Abstract
Mutation of p53 is the most common genetic change in human cancer, causing complex effects including not only loss of wild-type function but also gain of novel oncogenic functions (GOF). It is increasingly likely that p53-hotspot mutations may confer different types and magnitudes of GOF, but the evidences are mainly supported by cellular and transgenic animal models. Here we combine large-scale cancer genomic data to characterize the prognostic significance of different p53 mutations in human cancers. Unexpectedly, only mutations on the Arg248 and Arg282 positions displayed significant association with shorter patient survival, but such association was not evident for other hotspot GOF mutations. Gene set enrichment analysis on these mutations revealed higher activity of drug-metabolizing enzymes, including the CYP3A4 cytochrome P450. Ectopic expression of p53 mutant R282W in H1299 and SaOS2 cells significantly upregulated CYP3A4 mRNA and protein levels, and cancer cell lines bearing mortality-associated p53 mutations display higher CYP3A4 expression and resistance to several CYP3A4-metabolized chemotherapeutic drugs. Our results suggest that p53 mutations have unequal GOF activities in human cancers, and future evaluation of p53 as a cancer biomarker should consider which mutation is present in the tumor, rather than having comparison between wild-type and mutant genotypes.
Keywords: p53, mutation, gain-of-function, prognosis, cancer, CYP3A4
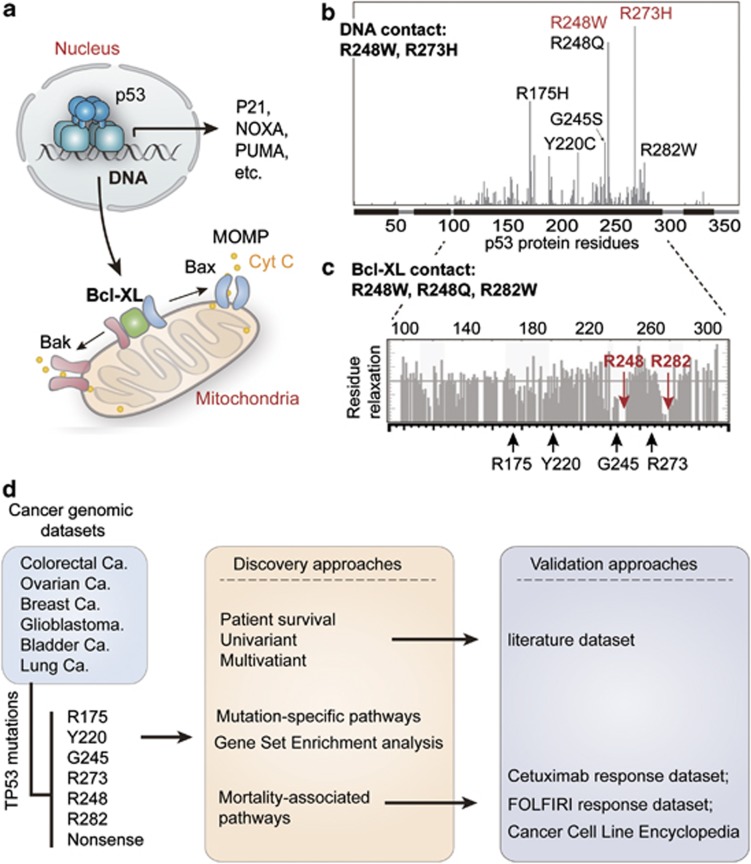
The TP53 gene, which resides on chromosome 17p13.1 and encodes the p53 protein, is the most frequent target for mutation in human cancers, with over 50% of all observed tumors exhibiting mutation at this locus.1, 2 The tumor suppressive functions of wild-type p53 are often explained by its transcriptional functions in the nucleus.3 In response to oncogenic mutations or DNA damage, p53 triggers a transcriptional program of cell cycle arrest, DNA repair, senescence, apoptosis4, 5 and metabolic remodeling6 (Figure 1a). The vast majority (95%) of p53 mutations in human cancers are missense mutations, sitting within the DNA-binding domain (amino acids 102–292) with hot spots at codons R175, Y220, G245, R248, R249, R273 and R2827 (illustrated in Figure 1b). A well-established classification system for p53 mutation is based on its roles as a transcriptional factor. Mutations that occur in the p53-DNA-binding surface are defined as contact mutations (R273H and R248W), whereas mutations that cause conformational instability of the p53 protein are classified as a structural mutations (R175H, Y220C, G245S, R248Q, R249S, R282W, etc)8 (illustrated in Figure 1b).
Figure 1.
Classification of p53 mutations and study design. (a) The function of p53 in nucleus and mitochondria. In response to DNA damage, p53 functions as a transcription factor, triggering cell cycle arrest and apoptosis by transactivation of p21, Noxa, Puma, so on. In addition, the function of p53 in mitochondria is mediated by its interaction with Bcl-XL, which releases Bak/Bax and causes mitochondrial outer membrane permeabilization (MOMP)-induced leakage of cytochrome c (cyt c) and apoptosis. (b) Frequently-occuring (hotspot) p53 mutations and classification of p53 mutations according to its effects on DNA contact and protein structural stability. The x-axis indicates amino acid sequence of p53 protein, and the y-axis displays frequency of mutation in the TCGA data sets. DNA contact mutations are labeled in red, whereas structural mutations are marked in black. (c) The position of hotspot mutations in relation to the regions of p53 that contact Bcl-XL. The gray bar plot indicates relaxation of p53 residues while in contact with Bcl-XL (higher relaxation indicates noncontact residues) as determined by nuclear magnetic resonance previously.11 The mutated residues sitting in the contact regions (R248 and R282) are highlighted in red. (d) Schematic diagram representing the study processes. Cancer cases collected from the indicated data sets of The Cancer Genome Atlas (TCGA) were grouped according to p53 mutations. The patient survival was compared using univariant and multivariant models, and then validated using an independent data set extracted from literatures. The enriched (signature) pathways for different mutations were determined using microarray data of TCGA colorectal cancer data set. Finally, the effects of p53 mutation and expression of CYP3A4 in response to FOLFIRI and cetuximab were analyzed using the indicated data sets
In addition to its transcription-dependent functions, the p53 protein has also been found to function in mitochondria,9 where it binds to Bcl-XL and induces mitochondrial outer membrane permeabilization (MOMP) and apoptosis.10 According to a p53–Bcl-XL complex model determined by NMR chemical shift perturbation, the p53-hotspot mutations R248W, R248Q and R282W sit in the binding surface to Bcl-XL11, 12 (Figure 1c). It is currently unknown if these ‘Bcl-XL contact' mutations confer any specific cancerous phenotypes.
It is becoming increasingly evident that mutant p53 proteins not only lose their tumor-suppressor function but some acquire oncogenic gain-of-function (GOF).1, 7, 13, 14 The most compelling support for GOF comes from mice engineered to harbor some of the hot spot tumor-associated p53 mutations. When compared with heterozygous or null (p53+/− or p53−/−) mice, animals with one mutant allele equivalent to human R175H or R273H showed more spontaneous carcinomas, sarcomas and lymphomas. Although these mutants increased tumor metastasis, they do not shorten mouse survival time as compared with p53-null background.15, 16 It was also found that R248Q mutation could accelerate onset of all tumor types and shorten the survival of mice,17 thus suggesting that hotspot mutants may confer different type and magnitude of GOF effects. A recent in-depth study demonstrated that R273H mutant gains new function by activation of EGFR signaling via suppressing mir-27a expression.18 It is of great importance to clarify the prognostic significance of p53 mutations, as genes that display oncogenic activities may present useful information for cancer prognosis.19 Presumably owing to the insufficient number of cases per mutation, previous studies have focused on the comparisons between wild-type and mutant p53,20 or between large categories of p53 mutants.21, 22 To our knowledge, no existing empirical research addresses the effects of individual p53 mutations on human cancers.
In this study, we combine large-scale cancer genomic data to characterize the effects of each hotspot p53 mutation on human cancer prognosis. The potential pathways that may mediate drug resistance and cancer mortality were also analyzed to provide insight into the unequal mutant GOF effects in human cancers.
Results
Mutations on Arg282 and Arg248 residues associate with shorter patient survival
To assess the effects of different p53 mutations in cancer outcome, we firstly performed survival analysis on the MSKCC bladder cancer data set (JCO, 2013) in combination with The Cancer Genome Atlas (TCGA) patient cohorts that had higher frequency of p53 mutation and adequate patient outcome data. These include breast invasive carcinoma,23 colorectal cancer,24 glioblastoma,25 lung squamous cell carcinoma26 and ovarian serous cystadenocarcinoma23 (analysis flowchart shown in Figure 1d). These data sets include a total number of 2916 cases, and the mutation spectrum of TP53 gene was similar to that reported previously.27 The overall survival was compared between patient groups with different p53 status, namely nonsense (loss-of-function (LOF)) mutation group and hotspot (GOF) mutation group. To analyze if mutant p53 aggregation28 or abolished contact with DNA is associated with patient outcome, the patients were also classified according to the type of p53 mutations.
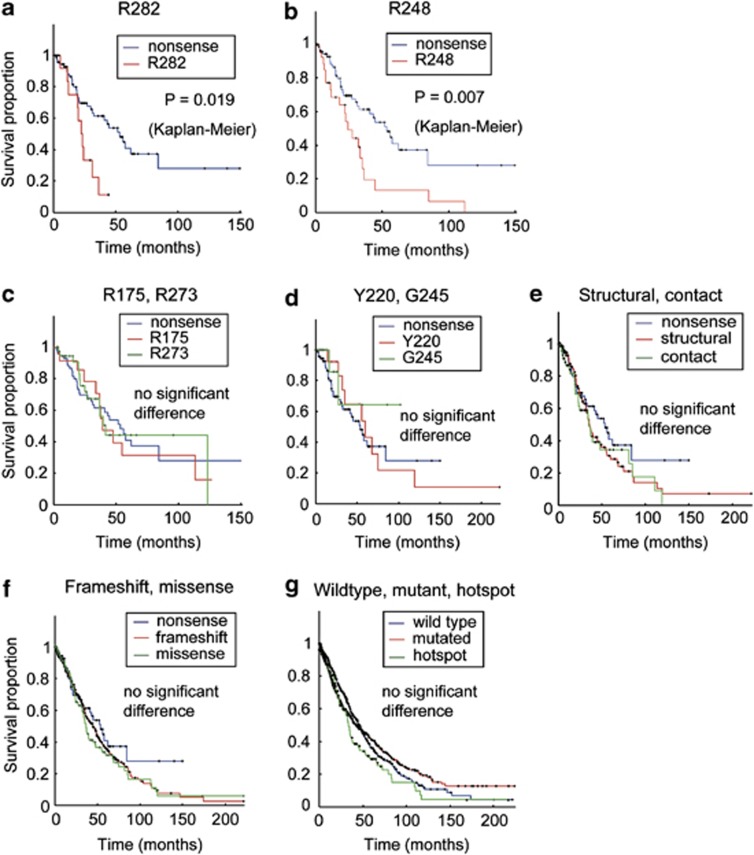
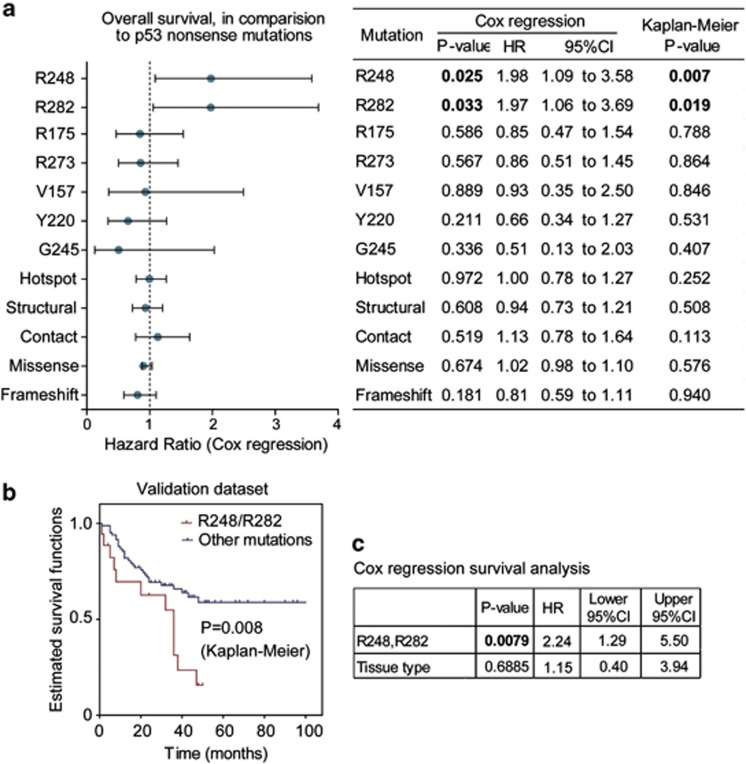
Kaplan–Meier survival analysis revealed that patients carrying p53 mutations on Arg248 and Arg282 residues had significantly shorter overall survival time than those carrying nonsense mutations (Figures 2a and b). Other frequently-occurring mutations at residues Y220, G245, R175 and R273 (Figures 2c and d) showed similar survival curves as nonsense mutations, and very few cases with the R249S mutation are available for further analysis. No difference in survival was found between structural, contact and nonsense mutations (Figure 2e). The frameshift, nonsense and missense mutations displayed comparable survival curves (Figure 2f), and even the comparison between wild-type and mutant p53 showed no significant difference in patient survival (Figure 2g). In the multivariate (Cox regression) analysis adjusting for cancer types, mutations on R282 and R248 residues showed ∼two fold higher hazard ratios than that of the nonsense mutations (Figure 3a).
Figure 2.
Univariate analysis on the effects of p53 mutations on cancer patient survival. Each hotspot mutation was compared with nonsense mutation using Kaplan–Meier survival analysis, and the P-values are indicated on the graph. Significantly shorter survival was found in patients baring p53 mutations on residues R282 (a) and R248 (b), but not on R175, R273 (c), Y220, and G245 (d). No significant difference was found between nonsense/structural/contact mutations (e), or between nonsense/frameshift/missense mutations (f). The wild-type and mutated p53 (including hotspots) showed no significant difference for overall patient survival (g)
Figure 3.
Multivariate survival analyses on the association between p53 mutations and patient survival. (a) The overall survival time of patients carrying indicated p53 mutations were respectively compared with those bearing nonsense mutations using Cox regression (including cancer type as covariant). The hazard ratios for different mutation types are plotted (round dots) with 95% confidence intervals. After adjustment for the factor of cancer types using Cox regression, the mutations on R282 and R248 positions showed ∼two fold higher hazard ratios (HR) than that of nonsense mtuations. The P-values for univariate analysis are also listed on the right (significant P-values shown in bold font). (b) Univariate (Kaplan–Meier) analysis on p53 mutant effects using independent validation data set. The detailed data and sources are shown in Supplementary Table 1. The overall survival curves of patients carrying R248Q/R248W/R282W mutations (plot in red) were compared with those bearing other mutations (blue plot). The P-value is indicated on the plot. (c) Multivariate (Cox regression) analysis on p53 mutant effects. After adjustment for cancer (tissue) types, the mutations on R248 or R282 positions showed significantly shorter survival (P=0.0079, hazard ratio =2.24)
Validation of mutant effects using an independent data set
We tested the association between R248/R282 mutations and cancer patient survival using an independent data set extracted from published literatures (data in Supplementary Table 1), and found these mutations are indeed associated with shorter patient survival (Figure 3b). Multivariate survival analysis adjusting cancer types also confirmed this finding (Figure 3c). In fact, mice carrying humanized mutation on R248 (but not R175 or R273) were also found with significantly shorter survival time,29 suggesting that frequently-occurring p53 mutations display unequal GOF effects in both human and mouse. Although the structural/contact classifications seem to be uncorrelated to patient survival, all the mortality-associated mutations are located in p53's binding surface to Bcl-XL (shown in Figure 1c), suggesting potential relevance of p53 mitochondrial apoptotic functions with cancer patient survival.
R248 and R282 mutations associate with drug metabolism enzymes
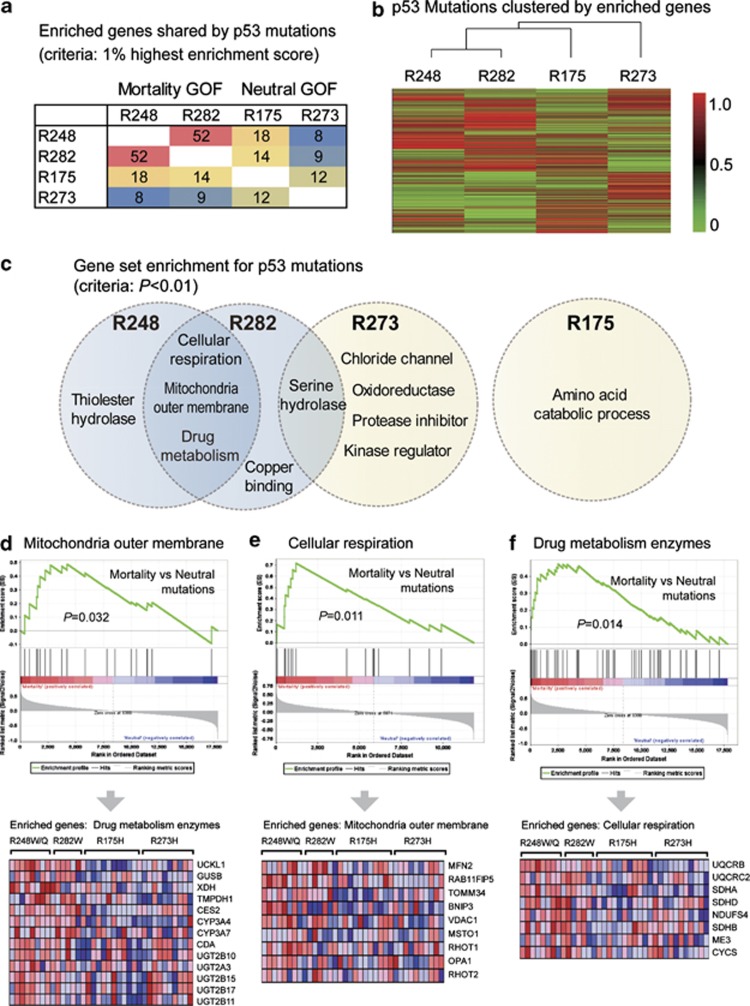
To probe the characteristic pathways associated with different p53-hotspot mutations, we selected the colorectal cancer (CRC) data set in TCGA as an example. The CRC cases contain more mortality-associated mutations (R248Q/W and R282W) and two intensively studied mutations (R175H and R273H), and thus were more suitable for further analysis. We performed Gene Set Enrichment Analysis (GSEA) to identify the specifically upregulated genes/pathways for each mutation as compared with nonsense mutations. When genes with top 1% enrichment scores were regarded as enriched genes (n=177, listed in Supplementary Table 2), R248W and R282W shared 52 commonly enriched genes. However, such similarity was not found between other mutations (Figure 4a). When mutations were classified according to their gene enrichment profiles, R248W and R282W fell into the same subgroup (Figure 4b). The mortality-associated mutations were both enriched in gene sets linked to mitochondria outer membrane, cellular respiration and drug metabolism enzymes (Figures 4c–f). The R273H mutation shared serine–hydrolase pathway with R282W, but R175H did not show significant similarity with other mutations (Figure 4c).
Figure 4.
Characteristic pathways of mortality-associated p53 mutations. (a) The number of enriched genes shared by different mutations. The colorectal cancer cases in TCGA were grouped according to p53 mutations, and Gene Set Enrichment Analysis (GSEA) was performed to identify upregulated genes/pathways for each mutation. The genes ranked as top 1% by enrichment scores were regarded as enriched genes (n=177), and the numbers of commonly enriched genes between the two mutations are shown. The mortality-associated mutations R248W and R282W shared 52 enriched genes, but such similarity is not found between other mutations. (b) Mutations are clustered according to their enrichment scores for each gene (enrichment profiles). Mortality-associated mutations show similar gene enrichment profiles and are clustered into one subgroup. (c) Enriched gene sets for different p53 mutations. Mortality-associated mutations (on R248 and R282 positions) shared the following enriched gene sets: (1) Mitochondrial outer membrane; (2) Cellular respiration; (3) Drug metabolism enzymes. Low similarity was found between other mutations. (d) GSEA plot (upper panel) and the expression of genes in the set ‘mitochondria outer membrane' (lower panel). (e) GSEA plot and gene expression in the pathway ‘cellular respiration'. (f) GSEA plot and expression of ‘drug metabolism enzymes' gene set
Mortality-associated P53 mutations induce CYP3A4 upregulation
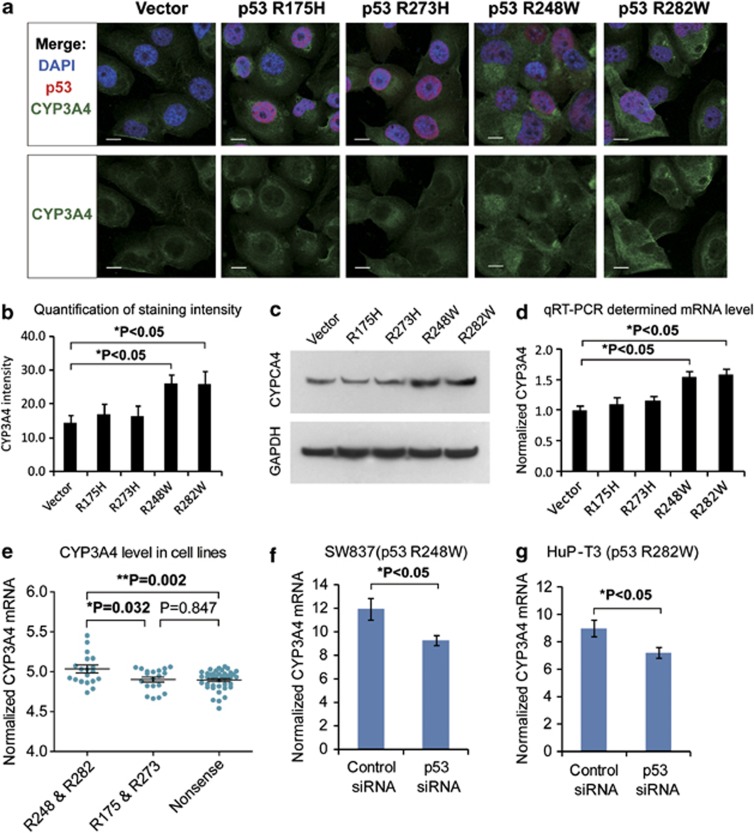
The GSEA analysis revealed that most of the drug metabolism enzymes in association with p53 R248/R282 mutations are responsible for the clearance of chemotherapeutic drugs. Among these, the most relevant one was cytochrome p450 (CYP3A4) , which is the main metabolizing enzyme for FOLFIRI (folinic acid/fluorouracil/irinotecan) regimen, etoposide, rapamycin and other antineoplastic drugs (Supplementary Table 3). We tested the effects of mutant p53 on CYP3A4 by ectopic expression of p53 mutants R175H, R273H and R282W in the H1299 cells (p53-null). Immunofluorescence and Western blot studies suggested that p53 R248 and R282 mutations induced higher expression of CYP3A4 protein (Figures 5a–c, full gel image for Wb shown in Supplementary Figure 1 online). Consistently, real-time quantitative reverse transcription PCR (qRT-PCR) detected increased expression of CYP3A4 mRNA in H1299 cells expressing the mortality-associated mutants (Figure 5d). When p53 mutants were expressed in another p53-null cell line SaOS2, increased CYP3A4 mRNA and protein levels were also observed for R248 and R282 mutations (Supplementary Figure S2). Further, we tested the association between p53 mutation and CYP3A4 expression in the Cancer Cell Line Encyclopedia (CCLE) data set, which contains drug response data for 639 human cancer cell lines with known genotypes.30 As expected, cancer cells bearing mortality-associated p53 mutations (R248, R282) displayed significantly higher level of CYP3A4 when compared with neutral GOF mutations (R175, R273) and nonsense LOF mutations (Figure 5e, data set in Supplementary Table 4). When the endogenous mutant p53 in SW837 (carrying p53 R248W) and HuP-T3 (p53 R282W) cells were suppressed by specific small interfering RNAs (siRNAs), the expression level of CYP3A4 also significantly decreased (Figures 5f and g). These results collectively demonstrate that mutant p53 R248W and R282W could induce the expression of CYP3A4 enzyme.
Figure 5.
Effects of p53 mutations on CYP3A4 expression. (a) Different p53 mutations were ectopically expressed in human H1299 cells (p53-null), and immunofluorescence was used to detect p53 (in red) and CYP3A4 (green). Cell nuclei were stained by DAPI in blue. Scale bars indicate 10 μm. (b) Statistical analysis of CYP3A4 fluorescence in cells expressing different p53 mutants. Data represent means±S.D. of four biological repeats. (c) Western Blot showing the level of CYP3A4 in H1299 cells expressing the indicated p53 mutants. Higher level of CYP3A4 protein expression was found in cells expressing p53 R248W and R282W mutations. (d) Quantitative RT-PCR detection of CYP3A4 mRNA expression in cells stably expressing different p53 mutations as indicated (data represent means±S.D., P<0.05, two-sided t-test). (e) CYP3A4 expression in cancer cells bearing different p53 mutations. The expression level of CYP3A4 was extracted using microarray data of Cancer Cell line Encyclopedia (CCLE) database. Then, the level of CYP3A4 was compared between cell bearing p53 mortality-associated mutations (on R248 or R282) or other mutations (on R175 R273) using Mann–Whitney test (P-value indicated). (f) The mRNA level of CYP3A4 in SW837 cells (bearing p53 mutation R248W) was measured by qRT-PCR. The CYP3A4 expression level decreased after mutant p53 was knocked down using specific siRNAs against p53 (data represent means±S.D., P<0.05, two-sided t-test). (g) The CYP3A4 expression level in HuP-T3 cells (carrying p53 mutation R282W) was determined by qRT-PCR. After p53 was suppressed by specific siRNAs, the CYP3A4 expression level also decreased (data represent means±S.D., P<0.05, two-sided t-test)
Cyp3a4 may mediate chemoresistance of P53 mortality mutations
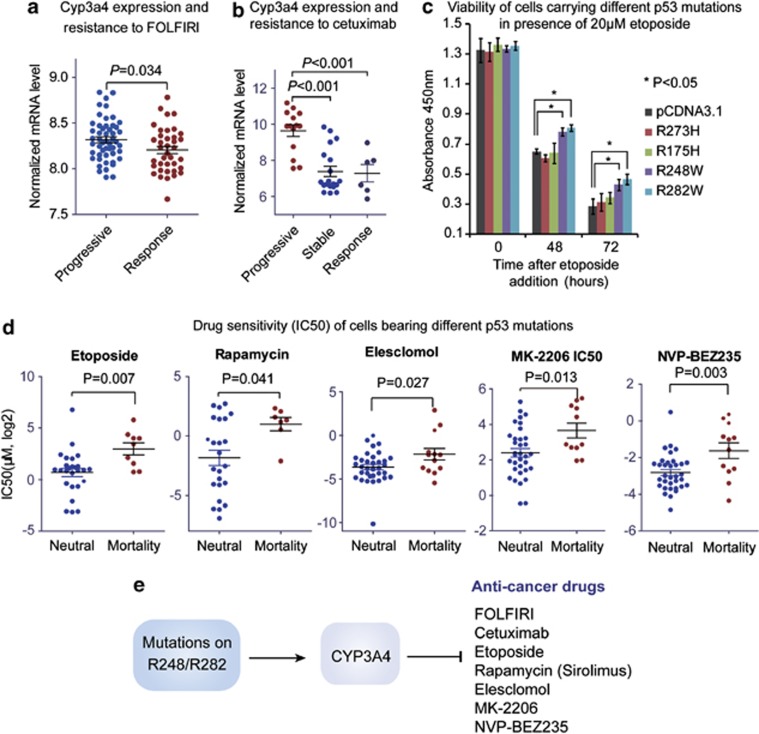
To clarify the relationship between CYP3A4 expression and resistance to FOLFIRI and cetuximab, we compared the expression levels of CYP3A4 in CRCs displaying different therapeutic responses (GEO data sets GSE13294 and GSE5851). In this context, higher level of CYP3A4 was found in nonresponse groups than in response groups treated by both drugs (Figures 6a and b). These results suggest that higher CYP3A4 level may contribute to the chemoresistance associated with p53 mortality mutations. To test the effects of p53 GOF mutations on cell response to chemotherapy, 20μM etoposide was added to H1299 cells stably expressing p53 mutations (R175H, R273H, R248W or R282W) or the control vector. As a result, cells bearing R248W and R282W mutations showed higher viability than the control group after incubation with etoposide for 48 h (Figure 6c). In support of these findings, cancer cells bearing mortality-associated mutations displayed increased inhibitory concentration (IC50) for etoposide, rapamycin, elesclomol, MK-2206 and NVP-BEZ235 (Figure 6d, Supplementary Table 5). Notably, all of the above drugs are substrates of CYP3A4, supporting our notion that CYP3A4 may have important roles in p53 mortality mutation-associated drug resistance (illustrated in Figure 6e).
Figure 6.
Mutant p53 confer chemoresistance by upregulating CYP3A4. (a) CYP3A4 is upregulated in CRC cases showing resistance to FOLFIRI regimen. The mRNA expression level of CYP3A4 was extracted from microarray data (GEO data set GSE13294), and subsequently compared between FOLFIRI-response cases and progressive cases using Mann–Whitney test (P-value indicated). (b) CYP3A4 is upregulated in CRC cases showing resistance to cetuximab. The mRNA expression level of CYP3A4 was obtained from microarray data (GEO data set GSE5851), and then compared between cetuximab-response, stable and progressive CRC cases using Mann–Whitney test (P-value indicated). (c) The effects of different p53 mutations on cell response to etoposide were determined by CCK-8 viability assay. The H1299 cells stably expressing p53 mutants were incubated with 20 μM etoposide, and cell viability was determined by CCK-8 assay. The x-axis indicates the time after etoposide addition, while the y-axis shows the readout of CCK-8 (absorbance at 450nm). Data represent mean± S.D. (*P<0.05, two-sided t-test). (d) Cancer cells bearing p53 mortality-associated mutations display resistance to multiple drugs metabolized by CYP3A4. The IC50 data for different drugs were obtained from CCLE data set and compared between cell lines carrying p53 mortality-associated mutations and other mutations (on R175 and R273) using Mann–Whitney test (P-value indicated). (e) Schematic diagram showing the proposed model for mortality-associated p53 mutations. The p53 GOF mutations on R248 and R282 induce the upregulation of CYP3A4, which in turn metabolizes most chemotherapeutic drugs
Discussion
Accumulating evidences suggest that p53 mutants display diverse oncogenic functions, and therefore should be considered as different proteins.1, 31 However, most existing evidences are based on cellular and transgenic animal models, and strong support from human cancers is awaited. By characterizing the effects of hotspot p53 mutations on cancer patient survival, we present the first evidence that p53 missense mutations display different patterns and strengths of GOF activity in human cancers.
Hotspot p53 mutations on two sites, namely R282 and R248 are associated with significantly shorter patient survival when compared with nonsense mutations. Previous studies have demonstrated that mice carrying mutations equivalent to human R175H and R273H showed no difference in survival time as compared with null mutation, although a broader tumor spectrum was observed.16 A recent study using humanized p53 mutant knock-in mice provided evidences that the R248Q mutation could significantly decrease survival time, but this effect was not found for G245S mutation.29 Our findings in human cancers are consistent with these results obtained from transgenic mouse models. To our knowledge, the GOF activity of R282W has not been characterized previously in mice.
Although each mutant p53 displayed unique set of enriched genes, a higher degree of similarity has been found between mortality-associated mutations. Importantly, we found that the p53 mutations on R248 and R282 could induce the expression of CYP3A4, which is one of the most important enzymes involved in the metabolism of chemotherapeutic drugs in the human body. Consistently, cancer cells bearing p53 R248/R282 mutations displayed resistance to multiple CYP3A4-metabolized antineuplastic drugs. These findings provide mechanistic insight into the mortality effect of a subset of p53 mutants.
In summary, we provide the first evidence for the unequal GOF effects of p53-hotspot mutations in human cancers, and probed the potential mechanisms by which R248/R282 mutations may lead to chemoresistance and mortality. These findings add an important piece of evidence to GOF effects of the p53 mutant, and suggest that p53 mutations have unequal prognostic significance in human cancers.
Materials and Methods
Plasmids construction
The pcDNA3-HA-p53 expression construct was constructed by inserting PCR-amplified cDNA sequences into pcDNA3 vector (Invitrogen, Carlsbad, CA, USA). All p53 mutations (R175H, R273H and R282W) were generated by site-directed mutagenesis PCR reaction using platinum PWO SuperYield DNA polymerase (Roche, Basel, Switzerland) according to the product manual. All plasmids were sequenced to confirm if the designed mutation is present, without any other unwanted mutation.
Cell culture and transfection
The human H1299 cells were maintained in Dmem medium (Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (Invitrogen) and cultured in a humidified incubator at 37 °C under 5% CO2. For transfection, cells were seeded into normal growth medium at 50% confluence in six-well tissue plates 24 h before transfection, and then transfected using FuGENE HD (Promega, Fitchburg, WI, USA) according to the product manual. Briefely, the transfection complex was made by 1 μg plasmid, 3 μl FuGENE HD and 100 μl media. Six hours after the complex was added to the cells, normal culture media was used to culture cells for additional 48 h, followed by gene expression analysis. siRNA for p53 has the sequence of 5′-GACUCCAGUGGUAAUCUAC-3′. Stable clones expressing different p53 mutations were selected with 600 μg/ml G418 for 12 weeks, and the expression of p53 was confirmed by immunofluorescence.
Quantitative real-time PCR analysis
Total RNA was extracted from H1299 cells using TRIzol reagent (Invitrogen). Reverse transcription was done using the First Strand cDNA Synthesis Kit (Takara, Shiga, Japan), according to the manufacturer's protocol. Quantitative PCR was performed using an ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) with SYBR Premix Ex Taq II (Takara). Quantification was calculated using the 2−ΔΔCT method and is presented as fold change. The expression of each target gene was normalized to that of 18S RNA. Primer sequences were: 18S (forward, 5′-CGGACAGGAT T GACAGATTGATAGC-3′, reverse, 5′-TGCC AGAG TCTCGTTCGTTATCG-3′); CYP3A4 (forward, 5′-AAGTCGCCTCGAAGATACACA-3′, reverse 5′-AAGGAGAGAACACTGCTCGTG-3′).
Immunofluorescence
The immunofluorescence assay was performed using a method described previously.32 The H1299 cells were plated into four-well chamber slides and transfected with different p53 mutations or the control vector. After stable expression clones were constructed, cells were fixed with 4% formaldehyde, permeabilized with 0.2% Triton X-100 and blocked in 3% BSA in PBS. The antibodies specific for CYP3A4 (Rabbit polyclonal, Santa Cruz, Dallas, TX, USA) or p53 (DO-1 mouse monoclonal, Santa Cruz) were used to label CYP3A4 and p53, respectively. Secondary antibodies (Alexa488-anti-rabbit and Alexa546-anti-mouse) were used to label CYP3A4, and p53, respectively. After staining with DAPI (1 : 10 000), the coverslips were added with antifade reagent (ProLong Gold, Invitrogen) and kept in the dark for 24 h. Images were acquired with a confocal fluorescence microscope (LSM710, Carl Zeiss, Oberkochen, Germany) using the same parameters: Lasers, 2.0% Pinhole, 60 μm; Gain, 405 nm, × 500; 488 nm, × 700; 543 nm, × 700; Digital Gain, × 1.0; Image averaging, × 4. Staining intensity of CYP3A4 was quantified using ZEN 2013 software package (Carl Zeiss) according to product manual.
Western blotting analysis
The methods for Western Blot have been described previously.32 Briefly, cells were harvested at 48 h after transfection, washed, and lysed with lysis buffer. Appropriate amounts of protein (30–50 μg) were resolved by electrophoresis in 8–12% Tris-glycine polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were blocked and then incubated for 1 h at room temperature with the appropriate primary antibody at dilutions specified by the manufacturer. They were next washed three times in 15 ml Tris-buffered saline with Triton (TBST) and incubated with the corresponding horseradish peroxidase-conjugated secondary antibody at 1 : 1000 dilution in TBST for 1 h. The membranes were washed three times for 5 min each with 15 ml TBST, then the bound secondary antibody was detected using an enhanced chemiluminescence system (Pierce Biotechnology, Rockford, IL, USA).
Cell viability assay
The protocol for cell viability assay has been described previously.33 Briefly, the MGC803 cells transfected with OCT1 vector, OCT1 vector+synbindin siRNA and control vector were seeded onto 96-well plates at 2500cells/well. Cell proliferation was measured using the Cell Counting Kit-8 (CCK-8, Dojindo, Kumamoto, Japan). At each time point, cells were incubated with 10 ml CCK-8 reagent per well (100 ml medium/well) for 30 min at 37 °C, 5% CO2. The absorbance was measured at 450 nm.
Cancer genomic and survival data
The genomic sequencing, gene expression profile and survival data analyzed in this study include MSKCC bladder cancer data set34 and all unrestricted data from TCGA, namely breast invasive carcinoma,23 CRC,24 glioblastoma,25 lung squamous cell carcinoma26 and ovarian serous cystadenocarcinoma.23 The published versions of these data sets include 2916 cancer cases with sequencing data and survival information.
Drug sensitivity profile of cells
We obtained CCLE data that include the sensitivity profiles of 639 human cancer cell lines to 131 anticancer drugs.30 The half maximal IC50 was used as a measure of the effectiveness of a drug on the cell lines. The mutation spectrum of TP53 in these cell lines, as well as the expression level of CYP3A4, were obtained from CCLE database.
Survival analysis
Survival analysis was carried out in R program using the ‘survival' package as described previously.35 In the Kaplan–Meier (log rank) survival test and Cox regression models, the censored status is indicated when the patient was still alive (or cancer-free) at the time of follow-up. The Cox regression model included cancer type as a covariant, and the P-value for mutation type is calculated after adjustment for the factor of cancer type. The hazard ratios and 95% confidence intervals were also determined for each mutation. The effects of different p53 mutations were compared with nonsense mutations as an indication of GOF effect.
GSEA
The GSEA package36 was used to determine if the members of a given gene set were generally associated with certain p53-hotspot mutations, and was therefore performed on all genes on the HG-U133 Plus 2 chip ranked by enrichment score from most negative to most positive. The maximum gene set size was fixed at 1500 genes, and the minimum size fixed at 15 genes. Thousand random sample permutations were carried out, and the significance threshold was set at FDR<0.01. If a gene set had a positive enrichment score, the majority of its members had higher expression in the tumor-bearing-specified p53 missense mutation than those with nonsense mutations, and the set was termed ‘enriched' in the specified hotspot mutation.
Clustering analysis
Different p53 hot spot mutations (R175H, Y220C, G245S, R248W, R248Q, R273H and R282W) were clustered according to their enrichment scores for each gene (based on GSEA analysis). The hierarchical clustering was carried out using the Cluster program (Pearson correlation, average linkage) and visualized by TreeView (EisenLab, Lawrence Berkeley National Laboratory, Berkeley, CA, USA).37
Acknowledgments
This project was supported by grants from the National Basic Research Program of China 973 Program (2010CB5293), the National High Technology Research and Development Program of China 863 Program (2012AA02A504), the Program for Innovative Research Team of Shanghai Municipal Education Commission and National Natural Science Foundation of China (30971330, 31371420, 81320108024) to JY Fang. The project was also supported by National Natural Science Foundation of China (81000861, 81322036, and 81272383), Shanghai ‘Oriental Scholars' project (2013XJ), Shanghai Science and Technology Commission ‘Pujiang Project' (13PJ1405900) and Shanghai Natural Science Foundation (12ZR1417900) to J Xu. The sponsors of this study had no role in the collection of the data, the analysis and interpretation of the data, the decision to submit the manuscript for publication, or the writing of the manuscript.
Glossary
- LOF
loss-of-function
- GOF
gain-of-function
- MOMP
mitochondrial outer membrane permeabilization
- TCGA
The Cancer Genome Atlas
- CCLE
Cancer Cell Line Encyclopedia
- GSEA
Gene Set Enrichment Analysis
- FOLFIRI
folinic acid-fluorouracil-irinotecan regimen
- IC50
half maximal inhibitory concentration
- CRC
colorectal cancer
- GEO
gene expression omnibus
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Author contributions
J Xu–study concept and design, acquisition of data wrote the paper; J Wang–acquisition of data, wrote the paper; Y Hu, J Qian, B Xu, H Chen, W Zou,–performed experiments and analyzed data; J-Y Fang–study design, wrote the paper.
Edited by G Raschellà
Supplementary Material
References
- Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- Xu J, Zhou X, Wang J, Li Z, Kong X, Qian J, et al. RhoGAPs attenuate cell proliferation by direct interaction with p53 tetramerization domain. Cell Rep. 2013;3:1526–1538. doi: 10.1016/j.celrep.2013.04.017. [DOI] [PubMed] [Google Scholar]
- Guo L, Liew HP, Camus S, Goh AM, Chee LL, Lunny DP, et al. Ionizing radiation induces a dramatic persistence of p53 protein accumulation and DNA damage signaling in mutant p53 zebrafish. Oncogene. 2013;32:4009–4016. doi: 10.1038/onc.2012.409. [DOI] [PubMed] [Google Scholar]
- Endo A, Moyori A, Kobayashi A, Wong RW. Nuclear mitotic apparatus protein, NuMA, modulates p53-mediated transcription in cancer cells. Cell Death Dis. 2013;4:e713. doi: 10.1038/cddis.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- Joerger AC, Fersht AR. The tumor suppressor p53: from structures to drug discovery. Cold Spring Harb Perspect Biol. 2010;2:a000919. doi: 10.1101/cshperspect.a000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Green DR. Dissecting p53-dependent apoptosis. Cell Death Differ. 2006;13:994–1002. doi: 10.1038/sj.cdd.4401908. [DOI] [PubMed] [Google Scholar]
- Follis AV, Chipuk JE, Fisher JC, Yun MK, Grace CR, Nourse A, et al. PUMA binding induces partial unfolding within BCL-xL to disrupt p53 binding and promote apoptosis. Nat Chem Biol. 2013;9:163–168. doi: 10.1038/nchembio.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagn F, Klein C, Demmer O, Marchenko N, Vaseva A, Moll UM, et al. BclxL changes conformation upon binding to wild-type but not mutant p53 DNA binding domain. J Biol Chem. 2010;285:3439–3450. doi: 10.1074/jbc.M109.065391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott AF, Apel IJ, Nunez G, Clarke MF. Bcl-XL protects cancer cells from p53-mediated apoptosis. Oncogene. 1995;11:1389–1394. [PubMed] [Google Scholar]
- Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011;18:1487–1499. doi: 10.1038/cdd.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Trinidad AG, Timpson P, Morton JP, Zanivan S, van den Berghe PV, et al. Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene. 2013;32:1252–1265. doi: 10.1038/onc.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Hanel W, Marchenko N, Xu S, Yu SX, Weng W, Moll U. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ. 2013;20:898–909. doi: 10.1038/cdd.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Cheng B, Miao L, Mei Y, Wu M. Mutant p53-R273H gains new function in sustained activation of EGFR signaling via suppressing miR-27a expression. Cell Death Dis. 2013;4:e574. doi: 10.1038/cddis.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradiso A, Scarpi E, Malfettone A, Addati T, Giotta F, Simone G, et al. Nuclear NHERF1 expression as a prognostic marker in breast cancer. Cell Death Dis. 2013;4:e904. doi: 10.1038/cddis.2013.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrendt SA, Hu Y, Buta M, McDermott MP, Benoit N, Yang SC, et al. p53 mutations and survival in stage I non-small-cell lung cancer: results of a prospective study. J Natl Cancer Inst. 2003;95:961–970. doi: 10.1093/jnci/95.13.961. [DOI] [PubMed] [Google Scholar]
- Vegran F, Rebucci M, Chevrier S, Cadouot M, Boidot R, Lizard-Nacol S. Only missense mutations affecting the DNA binding domain of p53 influence outcomes in patients with breast carcinoma. PloS One. 2013;8:e55103. doi: 10.1371/journal.pone.0055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Cuesta L, Oakman C, Falagan-Lotsch P, Smoth KS, Quinaux E, Buyse M, et al. Prognostic and predictive value of TP53 mutations in node-positive breast cancer patients treated with anthracycline- or anthracycline/taxane-based adjuvant therapy: results from the BIG 02-98 phase III trial. Breast Cancer Res. 2012;14:R70. doi: 10.1186/bcr3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S, et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol. 2011;7:285–295. doi: 10.1038/nchembio.546. [DOI] [PubMed] [Google Scholar]
- Hanel W, Marchenko N, Xu S, Xiaofeng YuS, Weng W, Moll U. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ. 2013;20:898–909. doi: 10.1038/cdd.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- Xu J, Wang J, Xu B, Ge H, Zhou X, Fang JY. Colorectal cancer cells refractory to anti-VEGF treatment are vulnerable to glycolytic blockade due to persistent impairment of mitochondria. Mol Cancer Ther. 2013;12:717–724. doi: 10.1158/1535-7163.MCT-12-1016-T. [DOI] [PubMed] [Google Scholar]
- Kong X, Qian J, Chen LS, Wang YC, Wang JL, Chen H, et al. Synbindin in extracellular signal-regulated protein kinase spatial regulation and gastric cancer aggressiveness. J Natl Cancer Inst. 2013;105:1738–1749. doi: 10.1093/jnci/djt271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer G, Al-Ahmadie H, Schultz N, Hanrahan AJ, Ostrovnaya I, Balar AV, et al. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J Clin Oncol. 2013;31:3133–3140. doi: 10.1200/JCO.2012.46.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun R, Finney R, Yan C, Chen QR, Hu Y, Edmonson M, et al. Discovery analysis of TCGA data reveals association between germline genotype and survival in ovarian cancer patients. PLoS One. 2013;8:e55037. doi: 10.1371/journal.pone.0055037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.