Abstract
This study investigated the association of copy number variants (CNVs) in type 2 diabetes (T2D) and T2D-associated end-stage renal disease (ESRD) in African Americans. Using the Affymetrix 6.0 array, >900,000 CNV probes spanning the genome were interrogated in 965 African Americans with T2D-ESRD and 1029 non-diabetic African American controls. Previously identified and novel CNVs were separately analyzed and were evaluated for insertion/deletion status and then used as predictors in a logistic regression model to test for association. One common CNV insertion on chromosome 1 was significantly associated with T2D-ESRD (p=6.17×10−5, OR=1.63) after multiple comparison correction. This CNV region encompasses the genes AMY2A and AMY2B, which encode amylase isoenzymes produced by the pancreas. Additional common and novel CNVs approaching significance with disease were also detected. These exploratory results require further replication but suggest the involvement of the AMY2A/AMY2B CNV in T2D and/or T2D-ESRD, and indicate that CNVs may contribute to susceptibility for these diseases.
Keywords: Copy number variation, African Americans, Diabetic nephropathy, End-stage renal disease, Genome-wide association study, Type 2 diabetes
Introduction
Diabetes is a complex and heterogeneous disease with a staggering global impact – most recent estimates indicate 346 million people worldwide suffer from this disease [1]. Type 2 diabetes (T2D) is the most common form of diabetes, accounting for more than 90% of cases, and occurs when peripheral tissue insulin resistance accompanies insufficient β-cell insulin production. While more than 80% of diabetes deaths occur in low- and middle-income countries [1], there remains an enormous diabetes burden in the United States as it is the seventh leading cause of death [2]. One of the most detrimental and expensive comorbidities of diabetes is kidney disease (nephropathy), which develops in ~40% of patients [3] and accounts for 10-20% of all-cause mortality [1]. The final stage of diabetes-associated nephropathy is end-stage renal disease (ESRD), a complication which occurs more frequently in African Americans than other American populations [4]. T2D and T2D-associated ESRD (T2D-ESRD) are likely influenced by complex interactions between environmental surroundings and lifestyle choices, highlighted by the significant increase in T2D over the last several decades [5]. In addition to environmental factors, numerous genetic loci have been identified as potential mediators of T2D and T2D-ESRD [6-8]. Though these have aided the understanding of the pathogenesis of these diseases, the combined impact of identified genetic contributors is low relative to the heritability of these diseases. While common genetic variants, specifically single nucleotide polymorphisms (SNPs), have been identified and explored in detail as possible mediators, copy number variation (CNV) is a relatively novel source of genetic variation that has only recently been reported as having a potential role in T2D in European-derived and Asian populations [9-11]. Additionally, prior studies of CNVs associated with T2D have not included African Americans [9-11].
Important for this study, recent reports have identified CNVs associated with related metabolic traits and disorders including BMI [12] and obesity [13-15]. CNVs associated with kidney disease (specifically glomerulonephritis) have also been reported [16]. Complex diseases with disparate incidence rates in different race groups are particularly interesting areas to focus attention, as gene dosage may impact disease manifestation [17]. Based on these observations we performed a CNV analysis of T2D and T2D-ESRD in an African American sample.
Materials and Methods
Subjects
A total of 965 African Americans with T2D-ESRD and 1029 non-diabetic African American controls were evaluated. Ascertainment criteria and recruitment methods have been described [18,19]. Recruitment and sample collection procedures were approved by the Institutional Review Board at Wake Forest School of Medicine and written informed consent was obtained from all participants. Case and control subjects were unrelated, self-described African Americans born in North Carolina, South Carolina, Georgia, Virginia or Tennessee. Subjects with T2D-ESRD were recruited from dialysis facilities (case subjects). T2D was diagnosed as developing diabetes at ≥25 years of age without prior diabetic ketoacidosis. Additionally, T2D-ESRD cases met at least one of the following criteria for inclusion: a) T2D diagnosed ≥5 years before initiating renal replacement therapy, b) background or greater diabetic retinopathy and/or c) ≥100 mg/dl proteinuria on urinalysis without other causes of nephropathy. Control subjects without a current diagnosis of diabetes or renal disease were recruited from the community and internal medicine clinics. DNA extraction was performed using the PureGene system (Gentra systems; Minneapolis, MN).
Genotyping and quality control
Genotyping was performed at the Center for Inherited Disease Research (CIDR) using 1μg of genomic DNA (diluted in 1X TE buffer and at 50ng/μl) on the Affymetrix Genome-wide Human SNP array 6.0 (Affy6.0). Genotyping and quality control have been previously described in depth [18,19].
Analysis
Principal components analysis (PCA) was performed utilizing all high-quality SNP data from the Affy 6.0; regions of high linkage disequilibrium and inversions were excluded [18,19]. Direct comparison of the PCA with FRAPPE [20] analysis of 70 ancestry informative markers (AIMs) resulted in a high correlation between PC1 and the AIMs (r2=0.87). The mean (± SD) African ancestry proportions in 965 T2D-ESRD cases and 1,029 controls were 0.80 ± 0.11 and 0.78 ± 0.11, respectively, as estimated by FRAPPE analysis [20]. PCA was also computed on intensity data to detect batch effects; intensity data were normalized using the software Partek [21] prior to CNV calling.
CNV data was analyzed using Affymetrix Genotyping Console 4.0 software with version 30 library and annotation files. This software implements the Birdsuite program tools to identify common, novel, and rare CNV [22]. Copy number (i.e., insertion or deletion) was estimated at all individual probes (novel CNV) and an a priori set of CNVs previously identified and described in McCarroll et al. [23] and implemented in Birdsuite [22]. Following software recommendations, common CNVs were called using the CANARY algorithm and novel CNVs were called using the hidden Markov models implemented in Birdseye [22]. For each method, probes or regions were categorized by copy number (CN) status with insertions defined as CN>2 and deletions defined as CN<2; probes with CN=0 were considered homozygous deletions. Insertions and deletions present in <5 individuals were discarded from further analyses. Following these criteria, for each of the 1,130 previously described CNV regions, 328 insertions and 941 deletions were deemed informative. For individual probes, 113,191 and 134,300 insertions and deletions, respectively, were deemed informative. CN status was coded as a binary variable indicating presence or absence of an insertion/deletion in each sample and used as the predictor in logistic regression models with covariates age, gender, and principal component 1 (PC1) (adjustment for admixture) to test for association with disease. Statistical significance, determined separately for insertions and deletions, was defined by Bonferroni correction. Genomic control inflation factors, based on a single probe when the detected CNV was estimated to extend over a region in which ≥1 probe identified the same CNV, and P-P plots were evaluated for evidence of systematic bias.
Results
Clinical characteristics of the study samples
Table 1 summarizes the clinical characteristics of the 1,994 study samples. Age at enrollment for T2D-ESRD cases was older than the non-diabetic controls; however, age at enrollment for controls was older than age at T2D diagnosis in the cases. The T2D-ESRD cases included a higher proportion of females; additionally, on average, all groups were overweight or obese at enrollment.
Table 1.
Clinical characteristics of study samples (values are presented as trait mean ± standard deviation).
| T2D-ESRD | Non-diabetic Controls | P-Value | |
|---|---|---|---|
| n | 965 | 1029 | |
| Female (%) | 61.2 | 57.3 | 0.076 |
| Age at Enrollment (years) | 61.6 ± 10.5 | 49.0 ± 11.9 | <0.0001 |
| Age at T2D diagnosis (years) | 41.6 ± 12.4 | — | <0.0001* |
| Age at ESRD diagnosis (years) | 58.0 ± 10.9 | — | |
| T2D to ESRD duration (years) | 16.2 ± 10.9 | — | |
| BMI (kg/m2) | 29.7 ± 7.0 | 30.0 ± 7.0 | 0.30 |
Comparison of Age at T2D diagnosis in T2D-ESRD cases to Age at Enrollment in Non-diabetic Controls.
Analysis
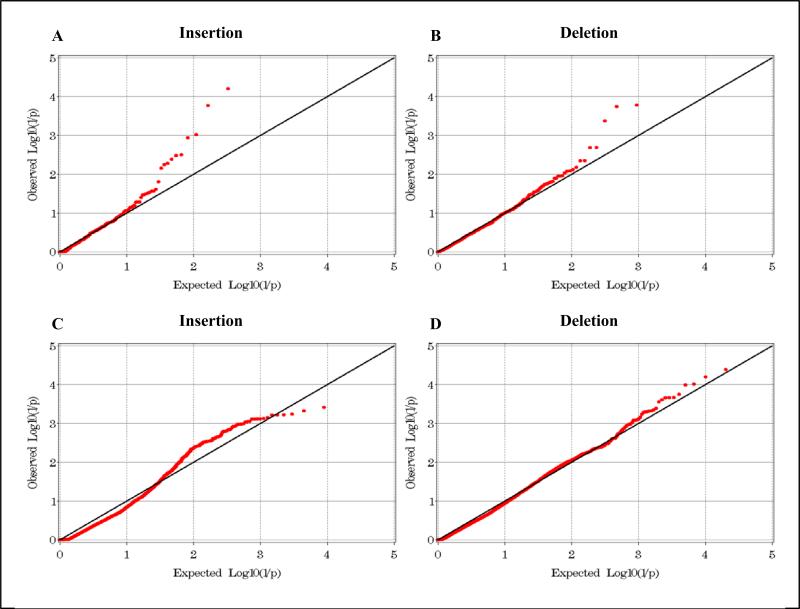
In the analysis of previously identified CNVs, 14,570 probes corresponding to 1,130 known CNV regions [23], including 328 insertions and 941 deletions, met QC parameters and were informative. Bonferroni-corrected thresholds of significance were P≤1.52×10−4 and P≤5.31×10−5 for previously identified insertions and deletions, respectively. Inflation factors (the mean of the X2 statistic) were computed based on informative regions; these were 1.13 and 0.98 for insertions and deletions, respectively. P-P plots for these suggest there was little evidence of a systematic bias (Figure 1A-B).
Figure 1.
P-P plots for insertion (A,C) and deletion (B,D) models of previously identified CNVs (A,B) and individual CNV probes (C,D).
One previously described CNV met a strict Bonferroni threshold of P≤1.52×10−4; this was an insertion on chromosome 1 spanning 50.15 kb; p=6.17×10−5, odd ratio (OR) =1.63 (95% CI 1.28-2.08). The insertion was common in cases (0.425) and controls (0.377). This region encompasses AMY2A and AMY2B, pancreatic amylase precursor genes which encode pancreatic amylase isoenzymes. Table 2 includes details of additional top scoring CNVs from this analysis.
Table 2.
Top CNV association information.
| hg18 Position∞ | Association | Frequency | Nearest Gene(s) | |||
|---|---|---|---|---|---|---|
| chr:start-end | P-value | OR (95% CI) | Cases | Ctrls | ||
| Known Insertions* | chr1:103910761-103960907 | 6.17x10−5 | 1.62 (1.28, 2.08) | 0.425 | 0.377 | RNPC3, AMY2B, LOC648740, AMY2A, AMY1A, AMY1C, AMY1B |
| chr12:36340414-36394320 | 1.68x10−4 | 0.59 (0.45, 0.78) | 0.172 | 0.242 | None within 500kb | |
| chr17:41521619-41719991 | 9.46x10−4 | 0.55 (0.39, 0.78) | 0.092 | 0.145 | KIAA1267, LOC644246, LRRC37A, ARL17B | |
| chr13:75007535-75015769 | 1.14x10−3 | 0.56 (0.40, 0.80) | 0.089 | 0.116 | COMMD6, UCHL3 | |
| chr15:28377089-28536721 | 3.12x10−3 | 0.44 (0.26, 0.76) | 0.897 | 0.892 | CHRFAM7A | |
| chr8:140673518-140675508 | 3.29x10−3 | 2.41 (1.34, 4.34) | 0.052 | 0.023 | KCNK9 | |
| Known Deletions* | chr5:180311316-180350709 | 1.64x10−4 | 1.55 (1.24, 1.95) | 0.418 | 0.353 | BTNL8, BTNL3, BTNL9 |
| chr1:187293847-187297986 | 1.78x10−4 | 3.44 (1.81, 6.67) | 0.057 | 0.041 | None within 500kb | |
| chr9:11957036-11965492 | 4.20x10−4 | 0.35 (0.19, 0.63) | 0.817 | 0.801 | None within 500kb | |
| chr12:11113633-11132799 | 2.03x10−3 | 0.7 (0.56, 0.88) | 0.534 | 0.595 | PRH1, TAS2R19, TAS2R31, TAS2R46, TAS2R43, TAS2R30 | |
| chr18:38133-68539 | 2.05x10−3 | 3.57 (1.59, 8.02) | 0.04 | 0.021 | ROCK1P1 | |
| Novel Insertions** | chr17:41700658-41700683 | 5.91x10−4 | 0.54 (0.38, 0.77) | 0.091 | 0.147 | KIAA1267, LOC644246, LRRC37A, ARL17B |
| chr7:142159095-142159120 | 3.81x10−4 | 3.11 (1.66, 5.83) | 0.062 | 0.017 | PRSS1, TRY6, PRSS2 | |
| chr8:12457161-12457186 | 5.71x10−4 | 0.49 (0.32, 0.73) | 0.058 | 0.093 | FAM66A, DEFB109P1, FAM90A25P, FAM86B2, LONRF1, MIR3926 | |
| chr12:34438874-34438899 | 2.14x10−3 | 3.27 (1.54, 6.98) | 0.041 | 0.019 | ALG10 | |
| chr10:135199054-135199079 | 2.35x10−3 | 0.49 (0.31, 0.77) | 0.051 | 0.072 | CYP2E1, SYCE1, SPRNP1 | |
| Novel Deletions** | chr14:21965715-21965740 | 4.03x10−5 | 0.07 (0.02, 0.24) | 0.003 | 0.026 | DAD1, ABHD4 |
| chr1:110027998-110028023 | 1.77x10−4 | 2.04 (1.41, 2.96) | 0.123 | 0.078 | GSTM4, GSTM2, GSTM1, GSTM5 | |
| chr7:142167293-142167318 | 2.15x10−4 | 0.6 (0.46, 0.79) | 0.169 | 0.259 | PRSS1, TRY6, PRSS2 | |
| chr2:71200196-71200221 | 8.11x10−4 | 0.34 (0.18, 0.64) | 0.022 | 0.048 | NAGK, MCEE, MPHOSPH10 | |
| chr3:145765721-145765746 | 1.49x10−3 | 0.17 (0.06, 0.50) | 0.008 | 0.013 | None within 500 kb | |
McCarroll et al. 2008.
Insertions analyzed=328, yielding a Bonferroni-adjusted threshold of P≤1.52×10-4; deletions analyzed=941, yielding a Bonferroni-adjusted threshold of P≤5.31×10-5.
Insertion probes analyzed=113191, yielding a Bonferroni-adjusted threshold of P≤4.42×10-7; deletion probes analyzed=134300, yielding a Bonferroni-adjusted threshold of P≤3.7×10-7. Association results reported are for the best probe in the region encompassed by the probes which span the reported physical position.
A by-probe analysis was performed to detect novel CNVs; 227, 287 individual probes passed QC metrics and were informative. These included 113,191 insertions and 134,300 deletions, corresponding to Bonferroni-corrected significance thresholds of P≤4.42×10−7 and P≤3.7×10−7 for insertions and deletions, respectively. Inflation factor calculations were 0.77 and 0.88 for insertions and deletions, respectively. P-P plots are shown in Figure 1C-D. No result from this analysis met strict Bonferroni-corrected significance thresholds; top results are shown in Table 2. The most significant insertion spanned 149.27 kb on chromosome 17; the best p-value for all probes in this region was 5.91×10−4, frequencies were 0.091 and 0.147 in cases and controls, respectively, and OR=0.54 (95% CI 0.38-0.77). Genes in this region include KIAA1267, LOC644246, LRRC37A (leucine rich repeat containing 37A) and ARL17B (ADP-ribosylation factor-like 17B). The most significant deletion spanned 247.16 kb on chromosome 14; the best p-value for all probes in this region was 4.03×10−5, frequencies were 0.003 and 0.026 in cases and controls, respectively, and OR=0.07 (95% CI 0.02-0.24). Genes nearest this region include DAD1 (defender against cell death 1) and ABHD4 (abhydrolase domain containing 4).
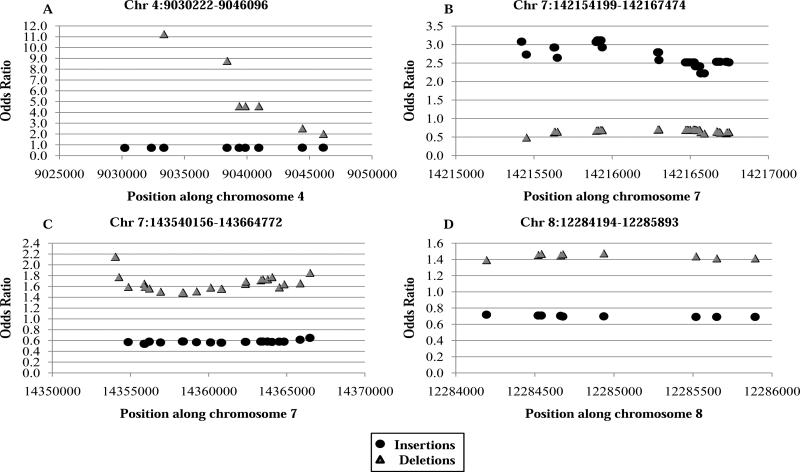
Further scrutiny of the probe-level data revealed four regions encompassed by three or more probes spanning >1 kb and where overlap of insertions and deletions occurred (Figure 2). Results trended towards association with disease susceptibility/protection. A rare overlapping region on chromosome 4 (Figure 2A) spanning 15.87 kb had an insertion p-value of 2.25×10−2, OR=0.74 and frequency=0.16 in cases and 0.19 in controls; the deletion p-value was 1.84×10−3, OR=11.25, frequency of 0.04 in cases and 0.02 in controls. The gene nearest this CNV was DEFB131, defensin beta 131, which belongs to the beta-defensin family and is highly expressed in the testis and moderately expressed in the prostate and small intestine [24]. Another overlapping region on chromosome 7 (Figure 2B) spanning 13.28 kb had an insertion p-value of 3.81x10−4, OR=3.11, case frequency=0.06 and control frequency=0.03. This CNV encompasses TRY6, which is thought to be a transcribed pseudogene that encodes a protein similar to trypsinogen [24]. Another overlapping region on chromosome 7 (Figure 2C) spaning 124.62 kb had an insertion p-value of 3.4510−3, OR=0.54, case frequency=0.07 and control frequency=0.10; the deletion p-value was 6.92×10−4, OR=2.15, case frequency=0.12 and control frequency=0.09. This CNV encompasses several genes, including five olfactory receptors. The fourth and more common overlapping region of interest occurred on chromosome 8 (Figure 2D); this region spans 1.70 kb and had an insertion p-value of 2.58×10−2, OR=0.60, case frequency=0.12, and control frequency =0.13; the deletion p-value was 1.56×10−2, OR=1.48, case frequency=0.16, control frequency=0.12. The gene nearest this CNV is predicted gene FAM66A, family with sequence similarity 66, member A.
Figure 2.
Plots of regions of overlapping CNVs (insertions and deletions) identified in by-probe CNV analysis. The OR of each probe is represented with annotation of insertion or deletion status.
Discussion
We performed a GWAS of common and novel CNV data obtained from the Affy 6.0 GWAS array in African Americans with T2D-ESRD. This array was designed to interrogate samples using 946K copy number probes, including 800,000 probes for uniform coverage across the genome and 140,000 additional probes for detection in regions of previously reported CNVs [23], and likely has better coverage of CNVs than other arrays utilized for CNV analyses in T2D. This allowed for two comprehensive analyses. First we examined previously identified common CNVs, a method that is useful for determining if known CNVs are associated with disease and if they exist in the population of interest. Secondly, we performed an unbiased genome-wide analysis based on individual probes, which theoretically enables the detection of undocumented CNVs. Both methods are particularly informative, as African Americans have not been included in prior studies that investigated the potential role of CNVs in T2D or T2D-ESRD.
A single CNV in a 50 kb region encompassing the AMY2A and AMY2B genes on chromosome 1 met Bonferroni-corrected thresholds of significance. AMY2A and AMY2B are pancreatic alpha amylase genes encoding secreted proteins that hydrolyze 1, 4-alpha-glucoside bonds in oligosaccharides and polysaccharides, catalyzing the initial step in digestion of dietary starch and glycogen. Both genes are expressed in the pancreas and AMY2B is also expressed in the liver. These characteristics suggest that the region is more likely associated with T2D risk. The region encompassed by this CNV contains more than 20 SNPs including several synonymous and nonsynonymous polymorphisms, intronic enhancers, and splice sites. Additionally, this CNV is within 50 kb of a previously identified area of common CNV in the salivary amylase gene AMY1, [25], although these have not been reported in disease studies. Our results suggest a potential role for disruption or duplication of these genes in the development of T2D and/or T2D-ESRD.
We additionally detected known and novel CNVs that occur at appreciable frequencies and approach statistically significant association with T2D/T2D-ESRD in this sample. These included four overlapping regions covering >1 kb where insertions and deletions trended towards association in opposite directions of effect. These regions likely represent true copy number events with a potential role in T2D and/or T2D-ESRD and warrant further investigation and replication.
A limitation of this report is that we compared cases with T2D-ESRD to non-nephropathy, non-diabetic controls. Therefore, association may be due to association with T2D or nephropathy. Unfortunately, appreciable numbers of African Americans with longstanding T2D lacking nephropathy or microalbuminuria are difficult to recruit due to the high prevalence of nephropathy in this population. Such individuals would comprise an ideal comparison group, useful for clarifying whether associations resulted from the presence of T2D or nephropathy. Additional limitations include the lack of replication in this study, which is required for the incorporation of this information into the current working model of the genetics of T2D/T2D-ESRD.
A previous study identified 1,362 CNVs in African Americans using the Affymetrix 500K array and detected two regions with significant frequency differences between African Americans and European Americans [26]. Using the Affy 6.0 array, we also detected that CNV events were common in this population. Another GWAS employing the Affy6.0 did not detect genome-wide significance between CNVs and anthropometric traits in African Americans [27]. Furthermore, a study that included African American children detected association between CNVs and childhood obesity [13]. While these represent efforts towards incorporating African Americans in studies of alternative genetic contributors to widely studied traits and diseases, there are currently no published reports investigating the role of CNVs in T2D in African Americans.
Conclusions
In summary, we observed a previously identified insertion on chromosome 1 associated with disease in a large African American sample with T2D-ESRD, which suggests a potential role for disruption or duplication of the encompassed genes in T2D and/or T2D-ESRD. Additionally, we detected that other common and novel CNVs were present at an appreciable frequency and may contribute to risk of T2D and/or T2D-ESRD in this population. These data are exploratory as CNVs represent a novel yet poorly understood form of genetic variation; validation is crucial for elucidation of additional mechanisms contributing to the etiology of these diseases.
Acknowledgements
Genotyping services were provided by the Center for Inherited Disease Research (CIDR), which is fully funded through a federal contract from the National Institutes of Health (NIH) to The Johns Hopkins University, contract number HHSC268200782096C. This work was supported by NIH grants K99 DK081350 (NDP), R01 DK066358 (DWB), R01 DK053591 (DWB), R01 HL56266 (BIF), R01 DK 070941 (BIF), R01 DK 084149 (BIF) and in part by a grant from the General Clinical Research Center of the Wake Forest School of Medicine, M01 RR07122.
Footnotes
Cooke Bailey JN, Lu L, Chou JW, Xu J, McWilliams DR, et al. (2013) The Role of Copy Number Variation in African Americans with Type 2 Diabetes-Associated End Stage Renal Disease. J Mol Genet Med 7: 61. doi:10.4172/1747-0862.1000061
References
- 1.WHO WHO Diabetes Fact sheet N°312. 2011 [Google Scholar]
- 2.CDC . National Diabetes Fact Sheet: national estimates and general information on diabetes and pre diabetes in the United States. Department of Health and Human Services, Centers for Disease Control and Prevention; USA: 2011. [Google Scholar]
- 3.de Boer IH, Rue TC, Hall YN, Heagerty PJ, Weiss NS, et al. Temporal trends in the prevalence of diabetic kidney disease in the United States. JAMA. 2011;305:2532–2539. doi: 10.1001/jama.2011.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.USRDS USRDS 2009 Annual Data Report. 2009 [Google Scholar]
- 5.Das SK, Elbein SC. The Genetic Basis of Type 2 Diabetes. Cell science. 2006;2:100–131. doi: 10.1901/jaba.2006.2-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prokopenko I, McCarthy MI, Lindgren CM. Type 2 diabetes: new genes, new understanding. Trends Genet. 2008;24:613–621. doi: 10.1016/j.tig.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeon JP, Shim SM, Nam HY, Ryu GM, Hong EJ, et al. Copy number variation at leptin receptor gene locus associated with metabolic traits and the risk of type 2 diabetes mellitus. BMC Genomics. 2010;11:426. doi: 10.1186/1471-2164-11-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bae JS, Cheong HS, Kim JH, Park BL, Kim JH, et al. The genetic effect of copy number variations on the risk of type 2 diabetes in a korean population. PLoS One. 2011;6:e19091. doi: 10.1371/journal.pone.0019091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sha BY, Yang TL, Zhao LJ, Chen XD, Guo Y, et al. Genome-wide association study suggested copy number variation may be associated with body mass index in the Chinese population. J Hum Genet. 2009;54:199–202. doi: 10.1038/jhg.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glessner JT, Bradfield JP, Wang K, Takahashi N, Zhang H, et al. A genome-wide study reveals copy number variants exclusive to childhood obesity cases. Am J Hum Genet. 2010;87:661–666. doi: 10.1016/j.ajhg.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walters RG, Jacquemont S, Valsesia A, de Smith AJ, Martinet D, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Li WD, Glessner JT, Grant SF, Hakonarson H, et al. Large copy number variations are enriched in cases with moderate to extreme obesity. Diabetes. 2010;59:2690–2694. doi: 10.2337/db10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 17.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonough CW, Palmer ND, Hicks PJ, Roh BH, An SS, et al. A genome-wide association study for diabetic nephropathy genes in African Americans. Kidney Int. 2011;79:563–572. doi: 10.1038/ki.2010.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer ND, McDonough CW, Hicks PJ, Roh BH, Wing MR, et al. A Genome-wide association search for type 2 diabetes genes in african americans. PLoS One. 2012;7:e29202. doi: 10.1371/journal.pone.0029202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 21.Partek: Next Generation Sequencing & Microarray Software.
- 22.Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 24.UCSC Genome Bioinformatics.
- 25.Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McElroy JP, Nelson MR, Caillier SJ, Oksenberg JR. Copy number variation in African Americans. BMC Genet. 2009;10:15. doi: 10.1186/1471-2156-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang SJ, Chiang CW, Palmer CD, Tayo BO, Lettre G, et al. Genome-wide association of anthropometric traits in African- and African-derived populations. Hum Mol Genet. 2010;19:2725–2738. doi: 10.1093/hmg/ddq154. [DOI] [PMC free article] [PubMed] [Google Scholar]