Abstract
The G1 kinase CDK4 is amplified or overexpressed in some human tumors and promotes tumorigenesis by inhibiting known tumor suppressors. Here, we report that CDK4 deficiency markedly accelerated lymphoma development in the Eμ-Myc transgenic mouse model of B lymphoma and that silencing or loss of CDK4 augmented the tumorigenic potential of Myc-driven mouse and human B cell lymphoma in transplant models. Accelerated disease in CDK4-deficient Eμ-Myc transgenic mice was associated with rampant genomic instability that was provoked by dysregulation of a FOXO1/RAG1/RAG2 pathway. Specifically, CDK4 phosphorylated and inactivated FOXO1, which prevented FOXO1-dependent induction of Rag1 and Rag2 transcription. CDK4-deficient Eμ-Myc B cells had high levels of the active form of FOXO1 and elevated RAG1 and RAG2. Furthermore, overexpression of RAG1 and RAG2 accelerated lymphoma development in a transplant model, with RAG1/2-expressing tumors exhibiting hallmarks of genomic instability. Evaluation of human tumor samples revealed that CDK4 expression was markedly suppressed, while FOXO1 expression was elevated, in several subtypes of human non-Hodgkin B cell lymphoma. Collectively, these findings establish a context-specific tumor suppressor function for CDK4 that prevents genomic instability, which contributes to B cell lymphoma. Furthermore, our data suggest that targeting CDK4 may increase the risk for the development and/or progression of lymphoma.
Introduction
MYC oncoproteins function as transcription factors that coordinate the expression of a large cast of genes that direct cell metabolism, growth, and division. Elevated MYC levels are a hallmark of rapidly dividing human malignancies, and this has major effects on cell physiology, provoking hyperproliferative and DNA damage responses (DDRs) as well as apoptosis (1, 2). In the Eμ-Myc transgenic mouse, a model of human B cell lymphoma (3), bypass of these three checkpoints accompanies malignant transformation. First, MYC activates a DDR in premalignant B cells (4), perhaps via MYC’s effects on unscheduled firing of DNA replication origins (5). Second, MYC induces the Arf tumor suppressor that inactivates the E3 ubiquitin ligase MDM2, leading to p53 stabilization and p53-dependent apoptosis (6, 7). Third, MYC triggers p27Kip1 destruction by the SCFSkp2 complex by inducing Cks1 expression as well as the expression of cyclin D2 (Ccnd2), which sequesters p27Kip1, and both responses likely contribute to the hyperproliferative response of MYC (8).
c-MYC directly induces the transcription of Cdk4 (9), a G1 serine/threonine kinase (10, 11) that is required for tumorigenesis in some contexts (12–14). CDK4 activation requires binding to the regulatory cyclin subunits cyclin D1, -D2, or -D3. CDK4:cyclin D complexes phosphorylate and inactivate the retinoblastoma (Rb) tumor-suppressor protein, releasing E2F transcription factors that regulate genes necessary for entry and progression through the S phase (15). Accordingly, CDK4 inhibitors block the activity of CDK4:cyclin D complexes and the proliferation of some cell types (15).
Given these facts, it has long been thought that CDK4 functions as an oncogene via its ability to suppress the functions of Rb or of the SMAD3 or FOXM1 tumor suppressors (16, 17). Indeed, some tumor types, for example mantle cell lymphoma, present with amplified CDK4 or CCND1 (cyclin D1) (18, 19). Further, loss-of-function mutations in the CDK inhibitors p16Ink4a and p15Ink4b occur in tumor types that display elevated CDK4:cyclin D activity (15, 20). In addition, loss or knockdown of CDK4 or cyclin-D1 disables RAS- and Neu-induced tumorigenesis (14, 21, 22), and a mutant form of cyclin D1 that binds to, but cannot activate, CDK4 also impairs Neu-induced tumorigenesis (23). Moreover, the CDK4:cyclin D2 complex is necessary for tumorigenesis provoked by Apc loss, which is MYC dependent (24). Finally, melanoma cells are highly reliant on CDK4, or on the related kinase CDK6, to suppress senescence, making them particularly susceptible to CDK4/6 inhibition (17).
One of the conundrums in cancer biology is that there are context-specific contributions of the Rb pathway in harnessing malignancy. For example, cyclin D1 or Cdk4 loss does not impair MYC- or WNT-driven breast adenocarcinoma (14) or tumor development in p53-deficient mice (25). Further, there is no selection for Rb loss during Myc-driven lymphomagenesis in Eμ-Myc transgenic mice (7), and selective inactivation of Ink4a does not affect MYC-driven lymphoma development (26).
Though DNA damage dampens CDK activity, for example, via p53-directed activation of the general CDK inhibitor p21Cip1, overall CDK activity also controls the DDR, where CDKs are necessary to repair double-strand breaks and for optimal activation of checkpoints (27). Given the necessary role of CDK4 downstream of MYC in epithelial cell tumorigenesis (12) versus the contrasting roles of MYC and CDKs in controlling the DDR (4, 28), we tested whether Cdk4 loss would affect Myc-driven lymphomagenesis. Surprisingly, we found that CDK4 deficiency markedly accelerates the onset of Myc-driven lymphoma, that loss or silencing of CDK4 augments the tumorigenic potential of extant lymphoma, and that CDK4 expression is suppressed in many subtypes of human B cell lymphoma. Further, we show that the lymphoma-promoting effects of this CDK4 deficiency are associated with loss of a CDK4/FOXO1/RAG1/RAG2 pathway that controls genomic stability.
Results
CDK4 deficiency accelerates Myc-driven lymphomagenesis.
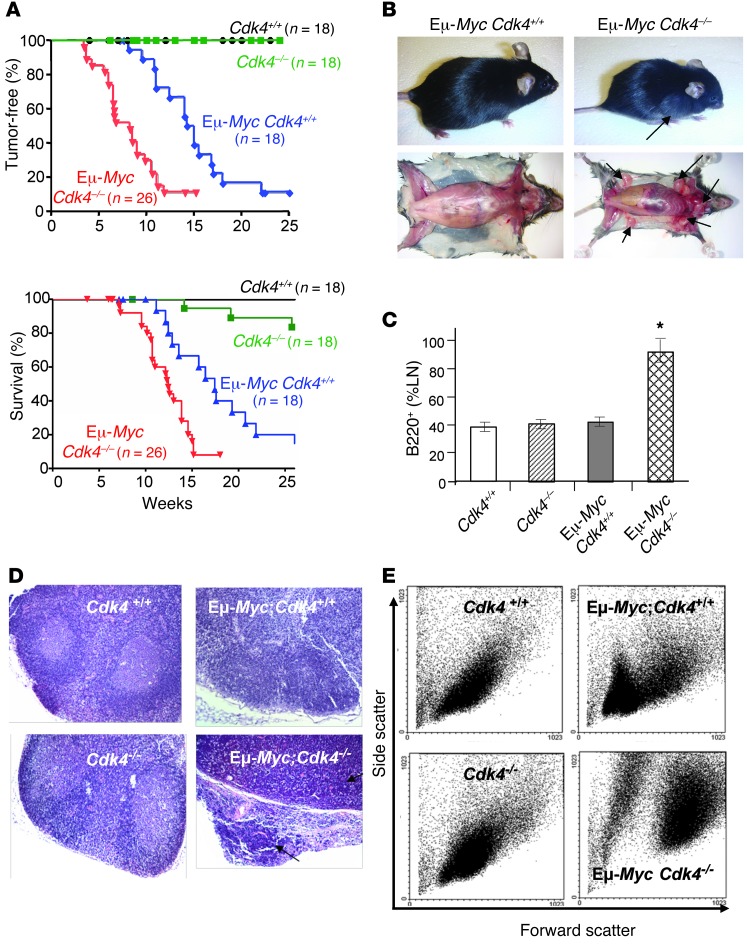
CDK4 deficiency is associated with rather late-onset hyperglycemia and type 2 diabetes that can lead to lethality in some animals, but these animals are not tumor prone (11, 13, 29). We therefore tested the potential role of CDK4 in Myc-driven lymphoma by crossing Cdk4+/– mice with Eμ-Myc transgenic mice (C57Bl/6). Eμ-MycCdk4+/– F1 offspring were then bred with Cdk4+/– mice to generate the three desired transgenic cohorts, Eμ-MycCdk4+/+, Eμ-MycCdk4+/–, and Eμ-MycCdk4–/– mice (Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI63139DS1), and these animals were then monitored daily for tumor onset and signs of morbidity. As expected, lymphomas were not observed in Cdk4+/+ or Cdk4–/– littermates. Eμ-MycCdk4+/+ mice displayed a typical course of lymphoma onset and survival, with a mean mortality of 18 weeks (Figure 1A). Surprisingly, Eμ-MycCdk4–/– mice had an accelerated course of lymphoma onset and corresponding reductions in lifespan (mean mortality 11 weeks, Figure 1A). Indeed, by 7 weeks of age, most Eμ-MycCdk4–/– mice had enlarged LNs (Figure 1B) that were composed almost exclusively of large immature B220+ B cells, whereas this was not evident in LNs from age-matched Eμ-MycCdk4+/+ littermates (Figure 1, B, C, E, and Supplemental Figure 1A). Eμ-MycCdk4+/– mice displayed an intermediate phenotype, with a mean survival of 13 weeks (Supplemental Figure 2A). Notably, Eμ-MycCdk4+/– lymphomas did not display loss of the remaining wild-type Cdk4 allele, and they expressed approximately half the level of CDK4 protein (Supplemental Figure 2B). Thus, CDK4 deficiency accelerates Myc-driven lymphomagenesis, and CDK4 functions as a modifier of lymphoma development.
Figure 1. CDK4 deficiency accelerates MYC-induced lymphomagenesis.
(A) Kaplan-Meier tumor-free mice (top) and survival (bottom) for Cdk4+/+, Cdk4–/–, Eμ-MycCdk4+/+, and Eμ-MycCdk4–/– littermates. Time to appearance of palpable cervical and peripheral LNs (>5 mm in at least one dimension) is shown for Eμ-MycCdk4+/+ and Eμ-MycCdk4–/– littermates. n = number of mice in each cohort. Statistical evaluation of tumor onset is based on the Mantel-Cox test for comparison of the Kaplan-Meier event-time format and on an unpaired Student’s t test for comparison of means and SDs. P = 0.0002 for E-MycCdk4–/– versus Eμ-MycCdk4+/+ mice. (B) Eμ-MycCdk4–/– mice developed rapid-onset lymphoma. Upper right panel: the majority of 7-week-old Eμ-Myc Cdk4–/– mice were moribund and had enlarged LNs (arrows, lower right panel). At this juncture, the LNs of Eμ-MycCdk4+/+ littermates were normal. (C) Approximately 90% of cells from the enlarged LNs of 7-week-old E-MycCdk4–/– mice were B220+ cells, whereas LNs from Cdk4+/+, Cdk4–/–, and Eμ-MycCdk4+/+ mice only had 34%–40% total B220+ B cells. Results are representative of 8 different mice in each cohort; mean ± SD, P = 0.0037. FACS analyses are shown in Supplemental Figure 1A. *P < 0.05. (D) H&E staining of LNs from 7-week-old Cdk4+/+ (upper left), Cdk4–/– (lower left), Eμ-MycCdk4+/+ (upper right), and Eμ-MycCdk4–/– (lower right) littermates. Enlarged LNs of Eμ-MycCdk4–/– mice had lymphomas with disseminated characteristics (black arrows) versus normal LN architecture in Cdk4+/+, Cdk4–/–, and Eμ-MycCdk4+/+ littermates. Original magnification, ×40. (E) FACS scatter plots showing that cells of LNs from 7-week-old Eμ-MycCdk4–/– mice were composed of a homogeneous population of large (B220+) cells.
Histological analyses revealed that the tumors arising in the LNs of Eμ-MycCdk4–/– mice were disseminated B cell lymphomas (Figure 1, C and D, and Supplemental Figure 1B). In contrast, normal LN architecture was present in 7-week-old Cdk4+/+, Cdk4–/–, and Eμ-MycCdk4+/+ mice (Figure 1D). We found that the lymphoma cells arising in the LNs of Eμ-MycCdk4–/– mice were large (Figure 1E) and were immature and mature B cell lymphomas typical of those found in Eμ-Myc transgenic mice (ref. 30, Supplemental Figure 1B, and Supplemental Table 2). Thus, CDK4 deficiency accelerates the onset of B lymphomas that typify Eμ-Myc mice.
CDK4 deficiency augments genomic instability of MYC-expressing B cells.
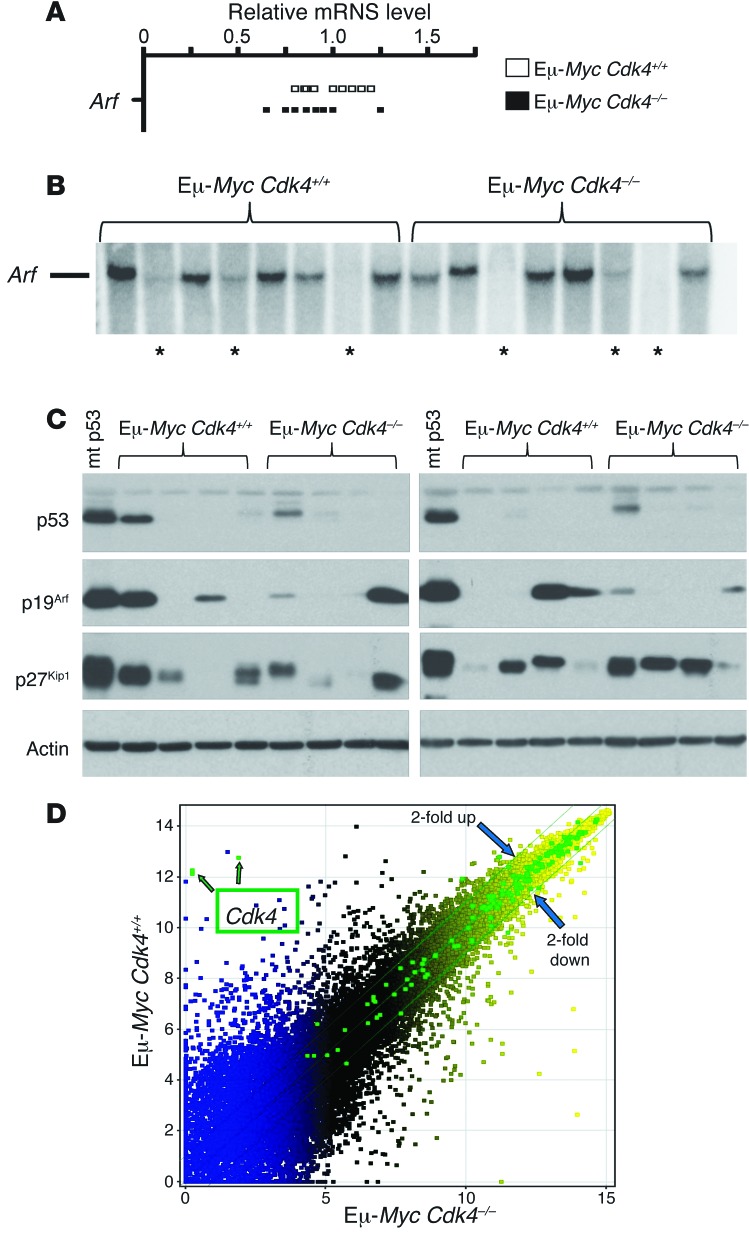
The hyperproliferative response of premalignant Eμ-Myc B cells relies at least in part on the destruction of p27Kip1 (8) and this response is offset by the induction of ARF/p53-dependent apoptosis (6). Accordingly, loss-of-function mutations in ARF or p53 are a hallmark of Eμ-Myc lymphoma (6, 7, 31). We found that the CDK4 deficiency had no effect on the elevated levels of Arf transcripts in Eμ-Myc B cells (Figure 2A) and that the apoptotic index of Eμ-MycCdk4+/+ and Eμ-MycCdk4–/– premalignant B cells was similar (Supplemental Figure 3). Further, the frequency of Arf deletion in Eμ-MycCdk4–/– versus Eμ-MycCdk4+/+ lymphoma was similar (3 of 8 for each cohort) (Figure 2B), and CDK4 deficiency had no effect on the alteration of p53, p19Arf, or p27Kip1 protein levels that typify Eμ-Myc lymphomas (Figure 2C), in which p27Kip1 levels are often reduced (8) and those of p19Arf and p53 are induced due to alterations in the ARF/MDM2/p53 circuit (6). As expected, sequencing established that high levels of p53 protein in some lymphomas (2 of 9 for each cohort) were due to hotspot missense mutations (R172H or R277H). Thus, CDK4 deficiency does not affect the ARF-p53 checkpoint in Eμ-Myc B lymphoma cells.
Figure 2. CDK4 deficiency does not affect the expression of Arf in Eμ-Myc B cells, alterations in the expression of p27Kip1, or inactivating mutations in p53 or Arf in Eμ-Myc lymphomas; nor does it affect direct MYC target genes.
(A) Relative levels of Arf mRNA in Eμ-MycCdk4+/+ and Eμ-MycCdk4–/– B cells were determined. Each rectangle shows levels in individual samples. (B) Biallelic deletions in Arf as detected by Southern blotting (asterisks) were evident in a similar fraction of Eμ-MycCdk4+/+ and Eμ-MycCdk4–/– lymphomas. (C) Levels of p53, p19Arf, p27Kip1, and actin proteins in the indicated representative lymphomas were determined (“mt p53” is a lysate from a previous Eμ-Myc lymphoma known to bear mutant p53). Lymphomas expressing high levels of p53 were found to harbor dominant-negative hotspot mutations in p53 by sequencing cDNA prepared from these tumors (data not shown). Tumors expressing high levels of p19Arf protein also have alterations in the ARF/p53 pathway (6). (D) Scatter Plot of all genes expressed in Eμ-MycCdk4+/+ and Eμ-MycCdk4–/– lymphomas are shown. Genes marked as green squares are direct MYC target genes in B cells (145 probesets that are upregulated in premalignant Eμ-Myc B cells versus wild-type B cells; ref. 50). Note that most direct MYC targets, other than CDK4 itself, were not affected by the CDK4 deficiency (differences are nearly all less than 2-fold).
To confirm that there were no overt effects of CDK4 deficiency on Myc transgene expression or function, we assessed c-MYC protein levels in Eμ-MycCdk4–/– versus Eμ-MycCdk4+/+ B cells and expression of bona fide MYC transcription targets. CDK4 loss did not affect the elevated levels of c-MYC protein in Eμ-Myc B cells or tumors (Supplemental Figure 4A), and expression profiling of Eμ-MycCdk4–/– versus Eμ-MycCdk4+/+ lymphoma demonstrated that CDK4 deficiency did not overtly affect the direct MYC targets that have been defined in human B cells (ref. 32 and Figure 2D).
The lymphoma-promoting effects of CDK4 deficiency could be due to effects on the hyperproliferative response manifest in Eμ-Myc B cells (8) or on possible autophagy (33) or senescence (34) checkpoints, particularly given the effects of the CDK4/Rb pathway on cell proliferation and on repression of senescence in melanoma (17). However, we observed no differences in the S phase indices of premalignant Eμ-MycCdk4+/+ and Eμ-MycCdk4–/– B cells (Supplemental Figure 4B) or in the expression of Rb, p16Ink4a, or D-type cyclins in the lymphomas that arose in these mice (data not shown). We also detected no differences in the levels of p62/sequestrin, a bona fide target of the autophagy pathway, or in the levels of senescence-associated β-galactosidase (Supplemental Figure 5). Finally, there were no compensatory changes in the expression of the related kinase CDK6 in CDK4-deficient B cells, which were comparable in Cdk4+/+, Cdk4–/–, Eμ-MycCdk4+/+, and Eμ-MycCdk4–/– B cells and in the lymphomas that arose in Eμ-MycCdk4+/+, Eμ-MycCdk4+/–, and Eμ-MycCdk4–/– mice (Supplemental Figure 2B and data not shown).
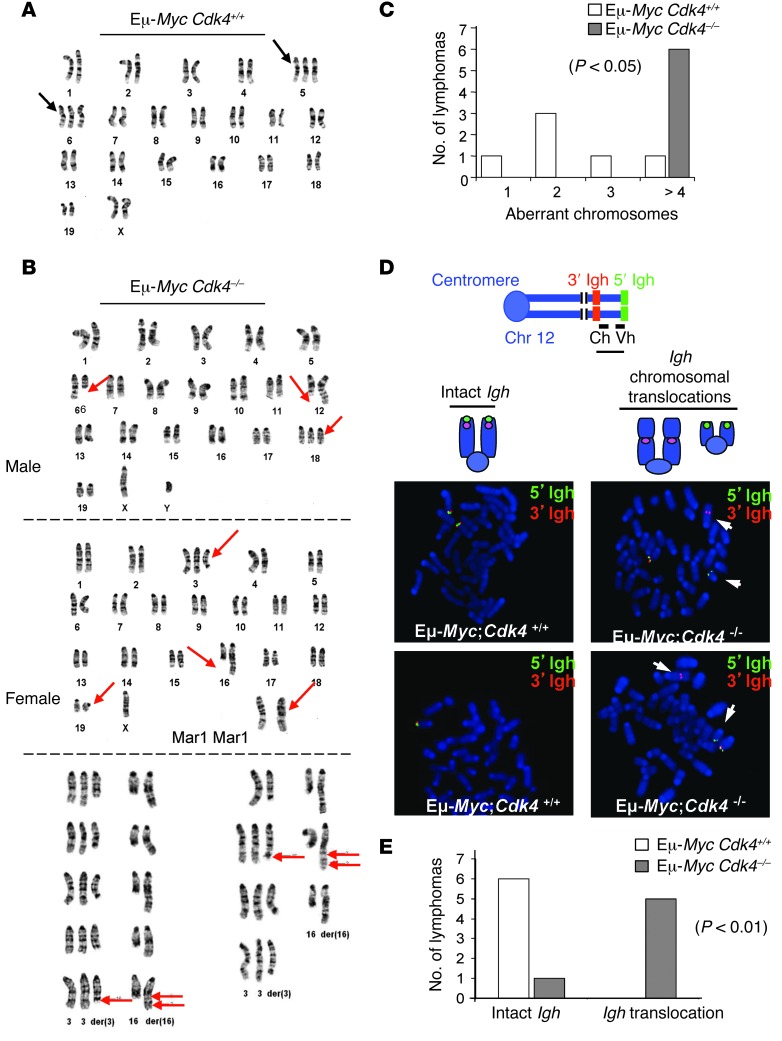
Given these findings and the roles of overall CDK activity in controlling the DDR (4, 28), we assessed the effects of CDK4 deficiency on genomic instability in Eμ-MycCdk4–/– versus Eμ-MycCdk4+/+ lymphomas. As noted previously (3, 35), many Eμ-MycCdk4+/+ B lymphomas exhibited trisomy of chromosome (chr) 6 and/or chr 5, indicating frequent nonrandom chromosomal aberrations (Figure 3, A and C). Of note, we found that Eμ-MycCdk4–/– lymphomas had multiple alterations, including a t(6C2;12F12) translocation, frequent trisomy of chr 18, loss of chr X, and complex translocations and rearrangements of chr 3 and chr 16 (Figure 3, B and C, and Supplemental Table 3). FISH analyses (36, 37) confirmed that there were marked increases in genomic instability (e.g., the Igh translocations) in Eμ-MycCdk4–/– versus Eμ-MycCdk4+/+ lymphomas (Figure 3, D and E). We also assessed the genomic alterations in these tumors (e.g., cleavage at fortuitous recombination signal sequences [RSSs] within the Bcl11b gene) by genomic PCR analyses, and Eμ-MycCdk4–/– lymphomas also had markedly increased numbers of fortuitous recombination events within Bcl11b lymphomas compared with those within Eμ-MycCdk4+/+ lymphomas (Supplemental Table 4, A and B). Finally, these alterations were associated with the induction of select proto-oncogenes in Eμ-MycCdk4–/– versus Eμ-MycCdk4+/+ lymphomas, including Wnt16, c-Raf, Tcf3, and Tcf4, and with increased levels of somatic hypermutation (SHM) targets in B cell tumors, including Pax5 and Bcl6 (Supplemental Figure 6). Thus, CDK4 deficiency leads to lymphomas that harbor rampant genomic alterations, and this is associated with dysregulation of at least some cancer-associated genes.
Figure 3. CDK4 deficiency augments genomic instability of Myc-driven lymphoma.
(A) Karyotype of E-MycCdk4+/+ lymphoma. Trisomy of chr 6 and/or chr 5 was evident in most lymphomas. (B) Abnormal karyotypes are a hallmark of Eμ-MycCdk4–/– lymphomas, including deletions of chr 6 at region C2, translocation of chr 6 and chr 12 with breakpoints (6;12)(C2;F1), trisomy of chr 18, loss of chr X, deletions of chr 3 and chr 16 at regions F2.2 and 16, respectively, translocation between chr 3 and chr 16, and the der(16) with [+der(3)t(3;16)(F2.2;C2),der(16)t(3;16) (F2.2;C2)ins(16;?)(C2;?)]. Karyotypes shown are representative of 20 cells for each lymphoma. (C) Statistical analysis of genomic instability in Eμ-MycCdk4–/– versus Eμ-MycCdk4+/+ lymphomas. The numbers of aberrant chromosomes (translocation, partial genomic loss and gain) per lymphoma are shown for Eμ-MycCdk4–/– and Eμ-MycCdk4+/+ lymphomas. Eμ-MycCdk4+/+ lymphomas (n = 6) with 0, 1, 2, and greater than or equal to 3 aberrant chromosomes are indicated. All Eμ-MycCdk4–/– lymphomas (n = 6) had more than 3 aberrant chromosomes (Supplemental Table 3). (D) FISH analyses of Eμ-MycCdk4–/– lymphomas. Metaphase samples of B220+ cells from Eμ-MycCdk4+/+ and Eμ-MycCdk4–/– lymphomas were assessed by FISH with 5′ (green) and 3′ (red) Igh probes that flank the 2Mb Igh locus on chr 12 (37). An intact Igh locus had colocalized red and green signals, while a broken locus had split red and green signals. FISH analyses confirmed complex translocations in Eμ-MycCdk4–/– lymphomas. Original magnification, ×1,500. Ch, constant (C) region of Igh locus; Vh, variable (V) region of Igh locus. (E) Statistical analysis of translocated versus wild-type Igh loci in Eμ-MycCdk4–/– and Eμ-MycCdk4+/+ lymphomas (n = 6, P < 0.01).
A CDK4/RAG1/2 pathway controls genomic stability and tumorigenesis in Eμ-Myc B cells.
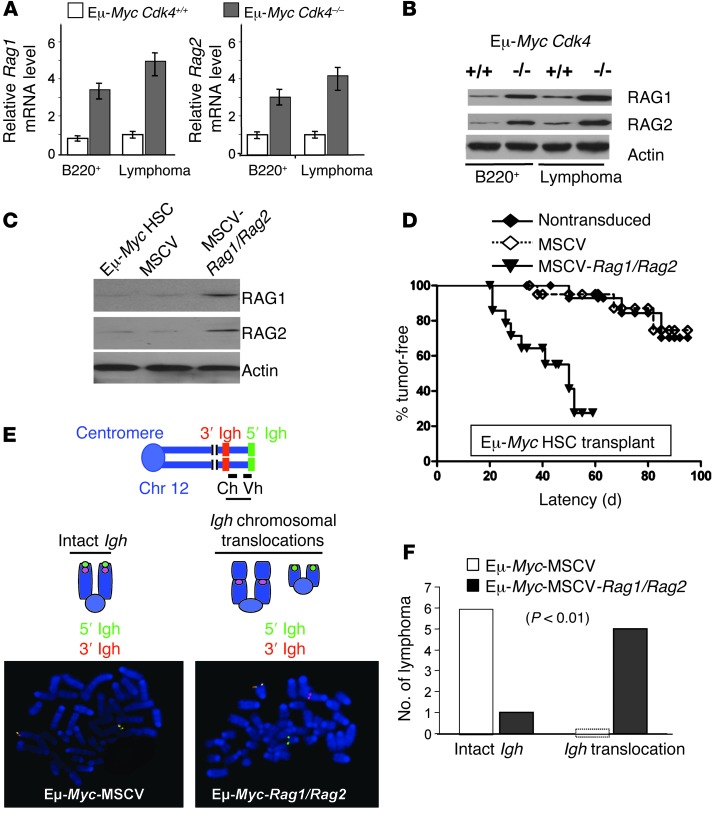
To determine the mechanism by which CDK4 deficiency renders Eμ-Myc B cells susceptible to genomic alterations, we assessed factors known to control genomic instability in lymphoid cells. Elevated levels of the lymphocyte-specific recombinase complex composed of RAG1 and RAG2, which are physically linked in the genome and are coordinately expressed (38, 39), are sufficient to drive aberrant translocations, recombination, and rearrangements of chromosomes that promote lymphomagenesis (40, 41). Notably, RAG1 and RAG2 mRNA and protein levels were selectively elevated in premalignant and neoplastic Eμ-MycCdk4–/– B cells (Figure 4, A and B).
Figure 4. CDK4 controls RAG1 and RAG2 expression in Eμ-Myc B cells, and forced coexpression of RAG1 and RAG2 is sufficient to augment the development of Myc-driven lymphoma.
(A) Rag1 and Rag2 mRNA levels in Eμ-MycCdk4–/– and Eμ-MycCdk4+/+ premalignant bone marrow B220+ and lymphoma cells (n = 5 mean ± SD, P < 0.005, respectively). (B) RAG1 and RAG2 protein levels in Eμ-MycCdk4–/– and Eμ-MycCdk4+/+ premalignant B220+ and lymphoma cells (data shown are representative of analyses of five cohorts of Eμ-MycCdk4–/– and Eμ-MycCdk4+/+ B220+ B cells and lymphomas). (C) Overexpression of RAG1 and RAG2 in Eμ-Myc HSCs. HSCs from E13.5–E15.5 Eμ-Myc fetal livers were transduced with MSCV-IRES-Puro (MSCV) control virus or were cotransduced with MSCV-Rag1-IRES-Puro and MSCV-Rag2-IRES-Hygro (MSCV-RAG1/RAG2) retroviruses. Lysates from these HSCs were analyzed by Western blotting. (D) Enforced coexpression of RAG1 and RAG2 accelerates lymphoma development. 3 × 106 HSCs (nontransduced, MSCV, or MSCV-Rag1/Rag2-HSCs; n = 15) were transplanted into lethally irradiated congenic recipients that were then followed daily for lymphoma onset (P < 0.001). (E) FISH analyses of lymphomas arising in recipient mice engrafted with Eμ-Myc HSCs transduced with MSCV control retrovirus or cotransduced with MSCV-RAG1/RAG2-expressing retroviruses. Metaphase cells from each cohort were assessed by FISH with 5′ (green) and 3′ (red) Igh probes. An intact Igh locus had colocalized red and green signals, while a broken, translocated locus had split red and green signals. Original magnification, x1,500. (F) Statistical analysis of Igh translocations in the two cohorts of lymphomas, Eμ-Myc-MSCV and Eμ-Myc-MSCV-RAG1/RAG2 (n = 6, P < 0.01).
Given the established roles of the RAG1/RAG2 complex in driving chromosomal alterations, we tested whether enforced coexpression of RAG1 and RAG2 was sufficient to provoke genomic instability and accelerate lymphoma onset in a fetal liver–derived Eμ-Myc HSC transplant model (20) and whether it was sufficient to augment the tumorigenic potential of extant Eμ-Myc lymphoma. Notably, lethally irradiated congenic recipients transplanted with Eμ-Myc HSCs cotransduced with MSCV-RAG1– and MSCV-RAG2–expressing retroviruses had a greatly accelerated course of lymphoma onset (Figure 4, C and D). As predicted, we found that RAG1/RAG2-expressing tumors had markedly increased numbers of genomic alterations (translocations, genomic deletions, and cleavage events at bona fide and fortuitous RSSs) that were associated with enhanced recombination at Igh and Bcl11b loci compared with the later-onset lymphomas arising in recipients transplanted with Eμ-Myc HSCs that were transduced with control MSCV virus (Figure 4, E and F, and Supplemental Table 4, C and D).
To further test whether RAG1 and RAG2 overexpression was sufficient to augment the tumorigenic potential of extant Eμ-Myc lymphoma, we cotransduced a randomly chosen tumor with MSCV-Rag1-IRES-Puro and MSCV-Rag2-IRES-Hygro viruses or with control MSCV-Puro virus. Syngeneic C57Bl/6 mice were then transplanted i.v. with 1 × 106 lymphoma cells via the tail vein, and the recipient mice were followed for their course of disease and overall survival. Notably, enforced RAG1 and RAG2 coexpression in extant Eμ-MycCdk4+/+ lymphoma induced more rapid disease in these recipients than in those transplanted with lymphoma transduced with MSCV-IRES control virus (Supplemental Figure 7). Again, the accelerated course of disease in recipients bearing RAG1/RAG2-expressing lymphomas was associated with marked increases in genomic instability (translocations, deletions, etc.; data not shown). Thus, enforced expression of RAG1 and RAG2 is sufficient to accelerate the onset and augmented tumorigenic potential of Myc-driven lymphoma, and this is associated with marked increases in genomic instability.
CDK4 controls Rag1 and Rag2 transcription via FOXO1.
To define the mechanism by which CDK4 loss controls RAG1 and RAG2 expression, we assessed known regulators of Rag1/Rag2 transcription. The FOXO family transcription factors FOXO1 and FOXO3a were candidates, as FOXO1 directly induces Rag1 and Rag2 transcription in B cells (39, 42, 43), and FOXO3a controls Arf transcription in Eμ-Myc B cells (44). Notably, levels of FOXO1, but not FOXO3a, were selectively elevated in premalignant and neoplastic Eμ-MycCdk4–/– compared with Eμ-MycCdk4+/+ B cells (Figures 5A and Supplemental Figure 8A).
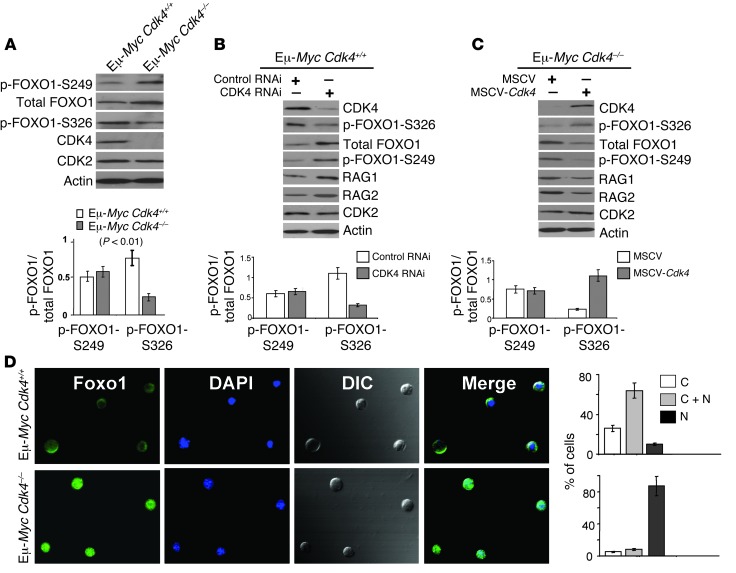
Figure 5. CDK4-dependent control of the levels and phosphorylation of FOXO1 in Eε-Myc B cells.
(A) Levels of the indicated proteins were determined by immunoblotting lysates of Eμ-Myc Cdk4+/+ versus Eμ-Myc Cdk4–/– lymphomas. Right: ratio of p-FOXO1/total FOXO1 (n = 5 for each cohort). p-FOXO1-S326 levels were markedly reduced in Eε-Myc Cdk4–/– lymphomas (P < 0.01). (B) CDK4 silencing affected the levels and phosphorylation of FOXO1 and RAG1 and RAG2 expression in Eε-Myc Cdk4+/+ lymphomas. Bottom: ratio of p-FOXO1/total FOXO1. Note the marked reduction in p-FOXO1-S326 following knockdown of CDK4 in malignant Eε-Myc Cdk4+/+ B cells (P < 0.05, n = 5). (C) Restoring CDK4 expression in Eμ-MycCdk4–/– lymphoma cells affected the levels and phosphorylation of FOXO1 and RAG1 and RAG2 expression. Bottom: ratio of p-FOXO1/total FOXO1. Note the marked increase in p-FOXO1-S326 following restoration of CDK4 in malignant Eμ-Myc Cdk4–/– B cells (P < 0.05, n = 5). (D) Increased levels of nuclear FOXO1 are a hallmark of Eμ-Myc Cdk4–/– lymphoma. Left panels: FOXO1; middle left panels: DAPI; middle right panels: light microscopy; right panels: merge. Quantification of FOXO1 localization is shown at right: C, cytoplasm; N, nucleus; C+N, cytoplasm and nucleus. The mean of triplicate experiments is shown (n = 100 Eμ-Myc Cdk4–/– and Eμ-Myc Cdk4+/+ lymphoma cells). Original magnification, ×600.
Proteasome-directed turnover of FOXO proteins is regulated by phosphorylation of select residues (e.g., Ser249 [S249] and S329 [mouse S326] of FOXO1 and S253 and S318/S321 of FOXO3a) that trigger their nuclear export (45-47). Indeed, we observed marked reductions in phosphorylated FOXO1-S326 (p-FOXO1-S326) in premalignant and neoplastic Eμ-MycCdk4–/– versus Eμ-MycCdk4+/+ B cells (Figure 5A). These effects on FOXO1 levels and phosphorylation were specifically linked to CDK4 status as: (a) there were no differences in CDK6 levels in wild-type versus CDK4-deficient Eμ-Myc B cells (Supplemental Figure 2B); (b) stable knockdown of CDK4 in Eμ-MycCdk4+/+ lymphoma cells led to increases in total FOXO1, yet reductions in p-FOXO1-S326 (Figure 5B); and (c) restoration of CDK4 expression in E-MycCdk4–/– lymphoma led to reductions in total FOXO1 levels and to corresponding increases in p-FOXO1-S326 levels (Figure 5C). In contrast, Cdk4 status did not affect p-FOXO1-S249 levels (Figure 5, A–C), nor did it affect p-S253- or p-FOXO3-aS318/S321 levels (Supplemental Figure 8A). Thus, FOXO1 expression is selectively controlled by CDK4 in MYC-expressing B cells, and increased FOXO1 levels in Eμ-MycCdk4–/– B cells are associated with decreased phosphorylation of FOXO1-S326.
Given these findings, we tested whether FOXO1 was a CDK4 substrate. Indeed, a consensus CDK4 site was conserved in FOXO1 (Supplemental Figure 8B), and in vitro kinase assays demonstrated that recombinant cyclin D1/CDK4 phosphorylates human FOXO1 at S329 (Supplemental Figure 8C). Further, in Eμ-MycCdk4–/– versus Eμ-MycCdk4+/+ lymphomas engineered to express a human GFP-FOXO1 fusion protein, we observed marked reductions in p-S329 GFP-FOXO1 levels (Supplemental Figure 8D). Finally, CDK4-mediated phosphorylation of FOXO1 at S326/S329 promoted FOXO1 cytoplasmic localization, as nuclear FOXO1 levels were elevated in Eμ-MycCdk4–/– lymphoma (Figure 5D), and mutation of FOXO1 S329 to alanine (FOXO1-S329A) increased nuclear GFP-FOXO1 levels in Eμ-MycCdk4+/+ lymphoma engineered to express this fusion protein (Supplemental Figure 8E). Thus, FOXO1 is a substrate of CDK4, and CDK4 deficiency is associated with increased levels of nuclear FOXO1.
To test whether the CDK4/FOXO1 circuit controls Rag1/Rag2 expression in MYC-expressing B cells, we assessed the effects of knockdown or overexpression of CDK4 in Eμ-Myc lymphoma cells. Stable knockdown of CDK4 in Eμ-MycCdk4+/+ lymphoma cells increased levels of RAG1 and RAG2 (Figure 5B), whereas restoring CDK4 expression in Eμ-MycCdk4–/– lymphoma reduced levels of RAG1 and RAG2 (Figure 5C). Thus, CDK4 suppresses FOXO1, which normally induces Rag1 and Rag2 expression (39, 42, 43).
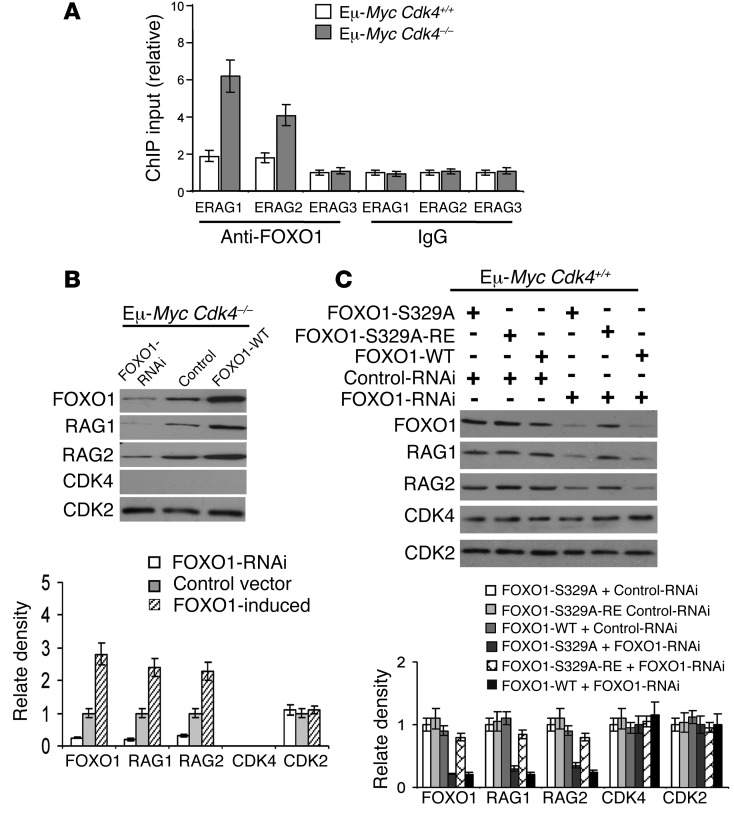
Since Rag1 and Rag2 are physically linked in the genome and are coordinately expressed and their transcription is directly regulated by FOXO1 (39, 42, 43), we tested whether the increases in RAG1 and RAG2 levels in the malignant B cells of Eμ-MycCdk4–/– mice were FOXO1 dependent. As predicted, ChIP analyses demonstrated FOXO1 binding to enhancers in the Rag1/Rag2 locus (ERAG1 and ERAG2) that harbor FOXO1 binding element motifs (refs. 39, 43, 48, and Figure 6A), and revealed that FOXO1 binding was augmented in Eμ-MycCdk4–/– B cells (Figure 6A). Moreover, we found that FOXO1 knockdown in Eμ-MycCdk4–/– lymphoma cells reduced RAG1 and RAG2 levels, whereas FOXO1 overexpression augmented RAG1 and RAG2 levels (Figure 6B). Further, enforced FOXO1-S329A expression in Eμ-MycCdk4+/+ lymphoma increased RAG1 and RAG2 levels, and reductions of RAG1 and RAG2 following FOXO1 knockdown were rescued by a FOXO1-S329A wobble mutant resistant to FOXO1 siRNA (Figure 6C). Thus, FOXO1 controls CDK4-dependent regulation of Rag1 and Rag2 transcription in Eμ-Myc B cells.
Figure 6. CDK4 deficiency induces Rag1 and Rag2 transcription via FOXO1 in Eμ-Myc lymphoma cells.
(A) ChIP analyses of FOXO1 binding to enhancers in the Rag1/Rag2 locus. ChIP was performed with anti-FOXO1 antibody or an isotype IgG control antibody, followed by qPCR analysis of FOXO1 binding to regulatory elements (ERAG1-ERAG3) in the Rag1/Rag2 locus (39, 43, 48, 65) in Eμ-Myc Cdk4–/– versus Eμ-Myc Cdk4+/+ lymphoma. Results are the fold enrichment in immunoprecipitates of FOXO1 antibody relative to control antibody (mean and SD of triplicate ChIP; P < 0.01). (B) Control of RAG1 and RAG2 expression by CDK4 is FOXO1 dependent. Eμ-Myc Cdk4–/– lymphoma cells were transfected with siRNA targeting FOXO1 (FOXO1-RNAi), a control siRNA (Ctrl RNAi), or a vector driving FOXO1 expression. Levels of FOXO1, RAG1, RAG2, CDK4, and CDK2 proteins were determined by immunoblotting. Note that FOXO1 knockdown reduced RAG1 and RAG2 levels, whereas FOXO1 overexpression increased their levels. Results shown are representative of three separate analyses. (C) FOXO1-mediated induction of RAG1 and RAG2 in Eμ-Myc B cells requires phosphorylation of S329 in FOXO1. Eμ-Myc Cdk4+/+ lymphoma cells were cotransfected with siRNA targeting FOXO1 and vectors expressing FOXO1-S329A or a wobble mutant of FOXO1-329A (FOXO1-S329A-RE) resistant to the FOXO1 siRNA. Note that FOXO1 knockdown decreased RAG1 and RAG2 levels and that this was reversed by expression of FOXO1-S329-RE. Immunoblotting with CDK4 and CDK2 showed equal loading. Results are representative of three independent experiments. Graphs display the median ± SD (n = 3; mean ± SD, with error bar).
CDK4 loss augments the tumorigenic potential of Myc-driven lymphoma.
The effects of CDK4 deficiency on the development of Eμ-Myc lymphoma suggested that CDK4 might also affect tumorigenic potential. To test this notion, limiting numbers of lymphoma cells (1–3 × 105) from tumors arising in Eμ-MycCdk4–/– and Eμ-MycCdk4+/+ (C57Bl/6) littermates (three independent lymphomas from each cohort) were transplanted into 6- to 8-week-old Cdk4+/+ (C57Bl/6) recipients, which were followed for the onset of disease and overall survival. Notably, disease was more penetrant and accelerated in recipients transplanted with Eμ-MycCdk4–/– lymphoma (P < 0.001; Figure 7A). Thus, CDK4 loss augments the tumorigenic potential of Myc-driven lymphoma.
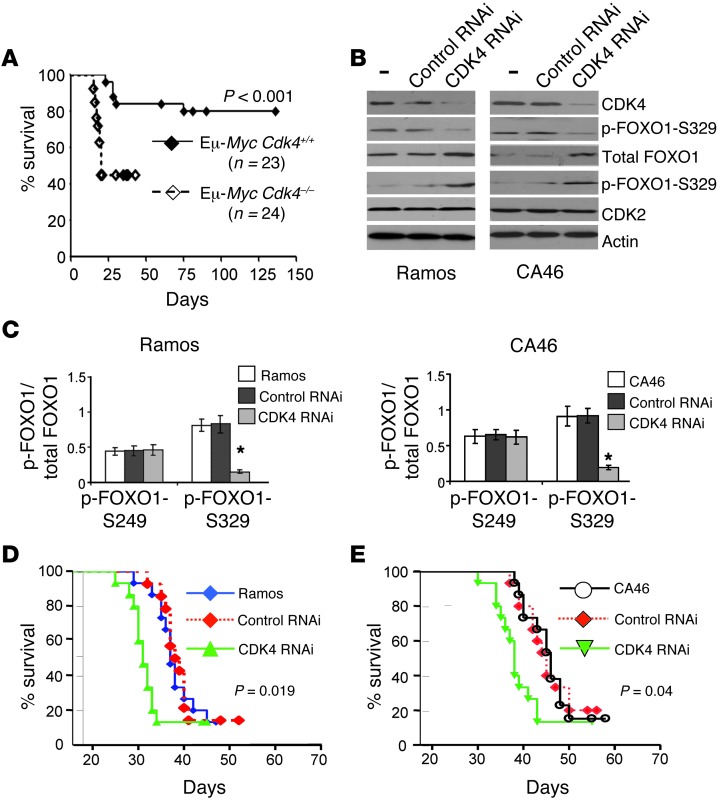
Figure 7. CDK4 deficiency augments the tumorigenic potential of Myc-driven lymphoma.
(A) B220+ cells (3 × 105) from individual Eμ-MycCdk4+/+ or Eμ-MycCdk4–/– lymphomas were transplanted i.v. via the tail vein into 6- to 8-week-old syngeneic C57Bl/6 recipient mice. Note the marked differences in the tumorigenic potential of Eμ-MycCdk4–/– versus Eμ-MycCdk4+/+ lymphomas (P < 0.001). n = number of mice per group. A Student’s t test was used for statistical analysis. Horizontal lines indicate mouse survival after inoculating lymphoma cells. (B) Knockdown of human CDK4 in human BL cells: Ramos (left) and CA46 (right) increased levels of FOXO1, yet led to decreases in p-FOXO1-S329 expression levels. There were no changes in CDK2 or actin levels following CDK4 knockdown. (C) The effects of CDK4 knockdown on the ratio of total FOXO1/p-FOXO1-S329 and p-FOXO1-S249 were determined for Ramos (left) and CA46 (right) BL cells. P < 0.05. (D and E) Knockdown of human CDK4 augments the tumorigenic potential of Ramos (D) and CA46 (E) lymphoma cells. Ramos or CA46 BL cells with stable CDK4 knockdown or cells harboring control RNAi were transplanted via the tail vein into 6- to 8-week-old nude recipients that were then followed for lymphoma onset. Statistical analyses were performed as above. n = 12 mice per group (P = 0.019 and P = 0.04, respectively).
Though we suspected that the accelerated disease in CDK4-deficient Eμ-Myc was associated with their inherently high genomic instability (Figure 3), we reasoned that CDK4 deficiency could have other effects on the tumor cells and tumorigenesis. To test this, we reintroduced CDK4 into Eμ-MycCdk4–/– mouse lymphomas by infecting these tumors with MSCV-CDK4-IRES-Puro virus (or with control MSCV-IRES-Puro virus; Supplemental Figure 9A). These CDK4-overexpressing tumors were then transplanted into syngeneic recipients that were followed for disease onset and overall survival. Eμ-MycCdk4–/– lymphomas transduced with CDK4-expressing retrovirus had a tumorigenic potential similar to that of the lymphomas transduced with control retrovirus (Supplemental Figure 9B). Thus, the augmented tumorigenic potential of CDK4-deficient Eμ-Myc lymphomas is due to defects such as genomic alterations that are acquired during tumor progression and that cannot be undone. Similarly, forced overexpression of CDK4 did not significantly enhance the tumorigenic potential of human Burkitt lymphoma (BL) cell lines (Supplemental Figure 10).
Although most human B cell lymphomas that we analyzed expressed little to no CDK4 (see below), two human BL cell lines (Ramos and CA46) expressed modest levels of CDK4 (Figure 7B). We thus used these cells to test whether silencing CDK4 also augments the tumorigenic potential of human MYC-driven B cell lymphoma. Efficient stable knockdown of CDK4 was achieved in both BL cell lines (Figure 7B). Of note, CDK4 knockdown led to decreased levels of p-FOXO1-S329 (Figure 7, B and C). Importantly, silencing CDK4 augmented the tumorigenic potential of both human BL cell lines when we injected them into immunocompromised nude recipient mice (Figure 7, D and E). Further, knockdown of CDK4 in these human lymphomas also led to tumors with greater genomic alterations (Supplemental Figure 11, A and B).
CDK4 behaves as an oncogene in other contexts (14, 49). Therefore, we tested the effects of silencing CDK4 on the tumorigenic potential of human estrogen receptor–positive MCF7 breast cancer cells. Notably, we found that stable knockdown of CDK4 impaired the tumorigenic potential of MCF7 xenografts in transplanted immunocompromised nude mice (Supplemental Figure 11C). Thus, the effects of CDK4 on tumorigenesis are context specific and, in contrast to other scenarios, CDK4 functions in lymphoma to harness tumorigenesis.
Suppression of CDK4 and elevated expression of FOXO1 are manifest in human B cell lymphoma subtypes.
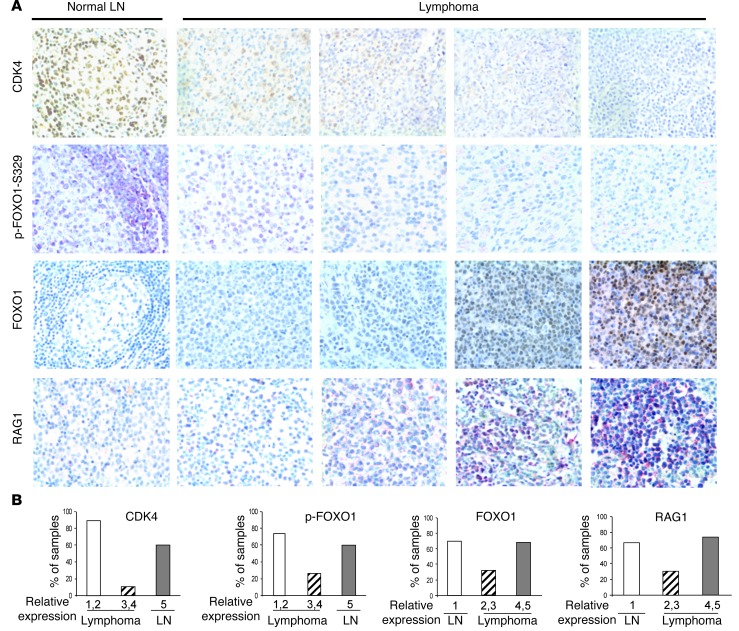
The profound effects of CDK4 deficiency on Myc-driven lymphoma in mice suggested that CDK4 might be altered in human B cell lymphoma. Indeed, immunohistochemical (IHC) analyses showed little to no expression of CDK4 protein in approximately 90% of the primary human B cell lymphomas tested (n = 125) when compared with the relatively high levels of CDK4 detected in normal LNs (n = 10) (Figure 8) and other tumor types (Supplemental Figure 12). Suppression of CDK4 levels was evident in most subtypes of non-Hodgkin lymphoma (NHL) including MALT, follicular lymphoma (FL), diffuse large B cell lymphoma (DLBCL), and BL (Supplemental Table 5). Furthermore, IHC analyses demonstrated that p-FOXO1-S329 levels were reduced and that total levels of FOXO1 were highly elevated in nearly 70% of human B cell lymphomas versus the levels expressed in normal LNs (Figure 8). Finally, we found that elevated levels of RAG1 in human B cell lymphomas were associated with reduced levels of CDK4 and were concordant with high levels of FOXO1 (Figure 8 and data not shown). These data suggest that a CDK4/FOXO1 pathway is also disabled in a significant proportion of non-Hodgkin B lymphomas.
Figure 8. CDK4 expression is suppressed, and FOXO1 and RAG1 expression is elevated in several subtypes of human B cell lymphomas.
(A) 125 cases of randomly collected patient B cell lymphoma samples from The Ohio State University Department of Pathology and US Biomax Inc., along with 10 normal LNs, were stained with antibodies specific to CDK4 (top panels), p-FOXO1-S329 (second panels), FOXO1 (third panels), or RAG1 (bottom panels). Original magnification, ×400. (B) For statistical analysis, the samples were graded on a scale of staining scores from 1 (lowest) to 5 (maximum), based on signal intensity for CDK4, p-FOXO1-S329, FOXO1, or RAG1. Approximately 90% of B cell lymphoma samples (111 of 125) had low or nondetectable levels of CDK4 (stages 1 and 2), and nearly 70% (87 of 125) of B cell lymphoma samples had low levels of p-FOXO1-S329 (stages 1 and 2). Accordingly, approximately 70% of these tumors (86 of 125) had elevated levels of total FOXO1 (stages 4 and 5), and approximately 64% (80 of 125) had elevated levels of RAG1 (stages 4 and 5). Among the lymphomas with elevated levels of FOXO1, more than 70% (61 of 86) had elevated levels of RAG1.
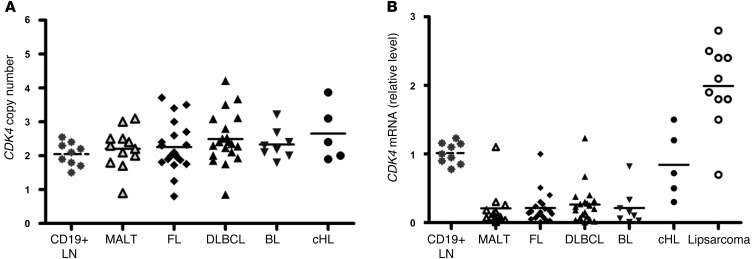
To address the mechanism by which CDK4 protein levels were suppressed in non-Hodgkin B cell lymphoma, we performed genomic CDK4 copy number analyses and quantitative RT-PCR (qRT-PCR) analyses of CDK4 transcript levels. We detected only about 4 of 60 lymphomas analyzed for copy number that showed a reduction to 1N, and although CDK4 amplification has been reported in human lymphoma and other tumor types, only 8 of these lymphomas had more than two copies of CDK4, and most of these tumors showed no overt amplification of CDK4 (Figure 9A). Strikingly, the reductions in CDK4 protein levels directly correlated with markedly reduced levels of CDK4 transcripts in the majority of human B cell lymphoma subtypes analyzed (Figure 9B). Thus, either CDK4 transcription or mRNA half-life is suppressed in many human non-Hodgkin B cell lymphomas.
Figure 9. CDK4 expression is repressed in subtypes of non-Hodgkin B cell lymphoma.
(A) The CDK4 gene copy number in human B cell lymphoma subtypes was determined by genomic qRT-PCR. Horizontal dotted line represents the mean CDK4 gene dosage detected in B cells from normal LN controls. Solid horizontal lines indicate the mean average copy number for the indicated lymphoma subtype. Most lymphomas (48 of 60 NHLs) were 2N. (B) CDK4 mRNA levels were suppressed in the majority of human non-Hodgkin B cell lymphomas. CDK4 transcript levels from the lymphomas assessed for the CDK4 copy number in A were determined by qRT-PCR. Values shown are relative to those of CDK4 transcripts expressed in normal B cells. Again, the dotted line represents the mean value of CDK4 mRNA in normal B cells, and solid lines represent the means found in the indicated subtypes of NHLs. Primary liposarcomas were also assessed for CDK4 transcript levels as controls.
Discussion
Given the established connections between c-MYC and CDK4 (9, 32) and that Cdk4 loss often impairs tumorigenesis, we predicted that Cdk4 deficiency would impair Myc-driven lymphomagenesis. Surprisingly, our studies revealed instead that Cdk4 loss accelerates Myc-driven lymphoma development and augments the tumorigenic potential of extant lymphoma. Furthermore, we discovered that CDK4 deficiency increases genomic instability in Eμ-Myc lymphomas, suggesting that this accelerates lymphomagenesis (4). Mechanistically, the effects of CDK4 deficiency are linked to the CDK4 substrate FOXO1 and to its transcription targets RAG1 and RAG2, which are associated with the aberrant chromosomal translocations that are manifest in Eμ-MycCdk4–/– lymphomas and which we show are sufficient to provoke genomic instability and to accelerate the development and augmented tumorigenic potential of Myc-driven lymphoma.
Our findings strongly support the notion that CDK4 harnesses Myc-driven lymphomagenesis despite the fact that it promotes and/or is required for tumorigenesis in other contexts (refs. 14 and 49 and Supplemental Figure 11B). Indeed, despite the fact that enforced cyclin D1 expression accelerates lymphoma development in Eμ-Myc transgenic mice (50), we show that loss of CDK4 accelerates the onset and augmented tumorigenic potential of Eμ-Myc lymphoma and that knockdown of human CDK4 also augments the tumorigenic potential of human BL cell lines. Finally, our analyses suggest that there is a strong selection for silencing CDK4 expression in several human B cell lymphoma subtypes, as many of these tumors express low levels of CDK4 mRNA and protein and the corresponding higher levels of FOXO1. These findings were surprising, given that there is no selection for Rb loss in the Eμ-Myc model (7) or effects of INK4a deficiency (26) and that CDK4 loss had no effect on senescence in premalignant B cells (Supplemental Figure 5B). However, our findings are generally consistent with previous studies showing that most human mantle cell lymphomas express very low levels of CDK4 protein and that only a small fraction of these tumors display an increased CDK4 gene copy number (only 4 of 62) (51). Further, others have reported that among 180 primary DLBCLs, there were only 21 recurrent copy number alterations (11.6%), which include copy gains of all possible genes, and that of these, there was a low frequency (only 13%) that had amplified CDK4 (52). Further, IHC analyses have shown that indolent FLs express no (12 of 21, 57%) or very low (9 of 12, 43%) levels of CDK4 and that 68% (15 of 22) of DLBCLs have no or low CDK4 levels (6 of 22 were negative and 9 of 22 had low levels of CDK4 (53).
Our findings have also revealed that CDK4 substrates other than Rb are important for tumorigenesis. Specifically, FOXO1 is now revealed as a CDK4 substrate that is important for lymphomagenesis, and FOXO1 is suggested to play oncogenic roles in this context by virtue of its ability to activate the expression of RAG1 and RAG2, which are sufficient to drive genomic instability and lymphomagenesis. Interestingly, recent reports have shown that FOXO1, FOXO3a, and FOXO4 can behave as oncoproteins in acute myeloid leukemia, but not in solid tumors (54), and that FOXO1 is highly expressed in NHLs (55). The latter findings are consistent with our studies (Figure 8), which show elevated FOXO1 expression in non-Hodgkin B cell lymphoma. Collectively, these findings suggest that the different roles of CDK4 and FOXO1 in tumorigenesis hinge on cell context.
We found that typical proliferative (p27Kip1) and apoptotic (ARF and p53) checkpoints of MYC-induced lymphoma were not affected by the CDK4 deficiency, nor were there effects on Myc transgene levels or on direct MYC transcription targets. However, we found that the effects on genomic instability were clearly manifest in Eμ-MycCdk4–/– lymphoma. Although alterations in cyclin E have been linked to genomic instability (56), we observed no effects of CDK4 loss on cyclin E levels in Eμ-Myc B cells (data not shown). However, there were profound effects on the FOXO1/RAG1/RAG2 pathway. Specifically, the data show that CDK4 phosphorylates FOXO1 at S326 (S329 in man) and that this modification is necessary for FOXO1 cytoplasmic localization, where it is destroyed by the proteasome (45–47). Accordingly, CDK4 loss leads to increased levels of nuclear FOXO1 in Eμ-Myc B cells. The effects of CDK4 on FOXO1 were selective, as we observed no changes in FOXO3a expression or phosphorylation in Eμ-MycCdk4–/– B cells. These findings are in accord with a lack of effect of CDK4 deficiency on Arf expression, as FOXO3a regulates Arf transcription in Eμ-Myc B cells (44). Thus, CDK4 targets FOXO1 through a mechanism independent of links between FOXO3a, ARF, and lymphomagenesis.
CDK2- or CDK1-dependent phosphorylation of FOXO1 at S249 blocks FOXO1 activity by provoking its cytoplasmic localization and destruction, and this response is disabled following DNA damage (45–47). However, we found that CDK4 status did not affect the levels of p-FOXO1-S249 in Eμ-Myc B cells. Rather, CDK4-directed phosphorylation of FOXO1 at S326 (human FOXO1 at S329) holds FOXO1 activity in check in lymphoma cells, and thus CDK4 loss leads to marked reductions of p-FOXO1-S326 or p-FOXO1-S329 and to increased levels of nuclear FOXO1 and induction of the FOXO1 targets Rag1 and Rag2, which are sufficient to provoke genomic instability and augment Myc-driven lymphomagenesis.
Precisely how CDK4 deficiency triggers genomic instability in MYC-expressing B cells is not resolved, but the data strongly suggest that this deficiency reflects aberrant activity of the RAG1/RAG2 complex, which can promote the formation of complex translocations coined “complicons” in the B cell milieu (40, 41, 57, 58). Although it has been reported that RAG1 deficiency also accelerates the development of lymphoma in the Eμ-Myc model (59), any genetic insult that provokes a progenitor B cell developmental blockade such as that seen with Rag1 deficiency (60) will trigger such a phenotype, leading to increased pools of highly proliferating, at-risk progenitor B cells (30). Indeed, genomic instability typifies Eμ-MycCdk4–/– B cell lymphoma, which selectively accumulates complex genomic abnormalities and is associated with the induction of several oncogenes that have been implicated in B cell malignancies. Collectively, increased genomic instability and/or aberrant expression of such targets would explain, at least in part, the accelerated course of lymphomagenesis manifest in Eμ-MycCdk4–/– transgenic mice as well as the increased tumorigenic potential of mouse and human lymphomas in transplant models.
Finally, it is important to note that anti-CDK4 strategies are being widely applied as anticancer therapies (49). Quite strikingly, the effects of CDK4 silencing are the opposite of those provoked by a small-molecule inhibitor that has been touted as a selective inhibitor of CDK4 and CDK6 (Supplemental Figure 13). In fact, given the limited profiling that has been done to date, it is unclear whether there are other kinases that are inhibited by these agents that then affect other critical factors needed to support lymphoma cell growth and survival (61–64). Along these lines, we note that it is striking that treatment of BL cells with these agents results in downregulation of steady-state levels of both CDK4 and CDK6, consistent with off-target effects. The effects of CDK4 deficiency on B cell lymphoma described herein indicate that the use of CDK4 suppression strategies in anticancer therapies needs to be carefully tailored, as such strategies may have untoward lymphoma-promoting effects.
Methods
Mice and tumor monitoring.
Cdk4+/– mice (C57Bl/6) were interbred with Eμ-Myc transgenic (C57Bl/6) mice. F1 Eμ-MycCdk4+/– mice were then backcrossed to Cdk4+/– mice to generate the desired cohort: Cdk4+/+, Cdk4+/–, Cdk4–/–, Eμ-MycCdk4+/+, Eμ-MycCdk4+/–, and Eμ-MycCdk4–/– mice. Mice were monitored daily for signs of morbidity and tumor onset, and log-rank survival tests were performed. Tumors were harvested immediately after euthanasia. Single-cell suspensions were obtained from the tumors and were analyzed by FACS; the remainder was snap-frozen in liquid nitrogen for DNA, RNA, and protein analysis. All animal-related procedures were performed in accordance with protocols approved by the IACUC of The Ohio State University.
Expression constructs.
The GFP-FOXO1-S329A construct was generated by site-directed mutagenesis of GFP-FOXO1 (provided by A. Bonni of Harvard University, Cambridge, Massachusetts, USA and S. Guo of Texas A&M Health Science Center, College Station, Texas, USA). The GST-FOXO1 and GST-FOXO1-S249A plasmids were also provided by A. Bonni. The RNAi-resistant FOXO1-RE plasmid was generated as described (45, 46).
Western blot analysis.
Lysates of cells and tissues were generated in SDS sample buffer (60 mM Tris-HCl at pH 6.8, 10% glycerol, 2% SDS, and 5% 2-mercaptoethanol). Equal amounts of lysate protein (50 μg) were separated on 8% to 12% SDS-PAGE gels, transferred to PVDF membranes (Amersham), and then incubated with primary antibodies specific to CDK4 (SC-260; Santa Cruz Biotechnology Inc.), CDK2 (2546; Cell Signaling Technology), CDK6 (SC-177; Santa Cruz Biotechnology Inc.), p-FOXO1 (S329) (ab52857; Abcam), RAG1 (NPP1-74190; Novus Biological), RAG2 (ab133609, i.e., Epitmics 5485-1 for mouse cells; Abcam); RAG2 (ab959955 for human cells; Abcam), p62 (SC-28359; Santa Cruz Biotechnology Inc.), p27Kip1 (610242; Transduction Labs), p53 (Ab-7; Oncogene Research), p19Arf (ab80; Abcam), FOXO1 (2880; Cell Signaling Technology), p-FOXO1-S249 (441245G; Invitrogen), FOXO3a (2497; Cell Signaling Technology), p-318/321-FOXO3a (9465; Cell Signaling Technology), or β-actin (A00702; Sigma-Aldrich). Blots were then rinsed in TBST and further incubated in peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies, respectively. Proteins were visualized using an ECL detection system (Pierce Biotech) and were exposed to film. All experiments were repeated in triplicate.
Eμ-Myc HSC transduction and transplantation.
Mouse Rag1 and Rag2 cDNA fragments were amplified with primers containing compatible restriction sites and then cloned into the MSCV-IRES-Puro and MSCV-IRES-Hygro retroviruses (Clontech), respectively. Retrovirus stocks were generated as described (13, 20). Days E13.5–E15.5 pregnant Eμ-Myc mice were humanely euthanized to obtain fetal livers, which were minced and cultured at approximately 3 × 106 cells/ml in media supporting HSC growth as described (13, 20). For transplantation, 6- to 8-week-old lethally irradiated C57Bl/6 recipient mice were reconstituted 6 hours later with approximately 3 × 106 viable fetal liver Eμ-Myc HSCs (control) or with Eμ-Myc HSCs transduced with MSCV control virus or cotransduced with MSCV-RAG1 and MSCV-RAG2 retroviruses by tail-vein injection. Lymphoma onset was defined as the occurrence of palpable cervical or peripheral LNs (at least 5 mm in diameter in one dimension). Statistical evaluation of tumor onset data was based on the log-rank (Mantel-Cox) test for comparison of the Kaplan-Meier event-time format and on an unpaired Student’s t test for comparison of the means and SDs. Three independent HSC transplantations were performed, and control and transduced Eμ-Myc HSCs were transplanted into 5 to 6 recipients.
Lymphoma transplantation assay.
Lymphoma cells (3 × 105) were injected into the tail veins of 6- to 8-week-old Cdk4+/+ female mice. Recipient animals were monitored for morbidity and tumor onset by palpating the LNs.
Expression profiling analyses.
The effects of CDK4 deficiency on direct MYC targets were assessed by comparing total gene expression profiles of Eμ-MycCdk4+/+ versus Eμ-MycCdk4–/– lymphomas with the 668 direct MYC target genes defined in human B cells (32). This gene list was imported into the wild-type B220+/precancerous Eμ-Myc B220+ gene chip robust multiarray averaging (GC-RMA) normalized database, and probesets were selected that were deferentially expressed (Student’s t test, P < 0.05, 2-fold change or higher) between wild-type B220+ and precancerous Eμ-Myc samples and that were expressed higher than the chip median in one or more samples. The resulting list was then imported into the Eμ-MycCdk4+/+and Eμ-MycCdk4–/– Affymetrix MAS5 normalized database. Microarray data were deposited in the Gene Expression Omnibus database (GEO GSE21683).
Statistics.
A Student’s t test was used to validate the significance of the observed differences. A P value less than 0.05 was considered statistically significant.
Additional methods are described in Supplemental Methods.
Supplementary Material
Acknowledgments
We thank H. Kiyokawa for providing Cdk4+/– mice; A. Bonni and S. Guo for the FOXO1 and FOXO1-S249A plasmids; F. Alt (Harvard University) for FISH probes; N. Heerema and J. Labanowska (The Ohio State University) for support and help with karyotype and FISH analyses; J. Ma, X. Bai, and G.L. Wang (The Ohio State University) for assistance with experimental techniques; X. Wang, H. Guan, Y. Jin, M. Gu, and L. Li (The Ohio State University) for assistance with sample analyses; M. Kernick (The Scripps Research Institute) for help with editing; and W.C. Chan (University of Nebraska Medical Center) and P. Carolyn (The Ohio State University) for critical reading and helpful comments. This research was supported by NIH grants CA113579 (to X. Zou) and CA076379 (to J.L. Cleveland), by NSFC grants 30571006 and 30671026 (to X. Zou), by monies from the State of Florida to Scripps Florida (to J.L. Cleveland), and by The Ohio State University MCC (to X. Zou).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article:J Clin Invest. 2014;124(4):1672–1684. doi:10.1172/JCI63139.
References
- 1.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8(12):976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 2.Soucek L, Evan GI. The ups and downs of Myc biology. Curr Opin Genet Dev. 2010;20(1):91–95. doi: 10.1016/j.gde.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams JM, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 4.Gorrini C, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448(7157):1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- 5.Dominguez-Sola D, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448(7152):445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 6.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13(20):2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13(20):2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller UB, et al. Myc targets Cks1 to provoke the suppression of p27Kip1, proliferation and lymphomagenesis. EMBO J. 2007;26(10):2562–2574. doi: 10.1038/sj.emboj.7601691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermeking H, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci U S A. 2000;97(5):2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsushime H, et al. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell. 1992;71(2):323–334. doi: 10.1016/0092-8674(92)90360-O. [DOI] [PubMed] [Google Scholar]
- 11.Rane SG, et al. Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in beta-islet cell hyperplasia. Nat Genet. 1999;22(1):44–52. doi: 10.1038/8751. [DOI] [PubMed] [Google Scholar]
- 12.Miliani de Marval PL, et al. Lack of cyclin-dependent kinase 4 inhibits c-myc tumorigenic activities in epithelial tissues. Mol Cell Biol. 2004;24(17):7538–7547. doi: 10.1128/MCB.24.17.7538-7547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou X, et al. Cdk4 disruption renders primary mouse cells resistant to oncogenic transformation, leading to Arf/p53-independent senescence. Genes Dev. 2002;16(22):2923–2934. doi: 10.1101/gad.1033002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Q, et al. Requirement for CDK4 kinase function in breast cancer. Cancer Cell. 2006;9(1):23–32. doi: 10.1016/j.ccr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18(22):2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 16.Matsuura I, Denissova NG, Wang G, He D, Long J, Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430(6996):226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 17.Anders L, et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell. 2011;20(5):620–634. doi: 10.1016/j.ccr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Italiano A, et al. Clinical and biological significance of CDK4 amplification in well-differentiated and dedifferentiated liposarcomas. Clin Cancer Res. 2009;15(18):5696–5703. doi: 10.1158/1078-0432.CCR-08-3185. [DOI] [PubMed] [Google Scholar]
- 19.Korz C, et al. Evidence for distinct pathomechanisms in B-cell chronic lymphocytic leukemia and mantle cell lymphoma by quantitative expression analysis of cell cycle and apoptosis-associated genes. Blood. 2002;99(12):4554–4561. doi: 10.1182/blood.V99.12.4554. [DOI] [PubMed] [Google Scholar]
- 20.Schmitt CA, et al. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell. 2002;109(3):335–346. doi: 10.1016/S0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- 21.Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411(6841):1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 22.Reddy HK, Graña X, Dhanasekaran DN, Litvin J, Reddy EP. Requirement of Cdk4 for v-H-ras induced breast tumorigenesis and activation of the v-ras-induced senescence program by the R24C mutation. Genes Cancer. 2010;1(1):69–80. doi: 10.1177/1947601909358105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landis MW, Pawlyk BS, Li T, Sicinski P, Hinds PW. Cyclin D1-dependent kinase activity in murine development and mammary tumorigenesis. Cancer Cell. 2006;9(1):13–22. doi: 10.1016/j.ccr.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 24.Finch AJ, Soucek L, Junttila MR, Swigart LB, Evan GI. Acute overexpression of Myc in intestinal epithelium recapitulates some but not all the changes elicited by Wnt/beta-catenin pathway activation. Mol Cell Biol. 2009;29(19):5306–5315. doi: 10.1128/MCB.01745-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padmakumar VC, Aleem E, Berthet C, Hilton MB, Kaldis P. Cdk2 and Cdk4 activities are dispensable for tumorigenesis caused by the loss of p53. Mol Cell Biol. 2009;29(10):2582–2593. doi: 10.1128/MCB.00952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413(6851):83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- 27.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431(7011):1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cerqueira A, Santamaria D, Martinez-Pastor B, Cuadrado M, Fernandez-Capetillo O, Barbacid M. Overall Cdk activity modulates the DNA damage response in mammalian cells. J Cell Biol. 2009;187(6):773–780. doi: 10.1083/jcb.200903033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsutsui T, et al. Targeted disruption of CDK4 delays cell cycle entry with enhanced p27(Kip1) activity. Mol Cell Biol. 1999;19(10):7011–7019. doi: 10.1128/mcb.19.10.7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen R, et al. Essential role of phospholipase C gamma 2 in early B-cell development and Myc-mediated lymphomagenesis. Mol Cell Biol. 2006;26(24):9364–9376. doi: 10.1128/MCB.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemann MT, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436(7052):807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeller KI, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103(47):17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maclean KH, Dorsey FC, Cleveland JL, Kastan MB. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest. 2008;118(1):79–88. doi: 10.1172/JCI33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reimann M, et al. Tumor stroma-derived TGF-β limits myc-driven lymphomagenesis via Suv39h1-dependent senescence. Cancer Cell. 2010;17(3):262–272. doi: 10.1016/j.ccr.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 35.Barna M, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456(7224):971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deriano L, et al. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature. 2011;471(7336):119–123. doi: 10.1038/nature09755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang JH, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460(7252):231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu W, et al. Coordinate regulation of RAG1 and RAG2 by cell type-specific DNA elements 5′ of RAG2. Science. 1999;285(5430):1080–1084. doi: 10.1126/science.285.5430.1080. [DOI] [PubMed] [Google Scholar]
- 39.Schulz D, et al. Gfi1b negatively regulates Rag expression directly and via the repression of FoxO1. J Exp Med. 2012;209(1):187–199. doi: 10.1084/jem.20110645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grundy GJ, et al. Initial stages of V(D)J recombination: the organization of RAG1/2 and RSS DNA in the postcleavage complex. Mol Cell. 2009;35(2):217–227. doi: 10.1016/j.molcel.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, et al. The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Adv Immunol. 2010;106:93–133. doi: 10.1016/S0065-2776(10)06004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol. 2008;9(6):613–622. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dengler HS, et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol. 2008;9(12):1388–1398. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bouchard C, Lee S, Paulus-Hock V, Loddenkemper C, Eilers M, Schmitt CA. FoxO transcription factors suppress Myc-driven lymphomagenesis via direct activation of Arf. Genes Dev. 2007;21(21):2775–2787. doi: 10.1101/gad.453107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang H, Regan KM, Lou Z, Chen J, Tindall DJ. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science. 2006;314(5797):294–297. doi: 10.1126/science.1130512. [DOI] [PubMed] [Google Scholar]
- 46.Yuan Z, et al. Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science. 2008;319(5870):1665–1668. doi: 10.1126/science.1152337. [DOI] [PubMed] [Google Scholar]
- 47.van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol. 2007;8(6):440–450. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- 48.Ochiai K, et al. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat Immunol. 2012;13(3):300–307. doi: 10.1038/ni.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 50.Nilsson JA, et al. Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell. 2005;7(5):433–444. doi: 10.1016/j.ccr.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 51.Hernandez L, et al. CDK4 and MDM2 gene alterations mainly occur in highly proliferative and aggressive mantle cell lymphomas with wild-type INK4a/ARF locus. Cancer Res. 2005;65(6):2199–2206. doi: 10.1158/0008-5472.CAN-04-1526. [DOI] [PubMed] [Google Scholar]
- 52.Monti S, et al. Integrative analysis reveals an outcome-associated and targetable pattern of p53 and cell cycle deregulation in diffuse large B cell lymphoma. Cancer Cell. 2012;22(3):359–372. doi: 10.1016/j.ccr.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Al-Assar O, et al. Transformed diffuse large B-cell lymphomas with gains of the discontinuous 12q12-14 amplicon display concurrent deregulation of CDK2, CDK4 and GADD153 genes. Br J Haematol. 2006;133(6):612–621. doi: 10.1111/j.1365-2141.2006.06093.x. [DOI] [PubMed] [Google Scholar]
- 54.Sykes SM, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011;146(5):697–708. doi: 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie L, et al. FOXO1 is a tumor suppressor in classical Hodgkin lymphoma. Blood. 2012;119(15):3503–3511. doi: 10.1182/blood-2011-09-381905. [DOI] [PubMed] [Google Scholar]
- 56.Loeb KR, et al. A mouse model for cyclin E-dependent genetic instability and tumorigenesis. Cancer Cell. 2005;8(1):35–47. doi: 10.1016/j.ccr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 57.Yin FF, et al. Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis. Nat Struct Mol Biol. 2009;16(5):499–508. doi: 10.1038/nsmb.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melek M, Gellert M. RAG1/2-mediated resolution of transposition intermediates: two pathways and possible consequences. Cell. 2000;101(6):625–633. doi: 10.1016/S0092-8674(00)80874-0. [DOI] [PubMed] [Google Scholar]
- 59.Nepal RM, Zaheen A, Basit W, Li L, Berger SA, Martin A. AID and RAG1 do not contribute to lymphomagenesis in Emu c-myc transgenic mice. Oncogene. 2008;27(34):4752–4756. doi: 10.1038/onc.2008.111. [DOI] [PubMed] [Google Scholar]
- 60.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68(5):869–877. doi: 10.1016/0092-8674(92)90030-G. [DOI] [PubMed] [Google Scholar]
- 61.Leonard JP, et al. Selective CDK4/6 inhibition with tumor responses by PD0332991 in patients with mantle cell lymphoma. Blood. 2012;119(20):4597–4607. doi: 10.1182/blood-2011-10-388298. [DOI] [PubMed] [Google Scholar]
- 62.Chiron D, et al. Induction of prolonged early G1 arrest by CDK4/CDK6 inhibition reprograms lymphoma cells for durable PI3Kdelta inhibition through PIK3IP1. Cell Cycle. 2013;12(12):1892–1900. doi: 10.4161/cc.24928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang X, et al. Prolonged early G(1) arrest by selective CDK4/CDK6 inhibition sensitizes myeloma cells to cytotoxic killing through cell cycle-coupled loss of IRF4. Blood. 2012;120(5):1095–1106. doi: 10.1182/blood-2012-03-415984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmitz R, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol. 2009;9(3):195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.