Abstract
Expression of apoptotic protease activating factor-1 (Apaf-1) gradually decreases during brain development, and this decrease is likely responsible for the decreased sensitivity of brain tissue to apoptosis. However, the mechanism by which Apaf-1 expression is decreased remains elusive. In the present study, we found that four microRNAs (miR-23a/b and miR-27a/b) of miR-23a-27a-24 and miR-23b-27b-24 clusters play key roles in modulating the expression of Apaf-1. First, we found that miR-23a/b and miR-27a/b suppressed the expression of Apaf-1 in vitro. Interestingly, the expression of the miR-23-27-24 clusters in the mouse cortex gradually increased in a manner that was inversely correlated with the pattern of Apaf-1 expression. Second, hypoxic injuries during fetal distress caused reduced expression of the miR-23b and miR-27b that was inversely correlated with an elevation of Apaf-1 expression during neuronal apoptosis. Third, we made neuronal-specific transgenic mice and found that overexpressing the miR-23b and miR-27b in mouse neurons inhibited the neuronal apoptosis induced by intrauterine hypoxia. In conclusion, our results demonstrate, in central neural system, that miR-23a/b and miR-27a/b are endogenous inhibitory factors of Apaf-1 expression and regulate the sensitivity of neurons to apoptosis. Our findings may also have implications for the potential target role of microRNAs in the treatment of neuronal apoptosis-related diseases.
Keywords: brain development, neuron, apoptosis, microRNA, fetal distress
Apoptosis occurs throughout the nervous system during development. It is estimated that at least half of the original cellular population is eliminated as a result of apoptosis in the developing nervous system because of the processes of the optimization of synaptic connections and removal of unnecessary neurons as well as pattern formation.1 Deletion of apoptotic genes from early development is lethal to the embryo.2, 3, 4 Interestingly, despite the ubiquitous expression of caspase-3 and caspase-9 throughout the organs of the body, the main pathology that arises from a deficiency in apoptosis is focused on the developing brain that reveals the essential physiological role of apoptosis in neural development. Pathologically, apoptosis and dysregulation of apoptosis also play roles in nervous system diseases, for example, infectious, degenerative, and neoplastic diseases of the nervous system.5 Among the multiple etiologies of perinatal brain injuries, hypoxia–ischemia is the most common and predominant.6 Hypoxia–ischemia is thought to be the final common end point for a complex convergence of events; some of these events are genetically determined and some are triggered by an in utero (but not necessarily intrapartum) stressor.7 Although apoptosis occurs after hypoxia–ischemia in both embryonic and adult stages, multiple studies have suggested that apoptosis plays a more prominent role in the embryo than in the adult after brain injuries caused by hypoxia–ischemia.8, 9, 10 This differential sensitivity of the brain to apoptosis may be explained, in part, by the differential age-dependent expression of apoptotic genes, mainly the apoptotic protease activating factor-1 (Apaf-1) gene.11
Apaf-1 is a component protein of the apoptosome. In the intrinsic apoptosis pathway, cytochrome c is released from the mitochondria to the cytosol and binds to Apaf-1 protein. Binding of cytochrome c to Apaf-1 allows the recruitment and activation of caspase-9 within the apoptosome.12 Deficiency in Apaf-1 is lethal to mouse embryos. Homozygous mutants die at embryonic day 16.5, and their phenotype includes severe craniofacial malformations, brain overgrowth, persistence of the interdigital webs, and dramatic alterations of the lens and retina.13 High level of Apaf-1 expression in brain tumors elevates the sensitivity of the brain tumors to apoptosis induced by cytochrome c that reveals that the protein level of Apaf-1 may directly influence the sensitivity of brain cells to apoptotic stimuli.14 Therefore, as a key apoptotic protein that may decide the apoptotic fate of cells, Apaf-1 expression is tightly regulated. Previous studies have reported that the expression of Apaf-1 decreases in rat cerebral cortex during development, and this would explain the high sensitivity of the nervous system to apoptosis at the embryonic stage. However, the mechanism by which Apaf-1 expression is downregulated in the brain during development is still unknown.
MicroRNAs (miRNAs) are a group of endogenous noncoding RNAs that consist of 18 to 25 nucleotides. The miRNAs play an important role in regulating gene expression at the posttranscriptional level by binding to complementary sites on target mRNAs that either block mRNA translation or trigger mRNA degradation.15, 16 The diversity of miRNAs and the multiple genes that are targeted by every miRNA provide miRNAs with versatile functions in the control of gene expression.17 Currently, miRNAs are thought to regulate the expression of most genes and, consequently, play regulatory roles in a wide variety of physiological and pathological cellular processes.18 In the nervous system, miRNAs have temporally and spatially specific expression patterns during the development of the brain19, 20, 21 and thus contribute to the processes of determining neuronal cell identities and specific functions.22, 23 In nervous system diseases, miRNAs may be dysregulated and influence the pathological progress and outcomes. Changes in the miRNA profile of the adult brain during hypoxia–ischemia have been reported. For example, in microglia cells, hypoxia causes the upregulation of FasL expression and the downregulation of miR-21 expression during hypoxia-induced microglial activation.24 Doeppner et al.25 reported that miR-124 promotes neuronal survival under ischemic conditions via Usp14-dependent REST degradation. However, the alterations in miRNAs during perinatal hypoxic–ischemic encephalopathy are still largely unknown.
In the present study, we tested the hypothesis that miRNAs may regulate Apaf-1 expression in the mouse cortex during development and in neurological diseases. We found miR-23a/b and miR-27a/b encoded by the miR-23a-27a-24 and miR-23b-27b-24 clusters that participated in the regulation of Apaf-1 expression. With various methods, we showed that the expression of miR-23-27-24 clusters gradually increased in the mouse cortex in a manner that was inversely correlated with the pattern of Apaf-1 expression. Furthermore, in a hypoxia-induced neuronal apoptosis mouse model, overexpression of the miR-23b and miR-27b clusters inhibited the neuronal apoptosis induced by intrauterine hypoxia. For the first time, we discovered that miRNAs regulate the sensitivity of neurons to apoptosis during development and hypoxia-induced brain injuries.
Results
Apaf-1 gene expression decreases during development in the mouse cerebral cortex
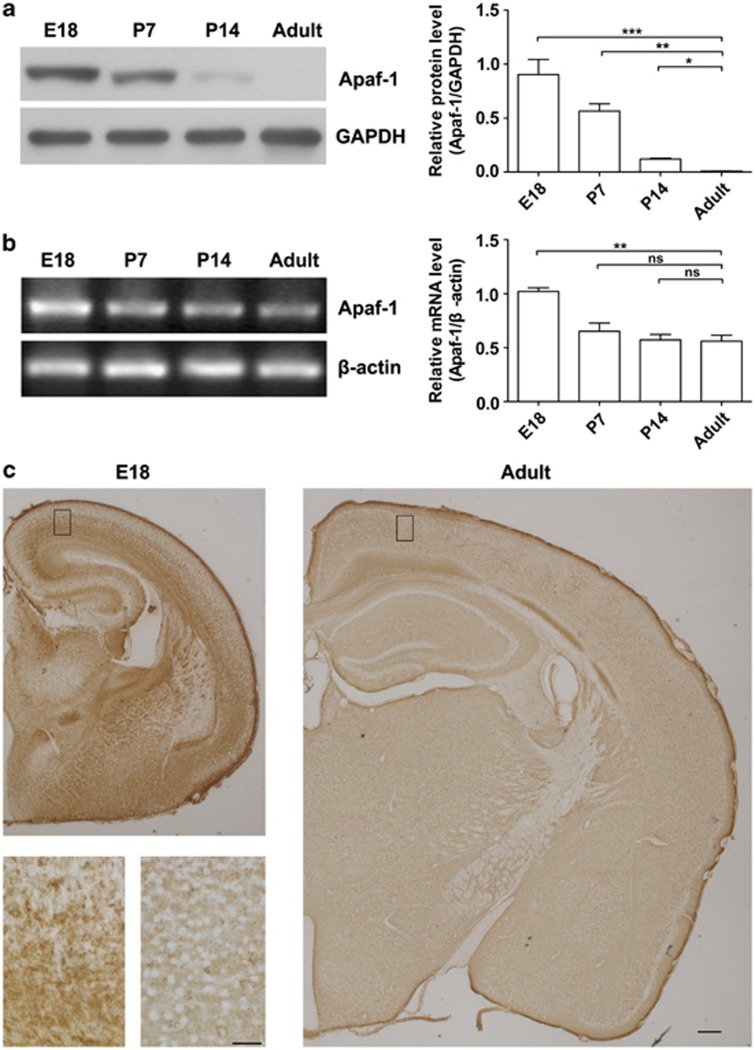
Previous studies have reported that the expression of Apaf-1 decreases during development in the rat cortex.11 Thus, we initially examined the expression of Apaf-1 protein and mRNA in the mouse cortex during brain development in the present study. Four time points were chosen to represent the entire developmental process: E18, P7, P14, and the adult stage (P60). We found that the Apaf-1 protein levels decreased as cortical development progressed (Figure 1a). Similarly, Apaf-1 mRNA also decreased during brain development (Figure 1b). Furthermore, immunohistochemistry (IHC) stain showed that the neuronal cells had little signal of Apaf-1 protein in the adult stage, whereas Apaf-1 protein was expressed in the neurons at the E18 stage (Figure 1c). This differential expression of Apaf-1 is consistent with the decreases in susceptibility to apoptosis during mouse brain development that implies that expression of Apaf-1 might contribute to the susceptibility of neurons to apoptosis.
Figure 1.
Decreased Apaf-1 gene expression during brain development. (a) Representative western blots of Apaf-1 in cortical samples at different developmental stages (E18, P7, P14, and adult). Each lane was loaded with equal amounts of total protein from the mixed samples of five individuals (n=5, unpaired t-test, *P<0.05, **P<0.01, and ***P<0.001). (b) Semiquantitative RT-PCR detection of Apaf-1 mRNA in the total RNA from cerebral cortices at different developmental stages. Quantitative RT-PCR detection of Apaf-1 mRNA (n=5, unpaired t-test, NS, not significant, **P<0.01). Error bars represent the S.E.M. (c) IHC stain of Apaf-1 in E18 and adult brain tissues. Scale bar represents 250 μm; scale bar (insert) represents 50 μm
Upregulation of miR-23-27-24 clusters during development in mouse cortex is inversely correlated with the Apaf-1 expression pattern
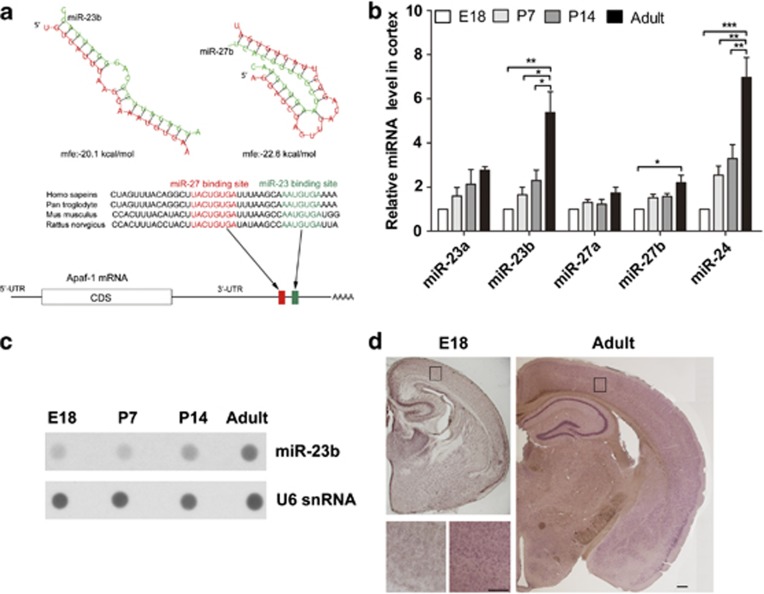
Compared with the downregulation of the mRNA level, the decrease of Apaf-1 protein level was more dramatic, and this indicates that Apaf-1 gene expression might be regulated at the posttranscriptional level. The miRNAs are a type of endogenous small RNA that suppresses gene expression at the posttranscriptional level. Using computer-aided algorithms,26 we found that four (miR-23a, miR-23b, miR-27a, and miR-27b) of the miR-23-27-24 clusters had conserved putative binding sites for Apaf-1 mRNA on 3′-UTR (Figure 2a). The miR-24 had less conserved binding sites. In the mouse genome, the mmu-miR-23a-27a-24-2 cluster is located on chromosome 8, and the mmu-miR-23b-27b-24-1 cluster is located on chromosome 13. We examined the levels of miR-23a/b, miR-27a/b, and miR-24 expression at the same developmental time points. The results from quantitative reverse transcription-PCR (qRT-PCR) experiments showed that expression of the miR-23-27-24 clusters clearly increased over these time points. The miR-23b expression gradually increased during maturation of the brain, and the miR-23b transcript levels were approximately fivefold greater in adult mouse cortices than in the cortices of the E18 pups. The miR-27b also exhibited dramatically increased expression in the adult stage. The miR-23a and miR-27a also showed gradually increases in expression, but these increases were relatively mild compared with that of the miR-23b-27b cluster. The miR-24 had a significantly increased expression pattern during development (Figure 2b). To further corroborate the qRT-PCR results, a RNA dot blotting assay and in situ hybridization staining were employed. As shown in Figure 2c, miR-23b levels were much higher in P60 adult mouse cortices than in E18 pup cortices. In contrast to IHC staining signal of Apaf-1, the in situ hybridization results revealed that miR-23b was abundantly expressed in the neurons and more miR-23b signals were detected in adult brain neurons (Figure 2d). Therefore, the expression patterns of the Apaf-1 gene and the miR-23-27 clusters were inversely correlated that indicates that the miR-23-27 clusters might inhibit Apaf-1 gene expression during brain development.
Figure 2.
The expression of miR-23-27 clusters increases during brain development. (a) Schematic of the Apaf-1 3′UTR indicating the locations of the miR-23 and miR-27 target sites that are conserved in vertebrates. The Apaf-1 3′UTR contains evolutionarily well-conserved sequences matched for the miR-23 and miR-27 families that were predicted by computer-aided algorithms. The free energies (mfes) of microRNA bindings were calculated by RNAHybrid software (BiBiServ, Bielefeld, Germany). (b) Quantitative RT-PCR detection of miR-23a, miR-23b, miR-27a, miR-27b, and miR-24 in cerebral cortex samples at different developmental stages (n=5, one-way ANOVA with Newman–Keuls multiple comparison test, *P<0.05, **P<0.01, and ***P<0.001). (c) Dot blot analyses of miR-23b and U6 snRNA in total RNA of the cerebral cortex. Each lane was loaded with equal amounts of total RNA extracted from the mixed samples of five individuals. (d) Expression of miR-23b in cortices of E18 and adult brain tissues detected by in situ hybridization. Scale bar represents 250 μm; scale bar (insert) represents 200 μm
MiR-23a/b and miR-27a/b suppress Apaf-1 expression at the posttranscriptional level
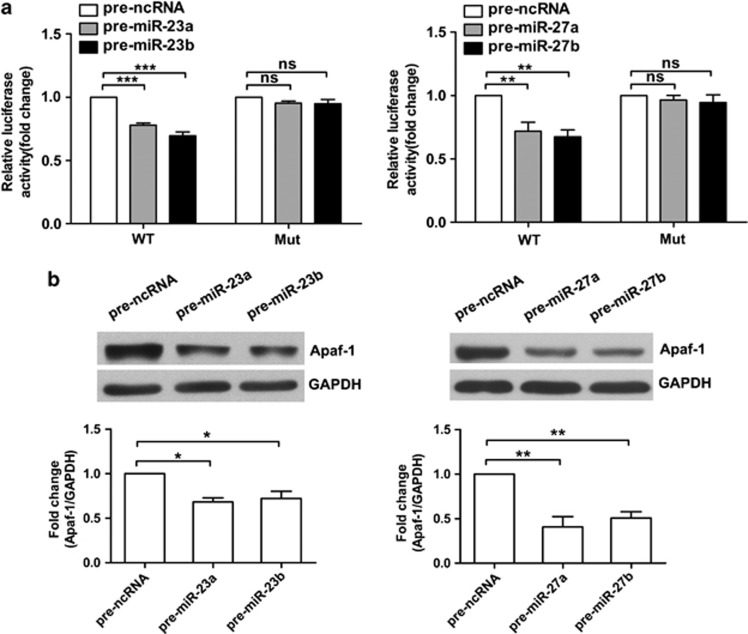
Gene expression analysis showed that the expression patterns of the miR-23-27-24 clusters were inversely correlated with that of the Apaf-1 gene. To confirm that these miRNAs actually suppress Apaf-1 gene expression, we performed the following studies. A 507-bp-length DNA that contains the putative binding sites for miR-23a/b, miR-27a/b, and miR-24 was cloned from the 3′-UTR of Apaf-1 mRNA to the pMIR-REPORT Luciferase plasmid. Then, luciferase assays were performed in HEK293T cells. Overexpression of miR-23a/b or miR-27a/b significantly suppressed luciferase activity. Conversely, after introducing mutations to seven sites in the predicted miRNA binding site on 3′-UTR of the plasmid, the inhibitory effect of overexpression of miR-23a/b or miR-27a/b was abolished (Figure 3a). However, overexpression of miR-24 had little effect on luciferase activity (Supplementary Figure 1a). Furthermore, western blotting analyses showed that Apaf-1 protein levels were decreased after cells received miR-23a/b or miR-27a/b (Figure 3b). Overexpression of miR-24 had little effect on Apaf-1 expression (Supplementary Figure 1b). These results indicate that miR-23a/b and miR-27a/b of miR-23-27-24 clusters suppressed Apaf-1 gene expression.
Figure 3.
The miR-23a/b and miR-27a/b suppress Apaf-1 protein expression. (a) HEK-293T cells were transfected with luciferase reporter plasmids carrying wild-type (WT) or mutant (Mut) Apaf-1 3′-UTRs. The luciferase activities were examined in the presence of pre-miR-23a/b (left) or pre-miR-27a/b (right) (n=5, unpaired t-test, NS, not significant, **P<0.01 and ***P<0.001). (b) Apaf-1 protein level in HEK-293T cells 48 h after transfection with pre-miR-23a/b or pre-miR-27a/b (n=5, unpaired t-test, *P<0.05 and **P<0.01)
Hypoxia causes increased neuronal apoptosis and increased Apaf-1 protein expression
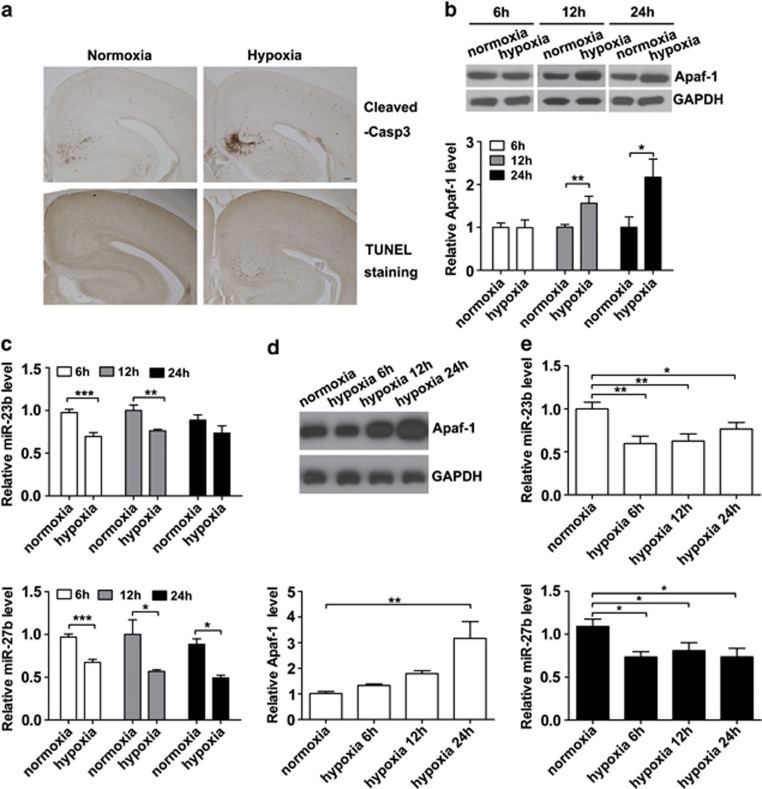
Physiologically, sporadic apoptotic cell death provides a selective mechanism that removes surplus or unwanted neurons during the process of brain development. However, apoptosis is uncommon in the mature mammalian brains under normal physiological conditions.27 Neuronal apoptosis also occurs in cortical neurons in many pathological conditions. Reduced oxygen supply (hypoxia) during the pre- and perinatal periods often leads to neonatal brain damage. Indeed, perinatal hypoxic–ischemic encephalopathy is a significant cause of mortality and morbidity in infants and young children, and apoptosis is thought to play an important role in perinatal hypoxic–ischemic encephalopathy.28 In a fetal distress mouse model, we observed a dramatic increase in apoptotic neurons in the E19.5 pup cortex exposure to hypoxia (Figure 4a). We examined whether Apaf-1 protein levels changed in hypoxia-treated cortices. As shown in Figure 4b, the expression of Apaf-1 protein was significantly increased at 12 and 24 h after 6 h of hypoxia treatment. Meanwhile, the expression of the miR-23b and miR-27b decreased before alterations in Apaf-1 expression at the 6-h time point after hypoxia treatment (Figure 4c). In primary cortical neurons, anaerobic treatment also caused increases in Apaf-1 protein expression and decreases in miR-23b and miR-27b expression (Figures 4d and e). These results imply that miR-23b and miR-27b are downregulated during hypoxia that relieves the miRNA-mediated inhibition of Apaf-1 expression and increases the sensitivities of neurons to apoptosis.
Figure 4.
Apaf-1 protein levels are elevated and miR-23b-27b cluster is decreased in cortices of E19.5 pups after hypoxia. (a) Coronal brain sections of E19.5 mice were stained with a cleaved caspase-3 (Cleaved-Casp3) antibody or with TUNEL staining kit. More positively stained neurons were observed in the hypoxia group (n=3) compared with the normoxia control group (n=3). Scale bar represents 100 μm. (b) Representative western blot images for Apaf-1 in the cerebral cortex under conditions of normoxia or hypoxia for 6 h followed by different recovery times (no recovery, recovery 6 h, or recovery 18 h). Relative fold changes of Apaf-1 protein level were quantified by densitometry (n=5, unpaired t-test, *P<0.05 and **P<0.01). (c) Quantitative RT-PCR detection of miR-23b and miR-27b expression in the cerebral cortices of different groups (n=5, unpaired t-test, *P<0.05, **P<0.01, and ***P<0.001). (d) Representative western blot images for Apaf-1 in primary cortical neurons under conditions of normoxia or hypoxia for 6, 12, and 24 h (n=3, unpaired t-test, **P<0.01). (e) Quantitative RT-PCR detection of miR-23b and miR-27b expression in primary cortical neurons under the conditions of normoxia or hypoxia for 6, 12, and 24 h (n=6, unpaired t-test, *P<0.05 and **P<0.01)
Overexpression of the miR-23b and miR-27b attenuates neuronal apoptosis through suppression of Apaf-1
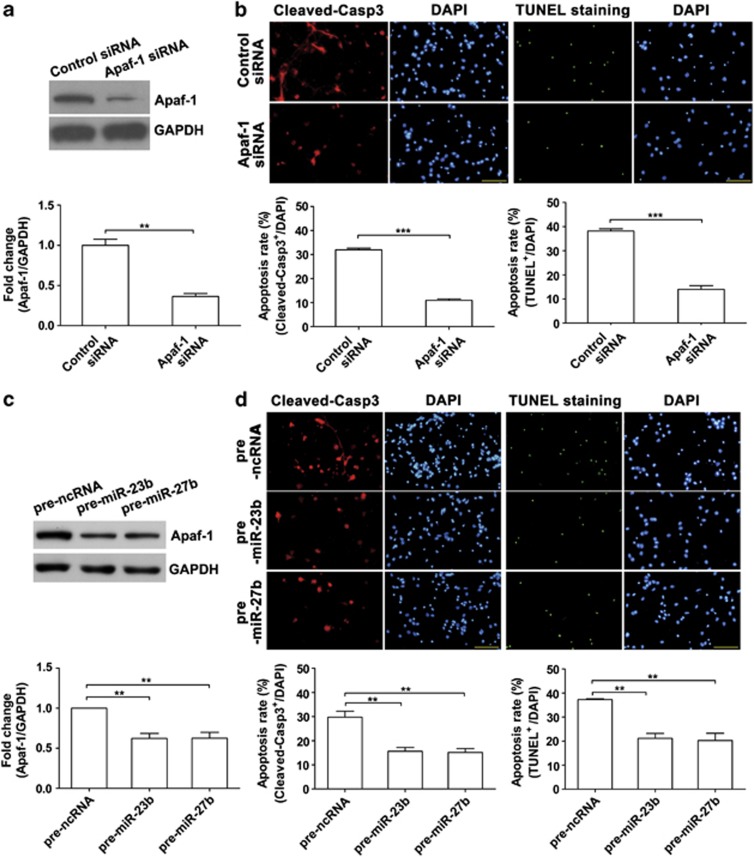
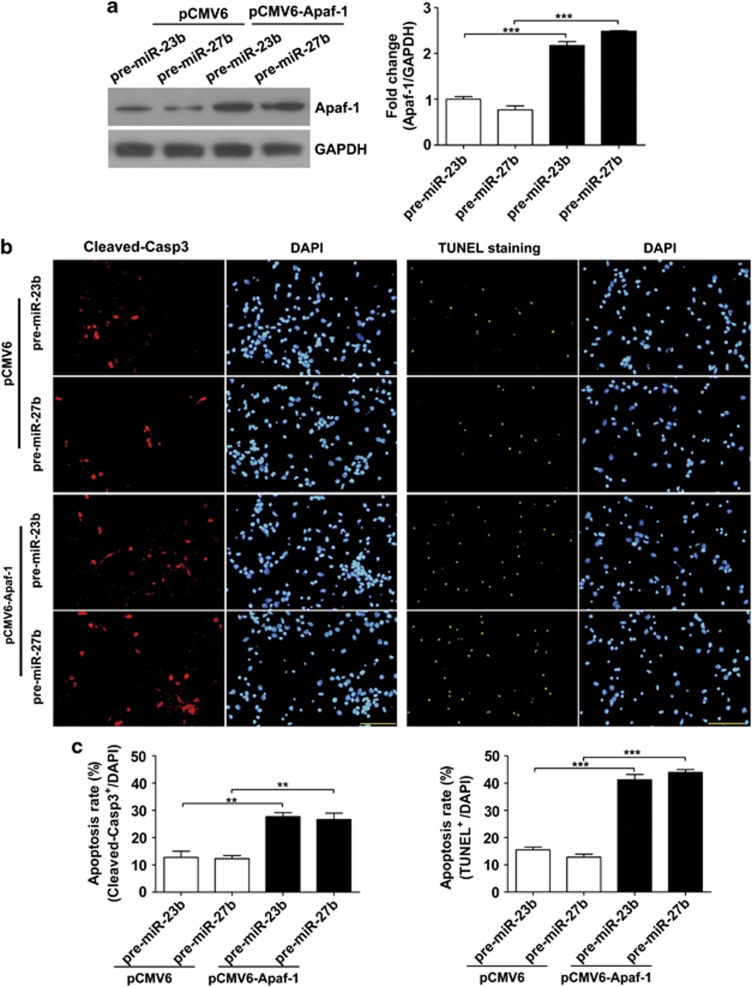
We investigated the role of Apaf-1 in neuronal apoptosis induced by hypoxia. A specific siRNA of Apaf-1 was employed to knock down Apaf-1 expression (Figure 5a). After the primary cortical neurons received the siRNA, these neurons received an anaerobic treatment (24 h), as we found that the cultured neurons were resistant to hypoxia treatment in vitro. We found that the neuronal apoptosis rate, calculated by cleaved caspase-3-positive neurons or terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) staining positive neurons respectively, was significantly decreased in siRNA group (Figure 5b). To examine the effects of miR-23b and miR-27b on hypoxia-induced neuronal apoptosis, we overexpressed miR-23b or miR-27b in primary cortical neurons, and these neurons received the anaerobic treatment (24 h). Western blot analyses showed that miR-23b or miR-27b significantly downregulated Apaf-1 protein levels (Figure 5c). Furthermore, the numbers of apoptotic neurons were reduced after the overexpression of miR-23b or miR-27b (Figure 5d). Next, we put an Apaf-1 expression vector into the neurons to elevate the Apaf-1 expression level that had received miR-23b or miR-27b precursor miRNAs to decrease Apaf-1 expression (Figure 6a). As shown in Figures 6b and c, elevated Apaf-1 expression caused a significant increase of neuronal death during hypoxia, and even miR-23b or miR-27b was also overexpressing. These results indicate that Apaf-1 is the main target of miR-23 and miR-27 during hypoxia, and through suppressing Apaf-1, miR-23a/b and miR-27a/b regulate the sensitivity of neuron to apoptosis.
Figure 5.
Overexpression of miR-23b or miR-27b represses Apaf-1 protein levels in primary cortical neurons and attenuates neuronal apoptosis caused by hypoxia. (a) Western blot analysis of Apaf-1 in primary cortical neurons 48 h after transfection with Apaf-1 siRNA. Relative amounts of Apaf-1 protein were quantified by densitometry (n=3, unpaired t-test, **P<0.01). (b) Immunofluorescence staining of Cleaved-Casp3 or TUNEL staining. Neurons were stained with Cleaved-Casp3 antibody or with TUNEL stain kit after hypoxia treatment (24 h) at DIV5. Statistical analysis of the percentage of Cleaved-Casp3-positive neurons or TUNEL-positive neurons to DAPI-stained cells (cell numbers=400–500, unpaired t-test, ***P<0.001). (c) Western blot analysis of Apaf-1 in primary cortical neurons 48 h after transfection with pre-miR-23b or pre-miR-27b. Relative amounts of Apaf-1 protein were quantified by densitometry (n=3, unpaired t-test, **P<0.01). (d) Immunofluorescence staining of Cleaved-Casp3 or TUNEL staining (cell numbers=600–700, unpaired t-test, **P<0.01). Scale bar represents 100 μm
Figure 6.
Overexpression of Apaf-1 re-activates the sensitivity of neurons overexpressing miR-23b/27b to apoptosis induced by hypoxia. (a) Western blot analysis of Apaf-1 in primary cortical neurons 48 h after co-transfection with plasmids (pCMV6 or pCMV6-Apaf-1) and miRNAs (pre-miR-23b or pre-miR-27b). Relative amounts of Apaf-1 protein were quantified by densitometry (n=3, unpaired t-test, ***P<0.001). (b) Immunofluorescence staining of Cleaved-Casp3 or TUNEL staining. Cultured cortical neurons co-transfected with plasmids (pCMV6 or pCMV6-Apaf-1) and miRNAs (pre-miR-23b or pre-miR-27b) were stained with Cleaved-Casp3 antibody or with TUNEL staining kit after hypoxia treatment (24 h) at DIV5. (c) Statistical analysis of the percentage of Cleaved-Casp3-positive neurons or TUNEL-positive neurons to DAPI-stained cells (cell numbers=700–800, unpaired t-test, **P<0.01 and ***P<0.001). Scale bar represents 100 μm
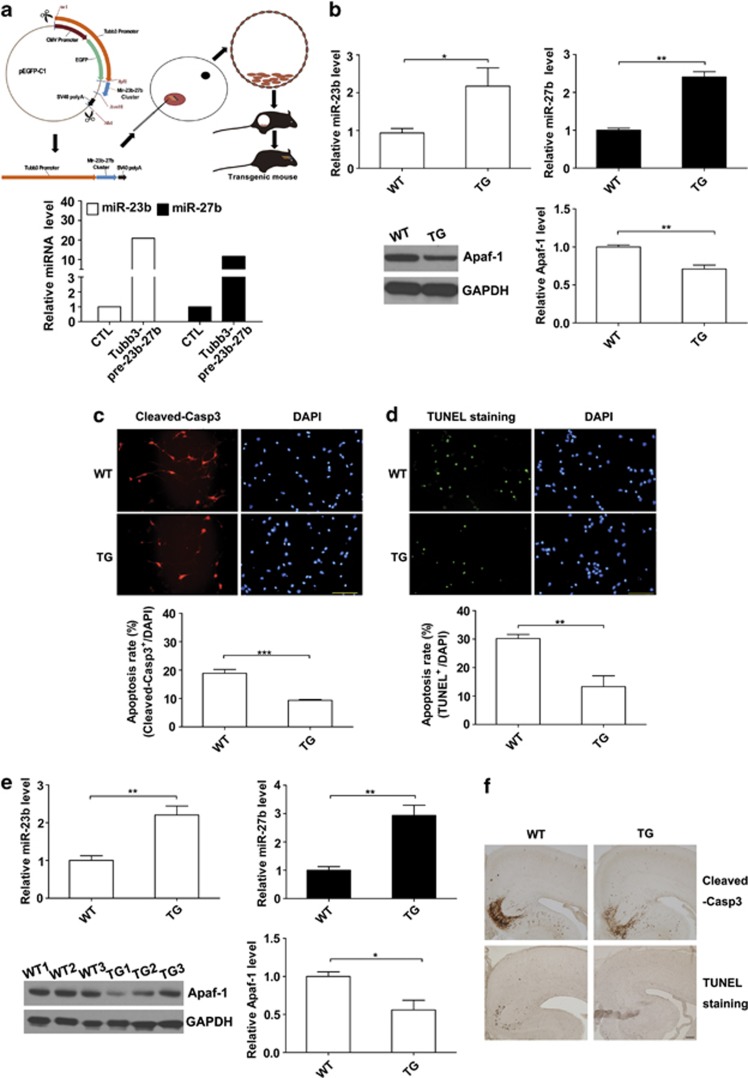
To further confirm the roles of miR-23b and miR-27b in neuronal apoptosis, we constructed neuronal-specific miR-23b-27b cluster transgenic mice. In these transgenic mice, the promoter of the tubulin βIII (Tubb3) gene was employed because of its high expression capability in the early embryonic brain stage (E9.5)29 that should allow for the ectopic overexpression of the miR-23b-27b cluster in developing cortices. The efficiency of the Tubb3 promoter was tested in primary neurons; miR-23b expression was ∼20-fold greater, and miR-27b expression was ∼10-fold greater after transfection of the miR-23b-27b expression plasmid (Figure 7a). Among the 10 founder transgenic mouse lines that were established (Supplementary Figure 2), we chose the line with the highest expression (the B line) and the respective wild-type (WT) mice as controls for further study. We found that the expression of miR-23b and miR-27b were significantly elevated in the transgenic mouse primary neuron cultures, and the expression of the Apaf-1 protein in transgenic mouse primary neurons was decreased (Figure 7b). Immunofluorescent staining of cleaved caspase-3 showed that the apoptotic rate was lower in transgenic mouse primary neurons than in WT mouse primary neurons (Figure 7c) after anaerobic treatment for 24 h. Similarly, TUNEL staining showed that the apoptotic rate was significantly lower in transgenic mouse primary neurons (Figure 7d). We further examined the expression levels of miR-23b and miR-27b in transgenic E19.5 mouse cortices. The miR-23b levels were elevated by ∼1.5-fold, and miR-27b levels were elevated by ∼2.5-fold in these transgenic mice. Consistently, the expression of Apaf-1 was reduced in the cortices of these mice (Figure 7e). After hypoxia treatment, the numbers of cleaved caspase-3-positive apoptotic neurons were reduced in the cortices of transgenic E19.5 pups. Similarly, TUNEL staining revealed that there were fewer apoptotic cells in the brains of miR-23b-27b transgenic mice (Figure 7f). Together, these results indicate that overexpression of the miR-23b-27b cluster alleviates neuronal apoptosis by inhibiting Apaf-1 in hypoxia-induced pathological conditions.
Figure 7.
The miR-23b-27b transgenic mice show more resistance to hypoxia-induced apoptosis. (a) A cartoon of generation of the miR-23b-27b cluster transgenic mice. The Tubb3 gene promoter was employed because of its high expression capability and specificity in neurons in the early embryonic stage. Quantitative RT-PCR detection of miR-23b and miR-27b in primary cortical neurons 24 h after transfection with the constructed plasmid. (b) Quantitative RT-PCR detection of miR-23b-27b (top) and western blot analysis of Apaf-1 protein levels (bottom) in cultured primary cortical neurons from wild-type (WT) mice and transgenic (TG) mice (n=3, unpaired t-test, *P<0.05 and **P<0.01). (c) Cultured cortical neurons isolated from E15.5 pups were stained with Cleaved-Casp3 antibody after hypoxia treatment (24 h) at DIV5 and counterstained with DAPI. Statistical analysis of the percentage of Cleaved-Casp3-positive neurons to DAPI-stained cells (cell numbers=400–500, unpaired t-test, ***P<0.001). Scale bar represents 100 μm. (d) Cultured cortical neurons were stained with TUNEL staining kit after hypoxia treatment. Statistical analysis of the percentage of TUNEL-positive neurons to DAPI-stained cells (cell numbers=400–500, unpaired t-test, **P<0.01). (e) Quantitative RT-PCR detection of miR-23b-27b (top) and western blot analysis of Apaf-1 protein levels (bottom) in the cerebral cortices from E19.5 mice (TG mice n=4 and WT mice n=6, unpaired t-test, *P<0.05 and **P<0.01). (f) Coronal sections of WT and transgenic neocortex at E19.5 were stained with Cleaved-Casp3 antibody after 6 h of hypoxia followed by a 6-h recovery or with TUNEL staining kit after a 18-h recovery. Scale bar represents 100 μm
Discussion
Apaf-1 is believed to be a key functional protein in physiological apoptosis during brain development and in pathological apoptosis related to CNS injury.11 In the present study, we found that the expressions of miR-23-27-24 clusters were elevated during brain maturation in a manner that was inversely correlated with Apaf-1 expression. The miR-23a/b and miR-27a/b could suppress Apaf-1 expression. Furthermore, the expression of the miR-23-27-24 was downregulated in brain tissue in a fetal distress mouse model. Forced expression of the miR-23b and miR-27b clusters suppressed Apaf-1 and alleviated neuronal apoptosis in primary cultured neurons and in E19.5 pup cortical neurons after hypoxia. These results show that miRNAs participate in the regulation of Apaf-1 gene expression and play important roles in physiological apoptosis during brain development and in pathological apoptotic processes caused by hypoxia.
The miRNAs modulate complex physiological and pathological processes by repressing the expression of multiple proteins. These inhibitory roles of miRNAs on gene expression are mediated by the imperfect base-match-guided binding of miRNAs to their target mRNAs, and it is this imprecise binding that allows each miRNA to target multiple mRNAs. Moreover, different miRNA binding sites may exist on the mRNA of a single gene. The miRNAs may act synergistically in cells of the tissue to enhance their suppressive effects on their common target genes; miRNAs can also function independently in the cells of different tissues. This versatile characteristic of miRNAs broadly extends the regulatory roles of miRNAs. We found that mmu-miR-23a/b and mmu-miR-27a/b had experimentally validated binding sites on the 3′-UTR of Apaf-1 mRNA, but not miR-24. In humans, hsa-miR-23a/b and hsa-miR-27a/b are predicted to be highest possible miRNAs that regulate Apaf-1 expression (miRanda-mirSVR and Targetscan). Very recently, Lian et al.30 have reported that the expression of miR-23a in human glioma tissues is significantly upregulated. Furthermore, they analyzed the results by using the luciferase reporter assay and western blot analysis in 293T cells and glioma cell line, respectively. This finding confirms that hsa-miR-23 suppresses human Apaf-1 expression. We found that the expression of each miRNA of miR-23-27-24 clusters increased during brain development, whereas the expression of Apaf-1 decreased during development. The inversely correlated expression patterns of Apaf-1 and the miR-23-27-24 clusters indicate that the miR-23a/b and miR-27a/b clusters function synergistically to repress Apaf-1 protein during the physiological development process.
Interestingly, according to the predictions of the miRanda-mirSVR algorithm,31 miR-23a/b also have putative binding sites for the human caspase-3 gene. Thus, the miR-23a/b and miR-27a/b may suppress multiple key proteins in the apoptosis pathway in human brain development. Moreover, it is likely that other miRNAs also synergistically suppress Apaf-1 and other proteins involved in apoptotic pathways.
Previous studies have noted that the sensitivity of the central nervous system to apoptosis is highest at the embryonic stage and gradually decreases during development. Physiologically, apoptosis is designed to remove unnecessary or unsuitable nerve cells that are overproduced during development. However, elevated sensitivity to apoptosis is a double-edged sword because embryonic neuronal cells are highly sensitive to various pathological injuries. Apoptosis may be the major pathway of cell death during the embryonic stage, and pathological neuronal apoptosis is more evident during the embryonic stage. Multiple studies have suggested that the role of neuronal apoptosis after hypoxia–ischemia in the neonate brain is more prominent than in the adult brain in which necrosis seems to be more prominent.7, 8, 9, 10 Inhibition of Apaf-1 signaling pathway can confer neuroprotection in rodent models of neonatal hypoxia–ischemia.32 The early forced expression of miR-23b-27b in transgenic mice allowed us to examine the inhibitory roles of miR-23b-27b on neuronal apoptosis induced by hypoxia. Our qRT-PCR results demonstrated that the expression of miR-23b and miR-27b increased significantly in transgenic mice during the embryonic stage. Importantly, the expressions of miR-23b and miR-27b were increased within the physiological range. IHC staining results showed that cleaved caspase-3-positive and TUNEL staining-positive neurons were significantly reduced in transgenic pups after hypoxia treatment. Therefore, the elevation of the expression of the miR-23a/b and miR-27a/b clusters may suppress the neuronal apoptosis induced by pathological injuries.
Apaf-1 protein levels are extremely low in adult brains, and the expressions of the miR-23-27 clusters are significantly higher in adults than in embryos. To further examine the roles of the miR-23-27 clusters on neuronal apoptosis in adults, it is needed to knock out or knock down the expression of the miR-23-27 clusters in future studies. However, both miR-23a-27a and miR-23b-27b clusters suppress Apaf-1 expression. Thus, double genome loci should be knocked out if we want to completely knock out the miR-23-27 clusters. The technical difficulty of knocking out each of these miRNAs hinders our ability to perform a loss-of function study. Perhaps, the miRNA sponge technique could be used to construct a functional miRNA knockdown transgenic mouse in the future.33 With this miR-23-27 sponge transgenic mouse, the roles of the miR-23-27 clusters in adults could be further explored.
Apaf-1 protein levels were elevated, whereas Apaf-1 mRNA levels did not change significantly after hypoxia (data not shown). Interestingly, miR-23 and miR-27 expression levels were downregulated earlier after hypoxia. These results suggest that hypoxia downregulated the expression of miR-23b and miR-27b, and this downregulation relieved the suppression of Apaf-1 protein. However, the mechanism by which the expression of the miR-23-27 clusters in neurons is regulated is still unknown, and the pathway by which hypoxia downregulates the expression of the miR-23-27 clusters is also unknown. Further studies are needed to explain the mechanism by which the miR-23b-27b cluster is downregulated by hypoxia.
Taken together, our finding suggests a potential molecular mechanism underlying age-dependent differences in which the miR-23-27 clusters normally repress Apaf-1 gene expression in the nervous system during development. In addition, the acute brain injury induced by hypoxia appears to suppress the expression of miR-23-27 clusters and leads to greater neuronal apoptosis. Upregulating the expression of miR-23-27 clusters may help to protect neurons from injury-induced apoptotic cell death during development.
Materials and Methods
Animals and reagents
C57BL/6J mice (from embryos to adults) were employed in the present study. All animals were handled in accordance with the NIH Guidelines for the Care and Use of Laboratory Animals and the protocols were approved by the Animal Care Committee of Nanjing University (Nanjing, China). All of the reagents used in this study were of analytical grade or molecular biology grade.
Cell culture, transfection, and hypoxia treatment in vitro
HEK293T cells were ordered from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Dissociated cortical neurons were prepared from E14.5–E15.5 cortices of C57BL/6J mice and cultured in Neurobasal medium (Life Technologies, Grand Island, NY, USA) containing 2% B27 (v/v, Life Technologies), 1 mM L-glutamine, 100 IU/ml penicillin, and 100 μg/ml streptomycin. B27 minus AO (Life Technologies) was substituted for B27 during the exposure of cortical neurons to hypoxia. HEK293T cells were transfected using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA), following the manufacturer's manual. Primary cortical neurons were transfected at DIV3 as described previously by An et al.34 To induce hypoxia in cultured neurons, anaeroPack bags (Mitsubishi Gas Chemical Company, Inc., MGC, Tokyo, Japan) were employed; cultured neurons at DIV5 were put into the bags, and the entire bags were incubated in a CO2 incubator for 6, 12, or 24 h.
RNA isolation and quantitative RT-PCR
Total RNA was extracted from tissues and the cultured cells using Trizol reagent (Invitrogen) according to the manufacturer's instructions. For quantitative RT-PCR analysis of mRNA, 1 μg of total RNA was reverse transcribed to cDNA with oligd(T) and Thermoscript (TaKaRa, Dalian, China). Real-time PCR was performed on an Applied Biosystems 7300 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) using SYBR green dye (Roche, Mannheim, Germany). The sequences of the primers used for gene amplification were as follows: Apaf-1 (forward): 5′-GTTGATGCTGTCATTATGTAGGC-3′ Apaf-1 (reverse): 5′-AGGTAAAAGGGGAAGTATGTGTT-3′ β-actin (forward): 5′-CTGTCCCTGTATGCCTCTG-3′ and β-actin (reverse): 5′-ATGTCACGCACGATTTCC-3′. Quantitative RT-PCR of mature miRNAs was performed using TaqMan miRNA probes (Ambion, Austin, TX, USA) according to the manufacturer's instructions. β-Actin and U6 snRNA were used for normalization in gene and miRNA expression studies, respectively. A relative fold change in expression of the target gene transcript was calculated with the equation 2−ΔCT.
Plasmid construction and luciferase reporter assay
A 507-bp segment of the 3′-UTR of Apaf-1 that contained the presumed miRNAs binding sites was amplified by PCR using mouse cDNA as a template with the following using primer sets: forward, 5′-GGACTAGTCACACTAGACAGGCACTCCACCG-3′ reverse, 5′-CCCAAGCTTGTTCAGCAAGGAAAGGGCCACAA-3′ (the SpeI and HindIII restriction sites were added for cloning). The PCR products were inserted into the p-MIR-report plasmid (Ambion). To test the binding specificity, we mutated the complementary site of miR-23a/b from AATGTGA to TTACACT and the complementary site of miR-27a/b from TACTGTGA to ATGACACT. For luciferase reporter assays, HEK-293T cells were cultured in 24-well plates, and each well was transfected with 0.2 μg of firefly luciferase reporter plasmid, 0.2 μg of β-galactosidase (β-gal) expression vector (Ambion), and 20 pmol of a precursor miRNA oligo using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. The β-gal vector was used as a transfection control. At 24 h post transfection, the cells were assayed using luciferase assay kits (Promega, Madison, WI, USA). The data depicted are representative of five independent experiments performed on different days.
MiRNA in situ hybridization
For tissue sections, brains of E18 and adult mice were dissected and fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) overnight at 4°C. Next, brains were cryoprotected in 15% sucrose and 30% sucrose in PBS and cut into 20-μm-thick coronal sections on a Leica cryostat after being embedding into an OCT compound. Brain cryostat sections were fixed in 4% PFA, pretreated with proteinase K, and hybridized with digoxigenin (DIG)-labeled Locked Nucleic Acid (LNA) probes (Exiqon, Woburn, MA, USA) in a 50% formamide hybridization mix at 45°C. Slides were then washed with 0.1 × SSC at 50°C. The DIG-labeled probes were detected by alkaline phosphatase (AP)-coupled anti-DIG (1 : 1000, Roche) followed by color development (substrate: BCIP/NBT, Invitrogen). The following probes were used: a probe for miR-23b (5′-GGTAATCCCTGGCAATGTGAT-3′) and a scrambled control probe (5′-GTGTAACACGTCTATACGCCCA-3′). The images were taken by an OLYMPUS IX-71 fluorescence inverted microscope (Olympus, Tokyo, Japan).
RNA dot blot
RNA (2.0 μg) was degenerated and transferred to a nylon membrane according to the standard protocol. The membrane was crosslinked and dried. The membrane was prehybridized in DIG Easy Hyb (Roche) for 30 min at 45°C and hybridized with DIG-labeled LNA probes (Exiqon) in a DIG Easy Hyb at 45°C for 12 h according to the manufacturer's instructions. After an extensive washing, the membrane was incubated with AP-coupled anti-DIG (1 : 1000, Roche) for 1 h at room temperature and detected with CDP-Star reagent (Roche).
Western blot
Samples of tissues and cultured cells were lysed in RIPA sample buffer and centrifuged at 12 000 × g for 10 min at 4°C. The supernatant fraction was collected, and the protein concentration was determined by a BCA assay (Pierce, Rockford, IL, USA). Proteins were fractionized on SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes. The membranes were blocked for 1 h, followed by an overnight incubation at 4°C with antibodies. After washes, membranes were incubated at room temperature for 1 h with the appropriate secondary antibody conjugated to horseradish peroxidase and detected with an enhanced chemiluminescence reagent (Cell Signaling Technology Inc., Danvers, MA, USA). The intensity of each band was scanned and quantified using BandScan software (Glyko Inc., Novato, CA, USA). The following antibodies were used: anti-Apaf-1 (rabbit, 1 : 500, Abcam, ab32372, Cambridge, MA, USA) and anti-cleaved caspase-3 (rabbit, 1 : 1000, Cell Signaling Technology Inc., 9661).
IHC and immunofluorescence staining
Coronal brain sections, 20-μm-thick, or primary cultured neurons were fixed in 4% PFA, incubated with PBS containing 0.3% hydrogen peroxide for 30 min, and blocked for 1 h at room temperature. Samples were incubated with primary antibodies at 4°C overnight, followed by incubation in a biotinylated secondary antibody for 2 h at room temperature. Antibody detection was amplified with a commercial catalyzed signal amplification kit (VECTASTAIN Elite ABC Kit; Linaris, Wertheim, Germany), according to the manufacturer's instructions. Reactions were visualized with DAB (Sigma, St. Louis, MO, USA). For immunofluorescence staining, incubation of Alexa Fluor 594-conjugated secondary antibody (1 : 1000, Invitrogen) was performed in a dark room at room temperature. Nuclei were counterstained with DAPI (Sigma) for 5 min and then washed in PBS.
siRNA and Apaf-1 overexpression plasmid
The siRNAs targeting mouse Apaf-1 (5′-GAUGGUCCUUGCAGUUGACAACAUA-3′, 5′-UAUGUUGUCAACUGCAAGGACCAUC-3′) and control siRNA were purchased from Invitrogen. Plasmid overexpressing Apaf-1 (pCMV6-Apaf-1; RC213433) and control plasmid (pCMV6; PS100001) were purchased from OriGene Technologies (Rockville, MD, USA).
Hypoxia treatment in vivo
Pregnant C57BL/6J wild-type mice (mated with male miR-23b-27b transgenic mice) were exposed to systemic hypoxia at the late stage of mouse gestation (gestation day 20; i.e., E19.5 for the pup). Mice were kept in continuous hypoxia with an inspired O2 fraction of 6% for 6 h (gas mixture: 6% O2, 94% N2; Hypoxic chambers, Coy Labs, Grass Lake, MI, USA). To enable adjustment to the hypoxic environment, O2 levels were gradually decreased from an O2 fraction of 21 to 6% in 2% steps every 10 min, as described previously.35 Controls were kept in the chamber under room air. After the incubation period, neonatal brains were dissected immediately or after a period of recovery (6 or 18 h). Cerebral cortices were isolated, frozen in liquid nitrogen, and stored at −70°C until protein and RNA extraction. For the IHC and TUNEL studies, the mice were allowed to recovery after the hypoxia period (6 h for cleaved caspase-3 staining and 18 h for TUNEL staining) before brain tissues were dissected and stained.
TUNEL assay
The brains of embryos were dissected and fixed in 4% PFA. Coronal brain sections were obtained, as described above. TUNEL assays were performed with an in situ apoptosis detection kit (KeyGEN BioTECH, Nanjing, China) according to the manufacturer's instructions.
Generation of miR-23b-27b transgenic mice
A schematic diagram of the transgenic construct is shown in Figure 7a. The mouse 3050-bp Tubb3 promoter was cloned by PCR from C57BL/6J mouse genomic DNA using the following primer sets: forward, 5′-GTTATTAATGTCGACAGGATGAGCTTTAAAATAG-3′ reverse, 5′-GAAGATCTGCTGACTTCACGCGGCTAGAGACGA-3′ (the AseI and BglII restriction sites were added for cloning). The mouse 613-bp miR-23b-27b cluster was cloned by PCR from C57BL/6J mouse genomic DNA using the following primer sets: forward, 5′-GAAGATCTTTCTAGAGCTAGCGAATTCCTCTC-3′ reverse, 5′-CGGGATCCGATCGCAGATCCTTCGCGGC-3′ (the BglII and BamHI restriction sites were added for cloning). These DNA fragments were subcloned into pEGFP-C1 plasmids (Clontech, Mountain View, CA, USA). The Nanjing Biomedical Research Institute of Nanjing University (NBRI, Nanjing University, China) provided the service to inject the miR-23b-27b transgenic construct into the pronuclei of C57/BL6 zygotes and implanted into pseudopregnant recipient females. Mice were genotyped by PCR using primers for miR-23b-27b cluster transgenic mouse (forward 5′-ATCTCGGTGCCGGTGCTGATGCT-3′ reverse 5′-GTCCGATTAGTGGATGTTTCTGTGG -3′).
Statistical analysis
Data are presented as mean±S.E.M. of at least three independent experiments. Direct comparisons were made using Student's t-tests and multiple group comparisons were made using one-way analyses of variance (ANOVA). Statistical significance was defined as P<0.05, 0.01, or 0.001 (indicated as *, **, or ***, respectively). The P-values of ≥0.05 were considered not significant (indicated as NS). Prism software 5.0 (GraphPad, Inc., La Jolla, CA, USA) was used for data analyses.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31100777 and 81170309 to Qipeng Zhang; 31000478 to Liang Li; 30988003 and 30871019 to Ke Zen; and 90608010 to Chen-Yu Zhang).
Author contributions
Qun Chen, Qipeng Zhang, Jie Xu, Hanqin Li, Liang Li, Susu Mao, and Fan Zhang planned and performed the experiments; Qun Chen and Qipeng Zhang analyzed the data and wrote the manuscript; Chenyu Zhang, Ke Zen, and Qipeng Zhang designed the experiments and revised the manuscript.
Glossary
- Apaf-1
apoptotic protease activating factor-1
- IHC
immunohistochemistry
- miRNA
microRNA
- Tubb3
tubulin βIII
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by D Bano
Supplementary Material
References
- Nijhawan D, Honarpour N, Wang X. Apoptosis in neural development and disease. Annu Rev Neurosci. 2000;23:73–87. doi: 10.1146/annurev.neuro.23.1.73. [DOI] [PubMed] [Google Scholar]
- Kuida K, Zheng TS, Na S, Kuan C, Yang D, Karasuyama H, et al. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature. 1996;384:368–372. doi: 10.1038/384368a0. [DOI] [PubMed] [Google Scholar]
- Hakem R, Hakem A, Duncan GS, Henderson JT, Woo M, Soengas MS, et al. Differential requirement for caspase 9 in apoptotic pathways in vivo. Cell. 1998;94:339–352. doi: 10.1016/s0092-8674(00)81477-4. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Kong YY, Yoshida R, Elia AJ, Hakem A, Hakem R, et al. Apaf1 is required for mitochondrial pathways of apoptosis and brain development. Cell. 1998;94:739–750. doi: 10.1016/s0092-8674(00)81733-x. [DOI] [PubMed] [Google Scholar]
- Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- Volpe JJ. Perinatal brain injury: from pathogenesis to neuroprotection. Ment Retard Dev Disabil Res Rev. 2001;7:56–64. doi: 10.1002/1098-2779(200102)7:1<56::AID-MRDD1008>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- McLean C, Ferriero D. Mechanisms of hypoxic-ischemic injury in the term infant. Semin Perinatol. 2004;28:425–432. doi: 10.1053/j.semperi.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Hu BR, Liu CL, Ouyang Y, Blomgren K, Siesjo BK. Involvement of caspase-3 in cell death after hypoxia-ischemia declines during brain maturation. J Cereb Blood Flow Metab. 2000;20:1294–1300. doi: 10.1097/00004647-200009000-00003. [DOI] [PubMed] [Google Scholar]
- Pulera MR, Adams LM, Liu H, Santos DG, Nishimura RN, Yang F, et al. Apoptosis in a neonatal rat model of cerebral hypoxia-ischemia. Stroke. 1998;29:2622–2630. doi: 10.1161/01.str.29.12.2622. [DOI] [PubMed] [Google Scholar]
- Sidhu RS, Tuor UI, Del Bigio MR. Nuclear condensation and fragmentation following cerebral hypoxia-ischemia occurs more frequently in immature than older rats. Neurosci Lett. 1997;223:129–132. doi: 10.1016/s0304-3940(97)13426-7. [DOI] [PubMed] [Google Scholar]
- Yakovlev AG, Ota K, Wang G, Movsesyan V, Bao WL, Yoshihara K, et al. Differential expression of apoptotic protease-activating factor-1 and caspase-3 genes and susceptibility to apoptosis during brain development and after traumatic brain injury. J Neurosci. 2001;21:7439–7446. doi: 10.1523/JNEUROSCI.21-19-07439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Cecconi F, Alvarez-Bolado G, Meyer BI, Roth KA, Gruss P. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell. 1998;94:727–737. doi: 10.1016/s0092-8674(00)81732-8. [DOI] [PubMed] [Google Scholar]
- Johnson CE, Huang YY, Parrish AB, Smith MI, Vaughn AE, Zhang Q, et al. Differential Apaf-1 levels allow cytochrome c to induce apoptosis in brain tumors but not in normal neural tissues. Proc Natl Acad Sci USA. 2007;104:20820–20825. doi: 10.1073/pnas.0709101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhila J, Sipila T, Icay K, Nicorici D, Ellonen P, Kallio A, et al. MicroRNA expression profiling reveals miRNA families regulating specific biological pathways in mouse frontal cortex and hippocampus. PLoS One. 2011;6:e21495. doi: 10.1371/journal.pone.0021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Jin P. Roles of small regulatory RNAs in determining neuronal identity. Nat Rev Neurosci. 2010;11:329–338. doi: 10.1038/nrn2739. [DOI] [PubMed] [Google Scholar]
- Sun AX, Crabtree GR, Yoo AS. MicroRNAs: regulators of neuronal fate. Curr Opin Cell Biol. 2013;25:215–221. doi: 10.1016/j.ceb.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Dong LY, Li YJ, Hong Z, Wei WS. miR-21 represses FasL in microglia and protects against microglia-mediated neuronal cell death following hypoxia/ischemia. Glia. 2012;60:1888–1895. doi: 10.1002/glia.22404. [DOI] [PubMed] [Google Scholar]
- Doeppner TR, Doehring M, Bretschneider E, Zechariah A, Kaltwasser B, Muller B, et al. MicroRNA-124 protects against focal cerebral ischemia via mechanisms involving Usp14-dependent REST degradation. Acta Neuropathol. 2013;126:251–265. doi: 10.1007/s00401-013-1142-5. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydar TF, Kuan CY, Flavell RA, Rakic P. The role of cell death in regulating the size and shape of the mammalian forebrain. Cereb Cortex. 1999;9:621–626. doi: 10.1093/cercor/9.6.621. [DOI] [PubMed] [Google Scholar]
- Northington FJ, Chavez-Valdez R, Martin LJ. Neuronal cell death in neonatal hypoxia-ischemia. Ann Neurol. 2011;69:743–758. doi: 10.1002/ana.22419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Geisert EE, Frankfurter A, Spano AJ, Jiang CX, Yue J, et al. A transgenic mouse class-III beta tubulin reporter using yellow fluorescent protein. Genesis. 2007;45:560–569. doi: 10.1002/dvg.20325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian S, Shi R, Bai T, Liu Y, Miao W, Wang H, et al. Anti-miRNA-23a oligonucleotide suppresses glioma cells growth by targeting apoptotic protease activating factor-1. Curr Pharm Des. 2013;19:6382–6389. doi: 10.2174/13816128113199990509. [DOI] [PubMed] [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Liang W, Hu X, Zhang W, Stetler RA, Vosler P, et al. Neuroprotection against hypoxic-ischemic brain injury by inhibiting the apoptotic protease activating factor-1 pathway. Stroke. 2009;41:166–172. doi: 10.1161/STROKEAHA.109.561852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trollmann R, Strasser K, Keller S, Antoniou X, Grenacher B, Ogunshola OO, et al. Placental HIFs as markers of cerebral hypoxic distress in fetal mice. Am J Physiol Regul Integr Comp Physiol. 2008;295:R1973–R1981. doi: 10.1152/ajpregu.00053.2008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.