Abstract
Granzymes are generally recognized for their capacity to induce various pathways of perforin-dependent target cell death. Within this serine protease family, Granzyme M (GrzM) is unique owing to its preferential expression in innate effectors such as natural killer (NK) cells. During Listeria monocytogenes infection, we observed markedly reduced secretion of macrophage inflammatory protein-1 alpha (MIP-1α) in livers of GrzM-deficient mice, which resulted in significantly impaired NK cell recruitment. Direct stimulation with IL-12 and IL-15 demonstrated that GrzM was required for maximal secretion of active MIP-1α. This effect was not due to reduced protein induction but resulted from heightened intracellular accumulation of MIP-1α, with reduced release. These results demonstrate that GrzM is a critical mediator of innate immunity that can regulate chemotactic networks and has an important role in the initiation of immune responses and pathogen control.
Keywords: NK cells, Granzyme M, MIP-1α, Listeria monocytogenes infection
Natural killer (NK) cells are lymphocytes of the innate immune system crucial for the defence against infection and cancer.1, 2 NK cells directly recognize infected cells and induce target cell death by the delivery of perforin (Pfp) and granzymes (Grzs), particularly GrzB.3 Besides this direct cytotoxic function, NK cells have an important regulatory role during inflammation.4, 5 They are embedded in a reciprocal crosstalk-network with immune cells such as macrophages, neutrophils and dendritic cells. This allows them to shape the immune response via the release of cytokines and chemokines.1, 6
In humans and rodents, three distinct Grz subfamilies exist. GrzA and K have trypsin-like activity; GrzB and H (Human) and B-G (rodent) exhibit chymotryptic activity, whereas GrzM processes its substrates at the carboxy side of long, hydrophobic side chain amino acids such as Met and Leu.7 Human, mouse and rat GrzMs share very similar substrate patterns and various structural characteristics.8, 9 In humans and rodents, GrzM is expressed constitutively and at high levels in NK cells, in subsets of innate T cells such as γδT cells10, 11 and in CD8+CD27−CD45RO− T effector cells.12 Recent studies have suggested that GrzM delivered by CD8 T cells can regulate Human Cytomegalovirus replication by targeting heterogeneous ribonucleotide particles.13 GrzM is able to induce Pfp-dependent target cell death in vitro, with some studies suggesting that this occurs independently of caspse,14, 15, 16 whereas others show a requirement of caspase activity.17, 18, 19 Interestingly, GrzM−/− mice have no defect of NK cell-mediated cytotoxicity, so the physiological relevance of these cell death pathways and GrzM's major biological role are still to be elucidated.
Our recent studies have demonstrated a critical role for GrzM in the regulation of LPS-induced inflammation and suggest that this Grz has functions other than direct cytotoxicity.20 We chose to determine the requirement of GrzM during Listeria monocytogenes infection where the pathophysiology of the inflammatory process is well characterized. L. monocytogenes is a Gram-positive bacillus causing severe, sometimes lethal food-borne, infections with a human case-fatality rate that can exceed 30%. Unless recognized and treated, Listeria infections can result in significant morbidity and mortality. Importantly, mouse models of Listeria infection have demonstrated a clear role for NK cells and APCs in the control of disease21, 22 where chemokine signaling networks are key components during the crosstalk that leads to effective immunity.
As part of this network, the chemokine macrophage inflammatory protein 1 (MIP-1α) belongs to the family of C-C chemokines, important for NK cell recruitment23, 24, 25 to the liver during MCMV infection.26, 27 While the major source of MIP-1α is thought to be macrophages,28, 29 NK cells can secrete MIP-1α in a cytokine-dependent manner.30, 31 Given our recent identification that GrzM can shape cross-talk during inflammation20 and the growing appreciation that Grzs are more than just inducers of apoptosis,20, 32 we sought to determine whether GrzM could shape immunity by regulating chemokine networks. In the absence of GrzM, we observed enhanced survival following Listeria challenge, and this was associated with a reduction in liver-specific NK cell recruitment, augmented effector function of NK cells in the liver and reduced liver pathology. We show that GrzM intrinsically regulates the release of Mip-1α from NK cells and that this is dependent upon IL-12/IL-15 signaling. Whereas GrzM and MIP-1α co-localise to cytotoxic granules, direct proteolytic processing of MIP-1α does not occur; rather GrzM is required for the efficient release of mature chemokine. These results demonstrate that GrzM can act as an immune stimulus and suggests its potential role as an intrinsic adjuvant.
Results
GrzM-deficient mice are less susceptible to L. monocytogenes infection
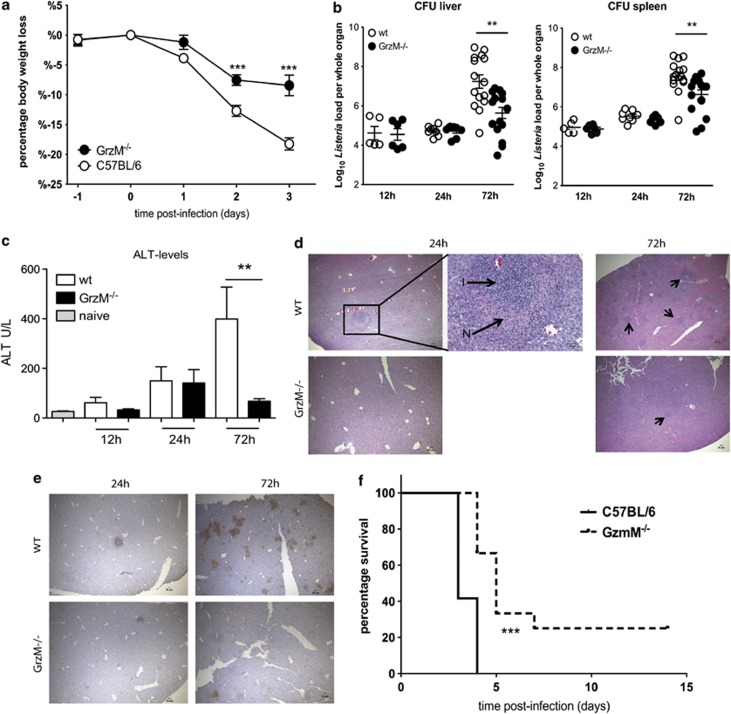
To evaluate the role of GrzM during microbial infection, WT and GrzM−/− mice were infected i.v. with 104 L. monocytogenes. During Listeria infection, WT mice lost an average of 12.5 and 20.0% of their initial body weight on days 2 and 3 post infection, respectively. In contrast, GrzM−/− mice were significantly less sensitive (P<0.0001) to bacterial infection and lost only 7.5% on day 2 and 8.4% on day 3 post infection (Figure 1a). In the first 24 h, there was no difference in the recovery of viable bacteria from the livers and spleens of WT or GrzM−/− mice (Figure 1b). However, by 72 h, the difference in the weight loss was clearly reflected in the bacterial burden of the recipients. GrzM−/− mice displayed significantly fewer viable bacteria in livers (P=0.0017) and spleens (P=0.0048) compared with WT mice.
Figure 1.
GrzM−/− mice are less susceptible to an infection with L. monocytogenes. (a–e) Female, age-matched WT and GrzM−/− mice were infected i.v. with 104 CFU of L. monocytogenes. (a) The recipients were monitored daily for weight loss and signs of illness. (b) After 12 h, on days 1 and 3 after infection mice were killed, livers and spleens were homogenized, serially diluted in PBS and plated on 5% sheep agar plates. Bacterial colonies were counted after 24 h of incubation at 37 °C. (c) Blood samples were taken 12, 24 and 72 h after infection and ALT levels were determined using the Architect plus analyser. (d) H & E staining of liver sections of WT and GrzM−/− mice 24 and 72 h after Listeria infection (N=necrotic foci, I=inflammatory infiltrations) (representative). Arrows indicate inflammatory foci. (e) TUNEL+ staining of WT and GrzM−/− mice 24 and 72 h after Listeria infection (representative). Pictures were taken with an Olympus BX-51, original magnification for images × 4 (NA 0.13, WD17 mm) or × 20 (NA 0.7, WD 0.65 mm). (f) The survival of recipients was monitored. Mice were killed when they had lost 20% of their initial body weight. Results are pooled (a) from two independent experiments with n=10, (b and c) from six independent experiments with n=5–15, (f) from two independent experiments, n=12
The reduced growth of viable bacteria in the organs of GrzM−/− mice coincided with significantly lower ALT levels (P=0.0069) in the serum 72 h after the infection, suggesting reduced hepatocellular damage (Figure 1c). Histological analysis of livers from infected WT and GrzM−/− mice after 24 and 72 h revealed multiple foci consisting of moderate to marked necrosis and a severe inflammatory mononuclear infiltrate in WT mice (Figure 1d). In contrast, the corresponding lesions in GrzM−/− mice were fewer and milder, with reduced necrosis and cellular infiltrate. TUNEL analysis showed massive apoptosis of hepatocytes in WT livers 72 h after infection (Figure 1e) where TUNEL+ cells were widely distributed and prevalent throughout the organs. In contrast, GrzM−/− organs contained far fewer TUNEL+ areas.
Overall, survival after Listeria infection was significantly enhanced (P<0.0001) for mice deficient in GrzM (Figure 1f). Collectively, these results showed a strong influence of GrzM on the immunopathology of L. monocytogenes infection and indicated a likely role for GrzM in the early inflammatory response.
Altered recruitment and function of GrzM−/− liver NK cells following L. monocytogenes infection
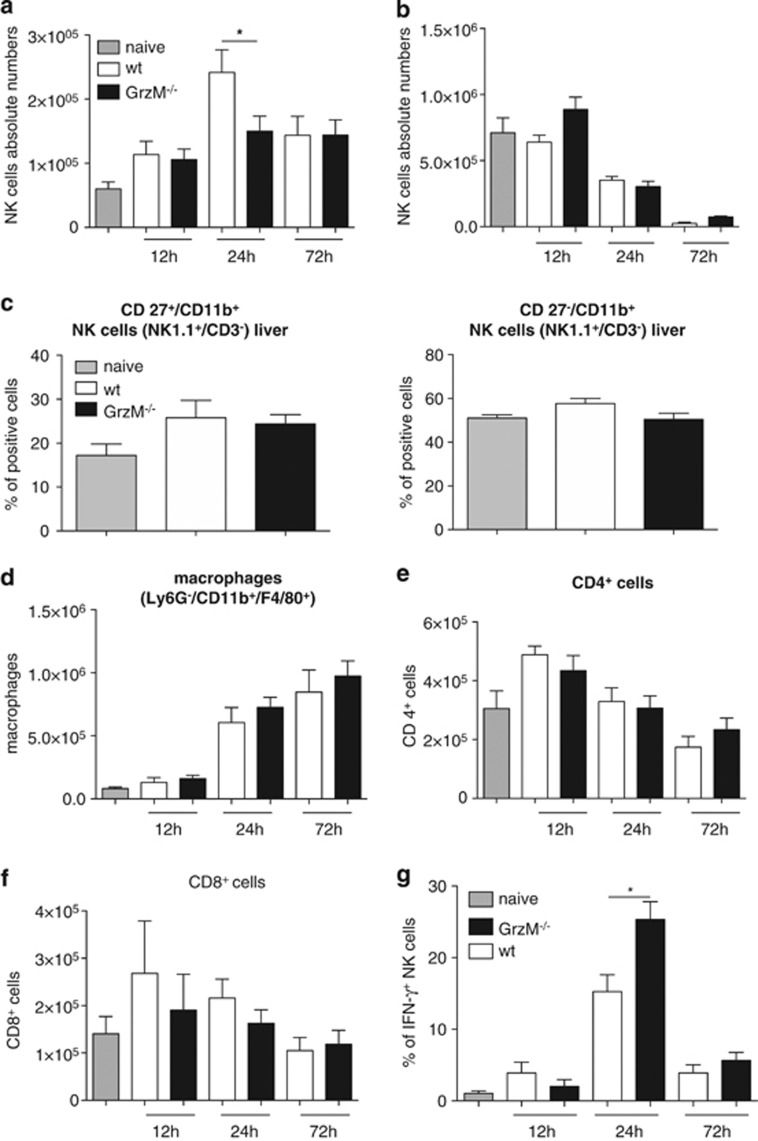
In order to expand our understanding of GrzM during L. monocytogenes-induced inflammation, the recruitment of NK cells to the liver and spleen was investigated. Twenty-four hours after infection, GrzM−/− mice displayed significantly reduced absolute numbers of NK cells (P=0.0459) in the livers (Figure 2a) but not in the spleens when compared with WT mice (Figure 2b). No effect on CD4+, CD8+ T cells and macrophages were observed in the spleen (Supplementary Figure 2). A more detailed investigation of liver NK cells 24 h after infection revealed comparable percentages of mature33, 34 NK cells (CD27+/11b+, CD27−/11b+) in WT and GrzM−/− mice (Figure 2c). Demonstrating the specificity of the response for NK cells, during the first 72 h of infection similar absolute numbers of CD4+, CD8+ T cells and macrophages were found in the livers of WT and GrzM−/− mice (Figures 2d–f).
Figure 2.
Recruitment and function of GrzM−/− NK cells is altered in the liver after L. monocytogenes infection. (a and b) Age-matched female WT and GrzM−/− mice were infected i.v. with 104 CFU of L. monocytogenes. At indicated time points after infection, mice were killed, livers (a) and spleens (b) were harvested, lymphocytes isolated and stained with mAbs reactive with NK1.1 and CD3 (shown are absolute numbers of NK cells), with (c) NK1.1, CD3, CD27 and CD11b, with Ly6G−/CD11b+/F4/80+ (d), CD4 (e), CD8 (f) or NK1.1, CD3 and IFN-γ (g) and analysed by flow cytometry. Results are pooled from six independent experiments, total n=5–8
Thus, the absence of GrzM resulted in a specific defect in liver NK cell recruitment following Listeria infection, which seemed counterintuitive to the improved bacterial clearance observed in GrzM−/− mice. As NK cell effector functional is strongly dependent on appropriate priming and activation,35, 36 we sought to determine any functional differences between NK cells in WT and GrzM−/− mice following Listeria challenge. In the livers, the frequency of NK cells positive for intracellular IFNγ was significantly increased (P=0.0115) in GrzM−/− mice at this time point (Figure 2g). Thus, the reduced infiltration of NK cells in GrzM-deficient mice correlated with increased activation of these cells, concomitant with the reduced bacterial burden.
Intrinsic capacity of GrzM−/− NK cells to secrete MIP-1α is impaired
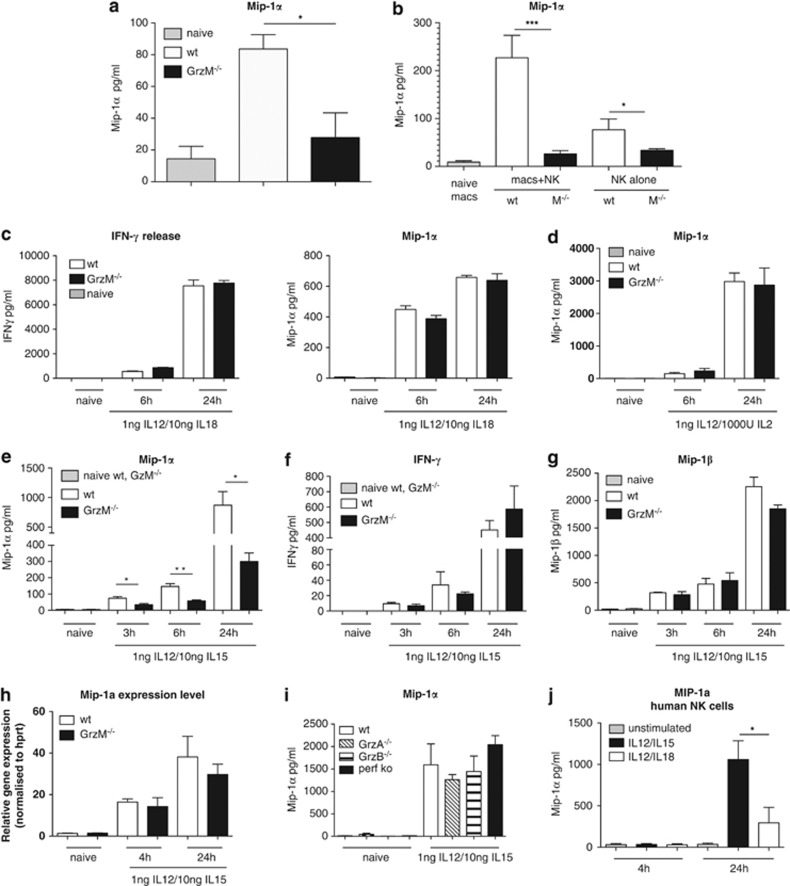
The chemokine MIP-1α is known to induce NK cell trafficking and migration to inflammatory sites.23, 25, 26 As we had observed fewer NK cells in the livers of Listeria-challenged GrzM−/− mice (Figure 2), we next sought to determine any effect of GrzM on MIP-1α production. To reveal whether GrzM deficiency influences the production and/or release of MIP-1α in vivo, the amount of chemokine secreted in bulk leukocyte cultures from livers of GrzM−/− and WT mice 24 h after L. monocytogenes infection was determined. We observed a significant reduction (P=0.0376) in MIP-1α produced by GrzM-deficient leukocytes compared with WT (Figure 3a). In order to determine whether GrzM delivery from NK cells to macrophages could alter MIP-1α production, we co-cultured NK cells purified from LPS-stimulated WT and GrzM−/− mice with bone marrow-derived macrophages and assessed MIP-1α production using cytometric bead array (CBA). We observed a significant decrease in MIP-1α (P=0.0003) produced in co-cultures of NK cells purified from GrzM−/− mice when compared with co-cultures containing WT NK cells (Figure 3b). Interestingly, we also observed a significant decrease in MIP-1α produced by purified GrzM−/− NK cells cultured alone (P=0.0357) when compared with WT NK cells (Figure 3b).
Figure 3.
Intrinsic capacity of GrzM−/− NK cells to secrete MIP-1α is impaired. (a) Age-matched female WT and GrzM−/− mice were infected i.v. with 104 CFU of L. monocytogenes. Twenty-four hours after infection, mice were killed, livers were harvested, liver cells were isolated, adjusted to 106 cells/ml and incubated at 37°C in a 96-well plate. After 6 h, supernatants of liver cells were taken and analysed by CBA for secreted MIP-1α. (b) WT and GrzM−/− mice were injected i.p. with 1 mg LPS. After 12 h, NK cells were enriched from spleens of treated or untreated (control) animals and co-incubated with naive bone marrow-derived macrophages at a ratio of 1 : 5 or were incubated without macrophages as a control for 4 h at 37°C. Subsequently, supernatants were harvested and analysed by CBA for MIP-1α as described above. Results are pooled from three independent experiments with (a) n=9 and (b) n=7. (c–h) NK cells were isolated from spleens of WT and GrzM−/− mice or (i) of GrzA−/−, GrzB−/− and pfp−/− mice and sorted to purity for NK1.1+/CD3-expressing cells by FACS (BD Aria II). NK cells (2 × 106 cells/ml) were left untreated as a control or stimulated with 1 ng IL-12/ml and 5 ng IL-18/ml (c), 1 ng IL-12/ml and 1000 IU/ml IL-2 (d) or 1 ng IL-12/ml and 10 ng IL-15/ml (e–i). After 6 and 24 h (c, d) or 3, 6 and 24 h (e–g) supernatants were taken and analysed by CBA for secreted IFN-γ and MIP-1α (c–f, i) or MIP-1α (g) (n=5–7). (h) After 4 and 24 h, the expression of mRNA coding for mouse MIP-1 was determined by real time PCR in WT and GrzM−/− NK cells (n=2). (j) Human NK cells were isolated from Buffy coats using the EasySep-negative enrichment kit. Cells (1 × 106) were left untreated, stimulated with 1 ng IL-12/ml and 10 ng IL-15/ml or with 1 ng IL-12/ml and 5 ng IL-18/ml. Supernatants were taken after 4 and 24 h and analysed by ELISA. Results are pooled from five independent experiments, n=5
Whereas the requirement for GrzM in MIP-1α release from macrophages was of interest, the decrease in MIP-1α production by activated NK cells from GrzM-deficient mice suggested an intrinsic role for GrzM in MIP-1α export. We next aimed to define the pathways by which GrzM could regulate this process intrinsically. NK cells from naive WT and GzM−/− mice were stimulated ex vivo with different combinations of IL-12p70, IL-15, IL-18 and IL-2. When stimulated with IL-12 (1 ng/ml) and IL-18 (5 ng/ml) or with IL-12 (1 ng/ml) and IL-2 (1000 U/ml) similar amounts of MIP-1α or IFN-γ were detected in the supernatant after 6 and 24 h (Figures 3c and d). In contrast, stimulation of NK cells with a combination of IL-12 (1 ng/ml) and IL-15 (10 ng/ml) led to significantly reduced amounts of secreted Mip-1α by GrzM−/− NK cells compared with WT NK cells after 3 h (P=0.0126), 6 h (P=0.0097) and 24 h (P=0.0417) (Figure 3e). This effect was specific for MIP-1α as comparable amounts of IFN-γ and MIP-1β were detected in the supernatants (Figures 3f and g). MIP-1α mRNA levels of WT and GrzM−/− NK cells were similarly upregulated after IL-12/IL-15 stimulation for 4 and 24 h, indicating that the reduction in MIP-1α secretion by GrzM−/− NK cells was not due to reduced gene transcription (Figure 3h). To exclude any contribution of other Grz or Pfp, NK cells isolated from pfp−/−, GrzA−/− and GrzB−/− mice were stimulated, and the supernatants were analyzed for secreted MIP-1α. Neither NK cells from pfp−/− mice, nor from GrzA−/− or B−/− mice, showed reduced MIP-1α secretion at any time point after IL-12/IL-15 stimulation (Figure 3i). Importantly, human NK cells secreted significantly higher amounts of MIP-1α (P=0.0137) after 24 h when stimulated with IL-12/IL-15 compared with stimulation with IL-12/IL-18 (Figure 3j).
GrzM and MIP-1α co-localize within the cytotoxic granules of primary human NK cells
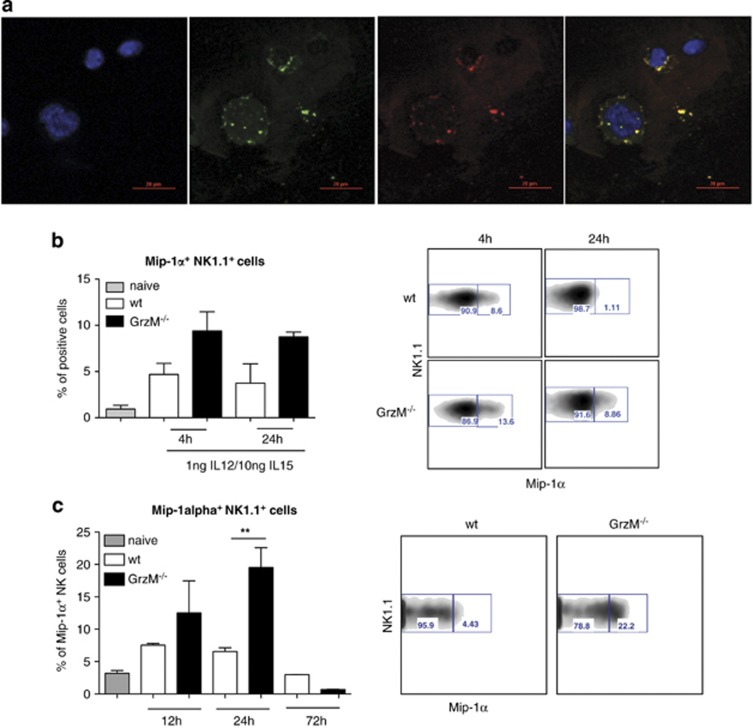
We next sought to assess whether GrzM has a role in the secretion of MIP-1α. Wagner et al.37 showed that HIV-specific cytotoxic CD8+ T cells co-localize GrzA and MIP-1α within the cytotoxic granules, and GrzA and MIP-1α are secreted together upon antigen-specific stimulation. To analyze how and where GrzM and MIP-1α interact within NK cells, confocal microscopy of naive human NK cells was performed. Our analysis revealed that in naive NK cells GrzM (stained green) and MIP-1α (stained red) are co-localized in the cytotoxic granules (Figure 4a). In naive NK cells, GrzM/MIP-1α-positive granules were distributed across the cytoplasm in small vesicles of relatively uniform size. Complementing these analyses, intracellular staining of MIP-1α in IL-12/IL-15-activated mouse NK cells revealed higher frequencies of MIP-1α+ GrzM−/− NK cells compared with MIP-1α+ WT NK cells at 4 and 24 h after stimulation (Figure 4b). A significant increase in the percentage of MIP-1α+ NK cells (P=0.0043) was also observed in the livers of GrzM−/− mice 24 h after the infection with Listeria (Figure 4c). Thus, GrzM and MIP-1α are in close contact within the cytotoxic granules of NK cells. Furthermore, our data suggest that IL-15 is a key cytokine regulating GrzM-specific release of MIP-1α from NK cells.
Figure 4.
GrzM and MIP-1α co-localize in the cytotoxic granules of NK cells. (a) Human NK cells were isolated from Buffy coats by using the EasySep-negative enrichment kit. NK cells were used immediately and spun onto slides and stained for Dapi (blue), GrzM (green) and MIP-1α (red). Data shown are representative of five experiments. Pictures were taken with a Nikon C2 confocal microscope. Original magnification × 40 (NA 1.3, WD 0.24 mm, oil). (b) NK cells were isolated from spleens of WT and GrzM−/− mice and sorted to purity for NK1.1+/CD3− expressing cells by FACS (BD Aria II). A total of 2 × 106 NK cells/ml were left untreated as a control or stimulated with 1 ng IL-12/ml and 10 ng IL-15/ml and treated with Golgi stop and Golgi plug for 1 h (left for further 3, 4 h time point) or 18 h (left for further 6, 24 h time point). After stimulation, cells were stained with mAbs reactive with NK1.1, CD3 and MIP-1α (n=3). (c) Age-matched female WT and GrzM−/− mice were infected i.v. with 104 CFU of L. monocytogenes. Twelve, twenty-four and seventy-two hours after infection with Listeria, mice were killed, livers were harvested, lymphocytes isolated and treated with Golgi stop and Golgi plug for 1 h, left for further 3 h and were stained with mAbs reactive with NK1.1, CD3 and MIP-1α (n=3–5)
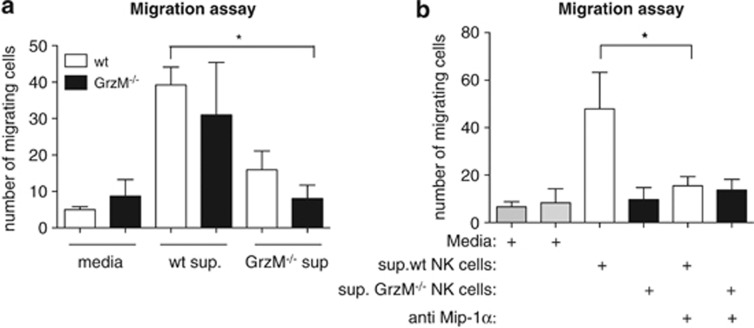
Impaired migration of NK cells towards MIP-1α secreted by GrzM−/− NK cells
We have shown that GrzM−/− NK cells secrete less MIP-1α after stimulation with IL-12/-15 or during Listeria infection, and that fewer NK cells are recruited into the livers of Listeria-infected GrzM-deficient mice. To evaluate whether these two observations are causally linked, chemotaxis assays were performed. Culture supernatants from WT and GrzM−/− NK cells stimulated for 4 h with IL-12 and IL-15 were added to the bottom chambers of transwell plates. Naive WT or GrzM−/− NK cells were incubated in the top chambers, and NK cell migration was assessed 4 h later. When we used supernatant from GrzM−/− NK cells, the migration of both GrzM−/− and WT NK (P=0.0138) cells was strongly impaired (Figure 5a). In contrast, WT and GrzM−/− NK cells showed a similar migratory capacity towards WT supernatant, indicating that the impaired migration of GrzM−/− NK cells is not a result of a defect in NK cell chemotaxis. When MIP-1α was neutralised, the migration towards WT supernatant of WT (P=0.0265) and GrzM−/− NK cells was reduced to background levels (Figure 5b). These data demonstrate that GrzM is required for optimal NK cell recruitment, and that, in turn, this recruitment is MIP-α-dependent.
Figure 5.
Migration of NK cells towards MIP-1α secreted by GrzM−/− NK cells is impaired. (a and b) Migration of naive WT and GrzM−/− NK cells towards supernatant of WT or GrzM−/− NK cells stimulated for (a) 4 h with IL-12/IL-15 or media or (b) supernatant treated with a neutralizing antibody against MIP-1α. Results are pooled from three experiments (n=6)
GrzM does not process/activate MIP-1α
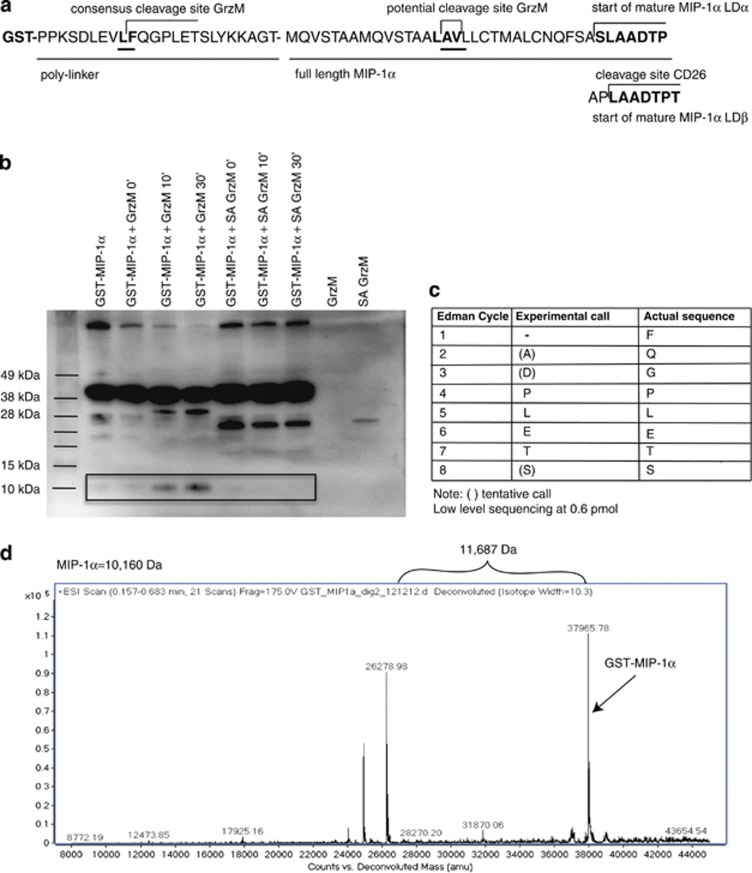
The ectodomain of plasma membrane-bound CD26, an aminodipeptidase, truncates MIP-1α by processing the N-terminus of the LDβ form after Proline.38, 39 Owing to the acidic environment of cytotoxic granules (pH 5.5), GrzM (a neutral serine protease) has suboptimal catalytic activity while being stored in that compartment, making it more likely that GrzM could process MIP-1α following secretion into the immune synapse.40, 41 This raises the possibility that GrzM might process MIP-1α following delivery to the APC, as macrophages do not synthesise GrzM.20 As MIP-1α contains a potential site for cleavage by GrzM (Leu-Ala-Val) within the signal peptide at position 15-16-17 (Figure 6a), it was necessary to formally exclude a direct effect of GrzM processing of MIP-1α. In order to do this, we utilised a GST fusion protein that incorporated full-length MIP-1α coupled to GST through a polylinker region. We incorporated a consensus cleavage site for human GrzM (Valine-Leucine-Phenylalanine) as a positive control (Figure 6a). Exposure of the fusion protein to low levels of active GrzM generated a protein product of ∼10 kDa, which was not produced by a catalytically inactive (Serine/Alanine-mutated) GrzM (Figure 6b). Protein purification and mass spectrometry established that the C-terminal cleavage product had a MW of 11 687 daltons (Figure 6d), which was well in excess of a product processed at the physiological site but corresponded to the mass predicted for full-length MIP-1α attached to a portion of the adjacent polylinker. N-terminal sequencing proved that processing of the GST fusion protein had taken place at the artificial site in the linker, but not within the MIP-1α polypeptide sequence, and specifically not at the potential GrzM consensus sequence (Figure 6c). This result clearly indicated that direct processing by GrzM is not responsible for MIP-1α activation or release.
Figure 6.
GrzM does not process MIP-1α. (a) Human recombinant full-length GST-tagged MIP-1α (sequence start mature MIP-1α: bold letters) was generated. The protein contains a GrzM consensus cleavage site in the polylinker region between GST and the start of full-length MIP-1α, as well as a potential cleavage site for GrzM in the full-length MIP-1α sequence. (b) This product was then incubated with low concentrations of human recombinant GrzM or a Serine/Alanine (S/A) mutant. The resulting products were used for immunoblotting (c) N-terminal sequencing and (d) mass spectrometry
Discussion
GrzM is not recognised as a principal mediator of apoptosis14 and is not essential for tumor control.42 Conversely, GrzM contributes to inflammatory processes observed during MCMV infection42 and LPS-induced endotoxicosis20; however, the mechanism is unclear. Our data now provide a molecular target for GrzM of biological relevance to inflammation. These studies demonstrate that GrzM is required for the optimal release of MIP-1α from NK cells. Importantly, MIP-1α release from GrzA−/− and B−/− NK cells was not affected, GrzM deficiency did not alter MIP-1β release, and the pathway was strictly dependent upon the activity of IL-12 and IL-15, indicating the strict specificity of this interaction. We have also demonstrated co-localisation of GrzM and MIP-1α in the cytotoxic granules of human NK cells. As MIP-1α is an important regulator of NK cell recruitment during inflammation,26, 27 our data suggest that GrzM-dependent MIP-1α production may auto-regulate the migration of NK cells, acting as an ‘internal-adjuvant' for the amplification of inflammation.
Twenty-four hours after L. monocytogenes infection, no difference was observed in the bacterial load from GrzM−/− and WT mice. This result indicated that GrzM had no influence on the initial infection of, or on the initial replication of the bacterium in, resident macrophages. However by 72 h, markedly fewer bacteria were recovered from the organs of GrzM−/− mice. This was a surprising outcome since GrzA and B deficiency reportedly resulted in increased pathogen burden.43, 44, 45 At later time points, GrzM−/− mice were less susceptible to Listeriosis when compared with their WT counterparts, exhibiting reduced liver damage, weight loss and considerably lower concentrations of pro-inflammatory serum cytokines (Supplementary Figure 1), all hallmarks of a ‘cytokine storm'-induced sepsis.46, 47, 48 These data suggested that GrzM might contribute to inflammation by amplifying the immune response, resulting in the high levels of cytokine production observed in WT mice. Indeed, co-incident with the reduced Listeria burden we observed a reduction in both serum cytokine levels and NK cell recruitment to the livers of GrzM-deficient mice.
As the function of NK cells is strongly dependent on appropriate priming, this observation suggested that the effects we observed in GrzM-deficient mice may arise from a more targeted immune response. After infection, NK cells are activated by cytokines and chemokines as well as by direct cell–cell contact.1, 49 These interactions result in activation of NK cells and production of pro-inflammatory cytokines. Conversely, NK cells can influence the behaviour of accessory cells in return, either by the secretion of cytokines, such as IFNγ, or by cytotoxicity50, 51 sculpting the subsequent immune response by altering the ability of the APC to continually stimulate inflammatory leukocytes. Hence, the reduced recruitment of NK cells to the liver in GrzM−/− mice could heavily influence crosstalk at the site of infection, resulting in a reduced inflammatory cytokine millieu. Our data suggest that, within the livers of GrzM−/− mice, the cellular immune-milieu and the resulting crosstalk is more beneficial for the host compared with the WT situation. This altered milieu results in the bacterial infection being contained more efficiently within the liver, preventing the development of aggressive systemic bacteremia.
MIP-1α is involved in the recruitment of NK cells to the liver27; however, it has been assumed that the source of this chemokine is tissue-resident APC activated via TLR signals.27, 52 Similar to other groups30, 31 we have found that NK cells can be a source of MIP-1α; however, its release is specifically dependent upon the activity of GrzM. Our data demonstrate that GrzM and MIP-1α co-localise in the cytotoxic granules, suggesting that GrzM may be a promising candidate to process or to truncate MIP-1α. The highly related murine and human MIP-1α are synthesized as 92 aa precursor proteins, which are matured by peptidases that cleave the hydrophobic signal sequence. As GrzM is a protease, our primary focus for the mechanism by which it increases MIP-1α output was to test for processing of MIP-1α by GrzM. Despite GrzM clearly being capable of cleaving at an accessible but artificial GrzM consensus sequence, no physiologically relevant interaction between GrzM and MIP-1α could be detected. Mouse and human GrzM exhibit very similar, although not identical, substrate specificities,53 as is the case for all Grzs. For example, both mouse and human GrzB cleave after selected aspartate residues but have different extended substrate preferences that reflect the different evolutionary pressures faced by the different species over many millions of years.54 Despite this, both are strongly pro-apoptotic, albeit they activate subtly different apoptotic pathways. All of the known functions of the granule exocytosis pathway are conserved across species, and as both mouse and human granules containing GrzM and MIP-1α, it is likely that a similar mechanism underpins MIP-1α release in both species.
A combination of cytokines is the key to inducing NK cell activation, as single agents do not stimulate NK to effector function.55, 56 This complex requirement is very relevant to both the clinical application of NK cells and to studies of NK cell regulation of adaptive immunity, as optimal NK cell activation is required in order to elicit the appropriate immune response. For example, overstimulation of NK cells results in a reduced CD8 T-cell response limiting the generation of functional memory CD8 T cells.57 It has been shown that the combination of IL-18 and IL-12 induce IFN-γ production, whereas IL-15/IL-12 induces IL-10, MIP-1α and TNF.35 Interestingly, optimal IFN-γ production requires priming via IL-18 and subsequent secondary stimulation (4 h after priming) by IL-12.58 A similar mechanism has been demonstrated for Pfp and GrzB upon IL-15 priming and subsequent IL-12 stimulation.59 The observation that GrzM-dependent MIP-1α production occurs only after IL-15 and IL-12 stimulation improves our understanding of GrzM biology and potentially the adoptive transfer of NK cells. Signal transduction following IL-15 stimulation has been extensively mapped, facilitating future studies that will identify the intersection between IL-15 and Granzyme M (GrzM). Using the appropriate NK cell stimulation to activate chemokine production and to amplify immune responses may prove useful for cellular therapies aimed at enhancing CD8 T-cell memory. Significantly, NK cell therapy is gaining traction in many clinical settings but the field remains young and the key components regulating the immune response are not well defined.60
Collectively, our study shows for the first time that GrzM can specifically regulate the release of the β-chemokine MIP-1α. Furthermore, we have demonstrated that the interaction between GrzM and MIP-1α is sensitive to cytokine stimulation combinations. These results demonstrate that GrzM can influence the recruitment of immune cells to inflammatory sites and that this interaction may be amenable to therapeutic targeting through specific cytokine stimulation of adoptively transferred NK cells.
Materials and Methods
Mice and bacterial infections
All experimental procedures and tissue/sample collections were approved by the Animal Ethics Committee at the Peter MacCallum Cancer Centre and the Department of Microbiology and Immunology of the University of Melbourne. B6.GrzM-deficient (GrzM−/−), B6.Pfp-deficient (pfp−/−) and GrzB-deficient (GrzB−/−) mice were generated42, 45, 61, 62 and maintained at the Peter MacCallum Cancer Centre under SPF conditions. Age-matched female wild-type (WEHI) and GrzM−/− mice were infected intravenously (i.v.) with 104 CFU of the L. monocytogenes EGD strain into the tail vein.22
Mice were weighed daily and monitored for signs of illness. Differences in the weight loss were calculated by two-way ANOVA test. Mice were considered to have succumbed to disease and were killed when they had lost 20% of their starting body weight. To study bacterial clearance, recipients were killed on pre-determined days after infection. Liver and spleen tissues were homogenized and the number of viable bacteria was calculated by serial dilution on blood agar plates, as described previously.22 For co-culture experiments, GrzM−/− and WT mice were injected with 1 mg per 30 g LPS intraperitonealy (i.p.) to induce inflammation and activate NK cells. Spleens of treated mice were harvested 18 h after the injection.
Histopathology
Formalin-preserved liver sections were processed and embedded in paraffin by standard techniques. Five-micrometer-thick longitudinal sections were stained with haematoxylin and eosin. TUNEL analysis was performed with the ApopTag Peroxidase In Situ Apoptosis detection kit (Millipore, Billerica, MA, USA) according to the manufacturer's instructions. Specimens were analysed with the Olympus BX-51 microscope at × 4 (NA 0.13, WD 17 mm) and × 20 (NA 0.7, WD 0.65) magnifications.
Serum alanine aminotransferase (ALT)
Blood was taken either from the retro-orbital sinus or by cardiac puncture at the indicated times, and serum ALT levels were measured using the Architect plus analyser (ci 4100, Abbott Park, Chicago, IL, USA), as an indicator of hepatic cell damage.
Flow cytometry
The following antibodies from e-Bioscience (San Diego, CA, USA) were used: CD8-PE-CY7 (clone 53–6.7), CD4- APC-Cy7 (clone RM4-5), NK1.1-PE (PK 136), CD3-Pacific Blue (17A2), CD69-FITC (H1.2F3), CD11b-APC-Cy7 (M1/70), Ly6G-PE (1A8), F4/80-APC (BM8), IFN-γ-FITC (XMG1.2), Mip-1α-APC (PFFM3), CD27-PeCy7 (LG.7F9). Single-cell suspensions of livers and spleens from WT and GrzM−/− mice were processed at indicated times after Listeria infection and stained for immune-cell subsets. For intracellular staining, the Cytofix/Cytoperm kit (554715; BD Biosciences Pharmingen, San Diego, CA, USA) from BD was used according to the manufacturer's instructions. For intracellular MIP-1α staining at 24 h, Golgi stop was added 6 h before cells were harvested.
Ex vivo stimulation of NK cells
Single-cell suspensions of spleen from naive WT, GrzM−/−, pfp−/− or GrzB−/−mice were enriched for NK cells (NK1.1+/CD3−) by negative selection with a mouse NK cell isolation kit II (Miltenyi Biotec, Bergisch Gladbach, Germany) and sorted to purity (BDAriaII, San Jose, CA, USA). Human NK cells were enriched from Buffy-coats (EasySep negative enrichment kit, Stemcell Technologies, Vancouver, BC, Canada; 19055). Mouse or human NK cells were stimulated with 1 ng IL-12/ml and 10 ng IL-15/ml, 1 ng IL-12/ml and 5 ng IL-18/ml, or 1000 U/ml IL-2 and 1 ng IL-12/ml for 3, 6 and 24 h or for 4 and 24 h. Cells were used for surface marker staining, intracellular staining or to extract RNA for RT-PCR; supernatants were harvested for CBA assays or were analysed by ELISA (R&D Systems, Minneapolis, MN, USA) according to the manufacturer's instructions.
Cytometric Bead array
The CBA Flex Systems Kit (BD Biosciences) was used to measure IFN-γ, MIP-1α and MIP-1β according to the manufacturer's instructions.
Semi-quantitative real-time PCR analysis for Mip-1α
NK cells enriched from single-cell suspensions of spleen cells from naive WT and GrzM−/− mice were stimulated with cytokines as described above. Total RNA was extracted by using the RNeasy mini kit according to the manufacturer's instructions (Qiagen, Hilden, Germany). Quantitative real-time PCR was performed on the ABI4900 light cycler, using cybergreen with high ROX for internal calibration (Agilent Technologies, Santa Clara, CA, USA). The following conditions were used for real-time PCR: 95 °C for 10 min, 40 cycles of 95 °C for 30 s, 60 °C for 30 s. A dissociation curve was determined to analyse primer specificity that was 95 °C for 15 s, 60 °C for 15 s and 95 °C for 15 s. All data were collected after the annealing step for each cycle and for the dissociation curve, after the annealing step and the final 95 °C denaturation step. Data from each sample were quantified relative to the housekeeping gene L32. All samples were run in triplicate and data reflect the normalised ΔΔCt (10000/2ΔΔ Ct) of the triplicate wells. Primers were as follows: mouse MIP-1α forward: 5′-GCCATATGGAGCTGACAC-3′ mouse MIP-1α reverse: 5′-TGCCTCCAAGACTCTCAG-3′.
Co-culture experiments
NK cells enriched from single-cell suspensions of spleen cells from naive or LPS-injected WT and GrzM−/− mice (2 × 105/ml) were co-cultured with naive bone marrow-derived macrophages (1 × 106/ml) at a ratio of 1 : 5. Cells were incubated 4 h at 37 °C in air containing 5% CO2. Culture supernatants were then analysed by CBA, as described above.
Cell migration assay
NK cells enriched from single-cell suspensions of spleen cells from naive WT and GrzM−/− mice were stimulated for 6 h with cytokines as described above, harvested and stored at −20 °C. On the following day, NK cells were enriched as described above from naive GrzM−/− and WT mice and adjusted to 2 × 106/ml. One hundred microliters of the enriched naive WT and GrzM−/− NK cells (2 × 105) were added into the top chamber of transwell membranes (Corning, 5 μm, CLS3421-48EA) over 600 μl of the defrosted, cell-free supernatant from the IL-12/IL-15-stimulated NK cells and incubated for 4 h. The transwells were then removed, and the plates fixed (MBF), permeabilized (0,1% Triton) and stained with DAPI (Invitrogen, Carlsbad, CA, USA) to facilitate counting of migrated cells. A neutralizing antibody against MIP-1α (Abcam, Cambridge, UK; ab 10383) was used as a specificity control for the response to MIP-1α.
Immunofluorescence
Human NK cells were enriched from Buffy-coats (EasySep negative enrichment kit, Stemcell Technologies, 19055). A total of 2 × 106 cells/ml were cytospun onto slides, air-dried, fixed in Bouins for 10 min, permealized with Triton for 5 min and blocked with 1% BSA in PBS for 30 min at 4 °C. Slides were incubated with goat anti-human MIP-1α (R&D Systems, AF-270-NA; 1 : 50) and mouse anti-human GrzM (1 : 500) for 45 min. After two washes with PBS slides were incubated with Alexa 488-conjugated anti-mouse IgG1 and Alexa 649 donkey anti-goat IgG for further 45 min (both diluted 1 : 1000; Molecular Probes, Grand Island, NY, USA). Control slides were incubated with only secondary antibodies or the mismatching antibodies. After staining, specimens were washed in PBS and a drop of Dapi anti-fade (Invitrogen) was used for nucleic staining, as well as to reduce photo-bleaching. Slides were examined by confocal imaging on a Nikon CS 2, original magnification × 40 (WD 0.24 mm oil) using the NIS software (Nikon Instruments Inc., Melville, NY, USA).
Western blot analysis
Recombinant full-length human GST-tagged MIP-1α, into which a processing consensus site for human GrzM (Valine Leucine Phenylalanine) was encoded in the polylinker region (Abnova, P01), was incubated with human recombinant GrzM or a S/A mutant (inactive) GrzM for the indicated times at 37 °C, or left on ice as a control. Products were separated on SDS-polyacrylamide gel and immunoblotting was performed as described previously using goat anti-human MIP-1α (R&D Systems, AF-270-NA) Blots were developed with ECL using the AP substrate CDP-Star (Applied Biosystems, Carlsbad, CA, USA).
N-terminal sequencing
GrzM-digested GST-MIP-1α was resolved by SDS-polyacrylamide gel electrophoresis and transferred to PVDF membrane, then stained with Coomassie brilliant blue G250. The band at ∼11 kDa was excised and subjected to N-terminal sequence analysis by Edman degradation (Australian Proteome Analysis Facility, Macquarie University, Sydney, Australia).
Mass spectrometry
Intact or GrzM-cleaved GST-MIP-1α were precipitated using methanol and chloroform to remove salts, according to the method of Wessel and Fluegge63 and dissolved in 2% formic acid for direct injection into an ESI-TOF mass spectrometer (Agilent Technologies 6220) with a constant flow of 0.25 ml/min 50% acetonitrile in 0.1% formic acid. Data were collected in positive ion mode and voltages used were capillary voltage −4000 V, fragmentor –250 V and skimmer −65 V. The acquisition scan range was 100–2000 m/z and the internal reference ions were 121.050873 and 922.009798. Acquisition was performed using the Agilent Mass Hunter Acquisition software version B.02.01 (B2116.30), and analysis was performed using Mass Hunter version B.03.01.
Statistical analysis
For statistical analysis, the data were subjected to the two-tailed Student's t-test, two-way ANOVA (weight loss experiments) or Mantel–Cox test (survival).
P-values: *P=<0.05, **P<0.005, ***P<0.0005.
Acknowledgments
We would like to thank J May for excellent technical assistance. This work was supported by a NH&MRC Project Grant (628302) and the NHMRC Program Grant in Cellular Microbiology (606788). MJS was supported by a NH&MRC Australia Fellowship. DMA was supported by an NHMRC CDF Level 1 (1028425).
Glossary
- NK cells
natural killer cells
- MIP-1α
macrophage inflammatory protein-1alpha
- Pfp
Perforin
- Grz
Granzymes
- CBA
cytometric bead array
- i.p.
intraperitonealy
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Ciliberto
Supplementary Material
References
- Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- Trinchieri G. Biology of natural killer cells. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapani JA, Smyth MJ. Functional significance of the perforin/granzyme cell death pathway. Nat Rev Immunol. 2002;2:735–747. doi: 10.1038/nri911. [DOI] [PubMed] [Google Scholar]
- Andrews DM, Estcourt MJ, Andoniou CE, Wikstrom ME, Khong A, Voigt V, et al. Innate immunity defines the capacity of antiviral T cells to limit persistent infection. J Exp Med. 2010;207:1333–1343. doi: 10.1084/jem.20091193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narni-Mancinelli E, Jaeger BN, Bernat C, Fenis A, Kung S, De Gassart A, et al. Tuning of natural killer cell reactivity by NKp46 and Helios calibrates T cell responses. Science. 2012;335:344–348. doi: 10.1126/science.1215621. [DOI] [PubMed] [Google Scholar]
- Newman KC, Riley EM. Whatever turns you on: accessory-cell-dependent activation of NK cells by pathogens. Nat Rev Immunol. 2007;7:279–291. doi: 10.1038/nri2057. [DOI] [PubMed] [Google Scholar]
- Rukamp BJ, Kam CM, Natarajan S, Bolton BW, Smyth MJ, Kelly JM, et al. Subsite specificities of granzyme M: a study of inhibitors and newly synthesized thiobenzyl ester substrates. Arch Biochem Biophys. 2004;422:9–22. doi: 10.1016/j.abb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Smyth MJ, Sayers TJ, Wiltrout T, Powers JC, Trapani JA. Met-ase: cloning and distinct chromosomal location of a serine protease preferentially expressed in human natural killer cells. J Immunol. 1993;151:6195–6205. [PubMed] [Google Scholar]
- Sayers TJ, Lloyd AR, McVicar DW, O'Connor MD, Kelly JM, Carter CR, et al. Cloning and expression of a second human natural killer cell granule tryptase, HNK-Tryp-2/granzyme 3. J Leukoc Biol. 1996;59:763–768. doi: 10.1002/jlb.59.5.763. [DOI] [PubMed] [Google Scholar]
- Krenacs L, Smyth MJ, Bagdi E, Krenacs T, Kopper L, Rudiger T, et al. The serine protease granzyme M is preferentially expressed in NK-cell, gamma delta T-cell, and intestinal T-cell lymphomas: evidence of origin from lymphocytes involved in innate immunity. Blood. 2003;101:3590–3593. doi: 10.1182/blood-2002-09-2908. [DOI] [PubMed] [Google Scholar]
- Sayers TJ, Brooks AD, Ward JM, Hoshino T, Bere WE, Wiegand GW, et al. The restricted expression of granzyme M in human lymphocytes. J Immunol. 2001;166:765–771. doi: 10.4049/jimmunol.166.2.765. [DOI] [PubMed] [Google Scholar]
- deKoning PJ, Tesselaar K, Bovenschen N, Colak S, Quadir R, Volman TJ, et al. The cytotoxic protease granzyme M is expressed by lymphocytes of both the innate and adaptive immune system Mol Immunol 201047903–911. [DOI] [PubMed] [Google Scholar]
- van Domselaar R, de Poot SA, Remmerswaal EB, Lai KW, ten Berge IJ, Bovenschen N. Granzyme M targets host cell hnRNP K that is essential for human cytomegalovirus replication. Cell Death Differ. 2013;20:419–429. doi: 10.1038/cdd.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JM, Waterhouse NJ, Cretney E, Browne KA, Ellis S, Trapani JA, et al. Granzyme M mediates a novel form of perforin-dependent cell death. J Biol Chem. 2004;279:22236–22242. doi: 10.1074/jbc.M401670200. [DOI] [PubMed] [Google Scholar]
- Lieberman J. The ABCs of granule-mediated cytotoxicity: new weapons in the arsenal. Nat Rev Immunol. 2003;3:361–370. doi: 10.1038/nri1083. [DOI] [PubMed] [Google Scholar]
- Smyth MJ, Street SE, Trapani JA. Cutting edge: granzymes A and B are not essential for perforin-mediated tumor rejection. J Immunol. 2003;171:515–518. doi: 10.4049/jimmunol.171.2.515. [DOI] [PubMed] [Google Scholar]
- Hu D, Liu S, Shi L, Li C, Wu L, Fan Z. Cleavage of survivin by Granzyme M triggers degradation of the survivin-X-linked inhibitor of apoptosis protein (XIAP) complex to free caspase activity leading to cytolysis of target tumor cells. J Biol Chem. 2010;285:18326–18335. doi: 10.1074/jbc.M109.083170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Xia P, Shi L, Fan Z. FADD cleavage by NK cell granzyme M enhances its self-association to facilitate procaspase-8 recruitment for auto-processing leading to caspase cascade. Cell Death Differ. 2012;19:605–615. doi: 10.1038/cdd.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Poot SA, Lai KW, van der Wal L, Plasman K, Van Damme P, Porter AC, et al. Granzyme M targets topoisomerase II alpha to trigger cell cycle arrest and caspase-dependent apoptosis. Cell Death Differ. 2013;21:416–426. doi: 10.1038/cdd.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony DA, Andrews DM, Chow M, Watt SV, House C, Akira S, et al. A role for granzyme M in TLR4-driven inflammation and endotoxicosis. J Immunol. 2010;185:1794–1803. doi: 10.4049/jimmunol.1000430. [DOI] [PubMed] [Google Scholar]
- Humann J, Lenz LL. Activation of naive NK cells in response to Listeria monocytogenes requires IL-18 and contact with infected dendritic cells. J Immunol. 2010;184 (9:5172–5178. doi: 10.4049/jimmunol.0903759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Strugnell R, Wijburg O, Brodnicki T. Measuring bacterial load and immune responses in mice infected with Listeria monocytogenes. J Vis Exp. 2011. p. 3076. [DOI] [PMC free article] [PubMed]
- Maghazachi AA, al-Aoukaty A, Schall TJ. C-C chemokines induce the chemotaxis of NK and IL-2-activated NK cells. Role for G proteins. J Immunol. 1994;153:4969–4977. [PubMed] [Google Scholar]
- Taub DD, Sayers TJ, Carter CR, Ortaldo JR. Alpha and beta chemokines induce NK cell migration and enhance NK-mediated cytolysis. J Immunol. 1995;155:3877–3888. [PubMed] [Google Scholar]
- Loetscher P, Seitz M, Clark-Lewis I, Baggiolini M, Moser B. Activation of NK cells by CC chemokines. Chemotaxis, Ca2+ mobilization, and enzyme release. J Immunol. 1996;156:322–327. [PubMed] [Google Scholar]
- Salazar-Mather TP, Lewis CA, Biron CA. Type I interferons regulate inflammatory cell trafficking and macrophage inflammatory protein 1alpha delivery to the liver. J Clin Invest. 2002;110:321–330. doi: 10.1172/JCI15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Mather TP, Orange JS, Biron CA. Early murine cytomegalovirus (MCMV) infection induces liver natural killer (NK) cell inflammation and protection through macrophage inflammatory protein 1alpha (MIP-1alpha)-dependent pathways. J Exp Med. 1998;187:1–14. doi: 10.1084/jem.187.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer M, von Stebut E. Macrophage inflammatory protein-1. Int J Biochem Cell Biol. 2004;36:1882–1886. doi: 10.1016/j.biocel.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Koch AE, Kunkel SL, Harlow LA, Mazarakis DD, Haines GK, Burdick MD, et al. Macrophage inflammatory protein-1 alpha. A novel chemotactic cytokine for macrophages in rheumatoid arthritis. J Clin Invest. 1994;93:921–928. doi: 10.1172/JCI117097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauriat C, Long EO, Ljunggren HG, Bryceson YT. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood. 2010;115:2167–2176. doi: 10.1182/blood-2009-08-238469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluman EM, Bartynski KJ, Avalos BR, Caligiuri MA. Human natural killer cells produce abundant macrophage inflammatory protein-1 alpha in response to monocyte-derived cytokines. J Clin Invest. 1996;97:2722–2727. doi: 10.1172/JCI118726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metkar SS, Menaa C, Pardo J, Wang B, Wallich R, Freudenberg M, et al. Human and mouse granzyme A induce a proinflammatory cytokine response. Immunity. 2008;29:720–733. doi: 10.1016/j.immuni.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 2009;113:5488–5496. doi: 10.1182/blood-2008-10-187179. [DOI] [PubMed] [Google Scholar]
- Silva A, Andrews DM, Brooks AG, Smyth MJ, Hayakawa Y. Application of CD27 as a marker for distinguishing human NK cell subsets. Int Immunol. 2008;20:625–630. doi: 10.1093/intimm/dxn022. [DOI] [PubMed] [Google Scholar]
- Fehniger TA, Shah MH, Turner MJ, VanDeusen JB, Whitman SP, Cooper MA, et al. Differential cytokine and chemokine gene expression by human NK cells following activation with IL-18 or IL-15 in combination with IL-12: implications for the innate immune response. J Immunol. 1999;162:4511–4520. [PubMed] [Google Scholar]
- Ferlazzo G, Munz C. NK cell compartments and their activation by dendritic cells. J Immunol. 2004;172:1333–1339. doi: 10.4049/jimmunol.172.3.1333. [DOI] [PubMed] [Google Scholar]
- Wagner L, Yang OO, Garcia-Zepeda EA, Ge Y, Kalams SA, Walker BD, et al. Beta-chemokines are released from HIV-1-specific cytolytic T-cell granules complexed to proteoglycans. Nature. 1998;391:908–911. doi: 10.1038/36129. [DOI] [PubMed] [Google Scholar]
- Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev. 2002;13:455–481. doi: 10.1016/s1359-6101(02)00045-x. [DOI] [PubMed] [Google Scholar]
- Proost P, Menten P, Struyf S, Schutyser E, De Meester I, Van Damme J. Cleavage by CD26/dipeptidyl peptidase IV converts the chemokine LD78beta into a most efficient monocyte attractant and CCR1 agonist. Blood. 2000;96:1674–1680. [PubMed] [Google Scholar]
- Anthony DA, Andrews DM, Watt SV, Trapani JA, Smyth MJ. Functional dissection of the granzyme family: cell death and inflammation. Immunol Rev. 2010;235:73–92. doi: 10.1111/j.0105-2896.2010.00907.x. [DOI] [PubMed] [Google Scholar]
- Henkart PA, Berrebi GA, Takayama H, Munger WE, Sitkovsky MV. Biochemical and functional properties of serine esterases in acidic cytoplasmic granules of cytotoxic T lymphocytes. J Immunol. 1987;139:2398–2405. [PubMed] [Google Scholar]
- Pao LI, Sumaria N, Kelly JM, van Dommelen S, Cretney E, Wallace ME, et al. Functional analysis of granzyme M and its role in immunity to infection. J Immunol. 2005;175:3235–3243. doi: 10.4049/jimmunol.175.5.3235. [DOI] [PubMed] [Google Scholar]
- Salti SM, Hammelev EM, Grewal JL, Reddy ST, Zemple SJ, Grossman WJ, et al. Granzyme B regulates antiviral CD8+ T cell responses. J Immunol. 2011;187:6301–6309. doi: 10.4049/jimmunol.1100891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullbacher A, Ebnet K, Blanden RV, Hla RT, Stehle T, Museteanu C, et al. Granzyme A is critical for recovery of mice from infection with the natural cytopathic viral pathogen, ectromelia. Proc Natl Acad Sci USA. 1996;93:5783–5787. doi: 10.1073/pnas.93.12.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebnet K, Hausmann M, Lehmann-Grube F, Mullbacher A, Kopf M, Lamers M, et al. Granzyme A-deficient mice retain potent cell-mediated cytotoxicity. EMBO J. 1995;14:4230–4239. doi: 10.1002/j.1460-2075.1995.tb00097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. The immunopathogenesis of sepsis. Nature. 2002;420:885–891. doi: 10.1038/nature01326. [DOI] [PubMed] [Google Scholar]
- Harrison C. Sepsis: calming the cytokine storm. Nat Rev Drug Discov. 2010;9:360–361. doi: 10.1038/nrd3162. [DOI] [PubMed] [Google Scholar]
- Remick DG. Pathophysiology of sepsis. Am J Pathol. 2007;170:1435–1444. doi: 10.2353/ajpath.2007.060872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz A, Stegmann KA, Riley EM. Activation of natural killer cells during microbial infections. Front Immunol. 2011;2:88. doi: 10.3389/fimmu.2011.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini C, Cassatella MA. The defensive alliance between neutrophils and NK cells as a novel arm of innate immunity. J Leukoc Biol. 2011;89:221–233. doi: 10.1189/jlb.0510250. [DOI] [PubMed] [Google Scholar]
- Thoren FB, Riise RE, Ousback J, Della Chiesa M, Alsterholm M, Marcenaro E, et al. Human NK Cells induce neutrophil apoptosis via an NKp46- and Fas-dependent mechanism. J Immunol. 2012;188:1668–1674. doi: 10.4049/jimmunol.1102002. [DOI] [PubMed] [Google Scholar]
- Zeng X, Moore TA, Newstead MW, Hernandez-Alcoceba R, Tsai WC, Standiford TJ. Intrapulmonary expression of macrophage inflammatory protein 1alpha (CCL3) induces neutrophil and NK cell accumulation and stimulates innate immunity in murine bacterial pneumonia. Infect Immun. 2003;71:1306–1315. doi: 10.1128/IAI.71.3.1306-1315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Poot SA, Westgeest M, Hostetter DR, Van Damme P, Plasman K, Demeyer K, et al. Human and mouse granzyme M display divergent and species-specific substrate specificities. Biochem J. 2011;437:431–442. doi: 10.1042/BJ20110210. [DOI] [PubMed] [Google Scholar]
- Kaiserman D, Bird CH, Sun J, Matthews A, Ung K, Whisstock JC, et al. The major human and mouse granzymes are structurally and functionally divergent. J Cell Biol. 2006;175:619–630. doi: 10.1083/jcb.200606073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapaque N, Walzer T, Meresse S, Vivier E, Trowsdale J. Interactions between human NK cells and macrophages in response to Salmonella infection. J Immunol. 2009;182:4339–4348. doi: 10.4049/jimmunol.0803329. [DOI] [PubMed] [Google Scholar]
- Walzer T, Dalod M, Robbins SH, Zitvogel L, Vivier E. Natural-killer cells and dendritic cells: "l'union fait la force". Blood. 2005;106:2252–2258. doi: 10.1182/blood-2005-03-1154. [DOI] [PubMed] [Google Scholar]
- Andrews DM, Estcourt MJ, Andoniou CE, Wikstrom ME, Khong A, Voigt V, et al. Innate immunity defines the capacity of antiviral T cells to limit persistent infection. J Exp Med. 2010;207:1333–1343. doi: 10.1084/jem.20091193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaix J, Tessmer MS, Hoebe K, Fuséri N, Ryffel B, Dalod M, et al. Cutting edge: priming of NK cells by IL-18. J Immunol. 2008;181:1627–1631. doi: 10.4049/jimmunol.181.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehniger TA, Cai SF, Cao X, Bredemeyer AJ, Presti RM, French AR, et al. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26:798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Parham P, Miller JS. NK cells—from bench to clinic. Biol Blood Marrow Transplant. 2012;18 (1 Suppl:S2–S7. doi: 10.1016/j.bbmt.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusel JW, Wesselschmidt RL, Shresta S, Russell JH, Ley TJ. Cytotoxic lymphocytes require granzyme B for the rapid induction of DNA fragmentation and apoptosis in allogeneic target cells. Cell. 1994;76:977–987. doi: 10.1016/0092-8674(94)90376-x. [DOI] [PubMed] [Google Scholar]
- Kagi D, Ledermann B, Burki K, Seiler P, Odermatt B, Olsen KJ, et al. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature. 1994;369:31–37. doi: 10.1038/369031a0. [DOI] [PubMed] [Google Scholar]
- Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.