Abstract
Structures of type-1 human immunodeficiency virus (HIV-1) reverse transcriptase (RT) have been determined in several forms, but only one contains an RNA/DNA hybrid. Here we report three structures of HIV-1 RT complexed with a non-nucleotide RT inhibitor (NNRTI) and an RNA/DNA hybrid. In the presence of an NNRTI, the RNA/DNA structure differs from all prior nucleic acid bound to RT including the RNA/DNA hybrid. The enzyme structure also differs from all previous RT–DNA complexes. As a result, the hybrid has ready access to the RNase H active site. These observations indicate that an RT–nucleic acid complex may adopt two structural states, one competent for DNA polymerization and the other for RNA degradation. RT mutations that confer drug resistance but are distant from the inhibitor-binding sites often map to the unique RT–hybrid interface that undergoes conformational changes between two catalytic states.
Keywords: RNase H, p51-p66 interface, drug resistance, bent and underwound helix
Type-1 Human Immunodeficiency Virus (HIV-1) is a lentivirus and the etiological agent of Acquired Immune Deficiency Syndrome (AIDS), a global pandemic for more than three decades. In addition to the RNA- or DNA-dependent DNA polymerase functions, the viral reverse transcriptase (RT) contains an RNase H activity, which hydrolyzes the RNA strand of an RNA/DNA hybrid1,2. Three of the four HIV-1 encoded enzymatic activities (protease, integrase and DNA polymerase) have been successfully targeted by antiviral drugs, but no inhibitor of RNase H has advanced to clinical trials. The viral DNA polymerase has the largest number of FDA-approved inhibitors for AIDS treatment (www.fda.gov), including nucleoside-analogs (NRTIs) and non-nucleoside RT inhibitors (NNRTIs)3,4. The success of these treatments has granted many HIV-infected individuals an active and fulfilling lifestyle. However, drug resistance continues to pose a major challenge, and new viral and host targets for drug development are needed.
HIV-1 RT consists of two polypeptide chains (p66 and p51) with both the DNA polymerase and RNase H activities residing in the p66 subunit5,6. RT is essential for copying and removing the viral RNA genome to make dsDNA before integration into the host genome7,8. The viral RNase H is homologous to cellular RNase H1 and contains four conserved carboxylates in its active site that coordinate the two Mg2+ ions necessary for catalysis9–11. To date, the majority of RNase H inhibitors target the Mg2+ ions in the active site11–14. Since the two-Mg2+-ion mechanism is used by all polymerases and many nucleases15,16, these RNase H inhibitors may suffer from a lack of specificity. However, viral RNase H differs from cellular RNases H1 in that it prefers a longer substrate (>18 bp) and fails to cleave the polypurine-track (PPT) sequence in the viral genome, thus leaving the PPT intact to prime the second DNA strand synthesis17–19. Regions outside of RNase H domain must contribute to substrate binding in RT and are therefore potential targets for HIV-1 RNase H inhibition.
A large number of HIV-1 RT structures have been reported, most of which are cocrystals of WT or mutant enzyme with NNRTIs13,14,20–24. There are more than twenty RT structures with ~20 bp nucleic acid substrates in binary or ternary complexes, representing the process of DNA synthesis or ATP-dependent excision of a chain terminator in the DNA-polymerase active site24. However, only one structure contains an RNA/DNA hybrid30. Unfortunately, none of the bound nucleic acid is captured in the RNase H active site in a catalytic configuration. These structures of HIV-1 RT can be grouped into two general states, namely the “closed” conformation when bound to a nucleic-acid and the “open” conformation with an NNRTI2.
Crystal structures of human and bacterial RNases H1 with an RNA/DNA hybrid in their active site have been determined9,10. Although the two active sites of RT have been proposed to act simultaneously19,31 because the RNase H primarily cleaves the RNA template 18–19 bp from the primer 3′ end bound to the polymerase domain, the nucleic acids cocrystallized with HIV-1 RT in the polymerization mode and the RNA/DNA hybrid in the RNase H active site brought in by superposition of the cellular RNase H1–substrate complex cannot be connected10. Even with severe bending, an RNA/DNA hybrid cannot be modeled to engage both the DNA polymerase and RNase H active sites10. Moreover, the anti-cooperativity of the two catalytic centers of RT and observations that NNRTIs can enhance the HIV-1 RNase H activity32–36 corroborate that DNA synthesis and RNA degradation occur sequentially on a single substrate. To determine how HIV-1 RNase H selects and binds its substrate and to open avenues for identifying new anti-HIV drug targets, we undertook structural analysis of HIV-1 RT complexed with non-PPT-containing RNA/DNA hybrids.
Results
Three crystals of HIV-1 RT with RNA/DNA hybrids
To favor binding of an RNA/DNA hybrid in the RNase H active site, we inhibited RT polymerase activity with an NNRTI (Nevirapine or Efavirenz) and included a non-instructive abasic analog (tetrahydrofuran) at the templating position of the RNA strand (Fig. 1). To avoid RNase H cleavage but preserve metal ion and nucleic acid binding, we replaced the catalytic carboxylate Asp498 with Ala (D498A) or Asn (D498N)9,10. By varying the length of the RNA/DNA duplex and single-strand overhang, we co-crystallized RT and the RNA/DNA hybrid in three different crystal lattices without chemical crosslinking or the necessity for a monoclonal antibody. A P3121 crystal form diffracted X-rays to 3.3Å resolution and contained wildtype RT, a nicked 22 bp hybrid and Efavirenz (WT22Efv) (Fig. 1a). With Nevirapine, R32 crystals containing D498N RT and a 25 bp hybrid (DN25Nvp) and P6122 crystals containing D498A RT and a 29 bp hybrid (DA29Nvp) diffracted X-rays to 4.85 – 5.0 Å (Supplementary Fig. 1a, b). These structures were determined and refined (Methods, Table 1). In all three cases, the electron density for RT and 21–22 bp of the RNA/DNA hybrids was unambiguous (Fig. 1b, Supplementary Fig. 1c–e). Efavirenz and Nevirapine were also visible in the WT22Efv and DN25Nvp crystals (Fig. 1b, Supplementary Fig. 1d), but the density for Nevirapine in the DA29Nvp structure was non-existing (Supplementary Fig. 1c), probably due to omission of the inhibitor in the crystal stabilization buffer. These protein structures are essentially identical with each other but very different from those complexed with DNA (Fig. 1a, c). As designed, the DNA polymerase active site is disengaged from catalysis. The p66 fingers subdomain is wide open and has few interactions with nucleic acid. The fingertip (residues 62–74), which interacts extensively with the template strand and an incoming nucleotide in the DNA synthesis mode, is disordered and not traceable here.
Figure 1.
HIV-1 RT complexed with an RNA/DNA hybrid and an NNRTI. (a) The WT22Efv structure. RT is shown as molecular surface with p66 in wheat and p51 in silver. The polymerase and RNase H active site are highlighted in yellow and cyan, respectively. CN stands for connection domain. The hybrid is shown as tube-and-ladder with RNA in red and DNA in blue. The nucleic acid sequence is shown in the same color scheme above. The “x” denotes the abasic tetrahydrofuran substitution, and black letters indicate bases that are not traceable. The nick in the RNA strand is indicated by a black arrow. (b) The RNA (red), DNA (blue) and Efavirenz (bright orange) in the WT22Efv structure are shown with the simulated annealing omit Fo-Fc map contoured at 4.0 σ. The purple arrowheads in (a) and (b) mark the unique bend in the hybrid that allows it to reach the RNase H active site. (c) The ternary complex structure 1RTD26 representing all previously reported RT–DNA complexes. It is shown with dsDNA in light and dark blue and dTTP in green. The RT–DNA crosslinking site (Q258C) is marked. Four α-helices, αA (p66 fingers), αK (p66 CN), αL (p51 CN), and αM (p51), are shown as cylinders to highlight different RT structures in (a) and (c). A white arrowhead indicates the absence of the bend in the DNA. (d) Pairwise superposition of hybrids in the three RT complexes. Each structure is color-coded as labeled.
Table 1.
Data collection and refinement statistics
| Data collection | |||
| Structure name | WT22Efv (4B30) | DA29Nvp (4B3P) | DN25Nvp (4B3Q) |
| Space group | P3121 | P6122 | R32 |
| Cell dimensions | |||
| a, b, c (Å) | 164.6, 164.6, 129.0 | 163.0, 163.0, 229.5 | 272.7, 272.7, 233.1 |
| Resolution (Å)* | 30–3.3 (3.45–3.3) | 50–4.85 (5.02–4.85) | 30.0–5.0 (5.09–5.00) |
| Rsym (%)* | 6.7 (71.6) | 15.9 (62.9) | 13.9 (87.1) |
| Mean I/σI* | 17.5 (2.3) | 14.7 (2.1) | 14.8 (1.7) |
| Completeness (%)* | 98.2 (99.9) | 99.7 (99.0) | 99.6 (100) |
| Redundancy* | 4.2 (4.2) | 11.6 (7.5) | 6.5 (6.8) |
| Refinement | |||
| Resolution (Å) | 30.0–3.3 | 46.0–4.8 | 30.0–5.0 |
| No. of reflections | 30184 | 9023 | 13005 |
| Rwork/Rfree | 27.5 / 29.5 | 36.4 / 40.4 | 36.1 / 40.7 |
| No. of atoms | |||
| Protein / nucleic acid | 7543 / 947 | 7284 / 878 | 7535 / 878 |
| Drug / Ca2+ | 21 / 2 | 0 / 0 | 20 / 0 |
| B-factors | |||
| Protein / nucleic acid | 80.6 / 81.8 | 188.9 / 193.2 | 149.0 / 149.8 |
| Drug / Ca2+ | 71.0 / 75.6 | 147.0 / - | |
| R.m.s. deviations | |||
| Bond distances (Å) | 0.008 | 0.008 | 0.008 |
| Bond angles (°) | 0.88 | 0.81 | 0.80 |
Highest resolution shell is shown in parenthesis.
The RNA/DNA hybrids in the WT22Efv and DN25Nvp structures are similar in register relative to RT but differed from that of DA29Nvp. In the first two crystals, the 3′-end of the DNA reached into the palm subdomain but is ~ 5Å removed from the polymerase active site, and the RNA 5′-overhang exits over the fingers subdomain as observed previously26 (Fig. 1a, b, Supplementary Fig. 1b). These two complexes therefore correspond to the DNA 3′-directed mode, also known as the primary RNase H cleavage11,19,37. In the DA29Nvp crystals, exit of the hybrid from the RNase H domain is blocked by a neighboring RT molecule (Supplementary Fig. 2a), and the 29 bp hybrid most likely extended through the gap between the p66 fingers and thumb subdomains (Supplementary Fig. 1a). The DA29Nvp structure may thus correspond to the end-independent mode of RNase H cleavage11.
The RNA/DNA hybrids in the WT22Efv and DA29Nvp crystals are both influenced by lattice contacts (Supplementary Fig. 2a, b) and the RNA strand is displaced from the RNase H active site by ~2Å. The DN25Nvp structure is suggestive of the potential chemistry of the RNase H reaction (Supplementary Fig. 2c, d), but the high-salt crystallization conditions and low resolution (5.0 Å) prevent unequivocal assignment of the active site configuration. The RNA/DNA hybrids in the three structures are overall in the A-form conformation and are similar but not identical. Perhaps because of a nick in the middle of the RNA strand in the WT22Efv structure, the hybrid appears susceptible to deformation (Fig. 1d). The half containing the 3′ end of DNA is superimposable with the hybrid in the DA29Nvp structure, and the other half is more similar to that in the DN25Nvp structure. Nevertheless the three RNA/DNA hybrids are much more similar to each other than all nucleic acids previously co-crystallized with RT (Fig. 1), and each bends by 20–30° when entering the RNase H domain (13–14 bp from the DNA 3′ end, Fig. 1, Supplementary Fig. 3). The following comparison uses the 3.3 Å WT22Efv structure as a representative.
Two distinct RT conformations in nucleic-acid complexes
In our RT–hybrid complexes, the protein is more similar to previously reported structures of apo-RT with an NNRTI (PDB: 1FK9, 3QIP)14,20,22 than RT–nucleic acid complexes (Fig. 2a–c, Supplementary Fig. 4), thus removing the distinction between “open” apo and “closed” nucleic acid-bound conformations. We found that the “open” structure of RT stabilized by NNRTIs is fully capable of binding RNA/DNA hybrid. The presence of the RNA/DNA hybrid in addition to an NNRTI is coupled with an expansion of RT (Fig. 2b, Supplementary Video 1). The ~2.5Å outward movement of the p66 fingers and RNase H domain is necessary to accommodate the hybrid. Concomitantly, the p66 thumb subdomain tilts toward the duplex, and the p51 thumb moves with the RNase H domain.
Figure 2.
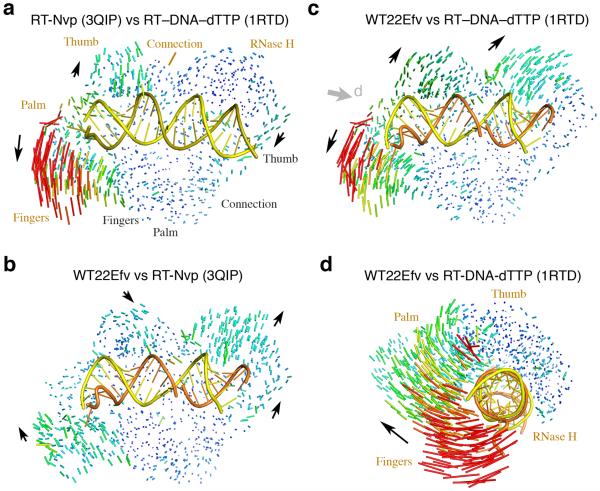
Structure comparison of HIV-1 RT. (a) An RT–DNA–dTTP ternary complex (1RTD26) versus an RT–NNRTI (3QIP14) complex. Differences are shown as a pairwise Cα-Cα vector map after superposition of p51. Increase in vector length is shown in blue (0.3 Å) to red (>8Å). The DNA duplex in 1RTD is shown as yellow and olive tube-and-ladder. (b) The WT22Efv structure versus 3QIP. The RNA (orange) and DNA (yellow) hybrid is also shown. Black arrows indicate the direction of movement from 3QIP to WT22Efv structure. (c) Comparison of WT22Efv with 1RTD after superposition of p51 as in (b). (d) Superposition of thumb, connection and RNase H in p66 between WT22Efv and 1RTD. The viewing angle is indicated by the grey arrowhead in panel c.
Structural changes are much more pronounced between our hybrid complexes and previously reported RT–nucleic acid complexes, e.g. 1RTD26, 2HMI38, 1HYS30 and 3KK227 (Fig. 2c, Supplementary Video 2). Based on difference-distance matrix analysis of each RT subunit (Supplementary Fig. 4), the thumb, connection and RNase H domain of p66 did not change to any appreciable degree compared to previously reported RT–nucleic acid complex structures (Supplementary Fig. 4b). After superposition of these three domains, the p66 fingers and palm in our complexes appeared to open up with a clockwise screw movement relative to the C-terminal RNase H, as if to extend and unwind the hybrid (Fig. 2d). Within p51 subunit the thumb underwent small movement relative to the other three subdomains (Supplementary Fig. 4b). We found that the most pronounced changes occur at the p66 and p51 interface, and the two subunits are ~2Å further apart in our hybrid complexes (Fig. 2c,d). Repacking of the two subunits has previously been observed when RT binds an NNRTI in the absence of nucleic acid substrate20,21,39,40, but it is more obvious in our RT–NNRTI–hybrid complexes (Fig. 2c). The changes at the p66-p51 interface are intimately linked to binding of the hybrid (see details below).
Structural comparison with the recently reported RT–DNA–NNRTI complex29 (PDB: 3V81) turned out to be not as revealing. Perhaps due to the crosslinking between RT and DNA and the procedure of soaking NNRTI into a preformed RT–DNA co-crystal, the protein structure of 3V81 is more similar to that of the RT–DNA–dNTP ternary complex than to that in either the RT–NNRTI complex or our WT22Efv structure (Fig. 3a).
Figure 3.

Structural comparison of WT22Efv and RT–DNA–Nvp (3V81)29. (a) Pairwise comparisons of RT in the 3V81 structure with RT complexed with Nevirapine (3QIP14), an RT ternary complex (1RTD26), and our WT22Efv (4B3O). The Cα-Cα vector maps are shown after superposition of p51 subunits and colored as in Fig. 2. Domain movements of 3V81 relative to others are indicated by black arrowheads. (b) Comparison of the entire RT–DNA–Nvp complex (3V81) with an RT–DNA–dNTP ternary complex (3KK227). Upon superposition of the nucleic acids, both of which are crosslinked to p66 via Q258C, not only are the DNAs highly similar, but also are the RT proteins except for the p66 fingers and palm subdomains as indicated by the grey arrowhead. The protein and DNA are color coded as indicated, and the NNRTI (Nvp) is shown as purple spheres.
A hybrid structure compatible with cleavage by RNase H
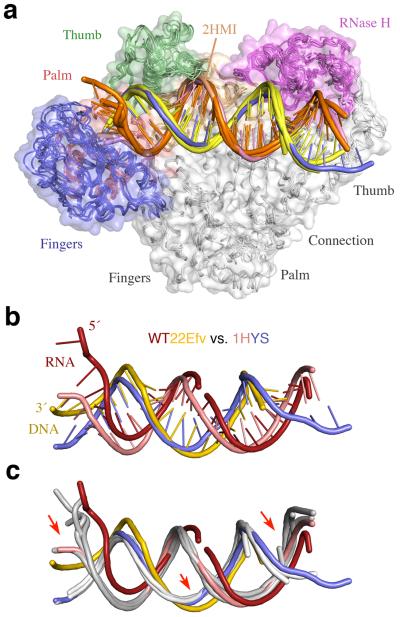
More than twenty structures of RT–nucleic acid complexes have been previously deposited in the PDB database, most of which were obtained after crosslinking the p66 thumb (via a Q258C substitution) to DNA (Fig. 1b)26,27,29. Only two structures of RT–nucleic acid complexes were obtained without the crosslink. One was co-crystallized with a monoclonal antibody bound to p51 (PDB: 2HMI)38, and the other with the PPT RNA/DNA hybrid (PDB: 1HYS)30. The DNA structures in the previously reported RT–DNA complexes (with or without an incoming dNTP, or even with an NNRTI29) are nearly identical (Fig. 3b, 4a). The only noticeable difference is that the DNA minor groove containing the crosslink is slightly wider than without (Fig. 4a). Surprisingly, the PPT-containing RNA/DNA hybrid (1HYS) superimposes well with every dsDNA co-crystallized with RT and better with all crosslinked DNA structures than uncrosslinked (2HMI), despite the presence of 2′-OHs.
Figure 4.
Structural comparison of nucleic acid complexed with RT. (a) Five previously reported RT–nucleic acid complexes (1RTD26, 1HYS30, 1T0552, 2HMI (uncrosslinked)38, and 3KJV27). The structures are shown after superposition of the p51 subunits. RT is shown as molecular surface with the fingers, palm, thumb, connection and RNase H domains color-coded blue, red, green, gold and magenta, respectively. The four DNA are depicted as orange template and yellow primer strands. The PPT RNA/DNA hybrid in 1HYS is shown in pink and blue. (b) Differences between the RNA/DNA hybrid in WT22Efv (red and yellow) and the PPT hybrid in 1HYS (pink and blue) after superposition of p51 subunits. (c) Comparison of the two hybrids and four DNAs (in light grey as in (a)). Red arrows highlight the main changes in WT22Efv.
The nucleic acid in our structures is considerably different from those reported previously and, more interestingly, from the PPT RNA/DNA hybrid of 1HYS (Fig. 4b, c). Although the sugar puckers in the previously reported PPT RNA/DNA were refined to have the A-like C3′-endo conformation, this hybrid duplex has a wide major groove and narrow minor groove with an average of 3.4Å rise and 34.5° twist per base pair, values typical of a B-form conformation. In contrast, our hybrid structure is mostly in the A-form with a wide minor groove and narrow major groove (Supplementary Fig. 3). The hybrids in our structures have two kinks. In addition to the typical 20–30° bend adjacent to the p66 thumb26, the hybrid is also bent by 20–30° when entering the RNase H domain near helix αM (Fig. 1a, b). Adjacent to this second bend, the RNA/DNA hybrid is underwound, which results in a widened major groove (Supplementary Fig. 3).
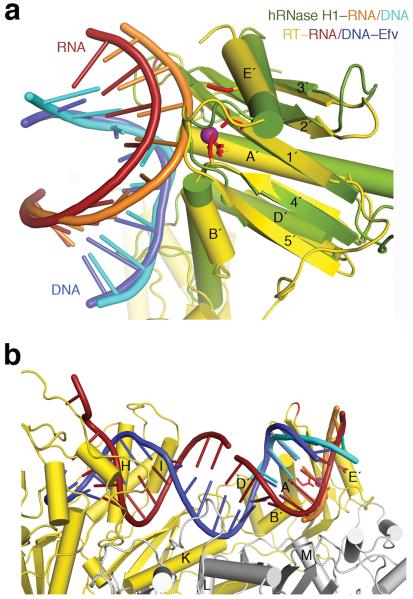
These changes in the hybrid (Fig. 4c) alter its position relative to the RNase H domain in our complexes. Five years ago to model an RNA/DNA in the RNase H active site based on the known RT–nucleic acid complex structures, the 3′-DNA end had to be relocated from the DNA polymerase catalytic center and the hybrid was bent in the middle towards the p51 subunit10. The hybrid structures in our RT complexes, however, can be readily connected with the RNA/DNA substrate co-crystallized with human RNase H1 after superposition of the homologous RNase H domains (Fig. 5, Supplementary Fig. 5). In the new structures the DNA 3′ end is indeed not positioned for polymerization. Yet the most interesting change was found in the middle of the 20 bp hybrid duplex, which is not as severely bent as proposed in order to place it for hydrolysis by the viral RNase H10, but is underwound with a widened major groove (Supplementary Video 3). Both the underwinding and the 20–30° kink observed here may not be possible for the polypurine track, thus resulting in no cleavage of PPT by RT17–19.
Figure 5.
The RNA/DNA hybrid structure is compatible with RNase H cleavage. (a) Comparison of WT22Efv with human RNase H1–substrate complex (2QK910). Human RNase H1 (green) is superimposed with the RNase H domain of HIV-1 RT (gold). The two hybrids are colored orange and cyan (human) and red and blue (WT22Efv). The RT active site is marked by the conserved carboxylates (red sticks) and the cation (purple sphere) in the hRNase H1 structure. Protein secondary structures are labeled according to convention24. (b) Compatibility with RNA degradation. Nucleic acids in the WT22Efv and human RNase H1 structures can be readily linked after superposition of the RNase H domain. p51 and p66 are depicted in silver and gold, respectively.
The RT-RNA/DNA interface and drug resistant mutations
The large conformational changes in the p66 thumb and fingers subdomains upon binding to NNRTI or nucleic acid have often attracted the most attention. Indeed in the WT22Efv structure, the presence of an NNRTI resulted in dramatic alterations of the p66 fingers (Fig. 2). With the altered nucleic acid structure near the fingers and thumb (Fig. 4b, c), the thumb (αH and αI) and the DNA polymerase “primer grip” (β12–β13) in p66 maintained their contacts with the RNA/DNA as in all RT–DNA complexes25. The structural changes that are coupled with the large conformational changes of the RNA/DNA hybrid and critical for RNA degradation, however, were found at the p51 and p66 subunit interface beyond the polymerase domain (Supplementary Video 2, 3).
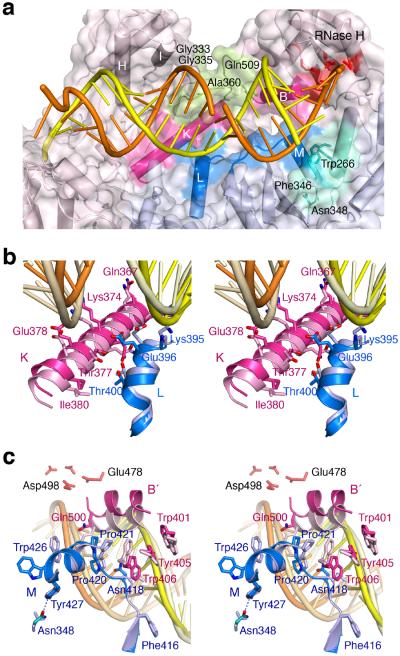
To accommodate the widened major groove of the RNA/DNA hybrid, two loops in p66, L18–K (the loop between β18 and αK, residues 356–364) and LB′–D′ (the loop between helices αB′ and αD′, residues 509–514) are re-configured to avoid steric clashes (Fig. 6a). To make these changes, an adjacent β turn (residues 333–336) that buttresses L18–K has two flexible glycine residues, Gly333 and Gly335. . Interestingly, mutations that introduce larger and polar side chains (G333D, G333E, G335C or G335D) as well as A360I/V and Q509L nearby have been implicated in NRTI and NNRTI drug resistance with a concomitant reduction of RNase H activity41 (36and reference therein). These mutations may prevent the conformational change needed to accommodate the altered RNA/DNA structure suitable for RNase H cleavage and thereby favor the polymerization-competent state.
Figure 6.
The altered p51–p66 subunit interface interacts with the RNA/DNA hybrid. (a) Overview of the WT22Efv structure with the altered subunit interface highlighted in blue (p51) and pink (p66). NNRTI and NRTI resistance mutations36 are found in the p66 connection and RNase H domain (pea green) that contacts the widened major groove. Residues surrounding Asn348 in p51 and forming van der Waals contacts with the RNA strand are colored teal. (b) A close-up stereo view of the interfacial helices between the p51 (blue) and p66 (pink) connection domains. The altered protein interface relative to 1RTD (an RT–DNA ternary complex, in lighter shades) is correlated with the change from DNA binding (pale yellow and wheat) to RNA/DNA hybrid (orange/yellow) binding. (c) A close-up stereo view of the back of (a). This shows the p51 C-terminus (blue) and helix αB′ (pink) in the WT22Efv structure and in 3KK227 (an RT–DNA ternary complex complete with helix αM in p51, in lighter shades). The hybrid in the WT22Efv (orange and yellow) and DNA in 3KK2 are also shown. In panels (b) and (c), the p51 subunits are superimposed.
Adjacent to the widened major groove in our WT22Efv structure, two areas of the altered p51-p66 subunit interface form new and substantial contacts with the minor groove of the hybrid (Fig. 6a). The first is between the two connection domains. Helix αL in p51 (residues 394–404) moves 1.5Å and helix αK in p66 (residues 364–384) moves 1.6Å in the opposite direction (Fig. 6b). As a result Gln394 and Lys395 of p51 and Lys366 of p66 directly contact the DNA strand. Not surprisingly, mutations K366R, T369I, E399D and A400T have been implicated in NRTI and NNRTI resistance36. The second is between helix αB′ in the RNase H domain (residues 499–508) and the p51 C-terminal helix αM (residues 422–430), which also move apart in our hybrid complexes (Fig. 6c). The extended segment comprising Phe416–Pro421 preceding αM first interacts with the DNA strand and then crosses the shallow minor groove, where the hybrid is bent, to form van der Waals contacts with the RNA strand three nucleotides from the scissile phosphate (Fig. 6a, c). The nearby tryptophan-repeat motif (residue 398–414) of p6642 concomitantly undergoes conformational changes (Fig. 6c), resulting in DNA binding by Trp406 and Gln407. The altered p51 C-terminal structure is stabilized by a hydrogen bond formed between Tyr427 and Asn348 (Fig. 6c). In previously reported RT–nucleic acid complexes, Asn348 is surrounded by hydrophobic sidechains and has no close contacts. The N348I mutation, which has often been found to confer drug resistance to both NRTI and NNRTI, exhibits reduced RNase H activity and altered cleavage patterns36,43,44.
p51 C-terminal helix αM (residues 422–430) is relatively flexible and has been modeled as random coil in earlier HIV-1 RT structures26 or when the last 13–15 residues in p51 (440 in total) are truncated24,25. In previously reported RT–nucleic acid complex structures, the p51 C-terminal residues are far removed from nucleic acid. The observation that deleting the C-terminal residues 427–440 of p51 alters RNase H activity of HIV-1 RT45,46 is surprising but can now be explained by the role of αM and residues preceding it in binding and orienting the nucleic acid substrate for RNA cleavage.
Discussion
The three RT–RNA/DNA hybrid structures described here reveal substantial differences in protein and nucleic acid conformation from those reported previously. While we cannot fully explain these alterations, inclusion of an NNRTI and the presence of an RNA/DNA hybrid without an artificial crosslinker are clearly contributing factors. These differences are consistent in the three different crystal forms, which differ in crystallization conditions, lattice contacts, hybrid lengths and NNRTI. It was observed previously that NNRTIs alter the p51-p66 interaction by both biochemical and structural methods14,20,39,40,42. It is interesting to note that the p51-p66 interface is rather hydrophilic and somewhat porous (Fig. 6, Supplementary Fig. 1e). The fact that NNRTI-induced conformational changes in the protein subunit interface and presence of the RNA/DNA hybrid allow the nucleic acid assume an orientation compatible with RNase H cleavage suggests that the structures observed here are functionally relevant.
Based on observations that (i), locking the nucleic acid substrate in the DNA polymerase active site reduces RNase H activity and (ii), disengaging DNA polymerase activity with an NNRTI enhances the viral RNase H33, we proposed that two active sites of HIV-1 RT cannot simultaneously engage in catalysis10. Gotte and co-workers suggested the opposite, based on observations that when nucleic acid is stabilized or presumably “locked” in the DNA polymerase active site, RNase H activity is still measureable, albeit greatly reduced31. Residual RNase H activity may, however, reflect transient dissociation of nucleic acid from the DNA polymerase active site rather than simultaneous catalysis. Analysis of concomitant polymerization and RNase H activities19 suggested that the two were coupled without substrate dissociation but did not exclude the sequential reactions.
The RT–RNA/DNA hybrid structures reported here support the proposal that the two active sites do not function simultaneously. Conformational flexibility in both the enzyme and RNA/DNA hybrid 47,48 are required to switch a nucleic acid substrate between the two catalytic centers. For polymerization, the thumb, palm and fingers play the essential role of substrate alignment as evidenced in all previous studies (Fig. 1b). However, to orient an RNA/DNA hybrid for RNA degradation, the connection domains of both subunits and the p51 thumb are required (Fig. 1a, 6a). NNRTIs inhibit polymerization by stabilizing the RT conformation incompatible for DNA synthesis, and NNRTI-resistant mutations often enhance the polymerization activity with a reduced RNase H activity 23,36. Mutations that confer drug resistance in the connection domain, which is distal from the NRTI and NNRTI-binding sites, have to act indirectly on drug binding, and were generically attributed to an “allosteric” effect in prior publications 35,36. Our RT–hybrid complex structures, however, show that the connection domains need to undergo conformational changes to orient the RNA/DNA and position the RNA strand for RNase H cleavage. Drug-resistant mutations in general tip the balance of substrate partitioning between the two active site in favor of DNA synthesis 36, which can be achieved directly by altering the drug-binding sites (e.g. Y181C, L74V or M184V) or “allosterically” by hindering the conformational changes in the connection domains (e.g. G333D, T369I or N348I) necessary for RNA degradation.
In contrast to cellular enzymes, HIV-1 RNase H depends on the rest of RT to bind an RNA/DNA hybrid and has a strong cleavage preference ~18 nucleotides upstream from the end of a hybrid marked by a 3′-end of DNA or a 5′ end of RNA 19,37,49. Of the ~1000Å2 enzyme–substrate interface, the DNA polymerase and RNase H active sites have been the primary target of anti-HIV drug development. Only recently vinylogous ureas, which bind at the subunit interface and inhibit RNase H activity, have been identified by high-throughput screening 46,50,51. The structures reported here reveal substantial contributions from the thumb and connection domains from both RT subunits in RNA/DNA binding thus providing an enlarged and underexplored target for future drug development.
Online Methods
Preparation of protein and nucleic acid substrates
Recombinant HIV-1 RT, a heterodimer of p66 and p51 subunits, was expressed in E. coli and purified as described previously 53. D498N and D498A mutations were introduced into the RNase H domain using QuikChange Kit (Stratagene) to eliminate RNase H activity. In addition, wildtype RT with a removable (His)6 tag was constructed by inserting a PreScission cleavage site between the tag and the p51 N-terminus. Briefly, His-tagged RT was purified using immobilized metal affinity and heparin-Sepharose chromatography. After removing the (His)6-tag by overnight proteolysis at 4°C in 50mM Tris·HCl (pH7.5), 150mM NaCl, 1mM EDTA and 1mM DTT, the protein solution was adjusted to 40mM NaCl and loaded onto a MonoS column. The tag-free RT was eluted with a linear NaCl gradient of 40–250mM. HPLC-purified RNA oligonucleotides were purchased from Dharmacon. DNA oligonucleotides (trityl-on) were purchased from the Facility for Biotechnology Resources at NIH and purified using an R3 reverse-phase column before detritylation. Hybrid substrates were annealed at a 1:1 ratio using a thermo cycler by heating at 75°C for 10 minutes and slow cooling to 20°C at a rate of 1°C/min.
Crystallization
Annealed RNA/DNA hybrids were mixed with HIV-1 RT and an NNRTI (Nevirapine or Efavirenz) at a 1.2:1:2 molar ratio in the buffer containing 10mM Tris·HCl (pH7.5), 50mM NaCl, 1mM DTT, 0.1mM EDTA and 5mM MgCl2 (for D498A/N mutant RT), or 5mM CaCl2 (for wildtype RT) and concentrated to ~15 mg/ml RT in the protein-nucleic acid complexes. Formation of a 1:1 complex of protein and hybrid substrate was confirmed by gel filtration chromatography and polyacrylamide gel electrophoresis (Supplementary Fig. 6). Crystals were grown by either the hanging or sitting drop vapor diffusion method at 4°C. Each complex, consisting of WT or mutant RT (Table 1), an NNRTI (Table 1) and an RNA/DNA hybrid (Fig. 1 and Supplementary Fig. 1), was mixed with reservoir solution at equal volume. The reservoir solution for the DA29Nvp crystals contained 50mM sodium cacodylate (pH6.5), 10mM MgCl2, 0.2M KCl, and 10% PEG4000 (w/v); for the DN25Nvp crystals it contained 1.8M (NH4)2SO4, 50mM Tris·HCl (pH8.5), and 25mM MgSO4; for the WT22Efv crystals it contained 0.1M sodium citrate (pH5.2), 0.1M CaCl2, and 7.5% PEG400 (v/v). Crystals were flash frozen in liquid nitrogen after sequential soaking in mother liquors with increasing concentrations of cryo-protectant, i.e. sucrose for DA29Nvp, glycerol for DN25Nvp, and PEG400 for WT22Efv.
Data collection, structure determination and refinement
X-ray diffraction data were collected at the beamline 22-ID, 22-BM and 23-IDB at the Advanced Photon Source, Argonne National Laboratory, Argonne, IL. Crystal forms in P3121 (WT22Efv), P6122 (DA29Nvp) and R32 (DN25Nvp) diffracted to 3.3Å, 4.8Å and 5.0 Å, respectively. Data were indexed and scaled using HKL2000 54 or XDS 55. Structures were solved by molecular replacement using the RT from the ternary complex (1RTD) and RT with NNRTI (1FK9) 22,26 with MolRep in CCP4 56. After placing the protein portion without any refinement, the density for RNA/DNA hybrid was clear in each case. All structures contained one heterodimeric HIV-1 RT and one RNA/DNA hybrid in each asymmetric unit.
Different refinement scenarios in PHENIX 57 were applied depending on data resolution, including grouped (rigid body) and restrained coordinate refinement, and restrained individual isotropic atomic displacement parameters refinement. Refinement was combined with bulk solvent correction and anisotropic scaling. Restraints on hydrogen bonds and planarity between base pairs of an RNA/DNA hybrid were custom-made and applied. Manual corrections were performed in COOT 58. Data-collection and refinement statistics are shown in Table 1.
The WT22Efv structure contains residues 4–556 of p66 (chain A) and residues 7–431 of p51 (chain B). Tracing of p66 is continuous except for the fingertip (residues 62–74), which is omitted due to the missing density in this region. Residues 216–230 in p51 are disordered. The RNA/DNA hybrid contains 24 out of 27 ribonucleotides and 22 out of 24 deoxynucleotides (Fig. 1a). The NNRTI Efavirenz is included (Fig. 1b). The protein has 95% residues in the favoured regions in Ramachandran plot. All seven outliers are associated with poorly defined electron densities. In the NA29Nvp structure, 519 residues of p66, 404 residues of p51, and 21 bp of RNA/DNA hybrid are built (Supplementary Fig. 1a). The protein has 94% residues in most preferred regions and 0.33% outliers in the Ramachandran plot. In the DN25Nvp structure 21 bp of RNA/DNA hybrid are traceable (Supplementary Fig. 1b), and Nevirapine is included. The RT has 95% residues in most preferred regions with no outliers.
Because these structures were solved by molecular replacement, to verify the structural models we calculated simulated annealing omit maps using Phenix.refine. Because the RNA/DNA consists of more than 10% of total diffraction matters, calculation of the simulated annealing of the entire hybrid was carried out in fragments to avoid distortion and assembled into a composite map. For the WT22Efv structures, DNA and RNA chains were split into three segments and simulated annealing Fo-Fc omit maps were calculated for each portion individually by setting the occupancy of the segment of nucleic acid to zero. The simulated annealing in phenix.refine was run with the parameter of ignore_zero_occupancy_atoms=False, which prevented the solvent mask from extending into the nucleic acid chain region. The composite omit maps thus generated clearly show the density for DNA and RNA at 4σ (Fig. 1b).
For low-resolution structures of DA29Nvp and DN25Nvp a similar approach was applied for DNA/RNA. However, the hybrids had to be split into smaller regions (3 nucleotides per run) to calculate simulated annealing omit maps. The simulated annealing omit maps countered at 2.5 sigma (DA29Nvp) and 3 sigma (DN25Nvp) clearly show the unbiased DNA/RNA path (Supplementary Fig. 1c, d).
Structural analysis
Proteins were superimposed using COOT 58, and standard deviations were computed using ALIGN 59. DDMP 60 was calculated using the algorithm written by P. Fleming (Johns Hopkins University). The pairwise Cα movement is displayed using ModeVector in PyMOL (www.PyMol.org). Nucleic acid structures were analyzed using 3-DNA 61 and Curve+ (gbio-pbil.ibcp.fr/cgi/Curves_plus) 62. All structural figures were prepared using PyMOL.
Supplementary Material
Acknowledgment
We thank D. Leahy and M. Nowotny for editing and critiquing the manuscript, C. Biertümpfel and Y. Zhao for help with data collection, and J.H.D. Cate for the ModeVector script to make Figure 2. The research was supported by the NIH Intramural AIDS Targeted Anti-viral Program (IATAP), and the intramural research programs of National Institute of Diabetes and Digestive and Kidney Diseases (W.Y., M.L. and L.T.) and National Cancer Institute (S. F. J. Le G. and J.T.M.).
Footnotes
Accession Codes Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank with accession codes 4B3O (WT22Efv), 4B3P (DA29Nvp) and 4B3Q (DN25Nvp).
Author Contributions J.T.M. prepared the HIV-1 RT mutants and proteins. L.T. made the RT–RNA/DNA–NNRTI complexes, grew the crystals and collected the diffraction data. M.L. refined the structures, and W.Y. carried out structure comparisons. W.Y. and S. Le G. originated the project. M.L., T.L., S. Le G. and W.Y. prepared the manuscript. All authors discussed the results and commented on the manuscript.
References
- 1.Varmus H. Reverse transcription. Sci Am. 1987;257:56–59. 62–54. doi: 10.1038/scientificamerican0987-56. [DOI] [PubMed] [Google Scholar]
- 2.Sarafianos SG, et al. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J Mol Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cihlar T, Ray AS. Nucleoside and nucleotide HIV reverse transcriptase inhibitors: 25 years after zidovudine. Antiviral Res. 2010;85:39–58. doi: 10.1016/j.antiviral.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 4.de Bethune MP. Non-nucleoside reverse transcriptase inhibitors (NNRTIs), their discovery, development, and use in the treatment of HIV-1 infection: a review of the last 20 years (1989–2009) Antiviral Res. 2010;85:75–90. doi: 10.1016/j.antiviral.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 5.di Marzo Veronese F, et al. Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science. 1986;231:1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]
- 6.Lightfoote MM, et al. Structural characterization of reverse transcriptase and endonuclease polypeptides of the acquired immunodeficiency syndrome retrovirus. J Virol. 1986;60:771–775. doi: 10.1128/jvi.60.2.771-775.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilboa E, Mitra SW, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 8.Whitcomb JM, Hughes SH. Retroviral reverse transcription and integration: progress and problems. Annu Rev Cell Biol. 1992;8:275–306. doi: 10.1146/annurev.cb.08.110192.001423. [DOI] [PubMed] [Google Scholar]
- 9.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metaldependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 10.Nowotny M, et al. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol Cell. 2007;28:264–276. doi: 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 11.Schultz SJ, Champoux JJ. RNase H activity: structure, specificity, and function in reverse transcription. Virus Res. 2008;134:86–103. doi: 10.1016/j.virusres.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw-Reid CA, et al. Inhibition of HIV-1 ribonuclease H by a novel diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid. J Biol Chem. 2003;278:2777–2780. doi: 10.1074/jbc.C200621200. [DOI] [PubMed] [Google Scholar]
- 13.Himmel DM, et al. Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site. Structure. 2009;17:1625–1635. doi: 10.1016/j.str.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lansdon EB, et al. Structural and binding analysis of pyrimidinol carboxylic acid and N-hydroxy quinazolinedione HIV-1 RNase H inhibitors. Antimicrob Agents Chemother. 2011;55:2905–2915. doi: 10.1128/AAC.01594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two- Mg2+-ion catalysis and substrate specificity. Mol Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Huber HE, Richardson CC. Processing of the primer for plus strand DNA synthesis by human immunodeficiency virus 1 reverse transcriptase. J Biol Chem. 1990;265:10565–10573. [PubMed] [Google Scholar]
- 18.Fuentes GM, Rodriguez-Rodriguez L, Fay PJ, Bambara RA. Use of an oligoribonucleotide containing the polypurine tract sequence as a primer by HIV reverse transcriptase. J Biol Chem. 1995;270:28169–28176. doi: 10.1074/jbc.270.47.28169. [DOI] [PubMed] [Google Scholar]
- 19.Gopalakrishnan V, Peliska JA, Benkovic SJ. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1992;89:10763–10767. doi: 10.1073/pnas.89.22.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 21.Hopkins AL, et al. Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors. J Med Chem. 1996;39:1589–1600. doi: 10.1021/jm960056x. [DOI] [PubMed] [Google Scholar]
- 22.Ren J, et al. Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase. Structure. 2000;8:1089–1094. doi: 10.1016/s0969-2126(00)00513-x. [DOI] [PubMed] [Google Scholar]
- 23.Ren J, Stammers DK. Structural basis for drug resistance mechanisms for non-nucleoside inhibitors of HIV reverse transcriptase. Virus Res. 2008;134:157–170. doi: 10.1016/j.virusres.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 24.Tu X, et al. Structural basis of HIV-1 resistance to AZT by excision. Nat Struct Mol Biol. 2010;17:1202–1209. doi: 10.1038/nsmb.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobo-Molina A, et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 27.Lansdon EB, et al. Visualizing the molecular interactions of a nucleotide analog, GS-9148, with HIV-1 reverse transcriptase-DNA complex. J Mol Biol. 2010;397:967–978. doi: 10.1016/j.jmb.2010.02.019. [DOI] [PubMed] [Google Scholar]
- 28.Meyer PR, Matsuura SE, Mian AM, So AG, Scott WA. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 29.Das K, Martinez SE, Bauman JD, Arnold E. HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism. Nat Struct Mol Biol. 2012;19:253–259. doi: 10.1038/nsmb.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarafianos SG, et al. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beilhartz GL, et al. HIV-1 reverse transcriptase can simultaneously engage its DNA/RNA substrate at both DNA polymerase and RNase H active sites: implications for RNase H inhibition. J Mol Biol. 2009;388:462–474. doi: 10.1016/j.jmb.2009.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hang JQ, et al. Substrate-dependent inhibition or stimulation of HIV RNase H activity by non-nucleoside reverse transcriptase inhibitors (NNRTIs) Biochem Biophys Res Commun. 2007;352:341–350. doi: 10.1016/j.bbrc.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 33.Radzio J, Sluis-Cremer N. Efavirenz accelerates HIV-1 reverse transcriptase ribonuclease H cleavage, leading to diminished zidovudine excision. Mol Pharmacol. 2008;73:601–606. doi: 10.1124/mol.107.038596. [DOI] [PubMed] [Google Scholar]
- 34.Shaw-Reid CA, et al. Dissecting the effects of DNA polymerase and ribonuclease H inhibitor combinations on HIV-1 reverse-transcriptase activities. Biochemistry. 2005;44:1595–1606. doi: 10.1021/bi0486740. [DOI] [PubMed] [Google Scholar]
- 35.Delviks-Frankenberry KA, et al. HIV-1 reverse transcriptase connection subdomain mutations reduce template RNA degradation and enhance AZT excision. Proc Natl Acad Sci U S A. 2008;105:10943–10948. doi: 10.1073/pnas.0804660105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delviks-Frankenberry KA, Nikolenko GN, Pathak VK. The “Connection” Between HIV Drug Resistance and RNase H. Viruses. 2010;2:1476–1503. doi: 10.3390/v2071476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peliska JA, Benkovic SJ. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992;258:1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- 38.Ding J, et al. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 A resolution. J Mol Biol. 1998;284:1095–1111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- 39.Hsiou Y, et al. Structure of unliganded HIV-1 reverse transcriptase at 2.7 A resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 40.Rodgers DW, et al. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brehm JH, Mellors JW, Sluis-Cremer N. Mechanism by which a glutamine to leucine substitution at residue 509 in the ribonuclease H domain of HIV-1 reverse transcriptase confers zidovudine resistance. Biochemistry. 2008;47:14020–14027. doi: 10.1021/bi8014778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tachedjian G, Orlova M, Sarafianos SG, Arnold E, Goff SP. Nonnucleoside reverse transcriptase inhibitors are chemical enhancers of dimerization of the HIV type 1 reverse transcriptase. Proc Natl Acad Sci U S A. 2001;98:7188–7193. doi: 10.1073/pnas.121055998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yap SH, et al. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med. 2007;4:e335. doi: 10.1371/journal.pmed.0040335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ehteshami M, et al. Connection domain mutations N348I and A360V in HIV-1 reverse transcriptase enhance resistance to 3'-azido-3'-deoxythymidine through both RNase H-dependent and -independent mechanisms. J Biol Chem. 2008;283:22222–22232. doi: 10.1074/jbc.M803521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cameron CE, Ghosh M, Le Grice SF, Benkovic SJ. Mutations in HIV reverse transcriptase which alter RNase H activity and decrease strand transfer efficiency are suppressed by HIV nucleocapsid protein. Proc Natl Acad Sci U S A. 1997;94:6700–6705. doi: 10.1073/pnas.94.13.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chung S, Miller JT, Johnson BC, Hughes SH, Le Grice SF. Mutagenesis of human immunodeficiency virus reverse transcriptase p51 subunit defines residues contributing to vinylogous urea inhibition of ribonuclease H activity. J Biol Chem. 2012;287:4066–4075. doi: 10.1074/jbc.M111.314781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perry JJ, Cotner-Gohara E, Ellenberger T, Tainer JA. Structural dynamics in DNA damage signaling and repair. Current opinion in structural biology. 2010;20:283–294. doi: 10.1016/j.sbi.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rambo RP, Tainer JA. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Current opinion in structural biology. 2010;20:128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerondelis P, et al. The P236L delavirdine-resistant human immunodeficiency virus type 1 mutant is replication defective and demonstrates alterations in both RNA 5'-end- and DNA 3'-end-directed RNase H activities. J Virol. 1999;73:5803–5813. doi: 10.1128/jvi.73.7.5803-5813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wendeler M, et al. Vinylogous ureas as a novel class of inhibitors of reverse transcriptase-associated ribonuclease H activity. ACS Chem Biol. 2008;3:635–644. doi: 10.1021/cb8001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chung S, et al. Structure-activity analysis of vinylogous urea inhibitors of human immunodeficiency virus-encoded ribonuclease H. Antimicrob Agents Chemother. 2010;54:3913–3921. doi: 10.1128/AAC.00434-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tuske S, et al. Structures of HIV-1 RT–DNA complexes before and after incorporation of the anti-AIDS drug tenofovir. Nat Struct Mol Biol. 2004;11:469–474. doi: 10.1038/nsmb760. [DOI] [PubMed] [Google Scholar]
- 53.Le Grice SF, Cameron CE, Benkovic SJ. Purification and characterization of human immunodeficiency virus type 1 reverse transcriptase. Methods Enzymol. 1995;262:130–144. doi: 10.1016/0076-6879(95)62015-x. [DOI] [PubMed] [Google Scholar]
- 54.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 55.Kabsch W. XDS. Acta Crystallographica Section D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.CCP4, The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 57.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cohen GH. ALIGN: a program to superimpose protein coordinates, accounting for insertions and deletions. J. Appl. Cryst. 1997;30 [Google Scholar]
- 60.Richards FM, Kundrot CE. Identification of structural motifs from protein coordinate data: secondary structure and first-level supersecondary structure. Proteins. 1988;3:71–84. doi: 10.1002/prot.340030202. [DOI] [PubMed] [Google Scholar]
- 61.Lu XJ, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.