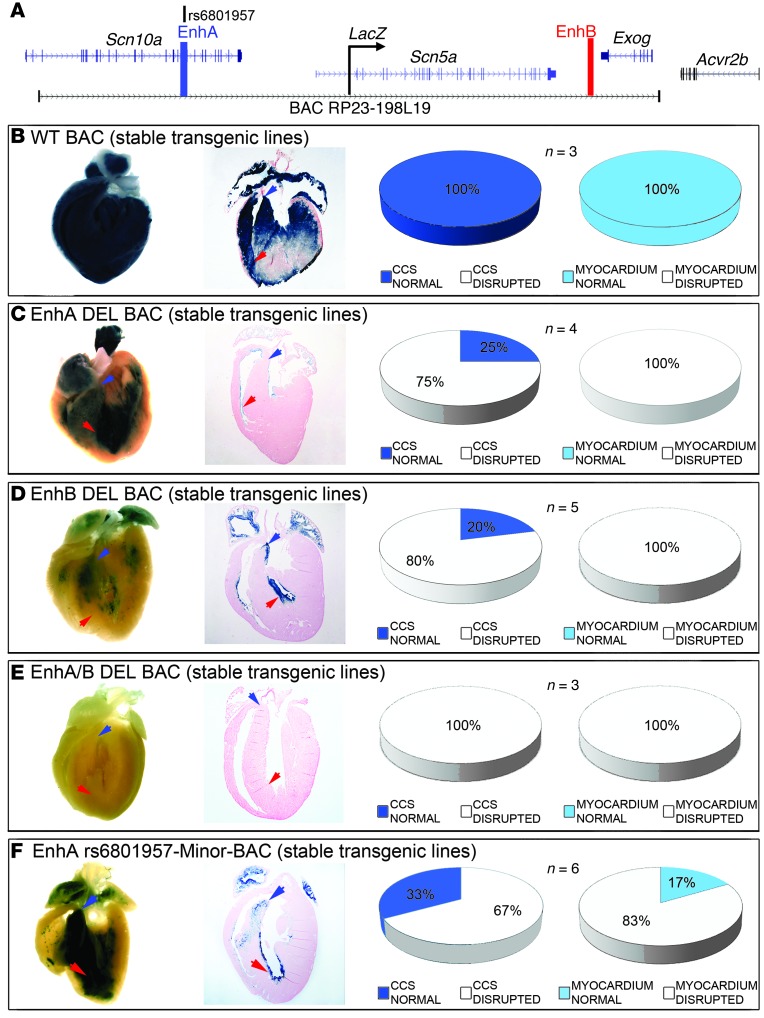
Figure 3. EnhA and EnhB are necessary for Scn5a cardiac expression.
(A) Modified murine BAC RP23-198L19 with LacZ inserted into the endogenous Scn5a translational start site. (B–F) Stable BAC transgenic lines, shown in whole-mount and cross-section histology; arrows indicate AV bundle (blue) and distal bundle branches (red). Pie charts show the distribution of LacZ expression in each genotype class. All studies were performed at 12 weeks of age; the number of independent transgenic lines examined is indicated (each analyzed in triplicate). (B) The WT enhancer demonstrated robust CCS and myocardial expression. (C) EnhA deletion eliminated proximal CCS and myocardial expression. (D) EnhB deletion eliminated distal CCS and myocardial expression. (E) Deletion of both enhancers eliminated CCS and myocardial expression entirely. (F) The minor allele at rs6801957 markedly altered Scn5a-LacZ expression, which was either entirely absent or absent from the atrioventricular bundle and confined to the distal ventricular septum in the majority of cases.
To investigate the necessity of EnhA and EnhB for Scn5a-LacZ reporter expression, we engineered deletions of EnhA (chr9:119,540,800–119,544,032), EnhB (chr9:119,378,051–119,379,479), or both from the BAC reporter and then evaluated Scn5a-LacZ expression in vivo. Removal of EnhA alone significantly abrogated Scn5a-LacZ: expression of LacZ was absent from ventricular myocardium in all independent adult and embryonic EnhA deletion BAC transgenics (Figure 3C, Supplemental Figure 2, G–O, and Supplemental Figure 4B). LacZ expression was disrupted in the CCS in the majority of EnhA deletion BAC transgenics. When present, LacZ expression was confined to the distal ventricular septum (Figure 3C, Supplemental Figure 2, G–O, and Supplemental Figure 4B). Removal of EnhB alone also substantially altered and reduced LacZ expression from the Scn5a locus (Figure 3D, Supplemental Figure 2, P–V, and Supplemental Figure 4C). Interestingly, the region of ventricular CCS Scn5a-LacZ expression that was maintained in EnhB deletion BAC transgenics (Figure 3D, Supplemental Figure 2, R–T, and Supplemental Figure 4C) was reciprocal to that maintained in EnhA deletion BAC transgenics (Figure 3C, Supplemental Figure 2, H–J, and Supplemental Figure 4B). Removal of both EnhA and EnhB together completely eliminated Scn5a-LacZ expression in all independent adult and embryonic transgenics (Figure 3E, Supplemental Figure 2, W–AC, and Supplemental Figure 4D). Taken together, these observations demonstrated that EnhA and EnhB are necessary and sufficient for in vivo Scn5a expression.
The SNP rs6801957, associated with conduction slowing in GWAS, leads to lower SCN5A expression in mice and in humans.
The observation that EnhA, which is located in Scn10a, was required for Scn5a expression suggested that common genetic variation affecting conduction system physiology and disease at SCN10A may modify SCN5A expression. A sentinel SNP located in SCN10A for conduction system physiology, rs6801957 (5), lies within EnhA (Figure 1A and Figure 3A). The major allele for rs6801957, G, establishes a conserved canonical T-box binding site (GGTGACAG) and promotes enhancer activity, whereas the minor allele A, associated with conduction slowing, disrupts a core nt in the T-box site (GGTAACAG) and decreases enhancer activity (21). This SNP is in strong LD with the nonsynonymous SCN10A SNP evaluated in prior studies (rs6795970, HapMap 2 CEU population, D′ = 1.0, r2 = 0.96, refs. 2, 3), with 2 other SNPs that implicated SCN10A in cardiac physiology in independent studies (rs6800541, HapMap 2 CEU population, D′ = 1.0, r2 = 0.96, ref. 4; rs6798015, HapMap 2 CEU population, D′ = 0.955, r2 = 0.84, ref. 1), and with the SNP that implicated SCN10A in Brugada syndrome (rs10428132, HapMap 2 CEU population, D′ = 1.0, r2 = 0.97, ref. 10) (Figure 1A).
We tested the allele-specific effects of the murine nt orthologous to SNP rs6801957 on Scn5a expression. The rs6801957 minor allele (A) was recombineered into the orthologous region of the Scn5a-LacZ BAC, and LacZ expression was analyzed in adult murine hearts (Figure 3F, Supplemental Figure 3, and Supplemental Figure 4E). The minor allele at rs6801957 substantially altered Scn5a-LacZ expression: expression of LacZ was markedly disrupted in the compact myocardium of the majority of independent transgenics (Figure 3F, Supplemental Figure 3, and Supplemental Figure 4E). LacZ expression was also significantly disrupted in the CCS of the majority of independent adult (4 of 6) and half of embryonic (8 of 16) transgenics (P = 0.04; Figure 3F, Supplemental Figure 3, and Supplemental Figure 4E). We concluded that the risk allele orthologous to SNP rs6801957 substantially decreased Scn5a expression and, conversely, that the major allele orthologous to SNP rs6801957 is necessary for normal CCS Scn5a expression.
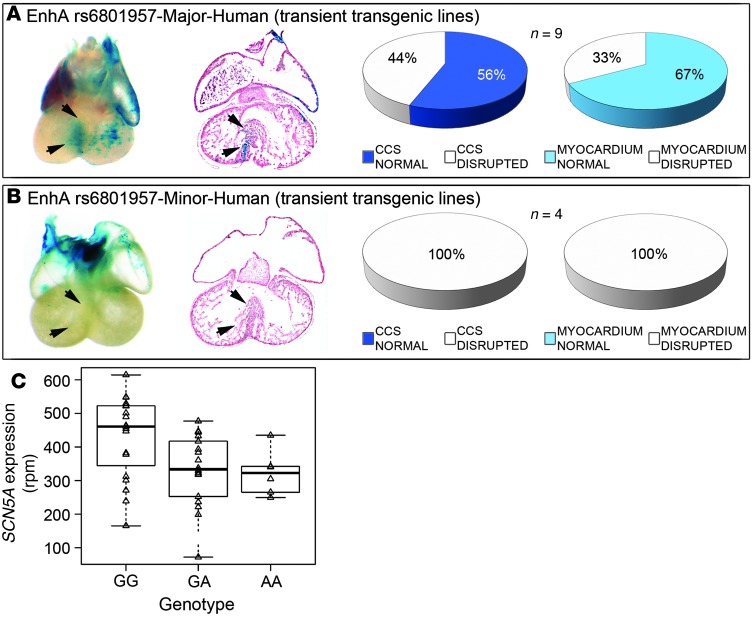
We next tested whether rs6801957 affects the sufficiency of human EnhA sequences to drive CCS expression in transgenic mice in vivo (Figure 4). Human EnhA with the major allele of rs6801957 was sufficient to drive Scn5a-LacZ expression in myocardium and the CCS in the majority of transient transgenic mouse embryos (Figure 4A). However, EnhA with the minor allele was insufficient for cardiac expression (P = 0.05; Figure 4B). We concluded that the major allele of SNP rs6801957 is necessary for normal CCS activity of EnhA.
Figure 4. SNP rs6801957 modulates EnhA activity.
Transient transgenic embryos at E14.5 harboring human 2.2-kb EnhA with major (A) and minor (B) allele of SNP rs6801957. Human EnhA with the major allele (n = 9) was sufficient to drive CCS expression (A), but human EnhA with the minor allele (n = 4) was not (B). (C) Correlation of SCN5A expression in human hearts with SNP rs6801957 genotype. SCN5A expression was assessed by RNA-seq in 42 human cardiac tissue samples, of which 19 were homozygous GG, 18 were heterozygous GA, and 6 were homozygous AA at locus rs6801957. Homozygous GG individuals had significantly higher SCN5A expression (428 ± 128 rpm) than GA (331 ± 104 rpm) or AA (322 ± 66 rpm) individuals. Each A allele reduced expression by 12% ± 6% (P = 0.01). A linear additive regression model was used to determine statistical significance. Lines within boxes represent median value; boxes represent interquartile range; whiskers represent 5th and 95th percentiles.
We hypothesized that rs6801957 influenced SCN5A expression in humans. Therefore, we evaluated SCN5A mRNA levels as a function of genotype at this SNP in 2 independent heart tissue bio-banks by RNA-seq and quantitative real-time RT-PCR (qRT-PCR). In the first set, GG samples (homozygous for the major allele associated with shorter PR and QRS intervals) expressed substantially more SCN5A mRNA (428 ± 128 normalized rpm; n = 19) than GA (331 ± 104 rpm; n = 18) or AA (322 ± 66 rpm; n = 6) tissue samples. In this set, each additional copy of the A allele reduced cardiac SCN5A expression by 12% ± 6% (additive genetic model, P = 0.01; dominant genetic model, P = 0.01). SCN10A was not appreciably expressed in any genotype. In the second set, samples homozygous for GG at rs6801957 expressed significantly more SCN5A RNA (0.060 ± 0.029 normalized expression; n = 33) than GA (0.048 ± 0.017 normalized expression; n = 37) or AA (0.050 ± 0.028 normalized expression; n = 17) tissue samples (additive genetic model, P = 0.11; dominant genetic model, P = 0.03). In this set, although the level of SCN5A mRNA did not increase linearly per copy of G allele, genotype at rs6801957 was significantly associated with SCN5A mRNA levels with the same direction of effect as in the first sample set. SCN10A variant rs6801957 had no detectable effect on SCN10A expression, although power to detect allele specific expression on SCN10A would be very poor, given its low expression in adult cardiac tissue.
Discussion
Our results unveil the genomic regulatory logic controlling Scn5a expression, with implications for interpreting cardiac GWAS and understanding the genomic basis of CCS function and disease. The physical associations between enhancers and promoters at the Scn10a-Scn5a locus were analyzed by 4C, a technique that provides a genome-wide view of all cis interactions (26, 37). Viewpoints from both EnhA and EnhB revealed interactions with the Scn5a and Scn10a promoter regions and, additionally, multiple contact points in and beyond Scn5a, fitting with ChIP-seq data. These contacts were confirmed when taking the promoters as viewpoint. Our findings indicate that the enhancers and promoters at this locus are all in close proximity, allowing the enhancers to simultaneously interact with and potentially regulate the transcription of both Scn5a and Scn10a. A conserved genomic architecture that enables enhancer sharing and coregulation of genes within an evolutionary conserved cluster has also been observed for the Irx and Hox gene clusters (38). Although the enhancers contact the Scn10a promoter, quantification of Scn10a transcript levels indicated that Scn10a transcription was not activated by these enhancers. Therefore, we focused our attention on the functionality of interactions between these enhancers and Scn5a. Using BAC transgenesis, an assay complementary to 4C contact mapping, we demonstrated the functionality of the interactions between EnhA and EnhB and Scn5a. The enhancers functioned in a modular pattern to determine Scn5a expression: EnhA, located in Scn10a, was required for proximal ventricular CCS expression; EnhB, located downstream of Scn5a, was required for distal ventricular CCS expression; and both enhancers were required for ventricular myocardium expression.
Our data indicate that the physiological role underlying common variants at SCN10A in cardiac rhythm control is not ascribed to the SCN10A gene product Nav1.8, but instead to the SCN5A gene product Nav1.5. SNPs identified by GWAS have modest effect sizes, an attribute inherent to their common frequency (39). This suggests that noncoding SNPs associated with human phenotypes by GWAS may cause expression alterations to genes with dosage-sensitive physiology. This paradigm held only for SCN5A, not SCN10A. SCN5A has an exquisitely dosage-sensitive relationship to ECG parameters and cardiac rhythm control. Heterozygous mutations in human SCN5A underlie numerous human conduction system diseases, including Brugada syndrome, long QT syndrome, atrial fibrillation, progressive cardiac conduction disease, and sudden cardiac death (14, 19, 20). Furthermore, noncoding variants within introns and downstream of SCN5A have also been associated with ECG parameters in GWAS. The mouse homozygous null phenotype is embryonic lethal, and mice haploinsufficient for Scn5a display a host of profound cardiac rhythm disturbances, including slowed cardiac conduction and a Brugada-like phenotype, similar to those associated with SNPs at neighboring SCN10A (2, 5). In contrast, whether the SCN10A product Nav1.8 plays a role in cardiac physiology remains controversial (11–13). The Scn10a knockout mouse displayed subtle cardiac ECG abnormalities, including modest PR interval shortening indicative of more rapid than normal conduction (2). This phenotype is at odds with the role of voltage-gated sodium channels in the rapid phase of myocardial depolarization, in which a decrease in channel function and concomitant sodium current would slow conduction (as observed by lowering Scn5a expression). Moreover, functional analysis of Scn10a indicated that the effects of pharmacologically blocking or genetically inactivating Scn10a are small, and provided conflicting evidence about whether Nav1.8 accelerates (5) or slows (2) conduction velocity. Expression analysis also supports a more substantial role for the gene product of Scn5a than that of Scn10a. During embryonic development, low levels of Scn10a expression can be qualitatively observed using in situ hybridization (21, 22) and qRT-PCR, demonstrating expression levels 4–5 times lower than those of Scn5a. However, as development progressed to adult stages, Scn10a expression remained low, whereas Scn5a transcript levels were induced to levels 100-fold greater than those of Scn10a. In humans, the expression discrepancy was 1,000-fold in favor of SCN5A over SCN10A. Although transcripts for both genes could be detected in heart tissue by PCR-based approaches, the confluence of data — including our SCN10A transcript quantification in adult heart tissue, our genotype-SCN5A expression correlation, and physiological observations of Scn10a knockout mice by others (2) — suggest that the contribution of Nav1.8 to cardiac physiology is minor. The absence of substantial effects in the Scn10a knockout, combined with the dramatic effects on electrophysiology observed for Scn5a knockout models (16, 18, 40), supports the contention that common genetic variation at the SCN10A locus affects cardiac physiology via dysregulation of SCN5A expression. Our observations do not formally rule out the possibility of other mechanisms affecting conduction physiology at the SCN10A-SCN5A locus. However, the totality of the evidence, including our present observations, strongly supports the relevance of the genomic mechanism we describe to cardiac conduction physiology and arrhythmia risk.
Mechanistically connecting noncoding genetic variations to their associated traits requires a commitment to understanding the functional biology of cis-regulatory elements, given that up to 85% of GWAS SNPs associated with human disease traits are noncoding (41, 42). Here, we presented an integrative and comprehensive experimental strategy to functionally link noncoding variants mapping within long-range cis-regulatory elements to their target genes. Combining high-resolution 4C strategies with engineering of BACs and mouse transgenic reporter assays, we demonstrated that the regulatory landscape of SCN5A extended into SCN10A and that the ECG-associated SNPs within SCN10A modified the function of an SCN5A enhancer. Using genomics and genetics data, we further demonstrated that these SNPs were associated with expression of SCN5A, but not SCN10A, in human hearts. Together, our data established SCN5A as the functional target of the noncoding variants within SCN10A associated with ECG parameters. The present work provides a genomic mechanism for the effect of common genetic variants at SCN10A on cardiac physiology and disease and provides a strong rationale for careful interrogation of the noncoding activity of GWAS variants prior to ascribing function to annotated genes. Our findings also suggest a genomic pathophysiological mechanism for Brugada syndrome, with the potential to refine diagnosis and implications for future therapeutic approaches.
Methods
Experimental animals
Generation of Scn5a-LacZ BAC transgenic mice was performed by the University of Chicago Transgenic Core Facility by pronuclear microinjection of DNA. Mice were maintained on a mixed genetic background and analyzed at E13.5, E14.5, and 12 weeks of age. All experiments used age-, gender-, and genetic strain-matched controls to account for any variations in data sets compares across experiments. Mice were bred and housed in specific pathogen–free conditions in a 12-hour light/12-hour dark cycle and allowed ad libitum access to standard mouse chow and water. Mice requiring medical attention were provided with appropriate veterinary care by a licensed veterinarian and were excluded from the experiments described. No other exclusion criteria existed.
4C template preparation
4C templates were prepared as previously described (43). In short, adult mouse hearts were isolated in ice-cold PBS. Single-cell suspensions were obtained by dissociation of tissue with IKA Ultra Turrax T5 FU, followed by dounce homogenization. Chromatin was cross-linked with 2% formaldehyde in PBS with 10% FCS for 10 minutes at room temperature, nuclei were isolated, and cross-linked DNA was digested with a primary restriction enzyme recognizing a 4-bp restriction site (DpnII), followed by proximity ligation. Cross-links were removed, and a secondary restriction enzyme digestion (Csp6I) was performed, followed again by proximity ligation. For all experiments, 200 ng of the resulting 4C template was used for the subsequent PCR reaction, of which 16 (total, 3.2 μg of 4C template) were pooled and purified for next-generation sequencing. The PCR products were purified using 2 columns per sample of the High Pure PCR Product Purification Kit (catalog no. 11732676001; Roche). The kit separates the PCR products larger than 120 bp from the adaptor-containing primers (∼75 and ∼40 nt in size). Similar results were obtained with products from a single PCR reaction (200-ng template).
4C-seq primer design
PCR primers were designed based on the following criteria. The size of the viewpoint fragment was at least 500 bp, to allow efficient cross-linking to other DNA fragments. The fragment end (the region between the primary and secondary restriction enzyme) was more than 350 bp, to allow efficient circularization during the second ligation step. Primers were designed to be maximally 20 nt in length. The strategy therefore produces sequencing reads (36 mers in this study) composed of the 4C primer sequence (20 nt, specific to a given viewpoint) followed by 16 nt that identify a captured sequence. The reading primer always hybridizes to, and ends at, the 3′ side of the first restriction recognition site. This design ensures analysis of only primary ligation events and provides sufficient sequence information to unambiguously identify most captured sequences. The nonreading primers, 18–20 nt in size, were designed at a distance ≤100 bp from the secondary restriction site. All primers had 35%–65% GC content and an optimal basic temperature of 55°C, ranging 45°C–65°C. Primers were checked against the mouse genome with MegaBLAST23 (settings, -p 88.88, -W 12, -e 1, -F T), which requires primers on the reading side to be matched uniquely in the genome and primers on the nonreading side to have a maximum of 3 perfectly matching BLAST high-scoring segment pairs (HSP). See Supplemental Table 1 for all primers used.
4C data analysis and statistics
4C templates were mixed and sequenced simultaneously in 1 Illumina HiSeq 2000 lane. The sequence tags generated by the procedure were prefixed by the 4C reading primer, which includes the DpnII restriction site sequence (see 4C-seq primer design). The 4C reading primer sequences were separated from multiplexed 4C-seq libraries, and the suffixes were extracted for further processing. Mapping and filtering of the sequence reads was done as previously described (26). The algorithm constructs a background model for remote intra- and interchromosomal contacts to correct for systematic biases that can occur during the 4C-seq experimental protocol. The algorithm is designed to use controls for sequencing errors and nonunique sequences while considering the high coverage (100×–100,000×) of fragment ends that are proximal to the viewpoint fragment. To normalize the interactions in close proximity to the viewpoint, the algorithm was used to calculate the median of normalized coverage for running windows of size 4 kb (depicted as black line) and sliding windows of 2–50 kb of linearly increasing size (depicted as color-coded multiscale diagrams). All median values represent enrichment relative to the maximum attainable 4-kb median value, whereas sliding windows represent enrichment relative to the maximum attainable 12-kb median value. The 20th and 80th percentiles were also computed and depicted as the green area around the 4-kb running windows (Supplemental Figure 1).
mRNA-seq and genotyping rs6801957
Ventricular tissue set 1.
Human left ventricular samples (n = 39) were obtained from patients of mixed ancestry who were given left ventricular assist devices. Discarded left ventricular tissue was obtained at the time of insertion of the device. Human right atrial samples (n = 4) were obtained as discarded tissue from patients who had cardiac surgery to repair their congenital heart disease lesion. The expression levels of Scn5a and Scn10a in adult mouse and human cardiac tissue were assessed by RNA-seq as described previously (30, 33–35). Samples were blinded to genotype at the SCN locus. Post-hoc analyses showed that there was no association between lane or batch and expression of SCN5A or SCN10A expression, independent of genotype. The rs6801957 genotype was assessed by dideoxy sequencing of PCR-amplified genomic DNA using standard procedures (44). rs6801957 sequences were amplified using the following primers: primer A, CACCTGGAGCTCCCTAAGA; primer B, GAGTCTGTAGCTCTCCCATAG. Primer A was used as sequencing primer. A linear additive regression model was used to determine statistical significance (45).
Ventricular tissue set 2.
Human left ventricular samples (n = 129) were collected from nonimplanted human donor hearts that were considered suitable for transplantation, yet not used due to logistical reasons. All heart samples used in this study were from individuals of self-reported European descent and were collected at centers in Szeged (gift from A. Varro, University of Szeged, Szeged, Hungary), Sydney (gift from C. dos Remedios, University of Sydney, Sydney, Australia), Miami (gift from N.H. Bishopric, University of Miami School of Medicine, Miami, Florida, USA), and Vanderbilt (gift from A.L. George, Vanderbilt University, Nashville, Tennessee, USA). Samples were flash-frozen and stored in liquid nitrogen. RNA and DNA were isolated from cardiac tissue following standard protocols. Manhattan distance hierarchical clustering using genome-wide SNP genotypic data confirmed a genetically homogeneous group. Genotype at rs6801957 was determined by means of a Taqman assay (Applied Biosystems). cDNA was prepared using Oligo-dT and Thermoscript First-Strand Synthesis System (Invitrogen) and used for qRT-PCR for determination of SCN5A transcript levels. All qRT-PCR assays were performed in triplicate. Expression values were normalized by dividing by the geometric mean of HPRT1 and TNNI3 mRNA expression values (46). For each set of triplicates, the mean, SD, and coefficient of variation were calculated. Sets with a coefficient of variation higher than 0.20 were left out of the analysis, as previously described (47), leaving a total of 87 subjects for analysis. The association between rs6801957 and SCN5A transcript levels (Supplemental Figure 5) was tested using an additive and dominant genetic model, taking gender, age, and center of origin as covariates. All calculations were performed in statistical programming language R, version 2.15.3.
BAC modification
Mouse BAC RP23-198L19 (mm9; chr9:119,351,100–119,598,458) was engineered in vitro using recombineering kits and protocols from Gene Bridges GmbH (Heidelberg). BAC RP23-198L19 was converted into an Scn5a transcriptional reporter (Scn5a-LacZ) by inserting a LacZ-ampicillin cassette, in frame, replacing the first exon of Scn5A using the Red/ET recombination kit and protocol from Gene Bridges (catalog no. K001). Primers used for generation of the recombineering cassette were as follows: Scn5a-LAI forward, CTTCCAGGCAGCCTGAGGAGAGCCTGTGCCCCCAGAAGCAGGATGAGAAGATGGCTCGCGATGATCCCGTCGT; Scn5a-LAI reverse, AGCCTCCTCCTCTGGCAGGCCCTCACGGCTCTCCTGTGAGGTGGCCGAACCTCGAGGCTAGCTCTAGAAGTCCAGC. Ampicillin-resistant colonies were PCR screened for homologous recombination using primers in the unaffected genomic region flanking the insertion and within the vector cassette, and the insertion junction was verified by sequencing using the following primers: Scn5A 5pI forward, CATCACACCCTGTGTTTGTCTC; Scn5A 5pI reverse, GGTACTATAGAAAGGGTCCAGGTCT; Scn5A 3pI forward, AGAAGATGGCAAACTTCCTGTTAC; Scn5A 3pI reverse, TGATTGGAATACAGATTAATGGTGA. Recombinant BACs were confirmed intact by restriction digest fingerprinting with PspXI (NEB).
EnhA and EnhB were deleted in the Scn5a-LacZ BAC through their targeted replacement, again using the Red/ET recombination kit and protocol from Gene Bridges (catalog no. K001). EnhA (chr9:119,540,801–119,544,032) was replaced with a spectinomycin resistance gene amplified from the iTol2-Amp plasmid using the following primers: Scn5a EnhADel forward, CTTAGGCAACCAGCCTGAATAGAAGCTGAAGCCACACCCAGCATTCCAGGGATAAAAATATATCATCATGCCTCCTC; Scn5a EnhADel reverse, GGCTCTTTGAAGAATCTGGCACAACTGTACATGGTCACTGCCTATCTGGTCACGTTAAGGGATTTTGGTCA. Insertion site location and fidelity was determined by PCR amplification and sequencing using the following primers flanking and internal to the junction: Scn5a EnhADelCheck5p forward, CAAAGGGCAGGTGAGAAGTC; Scn5a EnhADelCheck5p reverse, CGAACCGAACAGGCTTATGT; Scn5a EnhADelCheck3p forward, CACCAAGGTAGTCGGCAAAT; Scn5a EnhADelCheck3p reverse, CTCGGAGGAGCTTGTGTCAT. EnhB (chr9:119,378,052–119,379,479) was replaced with a kanamycin/neomycin resistance gene amplified from the rpsL-neo plasmid (Gene Bridges kit no. K002), using the following primers: Scn5a EnhBDel forward, TGAACTCAGCTTGTCAGGTTTGACAGCAAATACCGTTACCGGCCGAGCCAGGCCTGGTGATGATGGCGGGATC; Scn5a EnhBDel reverse, CCTGACTCTTGAGGTACATTCTTGCCCCCTTCCCCCCGTCCCCAACGTGATCAGAAGAACTCGTCAAGAAGGCG. The insertion site was checked with the following flanking/internal primers: Scn5a EnhBDelCheck forward, GCTTGTCAGGTTTGACAGCA; rpsL reverse, CAGACGAACACGGCATACTTTAC; Kan forward, ATCAGGATGATCTGGACGAAGAG; Scn5a EnhBDelCheck reverse, TGGCCTGACTCTTGAGGTACA.
Recombinant colonies were selected with appropriate antibiotic, and the BAC was confirmed intact by fingerprint analysis with PspXI.
SNP rs6801957, in EnhA, was converted from the major G allele to the minor A allele, without leaving an antibiotic resistance gene behind using the 2-step counter selection BAC modification kit (Gene Bridges kit no. K002). In the first step, a counter-selection cassette containing a kanamycin resistance and a streptomycin sensitivity gene was inserted into EnhA using primers EnhA MutCsHA1 (CAGAGTTCGTGTTCTTTACTCCCGGGAGGTGACACTCTGGCCTCGGCTGCGGCCTGGTGATGATGGCGGGATCG) and EnhA MutCsHA2 (CTAACAGCTGCTGCTATCAACTATCATTTTCGAGATTCCTTTGTCTGAGTTCAGAAGAACTCGTCAAGAAGGCG), resulting in the deletion of rs6801957. Recombinant colonies were selected for with kanamycin, and the insertion site was verified with the following primers: EnhA MutCsSpan forward, AATACACTGCGGGAGGTTTG; rpsL reverse, CAGACGAACACGGCATACTTTAC; Kan forward, ATCAGGATGATCTGGACGAAGAG; EnhA MutCsSpan reverse, CCTGGAGCCTTCTGATAACG. In the second step, the counter-selection cassette was replaced with a recombineering cassette consisting of an A nt flanked by homology arms of 205 and 290 bp, thereby restoring rs6801957, but with the minor allele. This recombination cassette was generated by first cloning the EnhA region into a pGL3 vector and converting the rs6801957 allele from G to A through traditional mutagenesis techniques, followed by PCR amplification from the EnhA MutCsSpan forward and reverse primers above and DpnI digestion to remove plasmid template. After recombination, colonies retaining the counter-selection cassette were selected against with streptomycin, and 5 of 5 sequenced colonies possessed the A allele. BACs were again checked for length and content using separate restriction digest fingerprints with NotI, PspXI, and EcoRV.
BAC DNA was prepared for pronuclear injection using Nucleobond PC20 Kit (Macherey-Nagel) from a fresh, overnight culture. An aliquot was checked for length and degradation via PFGE, and the remainder was dialyzed against PBS and submitted for pronuclear injection. Pronuclear injections were performed by the University of Chicago Transgenic Core Facility, supported by University of Chicago Cancer Center.
Human enhancer transgenics
Cloning of the human enhancer and testing in transient transgenics was performed as described previously (21).
Statistics
Values represent mean ± SD of the indicated number of measurements. Statistical significance was determined using 2-tailed Student’s t test or a linear additive regression model. A P value less than 0.05 was considered significant. No statistical method was used to predetermine sample size, and animal experiments were not performed in a blinded fashion. Mice were assigned at random to treatment groups for all mouse studies.
Study approval
All experiments were performed under an University of Chicago IACUC-approved protocol (ACUP no. 71737) and in compliance with US Public Health Service Policy on Humane Care and Use of Laboratory Animals.
Supplementary Material
Acknowledgments
We thank Nanette H. Bishopric, Alfred L. George, Andras Varro, and Cristobal dos Remedios for providing samples for expression analysis in human donor hearts. The study was funded by grants from the NIH (R01 HL114010, to I.P. Moskowitz and M.A. Nobrega), AHA (10PRE2670012, to D.E. Arnolds), the European Community’s Seventh Framework Programme contract (“CardioGeNet” 223463, to V.M. Christoffels), the Cardiovascular Onderzoek Nederland (PREDICT, to C.R. Bezzina), the German Foundation for Heart Research (F/09/10, to O. Toka), and the Federal Ministry of Education and Research (BMBF; FKZ01GI0601, to O. Toka).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article:J Clin Invest. 2014;124(4):1844–1852. doi:10.1172/JCI73140.
See the related Commentary beginning on page 1460.
References
- 1.Smith JG, et al. Genome-wide association studies of the PR interval in African Americans. PLoS Genet. 2011;7(2):e1001304. doi: 10.1371/journal.pgen.1001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers JC, et al. Genetic variation in SCN10A influences cardiac conduction. Nat Genet. 2010;42(2):149–152. doi: 10.1038/ng.516. [DOI] [PubMed] [Google Scholar]
- 3.Holm H, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42(2):177–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 4.Pfeufer A, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42(2):153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sotoodehnia N, et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet. 2010;42(12):1068–1076. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denny JC, et al. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation. 2010;122(20):2016–2021. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfeufer A, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41(4):407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho YS, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41(5):527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 9.Gudbjartsson DF, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448(7151):353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 10.Bezzina CR, et al. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet. 2013;45(9):1044–1049. doi: 10.1038/ng.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.London B. Whither art thou, SCN10A, and what art thou doing? Circ Res. 2012;111(3):268–270. doi: 10.1161/CIRCRESAHA.112.275032. [DOI] [PubMed] [Google Scholar]
- 12.Yang T, Atack TC, Stroud DM, Zhang W, Hall L, Roden DM. Blocking Scn10a channels in heart reduces late sodium current and is antiarrhythmic. Circ Res. 2012;111(3):322–332. doi: 10.1161/CIRCRESAHA.112.265173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verkerk AO, et al. Functional Nav1.8 channels in intracardiac neurons: the link between SCN10A and cardiac electrophysiology. Circ Res. 2012;111(3):332–343. doi: 10.1161/CIRCRESAHA.112.274035. [DOI] [PubMed] [Google Scholar]
- 14.Rook MB, Evers MM, Vos MA, Bierhuizen MF. Biology of cardiac sodium channel Nav1.5 expression. Cardiovasc Res. 2012;93(1):12–23. doi: 10.1093/cvr/cvr252. [DOI] [PubMed] [Google Scholar]
- 15.Stengl M. Experimental models of spontaneous ventricular arrhythmias and of sudden cardiac death. Physiol Res. 2010;59(suppl 1):S25–S31. doi: 10.33549/physiolres.932001. [DOI] [PubMed] [Google Scholar]
- 16.Leoni AL, et al. Variable Na(v)1.5 protein expression from the wild-type allele correlates with the penetrance of cardiac conduction disease in the Scn5a(+/–) mouse model. PLoS One. 2010;5(2):e9298. doi: 10.1371/journal.pone.0009298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Remme CA, et al. The cardiac sodium channel displays differential distribution in the conduction system and transmural heterogeneity in the murine ventricular myocardium. Basic Res Cardiol. 2009;104(5):511–522. doi: 10.1007/s00395-009-0012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papadatos GA, et al. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc Natl Acad Sci U S A. 2002;99(9):6210–6215. doi: 10.1073/pnas.082121299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilde AA, Brugada R. Phenotypical manifestations of mutations in the genes encoding subunits of the cardiac sodium channel. Circ Res. 2011;108(7):884–897. doi: 10.1161/CIRCRESAHA.110.238469. [DOI] [PubMed] [Google Scholar]
- 20.Tfelt-Hansen J, Winkel BG, Grunnet M, Jespersen T. Inherited cardiac diseases caused by mutations in the Nav1.5 sodium channel. J Cardiovasc Electrophysiol. 2010;21(1):107–115. doi: 10.1111/j.1540-8167.2009.01633.x. [DOI] [PubMed] [Google Scholar]
- 21.van den Boogaard M, et al. Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J Clin Invest. 2012;122(7):2519–2530. doi: 10.1172/JCI62613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnolds DE, et al. TBX5 drives Scn5a expression to regulate cardiac conduction system function. J Clin Invest. 2012;122(7):2509–2518. doi: 10.1172/JCI62617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abriel H. Cardiac sodium channel Na(v)1.5 and interacting proteins: Physiology and pathophysiology. J Mol Cell Cardiol. 2010;48(1):2–11. doi: 10.1016/j.yjmcc.2009.08.025. [DOI] [PubMed] [Google Scholar]
- 24.Hoogaars WM, et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21(9):1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.[No authors listed]. Effect of metoprolol CR/XL in chronic heart failure: Metoprolol CR/XL Randomised Intervention Trial in Congestive Heart Failure (MERIT-HF) Lancet. 1999;353(9169):2001–2007. [PubMed] [Google Scholar]
- 26.van de Werken HJ, et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat Methods. 2012;9(10):969–972. doi: 10.1038/nmeth.2173. [DOI] [PubMed] [Google Scholar]
- 27.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488(7409):116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blow MJ, et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42(9):806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457(7231):854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakabe NJ, et al. Dual transcriptional activator and repressor roles of TBX20 regulate adult cardiac structure and function. Hum Mol Genet. 2012;21(10):2194–2204. doi: 10.1093/hmg/dds034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delorme B, et al. Expression pattern of connexin gene products at the early developmental stages of the mouse cardiovascular system. Circ Res. 1997;81(3):423–437. doi: 10.1161/01.res.81.3.423. [DOI] [PubMed] [Google Scholar]
- 32.Miquerol L, et al. Architectural and functional asymmetry of the His-Purkinje system of the murine heart. Cardiovasc Res. 2004;63(1):77–86. doi: 10.1016/j.cardiores.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 33.DeLaughter DM, et al. Spatial transcriptional profile of the chick and mouse endocardial cushions identify novel regulators of endocardial EMT in vitro. J Mol Cell Cardiol. 2013;59:196–204. doi: 10.1016/j.yjmcc.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herman DS, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366(7):619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Islam S, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21(7):1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2(10):769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 37.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26(1):11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tena JJ, et al. An evolutionarily conserved three-dimensional structure in the vertebrate Irx clusters facilitates enhancer sharing and coregulation. Nat Commun. 2011;2:310. doi: 10.1038/ncomms1301. [DOI] [PubMed] [Google Scholar]
- 39.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322(5903):881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Probst V, et al. Haploinsufficiency in combination with aging causes SCN5A-linked hereditary Lenegre disease. J Am Coll Cardiol. 2003;41(4):643–652. doi: 10.1016/S0735-1097(02)02864-4. [DOI] [PubMed] [Google Scholar]
- 41.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods. 2007;4(11):895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 44.Montgomery KT, et al. Mutation detection using automated fluorescence-based sequencing. Curr Protoc Hum Genet. 2008;Chapter 7:Unit7.9. doi: 10.1002/0471142905.hg0709s57. [DOI] [PubMed] [Google Scholar]
- 45.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28(10):1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal Biochem. 2000;285(2):194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.