Abstract
Daily rhythms in animal behavior, physiology and metabolism are driven by cell-autonomous clocks that are synchronized by environmental cycles, but maintain ~24 hours rhythms even in the absence of environmental cues. These clocks keep time and control overt rhythms via interlocked transcriptional feedback loops, making it imperative to define the mechanisms that drive rhythmic transcription within these loops and on a genome-wide scale. Recent work identifies novel post-transcriptional and post-translational mechanisms that govern progression through these feedback loops to maintain a period of ~24 hours. Likewise, new microarray and deep sequencing studies reveal interplay among clock activators, chromatin remodeling and RNA Pol II binding to set the phase of gene transcription and drive post-transcriptional regulatory systems that may greatly increase the proportion of genes that are under clock control. Despite great progress, gaps in our understanding of how feedback loop transcriptional programs maintain ~24 hours cycles and drive overt rhythms remain.
Introduction
Organisms exposed to daily environmental cycles display diurnal rhythms in physiology, metabolism and behavior. These rhythms are generated and sustained by cell-autonomous circadian clocks, which help organisms anticipate predictable changes in the environment. They continue to operate in constant environmental conditions (i.e., free-run) with a period of about 24 hours. Genetic and molecular analysis of circadian clocks in Drosophila and mice revealed that the circadian timekeeping mechanism consists of interlocked transcriptional feedback loops, which drive rhythmic transcription of ‘clock genes’ that encode feedback loop components and ‘output genes’ that control physiological, metabolic and behavioral rhythms. Most clock genes are well conserved from insects to humans, and with few exceptions, play similar roles in the timekeeping mechanism.
Although transcriptional feedback loops were established as the molecular basis of circadian timekeeping more than 20 years ago [1,2], fundamental questions remain about the mechanisms by which these feedback loops sustain ~24 hours rhythm and drive rhythmic expression of output genes. Here we will review recent studies of clock protein synthesis and modifications that provide significant insight into post-transcriptional mechanisms that control feedback loop progression, and whole genome analysis of transcription, protein–DNA binding and chromatin modifications that shed new light on clock regulation of rhythmic gene expression.
The architecture of transcriptional feedback loops in animals
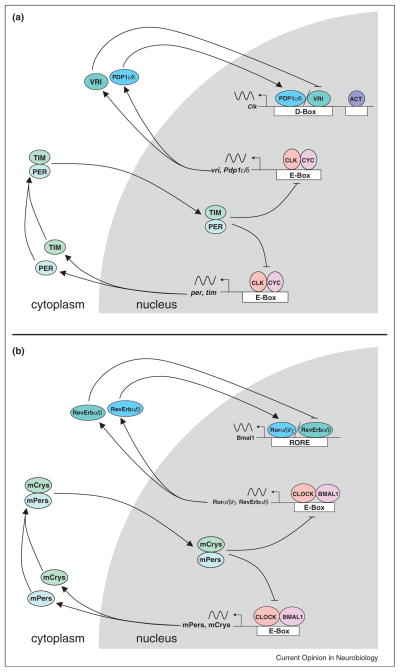
Transcriptional feedback loops that keep circadian time in animals have been largely derived from studies in Drosophila and mice. These feedback loops have recently been reviewed [3–5]; thus, we will present a sketch of their essential working parts (Figure 1). In both of these model systems, a pair of orthologous basic helix–loop–helix PER-ARNT-SIM (bHLH-PAS) transcription factors called CLOCK and BMAL1 (or its homologue NPAS2) in mammals and CLOCK (CLK) and CYCLE (CYC) in Drosophila form heterodimers that bind E-box regulatory elements to activate transcription of genes encoding their repressors, CRYPTOCHROME 1 and CRYPTOCHROME 2 (mCRYs) and PERIOD 1 and PERIOD 2 (mPERs) in mammals and PERIOD (PER) and TIMELESS (TIM) in Drosophila [6–10]. mPER–mCRY complexes in mammals and PER–TIM complexes in Drosophila accumulate in the cytoplasm, move into the nucleus, and then bind to and inactivate the CLOCK–BMAL1 and CLK–CYC activators, respectively, to repress transcription [11,12]. mPER–mCRY and PER–TIM are then degraded, which permits the activators to bind E-boxes and initiate the next cycle of transcription. The primary function of this ‘core’ feedback loop is to determine circadian period.
Figure 1.
Interlocked feedback loops that keep circadian time. Genetic architecture of the core and interlocked feedback loops of Drosophila (a) and mice (b). Gene, protein and regulatory element names are as defined in the text. Sinusoidal lines represent rhythmic mRNAs; arrows depict the synthesis, assembly and/or localization of clock proteins; blocked line denotes repression; gray background indicates events in the nucleus; white background indicates events in the cytoplasm.
CLOCK–BMAL1 and CLK–CYC also activate a second ‘interlocked’ feedback loop that controls rhythmic expression of activator genes (e.g., Bmal1 and Clk), which are transcribed in the opposite circadian phase as repressor genes (e.g., mPers/mCrys and per/tim) [13,14]. In mammals, this feedback loop is controlled by the nuclear hormone receptors Ror α/β/γ and RevErb α/β, which bind RevErbA/Ror-binding elements (RREs) to activate and repress Bmal1 transcription, respectively [15,16]. In contrast, this feedback loop is controlled by the basic leucine zipper (bZIP) transcription factor VRILLE (VRI) in flies, which binds D-box elements to repress Clk activation by PAR Domain Protein 1 δ/ε (PDP1 δ/ε) and other uncharacterized activators [17,18]. Both PAR bZIP and nuclear hormone receptors play major roles in animal physiology and metabolism. Their role in the clock represents a conserved element through which stability and precision of the clock is tied to the metabolic state of the animal.
The timing of feedback loop events during the daily environmental cycle is different in flies and mice. For example, per transcription in all fly tissues peaks around Zeitgeber Time (ZT) 15 (where ZT 0 is lights on and ZT 12 is lights off), whereas the mPers peak around ZT 6 in the ‘master’ brain pacemaker, called the suprachiasmatic nucleus (SCN), and 4–8 hours later in peripheral tissues [19]. This phase difference reflects the principle that light, the principal environmental cue, initially synchronizes the SCN clock, which then acts to synchronize peripheral clocks [20,21]. Light is able to synchronize SCN and Drosophila clocks because the accumulation of key repressor mRNAs and proteins in the core feedback loop is rate limiting; light-dependent degradation of TIM in Drosophila and induction of mPer1 transcription in mammals cause abrupt changes in the phase of the clock that ensure repressor levels are low in flies and high in mammals during daytime. The mechanisms that drive and interpret TIM degradation and mPer1 induction have been reviewed extensively [3,5,22]. Another essential function of these feedback loops is to drive expression of output genes that control overt rhythms, a topic we consider further below.
Mechanisms by which feedback loops maintain ~24 hours periods
The steps required for completing one cycle of the core feedback loop include activator binding to E-boxes, the transcription, RNA processing/cytoplasmic transport, translation, and nuclear localization of repressors, binding and inhibition of activators by repressors, and degradation of repressors. The time it takes to complete these steps should take much less than 24 hours, thus a net delay must be imposed to set the free-running period to ~24 hours. This ‘delay’ principle applies to Drosophila and mammalian systems alike, and the regulation of feedback loop processes common to both systems are remarkably similar [3–5], thus we will focus on Drosophila here for brevity and note important differences between these model systems.
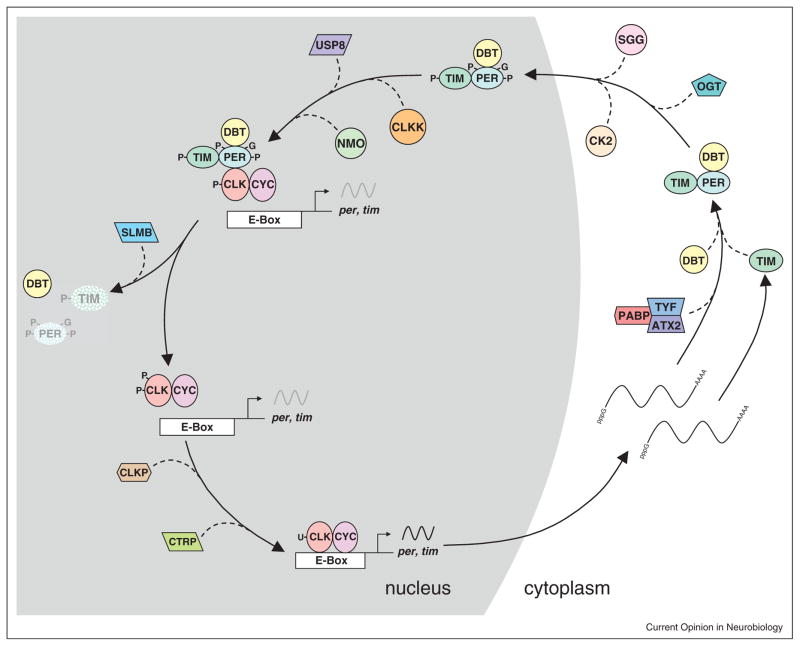
Several feedback loop processes are regulated at the post-translational level, including PER nuclear localization, transcriptional repression, and degradation (reviewed in [3]) (Figure 2). PER phosphorylation by SHAGGY (SGG)/glucose synthase kinase 3 (GSK3) and casein kinase II (CKII) promotes PER nuclear localization, and mutants in either kinase lengthen period [23–26]. This period lengthening suggests that SGG and CKII normally act to shorten period, indicating that this step is inherently slow and must be advanced to achieve a ~24 hours period. DOUBLE-TIME (DBT)/casein kinase I δ/ε (CKI) phosphorylates PER to promote transcriptional repression while decreasing PER stability in the nucleus, thus enhancing repression while limiting the time that repression can occur [27–29]. DBT/CKI, along with NEMO kinase, delays PER degradation in the nucleus by phosphorylating residues in the ‘per-short’ domain that includes the original period shortening perS mutant [29–31]. Phosphorylated PER is stabilized by TIM binding, which delays PER degradation until after TIM is destroyed after dawn [32,33]. Reduced PER–TIM binding, such as in the perL mutant [34], lengthens circadian period by increasing the time it takes PER to accumulate. Ultimately, PER phosphorylation by DBT/CKI at S47 forms a binding site for the E3 ubiquitin ligase SLIMB/β-TrCP, which targets PER for degradation via the ubiquitin–proteasome pathway [29]. Processes promoted by PER phosphorylation are counterbalanced by protein phosphatases, including PP2a and PP1, which dephosphorylate PER [35,36]. Phosphorylation-dependent regulatory mechanisms that delay PER degradation in the nucleus extend transcriptional repression for many hours, thereby delaying the core feedback loop. As in flies, clock protein phosphorylation governs the same feedback loop processes in mice, including nuclear localization, transcriptional repression and degradation (reviewed in [4]). Many kinases have the same specificity and function in the mammalian feedback loop (e.g., CKI phosphorylates the mPERs to promote their degradation in the nucleus), but clock components that serve a different function in mice are targeted by different kinases (e.g., AMPK targeting mCrys) [37].
Figure 2.
Regulatory events in the core loop of Drosophila that control 24 hours periodicity. Gene, protein and regulatory element names are as defined in the text. CLKK represents kinases that phosphorylate CLK; CLKP denotes phosphatases that dephosphorylate CLK; stippled proteins indicate degradation; black sinusoidal lines represent active transcription; gray sinusoidal lines represent repressed transcription; arrows depict the synthesis, assembly and/or localization of clock proteins; dashed lines denote the action of regulatory proteins; lines marked with pppG and AAAA depict mature mRNAs; P depicts phosphorylation, G indicates O-GlcNAcylation; U represents ubiquitylation; gray background indicates events in the nucleus; white background indicates events in the cytoplasm.
Phosphorylation is not the only post-translational modification of PER. Rhythmic glycosylation of cytosolic PER at S and T residues with O-linked N-acetylglucosamine (O-GlcNAc), which peaks around mid-night (e.g., ZT16–ZT20), acts to enhance PER stability and delay nuclear entry [38••]. Blocking the enzyme which O-GlcNAcylates PER, called O-GlcNAc transferase (OGT), shortens circadian period, which implies that PER O-GlcNAcylation imposes a delay in the core loop, perhaps by competing with phosphorylation-dependent PER nuclear entry and degradation. It is not clear how the delay in cytosolic PER accumulation (which generates the lag between per mRNA and protein accumulation) is controlled. Although a novel phosphorylation-dependent destabilization of cytosolic PER could generate such a lag, growing evidence indicates that this lag is mediated by regulated PER translation. Recent work shows that TWENTY-FOUR (TYF) is targeted to per mRNA via ATAXIN 2 (ATX2) RNA binding protein to form a complex with POLY-A BINDING PROTEIN (PABP) and promote PER translation [39••,40•,41••]. Since the role of this complex is to promote PER translation, it suggests that the lag in PER accumulation arises because per mRNA is difficult to translate or translation is repressed via a separate mechanism. Regulation of PER translation by TYF/ATX2/PABP complexes occurs only in brain pacemaker neurons [39••,40•,41••], suggesting that other mechanisms regulate the lag in PER accumulation in peripheral tissues.
As PER enters the nucleus, it binds to CLK–CYC and promotes CLK phosphorylation, transcriptional repression, and the release of CLK–CYC from E-boxes (reviewed in [3]). Although DBT/CKI plays a non-catalytic role in targeting CLK for phosphorylation, neither the kinases nor phosphorylation sites that inhibit CLK transcription and/or DNA binding have been identified. In contrast to CLK phosphorylation, CLK ubiquitylation peaks when CLK–CYC transcriptional activity is maximal from ZT10-14 [42•]. This rhythm in ubiquitylation is mediated by UBIQUITIN SPECIFIC PROTEASE 8 (USP8), which deubiquitylates CLK to downregulate CLK–CYC activity from ~ZT18-ZT4, thereby reinforcing PER-dependent repression [42•]. Once PER is degraded, CLK–CYC transcription is reactivated coincident with an increase in ubiquitylated CLK and a reduction in phosphorylated CLK, but the extent to which these processes contribute to delays that set the ~24 hours circadian period is not known. The increase in ubiquitylated CLK could be mediated by the Circadian TRIP (CTRIP) E3 ubiquitin ligase [43], ortholog of TRIP12 in mammals, whereas the reduction in phosphorylated CLK could result from CLK dephosphorylation or new CLK synthesis.
CLOCK phosphorylation correlates with mPER–mCRY binding and increased CLOCK phosphorylation [44], suggesting that similar mechanisms operate on mammals. However, BMAL1 phosphorylation, acetylation, sumoylation and ubiquitylation also control CLOCK–BMAL1 transcriptional activity [4], thus adding regulatory complexity compared to Drosophila, where CYC appears to be a permissive rather than an instructional factor [3]. Importantly, factors that mediate the post-translational modification of clock components are modulated by other signaling pathways and have other targets. Therefore, these steps also form nodes connecting the core clock with different signaling pathways. For example, fasting induced activation of AMPK in the mouse liver promotes mCRY degradation, thereby constituting a mechanism that integrates energy sensing with the core clock [37].
Regulation of rhythmic outputs via transcriptional feedback loops
Components of both the core and the interlocked loops are transcription regulators, so their action on other loci is the first step in generating overt rhythms. Here we will discuss how microarray and deep-sequencing approaches have revealed the extent of tissue-specific rhythms in chromatin state, factor binding, transcription and transcript abundance, and allude to novel post-transcriptional gene regulatory mechanisms. Since CLK–CYC and CLOCK–BMAL1 directly or indirectly initiate all circadian transcription in flies and mice, respectively, we will focus on these core regulators.
A genome-wide analysis of CLK, PER and RNA polymerase II (Pol II) binding in fly heads revealed CLK binding rhythms at >800 CLK target sites that peak at ~ZT14, followed by PER binding ~6 hours later [45•]. Only ~30% of rhythmically bound CLK targets showed rhythmic Pol II binding, but many of these genes were not previously detected as producing cycling mRNAs, likely because they represent a single RNA isoform and/or may have a limited expression pattern. Often the Pol II binding rhythm was not synchronous with that of CLK, implying that CLK–CYC binding can drive rhythmic expression in different circadian phases. Analysis of nascent and processed transcripts revealed rhythms in RNA editing, RNA splice variants, and non-coding RNAs that mediate ribosome biogenesis [46,47]. Important issues that arise from these studies are how CLK–CYC is targeted to specific genes and isoforms in different tissues, how the phase of Pol II binding is determined once activators bind, and how the clock regulates mRNA cycling at the post-transcriptional level.
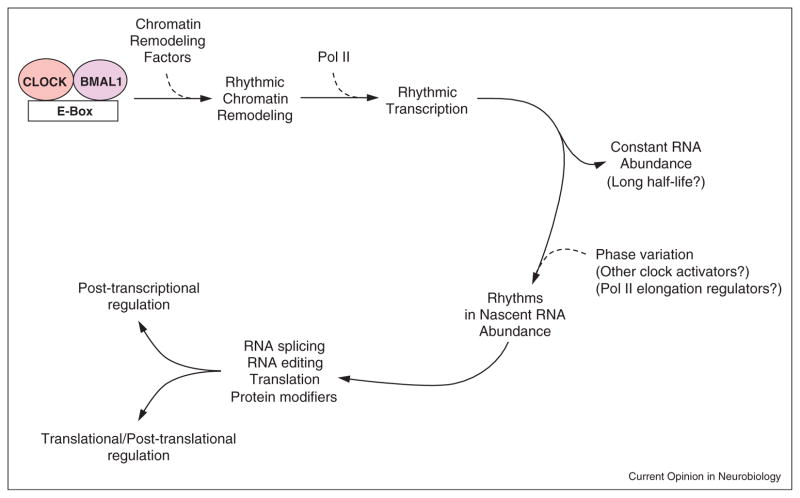
In mammals, global transcriptional regulation by components of the core loop also bear considerable similarity to that in insects (Figure 3). Integrative analyses of the dynamic chromatin environment and transcript abundance in mouse liver have revealed five major phases of circadian transcription [48••,49•,50••]. Maximum levels of CLOCK–BMAL1 complex along with p300 are detected around circadian time 8 (CT8, where CT12 corresponds to activity onset in constant darkness) when H3K9ac and H3K4me1 levels also peak. This marks the transcription activation phase, immediately followed by active transcription for the next 4–5 hours leading to maximum nascent transcript levels at ~CT14-16. Binding of mPER and mCRY repressors then marks the repression phase until ~CT22, after which accumulation of Ser5 phosphorylated Pol II marks the poised state of transcription initiation until ~CT2. Transcriptional de-repression must occur between CT2 and CT6 for the next round of activation to start. Robustly oscillating transcripts show coordinated rhythms in histone modifications and recruitment of clock components to their proximal regulatory sites [51••]. Beyond this generalized schema, locus specific regulation might produce transcription rhythms having different phases or magnitudes. These genomic studies also identified circadian oscillations in antisense RNA, non-coding RNAs, miRNA, RNA processing factors, and in ribosome biogenesis, thus offering mechanisms for generation of circadian rhythms at the post-transcriptional level [48••,50••,51••,52].
Figure 3.
Transcriptional regulation of circadian gene expression. Protein and regulatory element names are as defined in the text. Arrows, sequence of events triggered by CLOCK–BMAL1 binding to E-boxes; dashed lines, influence of transcription factors; chromatin remodeling factors, factors that alter chromatin structure; phase variation, regulation of nascent RNA cycling phase; parentheses, possible explanation for phenomena. See text for detailed description.
Recent deep-sequencing studies have produced some clues for tissue specific transcript oscillations. Transcriptionally silent loci show characteristic DNA and histone modification marks of silent transcription and a near absence of active marks. The expressed transcripts span several orders of dynamic range and roughly correlate with activation marks from proximal regulatory sites. The vast majority of cycling transcripts show oscillations with a peak to trough ratio of <10 fold; a small portion of the large transcript dynamic range. At the trough there are still detectable transcript levels and the loci are not completely devoid of activation chromatin marks [49•,51••]. This implies that tissue specific factors likely mark loci for basal transcription and clock components generate transcript oscillations. Such a dual mode of regulation likely explains the tissue specific nature of circadian outputs.
While these deep-sequencing approaches have revealed genome-wide rhythms in transcriptional regulation, some cautionary notes should be mentioned. As was seen earlier with micro-array studies, rhythmic transcript sets from the same organ (even those identified by the same groups) rarely overlap by >50%. Antibody quality, wet lab methods, and data analysis methods complicate these experiments. Hence, the lack of overlap between any two parameters may not be entirely due to biological differences. For example, statistical tests showed only a fraction of promoters with H3K4me3 oscillations also showing robust H3K27Ac rhythms, while visualization revealed a larger overlap [51••]. The peak phases of rhythmic H3K4me3 and H3K9Ac in one study were coincident while another study found them ~10 hours apart [48••,51••]. These discrepancies underscore the value of validating chromatin marks, factor binding and mRNA levels before detailed studies of individual genes commence.
Conclusions
Recent studies in Drosophila and mice have provided new insights into the nature of delays within the core feedback loop that generate a 24 hours period and the regulation of global rhythms in gene expression required for circadian timekeeping and driving overt rhythms. Although phosphorylation promotes nuclear localization and delays degradation of the PER and mPER repressors in the nucleus, new data in Drosophila show that O-GlcNAcylation of PER delays its nuclear localization and enhances its stability, possibly by competing with PER phosphorylation. CLK deubiquitylation by USP8 reinforces transcriptional repression by PER complexes, whereas CLK ubiquitylation and decreased phosphorylation may be involved in shifting CLK to a transcriptionally active state. TYF–ATX2–PABP complexes promote PER translation in brain pacemaker neurons, which suggests that inefficient PER translation accounts for the lag in cytoplasmic PER accumulation. Despite these advances in defining delays in the core loop, we do not know how PER complexes inhibit CLK–CYC, how CLK–CYC transcriptional activity is reactivated, the basis of inefficient PER translation in pacemaker neurons, whether translational regulation delays PER accumulation in other tissues, and the extent to which delays in the mammalian core loop are regulated by the same mechanisms.
Genome-wide analysis of transcript dynamics in their chromatin context has revealed novel gene regulatory mechanisms at the transcriptional, post-transcriptional and translational levels. CLK–CYC and CLOCK–BMAL1 activator binding promotes Pol II binding at different phases, indicating that additional factors regulate the phase of transcription. CLK–CYC and CLOCK–BMAL1 also regulate specific output genes in different tissues, which suggests that they combine with tissue-specific activators that permit circadian transcription of certain output genes. Rhythmic transcription extends to non-coding RNAs and enzymes that regulate gene expression at the post-transcriptional and translational levels. Given the potential for circadian regulation of gene expression at many different levels, it is likely that a much larger proportion of genes are under clock control than previously thought. However, this number will be hard to determine given the technical and biological variability.
Acknowledgments
This work was supported by National Institutes of Health grants DK091618 (SP), EY01607 (SP), and NS052854 (PEH).
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 2.Hardin PE, Hall JC, Rosbash M. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc Natl Acad Sci U S A. 1992;89:11711–11715. doi: 10.1073/pnas.89.24.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardin PE. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet. 2011;74:141–173. doi: 10.1016/B978-0-12-387690-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peschel N, Helfrich-Forster C. Setting the clock — by nature: circadian rhythm in the fruitfly Drosophila melanogaster. FEBS Lett. 2011;585:1435–1442. doi: 10.1016/j.febslet.2011.02.028. [DOI] [PubMed] [Google Scholar]
- 6.Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 7.Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 8.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 9.Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix–loop–helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci U S A. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 11.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 12.Lee C, Bae K, Edery I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK–CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol Cell Biol. 1999;19:5316–5325. doi: 10.1128/mcb.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glossop NR, Lyons LC, Hardin PE. Interlocked feedback loops within the Drosophila circadian oscillator. Science. 1999;286:766–768. doi: 10.1126/science.286.5440.766. [DOI] [PubMed] [Google Scholar]
- 14.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 15.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 16.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 17.Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- 18.Glossop NR, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron. 2003;37:249–261. doi: 10.1016/s0896-6273(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 19.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 21.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golombek DA, Rosenstein RE. Physiology of circadian entrainment. Physiol Rev. 2010;90:1063–1102. doi: 10.1152/physrev.00009.2009. [DOI] [PubMed] [Google Scholar]
- 23.Akten B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR. A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci. 2003;6:251–257. doi: 10.1038/nn1007. [DOI] [PubMed] [Google Scholar]
- 24.Ko HW, Kim EY, Chiu J, Vanselow JT, Kramer A, Edery I. A hierarchical phosphorylation cascade that regulates the timing of PERIOD nuclear entry reveals novel roles for proline-directed kinases and GSK-3beta/SGG in circadian clocks. J Neurosci. 2010;30:12664–12675. doi: 10.1523/JNEUROSCI.1586-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- 26.Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- 27.Nawathean P, Rosbash M. The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol Cell. 2004;13:213–223. doi: 10.1016/s1097-2765(03)00503-3. [DOI] [PubMed] [Google Scholar]
- 28.Kivimae S, Saez L, Young MW. Activating PER repressor through a DBT-directed phosphorylation switch. PLoS Biol. 2008;6:e183. doi: 10.1371/journal.pbio.0060183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiu JC, Vanselow JT, Kramer A, Edery I. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 2008;22:1758–1772. doi: 10.1101/gad.1682708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiu JC, Ko HW, Edery I. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell. 2011;145:357–370. doi: 10.1016/j.cell.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu W, Houl JH, Hardin PE. NEMO kinase contributes to core period determination by slowing the pace of the Drosophila circadian oscillator. Curr Biol. 2011;21:756–761. doi: 10.1016/j.cub.2011.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 33.Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 34.Gekakis N, Saez L, Delahaye-Brown AM, Myers MP, Sehgal A, Young MW, Weitz CJ. Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- 35.Fang Y, Sathyanarayanan S, Sehgal A. Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1) Genes Dev. 2007;21:1506–1518. doi: 10.1101/gad.1541607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sathyanarayanan S, Zheng X, Xiao R, Sehgal A. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell. 2004;116:603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- 37.Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38••.Kim EY, Jeong EH, Park S, Jeong HJ, Edery I, Cho JW. A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 2012;26:490–502. doi: 10.1101/gad.182378.111. This paper tests whether clock proteins within the core are O-GlcNacylated. The authors find that PER protein is rhythmically O-GlcNacylated, and that this modification both delays PER nuclear entry and stabilizes PER. These results reveal an important delay mechanism that functions in the cytosol to determine circadian period. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39••.Lim C, Allada R. ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science. 2013;340:875–879. doi: 10.1126/science.1234785. In this paper a proteomics strategy is taken to identify partners of TYF, which activates PER translation. This analysis reveals that ATX2 binds to TYF and PABP to activate PER translation in pacemaker neurons, which contrasts with previously characterized roles for ATX2 in translational repression. These studies provide a mechanistic basis for the TYF translational activation and a new role for ATX2 as a translation activator. [DOI] [PubMed] [Google Scholar]
- 40•.Lim C, Lee J, Choi C, Kilman VL, Kim J, Park SM, Jang SK, Allada R, Choe J. The novel gene twenty-four defines a critical translational step in the Drosophila clock. Nature. 2011;470:399–403. doi: 10.1038/nature09728. This paper used an overexpression screening approach to identify tyf. Genetic and molecular analysis showed that TYF promoted PER translation. This work was the first to identify a role for translational regulation within the Drosophila clock. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41••.Zhang Y, Ling J, Yuan C, Dubruille R, Emery P. A role for Drosophila ATAXIN-2 in the activation of PERIOD translation and circadian behavior. Science. 2013;340:879–882. doi: 10.1126/science.1234746. A genetic approach was taken to show that reduced ATX2 expression lengthened circadian period. Genetic, molecular and behavioral analysis of ATX2 revealed that it interacts with TYF and PABP in pacemaker neurons to promote PER translation. Along with Lim et al. [40•], this study provides a mechanistic basis for the TYF translational activation and a new role for ATX2 as a translation activator. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Luo W, Li Y, Tang CH, Abruzzi KC, Rodriguez J, Pescatore S, Rosbash M. CLOCK deubiquitylation by USP8 inhibits CLK/CYC transcription in Drosophila. Genes Dev. 2012;26:2536–2549. doi: 10.1101/gad.200584.112. This paper investigated the role of USP8 in the circadian clock because its mRNA was consistently identified as highly rhythmic. Genetic, molecular and behavioral studies show that USP8 deubiqityates CLK protein to reinforce PER-dependent transcriptional repression and that CLK ubiquitylation coincides with maximal transcription. This study is the first to show that a USP plays a role within the circadian timekeeping mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamaze A, Lamouroux A, Vias C, Hung HC, Weber F, Rouyer F. The E3 ubiquitin ligase CTRIP controls CLOCK levels and PERIOD oscillations in Drosophila. EMBO Rep. 2011;12:549–557. doi: 10.1038/embor.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 45•.Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, Tkachev S, Rosbash M. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev. 2011;25:2374–2386. doi: 10.1101/gad.178079.111. This paper used a ChIP-chip approach to map circadian recruitment of CLK, CYC, PER and Pol II to promoters in Drosophila heads. The experiments reveal temporally orchestrated recruitment of activator, repressor, and Pol II to the promoters of hundreds of genes, and identify RNA isoform-specific and tissue specific CLK targets. These results imply that CLK partner proteins contribute to gene-specific and tissue-specific circadian regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hughes ME, Grant GR, Paquin C, Qian J, Nitabach MN. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Res. 2012;22:1266–1281. doi: 10.1101/gr.128876.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez J, Tang CH, Khodor YL, Vodala S, Menet JS, Rosbash M. Nascent-Seq analysis of Drosophila cycling gene expression. Proc Natl Acad Sci U S A. 2013;110:E275–E284. doi: 10.1073/pnas.1219969110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48••.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. This paper used deep-sequencing method to quantify circadian patterns of histone modification, Pol II binding, clock component occupancy on DNA, and transcript levels in adult mouse liver. They found that the circadian transcriptional cycle consists of a poised state, a coordinated activation state and a repression state, each marked by distinct combinatorial histone modifications and protein DNA interactions. Only 22% of the rhythmic transcripts were driven by de novo transcription, suggesting a larger role of post-transcriptional steps in circadian outputs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49•.Le Martelot G, Canella D, Symul L, Migliavacca E, Gilardi F, Liechti R, Martin O, Harshman K, Delorenzi M, Desvergne B, et al. Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 2012;10:e1001442. doi: 10.1371/journal.pbio.1001442. This paper found that rhythmic Pol II recruitment at promoters rather than a rhythmic transition from paused to productive elongation underlies circadian transcription in the mouse liver. Modeling Pol II occupancy and transcript oscillations revealed a role of RNA half-life and post-transcriptional processing in circadian output regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife. 2012;1:e00011. doi: 10.7554/eLife.00011. This paper used a nascent-RNA sequencing approach and compared transcription to processed RNA and CLOCK–BMAL1 occupancy on promoters They found that the phase of CLOCK–BMAL1 binding does not determine the phase of transcript rhythm, and a prominent contribution of post-transcriptional processing to oscillations in processed transcripts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51••.Vollmers C, Schmitz RJ, Nathanson J, Yeo G, Ecker JR, Panda S. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 2012;16:833–845. doi: 10.1016/j.cmet.2012.11.004. This paper found circadian oscillations in chromatin marks characteristic of promoters and enhancers and transcript oscillations from proximal loci. Circadian rhythms in non-coding, miRNA and regulatory RNAs including an antisense Per2 transcript were identified. The amplitude of transcript oscillation paralleled binding of multiple clock components to proximal regulatory sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jouffe C, Cretenet G, Symul L, Martin E, Atger F, Naef F, Gachon F. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 2013;11:e1001455. doi: 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]