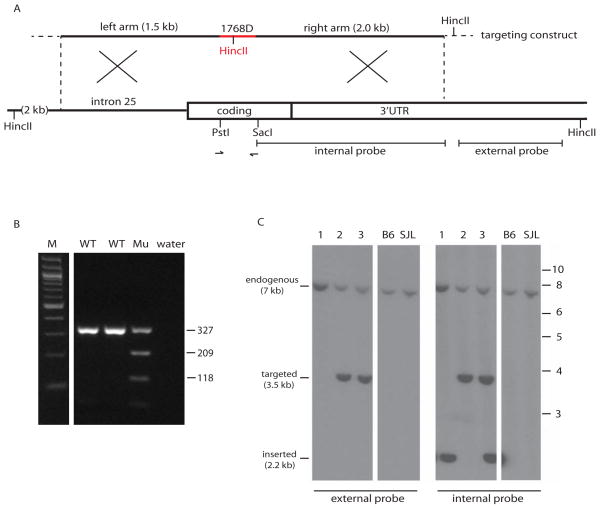
Figure 2. Structure of the targeting construct and genotype assays to detect targeted alleles and random insertions.
A. The 6.3 kb targeting construct was generated from a 320 bp synthetic fragment (red) containing 9 nucleotide substitutions (Figure 1B) plus flanking genomic fragments of 1.5 kb and 2.0 kb and the vector sequence of 3 kb (not shown). Arrows represent the PCR primers used to amplify a 327 bp fragment containing the targeted site for genotyping and sequencing. The indicated external probe was used for hybridization of Southern blots with HincII digested genomic DNA, generating a 3.5 kb HincII fragment from correctly targeted Scn8a loci. The internal probe was used to detect random insertions of the targeting construct by hybridization to the internal 2.2 kb HincII fragment generated from the HincII site in the vector. B. Detection of mice carrying the HincII site from the targeting vector by PCR amplification of the 327 bp genomic fragment (see A) followed by digestion with HincII. M, 100 bp ladder; WT, wildtype; Mu, mutant with the introduced HincII site. Fragment sizes in bp. C. Distinction between correctly targeted Scn8a and random insertion of the targeting construct by Southern blot of HincII digested genomic DNA followed by hybridization with the external probe (left) or the internal probe (right). Three representative genomic DNA samples are shown, containing a targeted allele, a random insertion, or both. MW markers in kb at right.