Abstract
Identifying a scalable, unbiased method for discovering which members of the human gut microbiota influence specific physiologic, metabolic and immunologic phenotypes remains a challenge. Here we describe a method in which a clonally-arrayed collection of cultured, sequenced bacteria was generated from one of several human fecal microbiota samples found to transmit a particular phenotype to recipient germ-free mice. Ninety-four bacterial consortia, of diverse size, randomly drawn from the culture collection, were introduced into germ-free animals. We identified an unanticipated range of bacterial strains that promoted accumulation of colonic regulatory T cells (Tregs) and expansion of Nrp1lo/− peripheral Tregs, as well as strains that modulated mouse adiposity and cecal metabolite concentrations using feature selection algorithms and follow-up mono-colonization. This combinatorial approach enabled a systems-level understanding of some of the microbial contributions to human biology.
Introduction
Characterizations of the structural configurations of human gut microbial communities are beginning to reveal differences between healthy individuals and those with various diseases (1–6). Experiments involving transplantation of intact uncultured microbiota from healthy humans to humans with colitis induced by Clostridium difficile or patients with metabolic syndrome have helped to establish a causal role for the microbiota in these disorders, and at the same time have provided proof-of-principle that the microbiota represents a therapeutic target for treating or preventing disease (6,7).
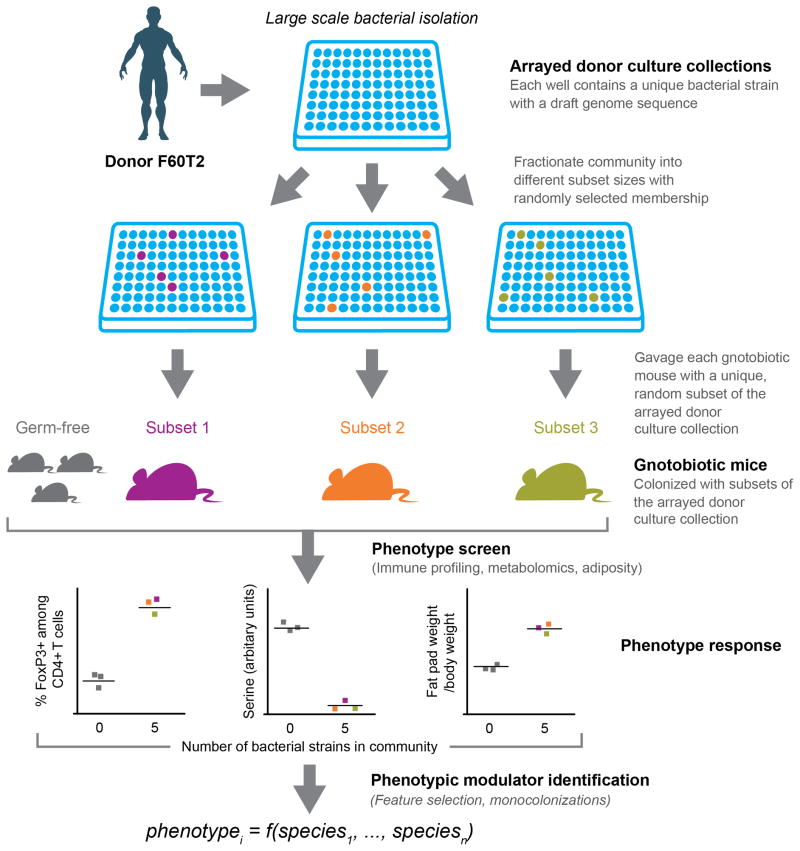
Transplantation of intact uncultured human gut microbiota samples from human donors with various physiologic or disease states, or cultured members of the microbiota, to germ-free mice provides an opportunity to identify specific microbial species that may influence the physiologic, metabolic and immunologic properties of humans (8–12). One challenge has been to develop a scalable, unbiased approach for identifying human gut bacterial strains that modulate phenotypic variation in recipient mice. Here we describe such a method (Fig. 1). It begins with a screen of gnotobiotic mice containing transplanted intact uncultured fecal microbiota from different human donors to identify transmissible phenotypes that can be attributed to each donor’s microbiota. We then generated a clonally-arrayed collection of cultured anaerobic bacteria in multi-well plates from a donor whose intact uncultured microbiota transmitted a phenotype of interest. Each well of the plate harbored a bacterial strain whose genome had been sequenced (13,14). The arrayed culture collection of bacteria was then randomly fractionated into subsets of various sizes. Each subset was gavaged into a germ-free animal, individually maintained in a sterile filter-topped cage, to observe the effect of the bacterial consortium on a host phenotype. By repeating this process across many subsets, the effect of each strain in the arrayed culture collection was assayed in the context of a diverse background of community memberships and sizes. Feature selection algorithms and follow-up experiments where mice were colonized with single strains (mono-colonization) were then used to identify the strains whose presence or absence best explained the observed phenotypic variation. We then identified bacterial strains that modulated several different host phenotypes: adiposity, intestinal metabolite composition, and the immune system.
Fig. 1. Overview of combinatorial ‘out-of-the-isolator’ gnotobiotics screen for identifying microbe-phenotype relationships.
Bacterial culture collections are generated from an intact uncultured (human) gut microbiota sample whose transplantation into gnotobiotic mice has already been shown to alter the animal’s physiologic, metabolic, immunologic or other properties. Each strain in the culture collection is present in a separate well of a multi-well plate. The arrayed collection of sequenced bacterial strains is then fractioned into random subsets of various sizes [shown are subsets (consortia) of five strains]. Each subset is gavaged into an individual germ-free animal maintained in a separate filter-top gnotobiotic cage to observe the effect of the subset on phenotypes of interest. By repeating this process across many subsets, the effect of each strain in the arrayed culture collection is assayed in the context of a diverse background of community memberships and sizes. Feature selection algorithms (for non-saturated phenotypes) and follow-up mono-colonization experiments (for phenotypes saturating at small community sizes) are used to identify the strains whose presence or absence best explains the observed phenotypic variation.
Results
Identifying human gut microbial communities that modulate phenotypic variation in gnotobiotic mice
We began by gavaging groups of 8–10 week old male germ-free C57BL/6J mice with human fecal microbiota samples obtained from five healthy US female donors (F57T2, F60T2, F61T1, F62T1, and F64T2 in table S1; one donor microbiota/group of mice; n=3–10 individually caged mice/group; each group maintained in a separate traditional flexible plastic film gnotobiotic isolator). Mice were fed a standard, autoclaved low-fat, high plant polysaccharide (LF-HPP) mouse chow. Two weeks after gavage, we defined the effects of the transplanted microbiota on regulatory T cells (Tregs) and adiposity.
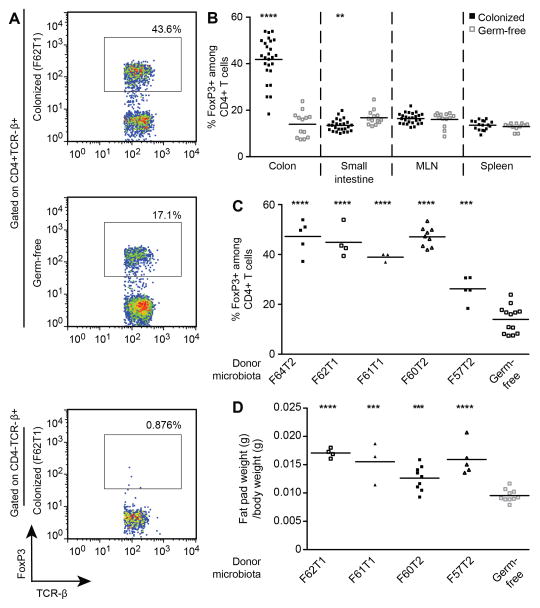
Tregs, which are key regulators of immune homeostasis in the gut and other tissues where they prevent inappropriate inflammatory responses (15), were measured in spleen, mesenteric lymph nodes, plus the small intestinal and colonic lamina propria by quantifying FoxP3 expression in CD4+ T cells using flow cytometry. We observed significant increases in the proportion of Tregs in the colonic lamina propria of mice colonized with the human donor microbiota compared to germ-free controls (Fig. 2A,B; table S2 which includes P-values), in keeping with a recent report using a single Japanese donor (16). This effect was observed with all five intact uncultured donor microbiota (Fig. 2C; table S2) in the context of recipient mice fed the LF-HPP chow, and additionally for donor F60T2 when tested on another diet with a more defined polysaccharide composition (table S2). The effect on Tregs was generally region-specific within the gut: the proportion of small intestine lamina propria Tregs in mice colonized with each of four different donor microbiota was not different from germ-free controls; it was modestly reduced in mice harboring microbiota from the one other donor, F60T2 (table S2; Fig. 2B). The effect was also organ-specific: no differences in the proportions of Tregs were found in the mesenteric lymph nodes in any of the colonized mice. Splenic Treg abundance was unchanged for two of the three donors where this tissue was assayed, and a modest increase was produced by one donor’s microbiota (F61T1) (Fig. 2B and table S2). These latter findings are consistent with a recent report that examined the effects of human gut microbiota on immune phenotypes outside of the colonic lamina propria (17). In addition to the effects on Tregs, these lean donor microbiota produced a 60 ± 17% (mean ± SD) increase in adiposity in recipient animals compared to germ-free controls, as judged by epididymal fat pad weight expressed as a proportion of total body weight (table S2; Fig. 2D).
Fig. 2. Intact uncultured human fecal microbiota drives increases in colonic Tregs and adiposity in recipient gnotobiotic mice.
Adult male germ-free C57BL/6J mice were gavaged with human fecal microbiota suspensions from one of five females living in the USA, or the mice were kept germ-free (see table S1 for donor metadata). Animals were fed a low-fat diet rich in plant polysaccharides. Two weeks after gavage, animals were sacrificed. Cells were prepared from the spleen, mesenteric lymph nodes plus small intestinal and colonic lamina propria (LP). FoxP3 expression was assessed in CD4+ T cells by intracellular flow cytometry. Epididymal fat pads were also harvested and weighed. (A) Representative flow cytometry plots showing FoxP3 expression in CD4+ T cells and CD4− T cells (staining control) isolated from the colonic lamina propria of mice containing a transplanted intact uncultured human fecal microbiota or CD4+ T cells from germ-free controls. The percentages shown represent frequency in the gate. (B) Frequency of FoxP3+ cells among CD4+ T cells in the indicated organs (data pooled from all recipient groups of mice, each harboring a transplanted microbiota from one of five donors, n=12–26 animals/group). (C) Frequency of FoxP3+ cells among colonic CD4+ T cells. Data are grouped according to the human microbiota donor rather than pooled (n=3–13 recipient mice/group). (D) Epididymal fat pat weight as a proportion of total body weight, measured at the time of sacrifice two weeks after gavage of the donor microbiota (n=3–10 recipient mice/group). Each point represents data from an individual mouse. Horizontal lines represent the mean. Statistical significance was determined using a two-tailed unpaired Student’s t-test. **, P<0.01; ***, P<0.001; ****, P<0.0001.
Given the phenotypic differences observed between germ-free mice and those colonized with the microbiota of each tested donor, we generated a clonally-arrayed collection of sequenced anaerobic bacterial isolates from the fecal microbiota of donor F60T2 (the rationale for selection of this donor was based on the fact that the effect size of her microbiota-transmitted phenotypes were representative of those produced by the other donors’ communities). We used whole genome sequence alignments to determine that the arrayed collection contained 17 unique strains (defined as isolates having ≥96% overall shared genome content) (13). These 17 strains represented the four most prominent bacterial phyla in healthy adult human gut microbiota (Bacteroidetes, Firmicutes, Actinobacteria, and Proteobacteria) (table S3).
We subsequently assessed the capacity of these cultured organisms to increase colonic Tregs and adiposity in recipient mice after a two-week colonization. Adult male germ-free mice fed a defined polysaccharide-rich diet were gavaged with the entire culture collection (i.e., all bacterial members together). We found that colonization with all members induced accumulation of a higher representation of Tregs among CD4+ T cells in the colonic lamina propria compared to germ-free controls [29.8±2.49% versus 19.5±1.87% (mean ± SEM), respectively, P=0.0099, two-tailed unpaired Student’s t-test; n=5 and n=15 mice, respectively; table S4]. Adiposity as judged by epididymal fat pad weight also slightly increased upon colonization relative to germ-free mice, although the difference was not significant [1.09±0.026% of total body weight versus 1.21±0.059% (mean ± SEM); P=0.051, two-tailed unpaired Student’s t-test; table S4]. These data suggested that bacterial strains capable of modulating these phenotypes were captured in the ‘personal’ culture collection.
A computational and experimental pipeline to identify microbial strains that modulate host phenotypes
Whereas we routinely build libraries of >40 bacterial strains from human gut microbiota samples (e.g., 13), we deliberately chose this smaller 17-member strain collection for two reasons: (i) it was capable of recapitulating the Treg and adiposity phenotype transmitted by the intact uncultured microbiota; and (ii) it was already sufficiently large to develop the experimental framework and computational tools for identifying effector strains that we had envisioned. Even with only 17 unique bacterial strains, identifying the set of bacterial strains isolated from donor F60T2 that modulated the observed immunologic and adiposity phenotypes represents a vast combinatorial challenge. For example, testing all possible subsets of five strains from the 17-member community with a single replicate (recipient germ-free mouse) would require >6000 communities and an equal number of mice. Testing all subsets of all possible sizes would require >100,000 communities. However, if we assume that many interactions between our gut microbes and our physiologic, metabolic and immunologic phenotypes are at least partially additive or involve only a few higher order interactions, the search to identify the causative groups can be reduced dramatically (18, 19). To illustrate this point, we first developed a probabilistic tool to simulate phenotypic responses, assuming an additive model. We generated a synthetic community of 20 strains in silico where any subset of the 20 organisms could be chosen to induce a phenotypic response (the ‘effector strains’). We modeled the effect size generated by each effector strain with a normal distribution: the mean was chosen to be 10 times the standard deviation (SD) based on the phenotypic effect size we noted in humanized gnotobiotic mice for both the increase in adiposity (mean/SD = 9.2) and the increase in proportion of Tregs (mean/SD = 9.4). Given that most of the intact uncultured donor microbiota in the gnotobiotic mouse screen had produced responses of similar magnitude for both Tregs and adiposity (Fig. 2), we reasoned that some host responses would become saturated. Therefore, we incorporated an option to introduce a maximum (saturation) value into the simulator, whereby the addition of more effector strains would not increase the effect size beyond a certain point. Given a definition of the number of effector strains, the effect size, and the standard deviation of the effect size, we simulated a population of phenotypic responses across all possible subset sizes from 0 to 20 (fig. S1 and fig. S2).
To generate empirical data to compare with our simulations, we sparsely sampled the combinatorial search space across all tested phenotypes by robotically fractionating the culture collection into random subsets containing anywhere from 1–17 strains (Fig. 1 and table S4). These subsets were then gavaged into 8–10 week old male C57BL/6J germ-free recipient mice (all mice fed the diet with defined polysaccharide composition). Non-colonized (germ-free) mice were included in all experiments as reference controls.
In order to increase the throughput of this approach, we developed and validated methods for screening each recipient mouse using filter top cages maintained on racks outside of, rather than within the flexible plastic film isolators typically used for gnotobiotic husbandry. This approach (‘out-of-the-isolator gnotobiotics’) allowed successful colonization without unwanted intrusion of environmental microbes over the course of a two-week experiment. Most community subsets were tested in a single mouse, while the mono- and bi-colonizations were tested with up to six replicates (i.e., 6 mice/subset community; 1 mouse/filter-topped cage; table S4). On average, each strain was tested in 46 different subsets [subset size 7.6 ± 3 (mean ± SD); total of 94 distinct subsets tested across 124 mice].
Two weeks after gavage, each mouse was sacrificed and a panel of phenotypic responses were measured that included (i) adiposity, (ii) short chain fatty acid concentrations [end products of microbial fermentation quantified by targeted gas chromatography-mass spectrometry (GC-MS) of cecal contents], (iii) other metabolites [identified by untargeted GC-MS and liquid chromatography (LC)-MS of cecal contents], and (iv) colonic lamina propria Tregs. At the time of sacrifice, fecal DNA was isolated and subjected to shotgun sequencing to quantify the representation of community members (20), while DNA yield per fecal pellet (ng DNA/mg stool) was used as a proxy for community biomass (21). Fifteen of the 17 bacterial strains tested were consistently detected in recipient mice (n=124 gnotobiotic animals surveyed). These 15 strains were the subject of all further analyses. Plots of fecal biomass versus subset size (1–15) revealed that biomass saturated with communities containing two or more strains, indicating that total biomass was constant across 90 of 94 (96%) of the consortia tested (fig. S3).
Identifying bacterial strains that influence gut metabolic phenotypes
The gut microbiota acts as a metabolic organ to harvest dietary nutrients that cannot be processed by the host alone, as well as nutrients generated by the host (e.g., mucosal glycans) (22). These nutrients can be converted into a diverse array of metabolites available to members of the microbiota as well as to the host. Most of this nutrient processing occurs in the form of fermentation in the distal gut (22). Therefore, we assayed cecal contents using targeted GC-MS to quantify concentrations of short-chain fatty acids (acetate, propionate, succinate, lactate, and butyrate), non-targeted GC-MS to measure 189 metabolites (primarily products of amino acid biosynthesis and carbohydrate metabolism), and non-targeted LC-MS to assay 973 metabolites/spectral features [the latter with negative electrospray ionization]. To focus our analysis on metabolites most likely generated or modified by the gut microbiota, we performed an unpaired two-tailed Student’s t-test to identify those whose concentrations were significantly different in mice colonized with subsets of the culture collection relative to germ-free controls (samples from 85–91 colonized mice and 16–21 germ-free mice were analyzed). In addition, we performed an F-test to identify metabolites whose concentrations displayed significantly different variance in the subset communities compared to germ-free mice (even though the two distributions may not have significantly different mean values). After false-discovery rate correction (FDR<0.01), we identified a total of 715 cecal metabolites/spectral features with significantly different mean or variance relative to these germ-free controls (table S5).
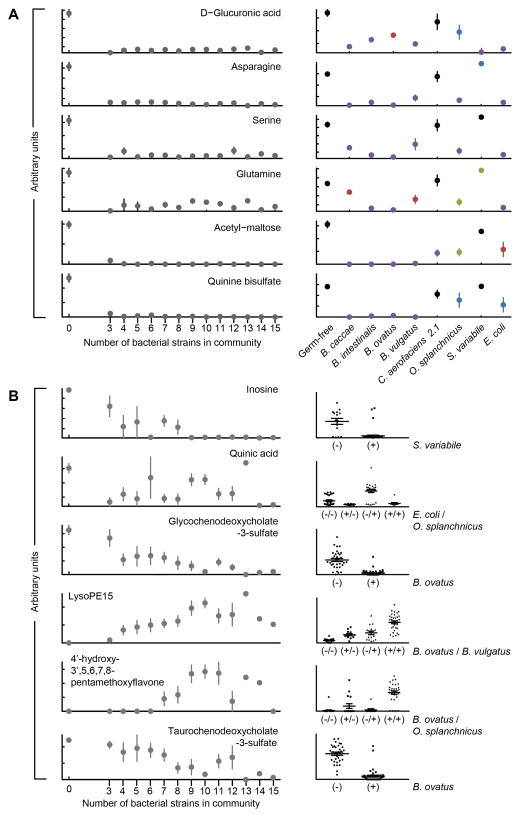
If a defined bacterial community has multiple strains that individually can saturate the effect size of a given phenotype, there will be a steep increase in the mean effect size at small community sizes that will saturate at all larger community sizes (fig. S1). Identifying the point at which the mean effect size has saturated can provide insights into the proportion of community members capable of perturbing that phenotype, and can inform follow-up experiments to identify the specific bacterial strains that mediate the phenotype. In an ideal system where sampling size is unlimited, follow-up experiments could focus on the subset size that produces the maximal entropy (information content) of the phenotype to generate the most informative data with the fewest number of animals: for example, in a 10-member community with only a single effector microbe, the maximum entropy would be a community size of five where the effector would be present in one-half of the communities. In practice, given the technical challenges of gnotobiotic husbandry, only a small number of animals can reasonably be sampled at each community size, so follow-up experiments can focus on consortia sizes that produce high phenotypic variance and avoid community sizes that are larger than the phenotype saturation point. The concentrations of more than 25% of measured cecal metabolites saturated at communities containing ≤3 members, suggesting that a large proportion of the strains in the culture collection were capable of significantly changing the concentrations of these compounds (e.g., Fig. 3A, left panel). Thus, identification of strains causally linked to these cecal metabolite concentrations in mice harboring more than three strains is likely not possible as the variation observed in the concentrations of each of the metabolites represents biological and measurement variation in the response rather than variation due to the addition or removal of species from the community (see right panel of Fig. 3A for an example of how, under these circumstances, follow-up mono-colonizations were used to identify strains capable of depleting a metabolite of interest).
Fig. 3. Bacterial strain-induced alterations in cecal metabolite concentrations.
(A) Metabolites whose levels in the cecum saturate at small community sizes (≤3 strains). A zero on the x-axis refers to germ-free controls. Saturation at low consortia sizes indicates that multiple strains are capable of modulating the metabolite. Mono-colonization (right portion of the panel) directly establishes that specific bacterial strains can alter the level of a given metabolite. Significance of the values are color coded: blue, P<0.05; red, P<0.01; green, P<0.001; purple, P<0.0001 as judged by an unpaired two-tailed Student’s t-test. Shown are mean values ± SEM. These examples represent a subset of the small-community saturated metabolites with high confidence identifications. Data for additional metabolites are presented in table S4. (B) Metabolites with more diverse patterns of changes in their levels as a function of community size reveal cases where fewer community members modulate the metabolite and instances where saturation requires the cumulative influence of multiple bacterial strains. For these metabolites, model-based approaches such as stepwise regression can be used to identify the strains that best explain the alterations in metabolite abundance. Mean values ± SEM are plotted. Each point in the right-hand portion of panel B represents a different community of 3–15 members. The x-axis groups the points into columns based on the strains identified by stepwise regression whose presence (+) or absence (−) in a community best explains the observed variation in metabolite level (P<0.001, F-test). The number of columns represents all possible combinations for presence/absence for the effector strains. The middle and upper/lower lines in each column denote the mean and SEM for all communities in that group.
For phenotypic responses that do not saturate or that only saturate at larger community sizes, the shape and variance of the community size versus effect size can once again provide information about the number of effector strains and how to identify them (fig. S2 and Fig. 3B). For phenotypes driven by a single effector microbe (i.e., only one of the 15 strains can induce a response), the mean effect size will linearly increase as the number of different strains in a subset is increased from 1 to all isolates in the culture collection. This linear increase occurs because at smaller community sizes the effector organism is less likely to be in the group than at larger community sizes. This becomes clear if we observe the data points themselves rather than the means, as the data points are drawn from a bimodal distribution with the number of points in each mode being driven by the subset size (fig. S2). As we simulate non-saturating systems with more than one effector strain with each strain generating the same effect size, this bimodality becomes multimodal where the number of modes is one more than the number of effector strains (fig. S2).
In practice, each effector strain can have a different effect size on the phenotype, which combined with the limited number of distinct communities of a given size that can be tested in mice, and the technical/biological variance of each phenotype, obscures identification of these modes. However, we can use feature selection to identify the microbes in each community whose presence or absence best explains the variation observed for a given phenotype. We modeled the concentration of each metabolite as a function of the community composition using the following equation:
where ymetabolite is the concentration of a given cecal metabolite, XstrainA… XstrainO are binary variables representing the presence (Xstrain = 1) or absence (Xstrain = 0) of each strain in the community, β0 is the estimated parameter for the intercept, and βstrainA … βstrainO are the linear regression estimated parameters reflecting the influence of each strain on the abundance of a given metabolite. For this analysis, we were interested in identifying metabolites whose concentration could be altered in the context of one bacterial community compared to another, rather than relative to germ-free controls. Therefore, we only included samples from mice harboring three or more strains (where biomass is saturated) in the model.
With this approach, we identified 42 metabolites where over half of the metabolite variation could be explained from the composition of the microbiota (mean R2 >0.5 with 10-fold cross validation; table S6). For each of these metabolites, which included bile acids, fatty acids, and amino acids, we used step-wise regression, as a feature selection method, to identify the community members that best explain the variation observed (Fig. 3B). As in our simulations with a single effector strain (fig. S2), we observed a bimodal distribution of cecal metabolite concentration where the two modes are largely explained by the presence or absence of the strain identified as influencing that metabolite (fig. S4). For example, the bile acids taurochenodeoxycholate-3-sulfate and glycochenodeoxycholate-3-sulfate were both significantly reduced when Bacteroides ovatus was present in a community (Fig. 3B; P-value = 7.50×10−21 and 6.58×10−22 respectively; two-tailed unpaired Student’s t-test). If we highlight the effect size distribution of the metabolite based on the presence/absence of B. ovatus, virtually all of the samples with low concentrations of these bile acids were obtained from mice harboring consortia containing B. ovatus (fig. S4A, black bars). These data could reflect direct consumption by the microbiota, the host or microbial-host co-metabolism.
These results illustrate how a model-based approach provides an opportunity to identify strains whose influence on a (metabolic) phenotype is robust to larger community composition; these identified strains could be used to generate mice with a microbiota designed to provide desired metabolic characteristics. The feature selection-based approach also identified additive metabolite-microbial community interactions involving more than one effector bacterial strain. One such example is our observation that concentrations of a lysophosphatidylethanolamine (LysoPE15) can be increased by either B. ovatus or B. vulgatus, but reaches maximal cecal concentrations when both strains were present in a community (Fig. 3B). Finally, this approach identified metabolite-microbiota interactions with more complex logic and non-linearities where alterations in metabolite abundance were dependent on the non-additive influence of multiple organisms. For example, quinic acid, a cyclic polyol obtained from plant-derived products, reached its highest levels when Odoribacter splanchnicus was present but Escherichia coli was not (i.e., the metabolite responded to the logic gate, ‘NOT E. coli AND O. splanchnicus’), while another metabolite (assignment, 4′-hydroxy-3′,5,6,7,8-pentamethoxyflavone) required both B. ovatus and O. splanchnicus to consistently reach high levels (i.e., ‘B. ovatus AND O. splanchnicus’; Fig. 3B, right panel).
Identifying strains that modulate host adiposity
Germ-free mice are known to be leaner than their colonized counterparts (23); this reflects a complex set of interrelationships, both known and yet to be characterized, between the gut microbiota, biotransformation of dietary components, and regulation of host metabolism (24, 25). The increase in the epididymal fad pad weight relative to total body weight appears to saturate at community sizes containing only 1–2 members (fig. S3), suggesting that a large proportion of the strains in the culture collection are capable of influencing host adiposity in this diet context. Therefore, to identify bacterial strains that increase adiposity, we focused our attention on mono-colonized mice. In addition to the five mono-colonizations performed in our initial screen (see above), we conducted seven more mono-colonizations in traditional gnotobiotic isolators with higher replication (n=4–11 animals/bacterial strain) (table S7). Overall, 8 of the 11 strains tested were able to produce a significant increase in adiposity two weeks after their introduction by oral gavage: they included five of the six Bacteroides strains surveyed (B. intestinalis, B. ovatus, B. vulgatus, B. caccae, and B. thetaiotaomicron) plus two other Bacteroidetes (Parabacteroides diastasonis, O. splanchnicus), and a member of Proteobacteria (E. coli) (mean percent increase for all significant strains, 21% ± 9 (SD); table S7 which includes P-values). The correlation between adiposity and fecal community biomass (table S8) for the mono-colonizations was below the threshold for significance (r = 0.49; P = 0.07, Pearson’s correlation). The finding that adiposity saturated at 1–2 strains in our combinatorial analysis of the 15 strains does not preclude the capacity of other gut microbes, microbe-microbe interactions, diet or host effects to shift this saturation point. For example, we recently characterized the microbiota of twins stably discordant for obesity: a culture collection generated from the microbiota of the obese co-twin transmitted greater increases in adiposity to recipient gnotobiotic mice than did the culture collections from the lean co-twin (9). Combinatorial gnotobiotics provides a way to screen multiple culture collections from lean and obese donors in order to identify strains that can change this saturation point.
Identifying strains that modulate the host immune system
Germ-free mice have marked defects in immune function relative to mice raised from birth with a mouse microbiota (conventionally-raised animals). These defects include decreased B and T cell (both αβ and γδ) function and impaired anti-microbial defense (26). Several research groups have identified gut microbes that have the capacity to shape different aspects of the intestinal and systemic immune systems, including differentiation of distinct CD4+ T cell effector subsets in the spleen (27) and small intestine (28, 29), boosting intestinal IgA production (30), and expanding the size and effector function of the CD4+ FoxP3+ regulatory T cell (Tregs) compartment (16, 31–34). Bifidobacterium infantis, Bacteroides fragilis and a consortium of Clostridia spp. isolated from humans have been shown to modulate intestinal Tregs (16, 33, 34) but as yet a systematic approach has not been applied to elucidate, in an unbiased fashion, the full scope of human-associated microbial species capable of modulating intestinal Treg responses.
During our phenotypic screens, we observed that as few as two different bacterial strains were sufficient to induce a level of colonic lamina propria Treg accumulation equivalent to that achieved with the complete uncultured microbiota [34.6±1.13% and 31.1±1.68% (mean ± SEM) for two-member and complete uncultured microbiota, respectively; P= 0.17 (two-tailed unpaired Student’s t-test); table S2]. Among the five mono-colonizatons in the initial screen, B. intestinalis produced the highest levels of colonic lamina propria Tregs [36.6% ± 4.39 (mean ± SEM) FoxP3+ among CD4+ T cells], and reached high densities of colonization. Collinsella aerofaciens, which colonized to low density, did not induce an increase in Tregs in this initial screen (table S9).
We performed follow-up mono-colonizations of adult male C57BL/6J mice with strains from the order Bacteroidales, including four members from the genus Bacteroides (B. caccae, B. thetaiotaomicron, B. vulgatus, and B. massiliensis), one from the genus Parabacteroides (P. distasonis), and one from the genus Odoribacter (O. splanchnicus) to determine if induction of Tregs was a general property of Bacteroides or if it extended across the order Bacteroidales. In addition, we colonized mice with E. coli, which reached high densities in mono-colonization (35). C. aerofaciens was used as a negative control (n=4–6 mice per treatment group per experiment). Tregs were measured in colonic lamina propria (Fig. 4 and table S9) as well as spleen and mesenteric lymph nodes (table S10) two weeks post-gavage.
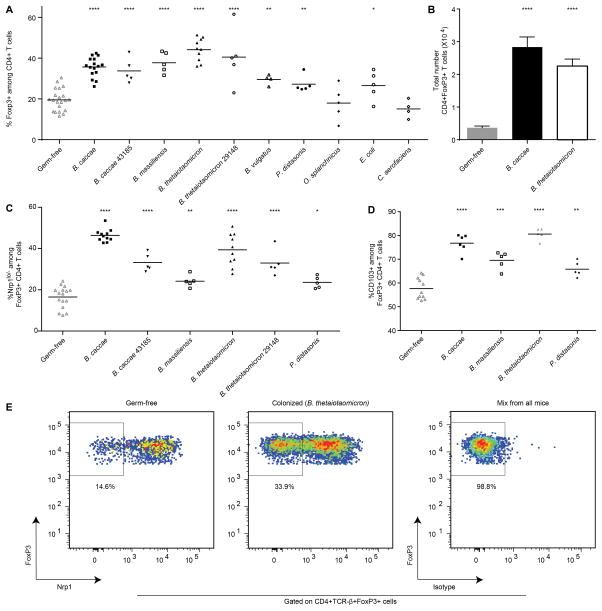
Fig. 4. Mono-colonization with select bacterial strains modulates colonic Tregs.
Adult male germ-free C57BL/6J mice were colonized with one bacterial strain as indicated, or were maintained as germ-free. Two weeks after gavage, mice were sacrificed. Cells from the colonic lamina propria were prepared and FoxP3, Neuropilin-1 (Nrp1) and CD103 expression were assessed in CD4+ T cells by flow cytometry. (A) Frequency of FoxP3+ cells among CD4+ T cells in germ-free versus mono-colonized mice (n=4–21 animals/group). (B) Total numbers of CD4+ FoxP3+ T cells for the indicated mono-colonized mice or germ free mice (n=10–16 animals/group). (C) Frequency of Nrp1lo/− cells among colonic FoxP3+ CD4+ T cells in germ-free versus mono-colonized mice (n=5–16 animals/group). (D) Frequency of CD103+ cells among colonic FoxP3+ CD4+ T cells recovered from germ-free or mono-colonized mice (n=5–11 animals/group). Horizontal lines in panels A, C and D represent the mean, and each point represents an individual mouse. Bars in B represent the mean and error bars show SEM. The statistical significance of difference observed between germ-free and colonized animals was determined using a two-tailed unpaired Student’s t-test. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. (E) Representative flow cytometry plots showing Nrp1 expression in FoxP3+ CD4+ T cells recovered from the colonic lamina propria of germ-free versus mono-associated animals. The percentages shown represent frequency in the gate. Isotype stain (control) was performed on a mixture of colonic lamina propria cells from all groups.
Each Bacteroides strain resulted in significant increases in the proportions of FoxP3+ Treg cells among CD4+ T cells in the colon lamina propria (Fig. 4A and fig. S5A; see table S9 for P-values). The extent of increase was not significantly correlated with the densities of colonization of these strains (P=0.26; Pearson’s correlation). Although mono-colonization with P. distasonis led to a significant increase in Tregs compared to germ-free controls, colonization with O. splanchnicus, which like P. distasonis is in the family Porphyromonadaceae, did not produce a significant effect (P=0.593, two-tailed unpaired Student’s t-test; these differences did not correlate with fecal biomass). E. coli also directed a significant increase in Tregs frequency (P=0.021, two-tailed unpaired Student’s t-test) while as expected, our negative control C. aerofaciens, did not lead to significant changes (Fig. 4A; table S9; P=0.132, two-tailed unpaired Student’s t-test). Further characterization of the response to B. caccae and B. thetaiotaomicron revealed that the observed relative increase in colonic Tregs was accompanied by absolute increases in the number of these cells, compared to germ-free controls (Fig. 4B; P=6.01×10−10 and 1.62×10−10 respectively, two-tailed unpaired Student’s t-test). Moreover, other gut-derived strains of each of these two species obtained from the American Type Culture Collection (ATCC) and thus representing different donors than the individual used to produce our culture collection, also drove significant increases in the proportions of colonic Tregs (Fig. 4A). Our culture collection did not contain any members of the Clostridia, which have recently been shown to be potent inducers of colonic Treg accumulation. However, these studies documented that mono-colonization with the majority of the Clostridia strains isolated did not lead to increases in Tregs (16).
To assess whether our results in C57BL/6J mice were generalizable to additional mouse strains, we colonized adult male germ-free NMRI mice with B. caccae, a potent inducer of Tregs (Fig. 4A), and sacrificed the mice two weeks later. B. caccae was able to drive significant increases in colonic Tregs demonstrating that the Treg response to this bacterial strain is conserved across two different mouse genetic backgrounds [20.59 ± 1.01% FoxP3+ Tregs among CD4+ T cells in NMRI animals colonized with B. caccae versus 10.96 ± 1.11 % in germ-free NMRI controls (mean ± SEM); n=10 mice/per group, pooled data from two independent experiments, P=4.88×10−6 (two-tailed unpaired Student’s t-test); table S9].
The intestinal microbiota can modulate several aspects of colonic Tregs besides expansion, such as altering their T-cell receptor (TCR) repertoire (36, 37) and favoring accumulation of peripherally derived Tregs (pTregs) over those that differentiate in the thymus (tTregs) (16, 31, 32, 37, 38). To determine whether the observed increases in Tregs represented accumulation of pTregs or tTregs, we assayed expression of neuropilin-1 (Nrp1) in FoxP3+ Treg cells [tTregs are Nrp1hi, while pTregs are Nrp1lo/−; (38, 39)]. These assays were conducted using C57BL/6J mice mono-colonized with B. caccae, B. massiliensis, B. thetaiotaomicron or P. distasonis from our culture library plus a strain of B. caccae and B. thetaiotaomicron obtained from the ATCC. Greater proportions of Nrp1lo/− cells among Tregs were documented under each colonization condition (Fig. 4C,E), indicating differentiation of pTregs locally rather than expansion of pre-committed tTregs. Upon colonization, greater proportions of Nrp1lo/− Tregs were also observed in the mesenteric lymph nodes but not in the spleen, likely reflecting the preferential exposure of cells in the mesenteric lymph nodes to gut-associated microbial antigens, although these differences were not equivalent to those seen in the colon (table S11). Colonization of germ-free C57BL/6J mice with B. caccae, B. thetaiotaomicron, B. massiliensis or P. distasonis also resulted in significantly increased proportions of colonic Tregs expressing the ∝E integrin CD103, suggesting a more activated phenotype (32, 40, 41) (Fig. 4D, fig. S5B).
Short chain fatty acids have recently been shown to represent a microbiota-derived molecular cue capable of promoting colonic Treg accumulation (42–44). Therefore, we measured the concentrations of acetate, propionate, butyrate, succinate and lactate in cecal contents from a subset of our mono-colonizations. Colonization with B. caccae, B. thetaiotaomicron, B. massiliensis or P. distasonis led to increases in acetate, propionate and succinate relative to germ-free controls (table S12) while butyrate was only significantly elevated above concentrations observed in germ-free controls in the case of mice colonized with P. distasonis.
Discussion
These results show that the capacity to induce Tregs and modify their phenotype is a characteristic of more effector strains than was appreciated previously. Our findings concerning the role of human gut bacteria in shaping features of the gut mucosal immune system complements and extends the elegant work by Atarashi et al (16). They used a single selective condition (chloroform treatment) to recover a group of 17 strains (all of which were described as members of the class Clostridia), from the human fecal microbiota of a single donor and showed the consortium was capable of expanding the colonic regulatory T cell compartment in gnotobiotic mice. The fact that we found this effector activity among gut species belonging to other bacterial phyla suggests that distribution of this functional capacity may be beneficial in ensuring that this tolerogenic cell type is consistently and persistently maintained in different microbial community and host contexts. The approach we describe allows systematic follow-up analyses of the extent to which the Treg response is affected by factors such as age at colonization or by different diets that produce abrupt and substantial alterations in microbiota configurations (45–47). Of interest, despite identifying members of different human gut bacterial phyla that shape the Treg response, our study and that of Atarashi et al revealed that intestinal short chain fatty acid concentrations increased upon colonization. Given the substantial amount of data supporting a role for short chain fatty acids in induction of Tregs (42–44), this suggests a common pathway by which different microbes converge to modulate this facet of the host immune system. The genetic manipulability of some of the strains identified in this report, notably the Bacteroides, affords an opportunity to test this and other hypotheses, and advance our knowledge about the molecular underpinnings of microbiota-Treg cross-talk.
As the field of human microbial ecology research moves from observational studies to hypothesis-driven experiments designed to directly test the contributions of the microbiota and its components to health, there is a growing need to develop and transition to a modernized set of Koch’s postulates (48) where the groups of microbes that modulate host phenotypic responses are identified along with the environmental factors (e.g. dietary) necessary for the response to be fully manifest. We have developed a platform for systematically identifying microbe-host phenotype interactions in different (human) donor microbiota using gnotobiotic mice that can represent different host genetic features, and different environmental conditions of interest. With the 17 strains in our culture collection, there were more than 100,000 possible combinations to search for effector strains. Using the mathematical and experimental strategies described, we only needed 100 combinations to identify multiple effector microbes for three very diverse biological responses (metabolic, adiposity, Tregs). This represents a 1000-fold reduction in the search space compared to what would be required theoretically. By testing these 100 combinations of microbes in an out-of-the-isolator gnotobiotic caging system rather than in traditional flexible film isolators, we overcame what would have been an insurmountable practical barrier to performing these studies for most groups. Our entire study could have been completed with a single flexible film isolator to generate the required germ-free mice. This feature suggests that our overall approach should be accessible to many investigators, since animal facilities with small numbers of gnotobiotic isolators already exist in numerous universities.
While identifying effector strains represents a critical first step in mechanistic analyses of how the gut microbiota impacts various facets of host biology, once such strains are identified, much additional work needs to be done. For example, numerous other important components of the intestinal immune system may also be affected by colonization with the strains we identified, including B cell class switching to IgA, macrophage/dendritic cell effector or migratory properties, and γδ T cell function. Another important goal is to identify the effector molecules produced by the identified effector strains and the host signalling pathways through which these molecules act. Using gnotobiotic mice genetically deficient in various components of the immune system (such as toll-like receptors or inflammasomes) and effector strains that are genetically manipulated (e.g., through whole genome transposon mutagenesis) represent ways for pursuing this goal. While additional elements of these mechanistic analyses will be dependent on the biological processes being interrogated, in principle, this platform can be applied to any microbiota-associated phenotype. Finally, our approach has therapeutic implications since it represents an enabling system for identifying and characterizing next generation probiotics, or combinations of pre- and probiotics (synbiotics).
Materials And Methods
Animal husbandry
All studies involving mice used protocols approved by the Washington University Animal Studies Committee. The development and implementation of ‘out-of-the-isolator’ gnotobiotic husbandry is detailed in Supplementary Results. Mice were fed one of two autoclaved mouse chows [one low in fat and rich in plant polysaccharides (B&K Universal, diet 7378000) or another with a more defined polysaccharide composition (Harlan Teklad, diet TD.2018S)] ad libitum. Animals were maintained under a strict 12-hour light cycle (lights on at 0600h).
Human fecal samples
De-identified fecal samples were collected as part of previous studies (5,9). Methods used for recruitment of subjects, obtaining informed consent, and for sample collection, de-identification and storage were approved by the Washington University IRB.
Human fecal microbiota transplants
Previously frozen fecal microbiota samples were prepared for transplantation into germ-free mice as previously described (9). Briefly, frozen (−80°C) fecal material was resuspended in sterile pre-reduced PBS-0.05% (w/v) cysteine-HCl at a concentration of 100 mg/mL by vortexing for 5 min in an anaerobic chamber (Coy Laboratory Products; atmosphere composed of 75% N2, 20% CO2, 5% H2). Large particulate material was allowed to settle by gravity for 5 min. The supernatant was placed in a glass vial with a crimped top to maintain the anaerobic environment after removal from the anaerobic chamber. Once transported to the gnotobiotic facility, and following introduction into the gnotobiotic isolator after sterilization of the outside surface of the vessel (Clidox-S; Pharmacal Research Laboratories), a 200 μl aliquot of the sample was introduced into recipient mice with a single oral gavage.
Clonally arrayed bacterial culture collections from human fecal samples
Methods for generating and sequencing the culture collection produced from a frozen fecal microbiota sample obtained from donor F60T2 are described elsewhere (9, 13).
Growth of bacterial strains for inoculation into germ-free animals
Strains from frozen stocks were grown as monocultures in an anaerobic chamber in modified Gut Microbiota Medium [GMM where short chain fatty acids were replaced with sodium acetate (final concentration 12.2 mM); ref. 14] for 2–3 d in deep well 96 well plates, or in 15 mL tubes. Strains were mixed together in various combinations, or maintained as a monoculture, and then combined with sterile glycerol (final concentration 15% v/v) to create stocks that were then placed in a glass vial with a crimped top (in the anaerobic chamber), and stored at −80°C until further use. Just prior to introduction to the gnotobiotic isolator, vials with these strains were thawed, and administered to each animal as a single 200 μL aliquot by oral gavage. All strains were isolated in-house as described above except B. caccae ATCC 43185 and B. thetaiotaomicron ATCC 29148 which were purchased from the ATCC.
Community Profiling by Sequencing (COPRO-Seq) of community composition
We have previously described methods for using short-read (50nt) shotgun sequencing of fecal DNA to quantify the relative and absolute abundances of members in defined consortia of sequenced human gut bacterial strains that had been introduced into germ-free recipients (20, 45). We applied the same approach (COPRO-Seq) to identify which strains in the collection were capable of consistently colonizing mice. Unique barcoded adapters were ligated onto each sequencing library, which was then size-selected (~350bp) to remove adapter dimers. To increase efficiency, barcoded sequencing libraries were pooled and amplified together. The purified amplicons were then sequenced on either an Illumina MiSeq or HiSeq 2000 instrument (paired-end reads; 50nt in one direction, 25nt in the other direction). Reads were split into samples using the barcode sequence (we required that both ends of the pair-end sequence have the same barcode in order to reduce the number of chimeras). Reads generated from each fecal sample were then mapped back to the sequenced genomes of members of the culture collection, as well as to the mouse genome, using bowtie (49), allowing up to one mismatch/read. The relative abundance of each strain was calculated as the fraction of ‘non-mouse’ sequencing reads mapping to each strain’s genome. We generated Illumina sequencing libraries from 1–2 fecal pellets taken from mice harboring each community. For libraries with >10,000 sequencing reads, we calculated the ratio between the median relative abundance for an organism when that organism was not gavaged into the mouse, divided by the median relative abundance when the organism was gavaged. For organisms whose median relative abundance was >10%, there were ~500x fewer reads in sample where the strain was absent than when it was present, thus providing an estimate of the maximum signal/noise ratio for the COPRO-Seq assay. We defined organisms capable of colonizing mice within the detection limit of our assay as those with a signal/noise ratio of >10. Only two of the strains in the 17-member arrayed culture collection, Ruminococcus albus and Ruminococcus bromii, fell below this threshold, with signal/noise ratios of 1.6 and 6.5 respectively.
For mono-colonization experiments, DNA prepared from one or two fecal pellets obtained at the time of sacrifice, was used to verify that only a single bacterial strain was present in the recipient mouse’s microbiota. The median number of sequencing reads mapping to the relevant strain in all mono-colonized animals (n=82) was >99.5% (table S13).
Simulating phenotypic responses as a function of community size
We developed a web application to simulate phenotypic responses with a probabilistic additive model (http://faithlab.mssm.edu/model_viz/). The application allows adjustment of community size, the number of effector microbes, the effect size (all effector microbes are assumed to have the same effect size), the standard deviation of the effect size, the sample size (how many different random communities to sample at a given subset size), and the effect size saturation (i.e., the maximum effect size summed across effector strains). Given a definition of these variables, the total effect size is calculated as max(sum effect size, effect size saturation), where sum effect size = effect size * number of effector strains. A final effect size for the community is then simulated by drawing a random sample from a normal distribution with the user-defined standard deviation and the mean set to the total effect size. This process is repeated for the requested sample size across all community subsets from zero to the total community size.
Isolation and characterization of immune cells
Isolation
Spleens and mesenteric lymph nodes were harvested and placed in ice-cold Dulbecco’s Phosphate Buffered Saline (DPBS; Gibco) containing 0.1% bovine serum albumin (BSA) w/v (Sigma). Single cell suspensions were prepared by passage through a cell strainer (70 μm-diameter pores; BD Falcon) and resuspension in DPBS/0.1% BSA. Splenocytes were pelleted by centrifugation at 453 × g for 5 min and supernatant was removed. Pellet was resuspended by gentle flicking of the tube and red blood cells were removed by addition of 700 μL of pre-warmed (37°C) “ammonium-chloride-potassium (ACK) Lysis” buffer (Gibco) to splenocytes followed by immediate vortexing. After a 3 min incubation at room temperature, ~10 mL of DPBS/0.1% BSA was added to halt the lysis, the cell suspension was passed through a cell strainer (70 μm-diameter pores), centrifuged at 453 × g for 5 min and the supernatant was removed. Spleen and mesenteric lymph nodes cell suspensions were stored on ice until further use.
Colonic lamina propria cells were isolated using a previously described protocol with modifications (50). Briefly, the colon was harvested immediately after sacrifice, and opened longitudinally with scissors. Luminal contents were removed by gentle scraping and the colon was trimmed of fat/mesentery. The remaining tissue was minced into small pieces (~2–3 mm3) and the fragments were placed on ice in ~10 mL DPBS/0.1% BSA until further processing. DPBS/0.1% BSA was aspirated and the tissue fragments were placed in 15 mL of ‘complete’ RPMI [RPMI 1640 (Gibco) containing heat inactivated fetal bovine serum (5% v/v; Gibco), penicillin (100 U/mL; CellGro), streptomycin (100 μg/mL; Gibco), L-glutamine (2.0 mM; CellGro), and HEPES (20 mM; CellGro)] plus ethylenediaminetetraacetic acid (EDTA; 5 mM; CellGro). The mixture was shaken at 180 rotations per minute (rpm) for 20 min (at 37°C) in an orbital shaker after which time the medium was aspirated. This step was repeated once and the colonic tissue was washed of excess EDTA by placing in 10 mL complete RPMI at room temperature for 5 min, after which time the medium was removed by aspiration. This step was repeated once, and the colonic tissue fragments were digested by placing in 10 mL complete RPMI supplemented with 0.3 mg/ml Collagenase Type VIII (Sigma) and 0.075 U/mL Dispase (BD Bioscience) for 60 min at 37°C with shaking at 180 rpm. Tissue fragments were vortexed and then allowed to settle briefly, and the supernatant was collected by passing through a cell strainer (BD Falcon; 100 μm pore diameter); an equal volume of complete RPMI with EDTA (5 mM) was used to wash the strainer and added to the harvested cell suspension to ensure that digestion was terminated. Cells were pelleted by centrifugation at 453 × g, the supernatant was discarded, the pellet was resuspended by flicking, and cells were stored on ice in 10–20 mL DPBS/0.1% BSA. The colonic tissue fragments that remained from this initial round of digestion were subjected to an identical second round of digestion and the resulting cells were pooled with those from the first digestion. No Percoll enrichment step was performed.
Flow cytometry
Cells were placed in a round bottom 96-well plate for staining. Cells were first incubated in DPBS/0.1% BSA containing anti-CD16/CD32 (Fc block (2.4G2); BD Pharmingen) at 4°C for 20 min to prevent non-specific binding of antibody in subsequent steps. Cells were pelleted by centrifugation at 453 × g for 5 min (4°C). The resulting supernatant was discarded and the pelleted cells were surface stained (with anti- CD4, TCR-β, CD103, Nrp1 or isotype control) by resuspending in DPBS/0.1% BSA containing the appropriate cocktail of fluorophore-conjugated antibodies. Cells were stored at 4°C in the dark for 20 min and were then washed by adding 200 μL of DPBS/0.1% BSA followed by centrifugation at 453 × g for 5 min. The supernatant was discarded, and the cell pellets were washed a second time with 250 μL of DPBS/0.1% BSA. Pellets were resuspended by vortexing and fixed in 100 μL Fixation/Permeabilization Buffer (eBioscience). Cells were stored for 16–40 h at 4°C in the dark. For intracellular staining, 200 μL of DPBS/0.1% BSA was added and the cells were centrifuged at 652 × g for 5 min. The supernatant was discarded, cells were resuspended by vortexing, and the step was repeated a second time. One hundred microliters of Permeabilization Buffer (eBioscience) supplemented with normal rat serum (Sigma; 2% v/v) was added and the cells were (i) mixed by pipetting, (ii) placed in the dark for 30–60 min at 4°C, (iii) centrifuged at 652 × g for 5 min, (iv) the resulting supernatant was discarded and the cells were resuspended in Permeabilization buffer containing anti-FoxP3 or isotype control antibody. Cells were subsequently mixed by pipetting and stored at 4°C for 30 min (or room temperature for 20 min).
Cells were washed by addition of 200 μL of Permeabilization buffer, centrifuged at 652 × g for 5 min, the supernatant was discarded, and the cells were resuspended by vortexing. This wash step was repeated twice, first with Permeabilization buffer and subsequently with DPBS/0.1% BSA. Cells were then resuspended in DPBS/0.1% BSA and analyzed using an LSR II, Canto II or Aria III flow cytometer (BD Biosciences). Data were compensated using FACSDiva (BD Biosciences) or FlowJo software (Tree Star), and then further analyzed using FlowJo software.
The following antibodies were used: CD4 PE-Cy7 (RM4-5; eBioscience), CD4 PE (GK1.5; eBioscience), CD4 APC (GK1.5; eBioscience), CD4 V500 (RM4-5; BD Horizon), FoxP3 eFluor-450 (FJK-16s; eBioscience), TCR-β PerCp-Cy5.5 (H57-597; BD Pharmingen), CD103 PE (M290; BD Pharmingen), and Nrp1 APC (FAB566A; R&D Systems). The appropriate fluorophore-conjugated isotype control antibodies were used where indicated. Cells were counted using a hemacytometer (American Optical Corp.).
Targeted quantification of short chain fatty acid concentrations in cecal contents using gas chromatography-mass spectroscopy
Methods for targeted quantification of short chain fatty acid concentrations are described in detail elsewhere (51). Briefly, cecal contents were homogenized with 10 μL of a mixture of internal standards (20mM acetic acid-13C2,D4, propionic acid-D6, butyric acid-13C4, lactic acid-3,3,3-D3 and succinic acid-13C4), 20μL of 33% HCl, and 1 mL diethyl ether. The mixture was centrifuged and the organic fraction was removed. The diethyl ether extraction was repeated. After combining the two ether extracts, a 60 μL aliquot was derivatized with 20 μL N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide (MTBSTFA) for 2 h at room temperature. Derivatized samples (1 μL) were injected with 15:1 split into an Agilent 7890A gas chromatography system, coupled with 5975C mass spectrometer detector (Agilent, CA). The quantity of each metabolite was determined from the spike-in internal standards and a calibration curve. Quantities were normalized to the initial weight of the sample.
Non-targeted analysis of cecal metabolites using GC-MS
Cecal contents were homogenized with 20 vol/wt of HPLC grade water. Homogenates were centrifuged (20,800 × g for 10 min at 4°C). A 200 μL aliquot of the supernatant was transferred to a clean tube and combined with ice-cold methanol (400 μL). The mixture was subsequently vortexed and centrifuged, and a 500 μL aliquot of the resulting supernatant, together with 10 μL of lysine-13C6,15N2 (2 mM), was evaporated to dryness using a speed vacuum. To derivatize the sample, 80 μL of a solution of methoxylamine (15 mg/mL in pyridine) was added to methoximate reactive carbonyls (incubation for 16 h for 37°C), followed by replacement of exchangeable protons with trimethylsilyl groups using N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) with a 1% v/v catalytic admixture of trimethylchlorosilane (Thermo-Fisher Scientific, Rockford, IL) (incubation for 1h at 70°C). Heptane (160 μL) was added and a 1 μL aliquot of each derivatized sample was injected into the GC/MS system. Metabolite identification was done by co-characterization of standards.
Untargeted analysis of cecal metabolites using ultra high performance liquid chromatography-mass spectrometry (UPLC-MS)
Frozen cecal samples were homogenized with 20 vol/wt of cold methanol and 1 vol/wt cysteine 13C6,15N2 (4 mM). Samples were subsequently incubated at −20°C for 1h and centrifuged 10 min at 20,800 × g. The resulting supernatant (300μL) was collected and dried in a SpeedVac at room temperature. Dried samples were resuspended in 100 μL of 95:5 water:ethanol, clarified for 5 min by centrifugation at 20,800 × g for 10 min at 4°C, and the supernatant was separated for UPLC-MS. Analyses were performed on a Waters Acquity I Class UPLC system (Waters Corp., Milford, MA) coupled to an LTQ-Orbitrap Discovery instrument (Thermo Fisher Corporation). Mobile phases used for negative ionization were (A) 5 mM ammonium bicarbonate in water and (B) 5 mM ammonium bicarbonate in a mixture of acetonitrile/water (95/5).
Statistical analysis
Unpaired Student’s t-tests (two-tailed) were used to determine statistically significant differences in adiposity and immune cell populations between germ-free animals and gnotobiotic animals colonized with one or more microbes. False-discovery rate (FDR) corrected t-tests were used to compare all 1167 metabolites concentrations in germ-free versus colonized animals was employed. Significant differences in metabolite variance between germ-free and colonized animals were determined by FDR corrected F-test. Linear regression with 10-fold cross-validation was used to model metabolite concentration as a function of community composition, while microbial strains that best explained the variation in concentration of each metabolite were identified with stepwise-regression.
Supplementary Material
fig. S1 – Simulating phenotype effect size and saturation as a function of microbial community size
fig. S2 – Simulating phenotype effect size as a function of microbial community size
fig. S3 - Phenotypic responses as a function of community membership size
fig. S4 – Modeling identifies individual bacterial strains that modulate cecal metabolite concentrations in complex communities
fig. S5 – Representative flow cytometry plots of FoxP3 and CD103 expression in the colon of mono-colonized and germ-free mice
table S1 - Human subject metadata
table S2 - Influence of donor microbiota on the proportion of regulatory T cells and on host adiposity
table S3 - Bacterial strains represented in clonally arrayed culture collection from donor F60T2
table S4 - Community composition and phenotypic responses in mice harboring subsets of the arrayed culture collection from donor F60T2
table S5 - Cecal metabolites with significantly different concentrations or variances between germ-free mice and mice colonized with subsets of bacterial strains from donor F60T2
table S6 - Modeling phenotypic variation as a function of community composition
table S7 - Differences in adiposity between germ-free mice and mice mono-colonized with bacterial strains from the donor F60T2 culture collection
table S8 - Differences in microbial biomass (DNA (ng)/wet weight of feces (mg)) between germ-free mice and animals mono-colonized with bacteria strains from the culture collection from donor F60T2
table S9 - Differences in the percentage of Treg cells (% FoxP3+ in CD4+ T cells) of the colonic lamina propria between germ-free mice and animals mono-colonized with bacterial strains from the donor F60T2 culture collection
table S10 - Differences in the percentage of Treg cells (% FoxP3+ in CD4+ T cells) in mesenteric lymph nodes and spleens between germ-free mice and animals mono-colonized (C57BL/6J) with bacterial strains from the donor F60T2 culture collection
table S11 - Differences in the percentage of peripheral Treg cells (Neuropilin1lo/− cells in CD4+FoxP3+ T Cells) in mesenteric lymph nodes and spleens harvested from germ-free versus mono-colonized C57BL/6J mice
table S12-Differences in short chain fatty acid concentrations in cecal contents between germ-free and mono-colonized C57BL/6J mice
table S13 - Median relative abundance of reads from strains used in mono-colonization experiments in recipient animal’s fecal DNA
Acknowledgments
We are grateful to David O’Donnell and Maria Karlsson for their help with gnotobiotic animal husbandry, Federico Rey for suggestions regarding procedures for out-of-isolator gnotobiotic husbandry, Jessica Hoisington-Lopez for assistance with DNA sequencing, Janaki Guruge for help with culturing anaerobic human gut bacteria, Marty Meier for his assistance with implementing robotic interfaces, and members of the Gordon lab for their many suggestions and support during the course of this work.
Funding
This work was supported by grants from the NIH (DK30292, DK70977, DK078669), and the Crohn’s and Colitis Foundation of America. P.P.A is the recipient of a Sir Henry Wellcome Postdoctoral Fellowship from the Wellcome Trust [096100].
Footnotes
Author Contributions
J.J.F, P.P.A, and J.I.G. designed experiments; J.J.F., P.P.A. and V.K.R. generated the data involving phenotyping of the immune system and adiposity measurements; J.J.F., P.P.A. and and J.C. generated the metabolomic data; J.J.F., P.P.A., and J.I.G. analyzed the data; J.J.F., P.P.A, and J.I.G. wrote the paper.
Competing Interests
JIG is co-founder of Matatu, LLC, a company that is characterizing the role of diet-by-microbiota interactions in defining health. The other authors declare that they have no competing interests.
“This manuscript has been accepted for publication in Science Translational Medicine. This version has not undergone final editing. Please refer to the complete version of record at www.sciencetranslationalmedicine.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.”
References
- 1.Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, Gougis S, Rizkalla S, Batto JM, Renenault P, Dore J, Zucker JD, Clement K, Ehrlich SD, Blottiere H, Leclerc M, Juste C, de Woulters T, Lepage P, Fouqueray C, Basdevant A, Henegar C, Godard C, Fondacci M, Rohia A, Hajduch F, Weissenbach J, Pelletier E, Le Paslier D, Gauchi JP, Gibrat JF, Louix V, Carre W, Maguin E, van de Guchte M, Jamet A, Boumezbeur F, Layec S ANR MicroObes consortium. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 2.de Vos WM, de Vos EA. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr Rev. 2012;70(Suppl 1):S45. doi: 10.1111/j.1753-4887.2012.00505.x. [DOI] [PubMed] [Google Scholar]
- 3.Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, Nielsen J, Backhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 4.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jorgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clement K, Dore J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker JD, Raes J, Hansen T, Bork P, Wang J, Ehrlilch SD, Pedersen O, Guedon E, Delorme C, Layec S, Khaci G, van de Guchte M, Vandemeulebrouck G, Jamet A, Dervyn R, Sanchez N, Maguin E, Haimet F, Winogradski Y, Cultrone A, Leclerc M, Juste C, Blottiere H, Pelletier E, LePaslier D, Artiguenave F, Bruls T, Weissenbach J, Turner K, Parkhill J, Antolin M, Manichanh C, Casellas F, Boruel N, Varela E, Torrejon A, Guarner F, Denariaz G, Derrien M, van Hylckama Vlieg JE, Veiga P, Oozeer R, Knol J, Rescigno M, Brechot C, M’Rini C, Merieux A, Yamada T MetaHIT consortium. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser EE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 7.Vrieze A, Van Nood E, Holleman F, Salojarvi J, Koote RS, Bartelsman JF, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, Derrien M, Druesne A, Van Hylckama Vlieg JE, Bloks VW, Groen AK, Heillig HG, Zoetendal EG, Stroes ES, de Vos WM, Hoekstra JB, Nieuwdorp M. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 8.Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Backhed HK, Gonzalez A, Werner JJ, Angenent LT, Knight R, Backhed F, Isolauri E, Salminen S, Ley RE. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, Liu J, Houpt E, Li MV, Holmes E, Nicholson J, Knights D, Ursell LK, Knight R, Gordon JI. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 13.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A. 2011;108:6252. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izcue A, Coombes JL, Powrie F. Regulatory lymphocytes and intestinal inflammation. Annu Rev Immunol. 2009;27:313. doi: 10.1146/annurev.immunol.021908.132657. [DOI] [PubMed] [Google Scholar]
- 16.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 17.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis d, Benoist C, Relman DA, Kasper DL. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Box GEP, Hunter JS, Hunter WG. Statistics for experimenters: design, innovation, and discovery. 2. Wiley-Interscience; Hoboken, N.J: 2005. p. xvii.p. 633. Wiley series in probability and statistics. [Google Scholar]
- 19.Hastie T, Tibshirani R, Friedman JH. The elements of statistical learning: data mining, inference, and prediction. 2. Springer; New York, NY: 2009. p. xxii.p. 745. Springer series in statistics. [Google Scholar]
- 20.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, Chervaux C, Knights D, Lozupone CA, Knight R, Duncan AE, Bain JR, Muehlbauer MJ, Newgard CB, Heath AC, Gordon JI. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudio NA, Muegge BD, Henrissat B, Hettich RL, Gordon JI. Effects of Diet on Resource Utilization by a Model Human Gut Microbiota Containing Bacteroides cellulosilyticus WH2, a Symbiont with an Extensive Glycobiome. PLoS Biol. 2013;11:e1001637. doi: 10.1371/journal.pbio.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 25.Delzenne NM, Neyrinck AM, Bäckhed F, Cani PD. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat Rev Endocrinol. 2011;7:639. doi: 10.1038/nrendo.2011.126. [DOI] [PubMed] [Google Scholar]
- 26.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Briddonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 29.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Isanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 33.O’Mahony C, Scully P, O’Mahony D, Murphy S, O’Brien F, Lyons A, Sherlock G, MacSharry J, Shanahan KB, O’Mahony L. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PLoS Pathog. 2008;4:e1000112. doi: 10.1371/journal.ppat.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reigstad CS, Hultgren SJ, Gordon JI. Functional genomic studies of uropathogenic Escherichia coli and host urothelial cells when intracellular bacterial communities are assembled. J Biol Chem. 2007;282:21259. doi: 10.1074/jbc.M611502200. [DOI] [PubMed] [Google Scholar]
- 36.Cebula A, Seweryn M, Rempala GA, Pabla SS, Mcindoe RA, Denning TL, Bry L, Kraj P, Kisielow L. Ignatowicz, Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497:258. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss JM, Bilate AM, Gobert M, Ding Y, Curotto de Lafaille MA, Parkhurst CN, Xiong H, Dolpady J, Frey AB, Ruocco MG, Yang Y, Floess S, Huehn J, Oh S, Li MO, Niec RE, Rudensky AY, Dustin ML, Littman DR, Lafaille JJ. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J Exp Med. 2012;209:1723. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yadav M, Louvet C, Davini D, Gardner JM, Martinez-Liordella M, Bailey-Bucktrout S, Anthony BA, Sverdrup FM, Head R, Kuster DJ, Ruminski P, Weiss D, Von Schack D, Bluestone JA. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med. 2012;209:1713. doi: 10.1084/jem.20120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huehn J, Siegmund K, Lehmann JC, Siewert C, Haubold U, Feuerer M, Debes GF, Lauber J, Frey O, Przybyiski GK, Niesner U, de la Rosa M, Schmidt CA, Brauer R, Buer J, Scheffold A, Hamann A. Developmental stage, phenotype, and migration distinguish naive- and effector/memory-like CD4+ regulatory T cells. J Exp Med. 2004;199:303. doi: 10.1084/jem.20031562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephens GL, Andersson J, Shevach EM. Distinct subsets of FoxP3+ regulatory T cells participate in the control of immune responses. J Immunol. 2007;178:6901. doi: 10.4049/jimmunol.178.11.6901. [DOI] [PubMed] [Google Scholar]
- 42.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013 doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 44.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, Deroos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013 doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333:101. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferreira RB, Willing BP, Finlay BB. Bringing Koch’s postulates to the table in IBD. Cell Host Microbe. 2011;9:353. doi: 10.1016/j.chom.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 49.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uhlig HH, Coombes J, Mottet C, Izcue A, Thompson C, Fanger A, Tannapfel A, Fontenot JD, Ramsdell F, Powrie F. Characterization of Foxp3+CD4+CD25+ and IL-10- secreting CD4+CD25+ T cells during cure of colitis. J Immunol. 2006;177:5852. doi: 10.4049/jimmunol.177.9.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rey FE, Gonzalez MD, Cheng J, Ahern PP, Gordon JI. Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc Natl Acad Sci U S A. 2013;110:13582–13587. doi: 10.1073/pnas.1312524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
fig. S1 – Simulating phenotype effect size and saturation as a function of microbial community size
fig. S2 – Simulating phenotype effect size as a function of microbial community size
fig. S3 - Phenotypic responses as a function of community membership size
fig. S4 – Modeling identifies individual bacterial strains that modulate cecal metabolite concentrations in complex communities
fig. S5 – Representative flow cytometry plots of FoxP3 and CD103 expression in the colon of mono-colonized and germ-free mice
table S1 - Human subject metadata
table S2 - Influence of donor microbiota on the proportion of regulatory T cells and on host adiposity
table S3 - Bacterial strains represented in clonally arrayed culture collection from donor F60T2
table S4 - Community composition and phenotypic responses in mice harboring subsets of the arrayed culture collection from donor F60T2
table S5 - Cecal metabolites with significantly different concentrations or variances between germ-free mice and mice colonized with subsets of bacterial strains from donor F60T2
table S6 - Modeling phenotypic variation as a function of community composition
table S7 - Differences in adiposity between germ-free mice and mice mono-colonized with bacterial strains from the donor F60T2 culture collection
table S8 - Differences in microbial biomass (DNA (ng)/wet weight of feces (mg)) between germ-free mice and animals mono-colonized with bacteria strains from the culture collection from donor F60T2
table S9 - Differences in the percentage of Treg cells (% FoxP3+ in CD4+ T cells) of the colonic lamina propria between germ-free mice and animals mono-colonized with bacterial strains from the donor F60T2 culture collection
table S10 - Differences in the percentage of Treg cells (% FoxP3+ in CD4+ T cells) in mesenteric lymph nodes and spleens between germ-free mice and animals mono-colonized (C57BL/6J) with bacterial strains from the donor F60T2 culture collection
table S11 - Differences in the percentage of peripheral Treg cells (Neuropilin1lo/− cells in CD4+FoxP3+ T Cells) in mesenteric lymph nodes and spleens harvested from germ-free versus mono-colonized C57BL/6J mice
table S12-Differences in short chain fatty acid concentrations in cecal contents between germ-free and mono-colonized C57BL/6J mice
table S13 - Median relative abundance of reads from strains used in mono-colonization experiments in recipient animal’s fecal DNA