Abstract
Inhibition of protein tyrosine phosphatase 1B (PTP1B) has been suggested as an attractive target to improve insulin sensitivity in different cell types. In the present work, we have investigated the effect of PTP1B deficiency on the response of human and murine macrophages. Using in vitro and in vivo approaches in mice and silencing PTP1B in human macrophages with specific siRNAs, we have demonstrated that PTP1B deficiency increases the effects of pro-inflammatory stimuli in both human and rodent macrophages at the time that decreases the response to alternative stimulation. Moreover, the absence of PTP1B induces a loss of viability in resting macrophages and mainly after activation through the classic pathway. Analysis of early gene expression in macrophages treated with pro-inflammatory stimuli confirmed this exacerbated inflammatory response in PTP1B-deficient macrophages. Microarray analysis in samples from wild-type and PTP1B-deficient macrophages obtained after 24 h of pro-inflammatory stimulation showed an activation of the p53 pathway, including the excision base repair pathway and the insulin signaling pathway in the absence of PTP1B. In animal models of lipopolysaccharide (LPS) and D-galactosamine challenge as a way to reveal in vivo inflammatory responses, animals lacking PTP1B exhibited a higher rate of death. Moreover, these animals showed an enhanced response to irradiation, in agreement with the data obtained in the microarray analysis. In summary, these results indicate that, although inhibition of PTP1B has potential benefits for the treatment of diabetes, it accentuates pro-inflammatory responses compromising at least macrophage viability.
Keywords: protein tyrosine phosphatase, cell viability, p53, radiation sensitivity
Protein tyrosine phosphatase 1B (PTP1B) is a ubiquitously expressed phosphatase that has emerged as a relevant modulator of signaling pathways initiated by the activation of the tyrosine kinase receptor superfamily.1 Clinically relevant, PTP1B is a critical node of insulin signaling due to its ability to dephosphorylate and inactivate the insulin receptor, thereby switching off insulin signaling.2, 3 Indeed, PTP1B-deficient mice are a unique model of insulin hypersensitivity due to enhanced insulin action.4, 5, 6 These mice are protected against diet4, 6 and age-induced obesity and insulin resistance linked to a low grade of chronic inflammation in white adipose tissue.7, 8 PTP1B is also involved in the control of immune cell signaling,9, 10 controlling cytokine signaling pathways by dephosphorylation of janus kinase 2 (JAK2), non-receptor tyrosine-protein kinase 2 (TYK2) and signal transducer and activator of transcription 5 (STAT5).11, 12 Moreover, it has been reported that interleukin-4 (IL-4) induced PTP1B mRNA in a phosphatidylinositol 3-kinase (PI3K)-dependent manner and enhanced PTP1B protein stability to suppress IL-4-induced STAT6 signaling.13
PTP1B-deficient mice exhibited an increase in monocyte/macrophages in the spleen and the bone marrow.14 This was due to a decreased threshold of response to macrophage colony-stimulating factor (M-CSF) by enhancing tyrosine phosphorylation of the activation loop of its receptor (M-CSF1R). In addition to lineage specification, PTP1B modulates activation of monocytes/macrophages. In fact, in the absence of PTP1B tissue macrophages display an activated phenotype assessed by the increased expression of CD80 that has a role in the maintenance of inflammation. However, this effect might require developmental adaptations in macrophages since short-term treatment of mice with the PTP1B inhibitor suramin protects against apoptotic liver damage induced by CD95 and against endotoxic shock mediated by tumor necrosis factor-α (TNF-α).15
In this work, we have characterized the effect of PTP1B deficiency on the early signaling in response to pro-inflammatory challenge in murine and human macrophages. Our data show that in the absence of PTP1B both classic and innate responses (M1 polarization) are overactivated at the time that alternative polarization (M2) is reduced.16, 17 Studies using in vivo models provide support to these enhanced pro-inflammatory responses. However, these data also suggest that the broad use of PTP1B inhibitors, although with potential benefits over the insulin signaling pathway, might exert undesirable effects on response to stressors of the immune system including the fine tuning of the pro-inflammatory and pro-resolution balance.
Results
PTP1B deficiency results in enhanced M1 responses and attenuated activation by M2 stimuli
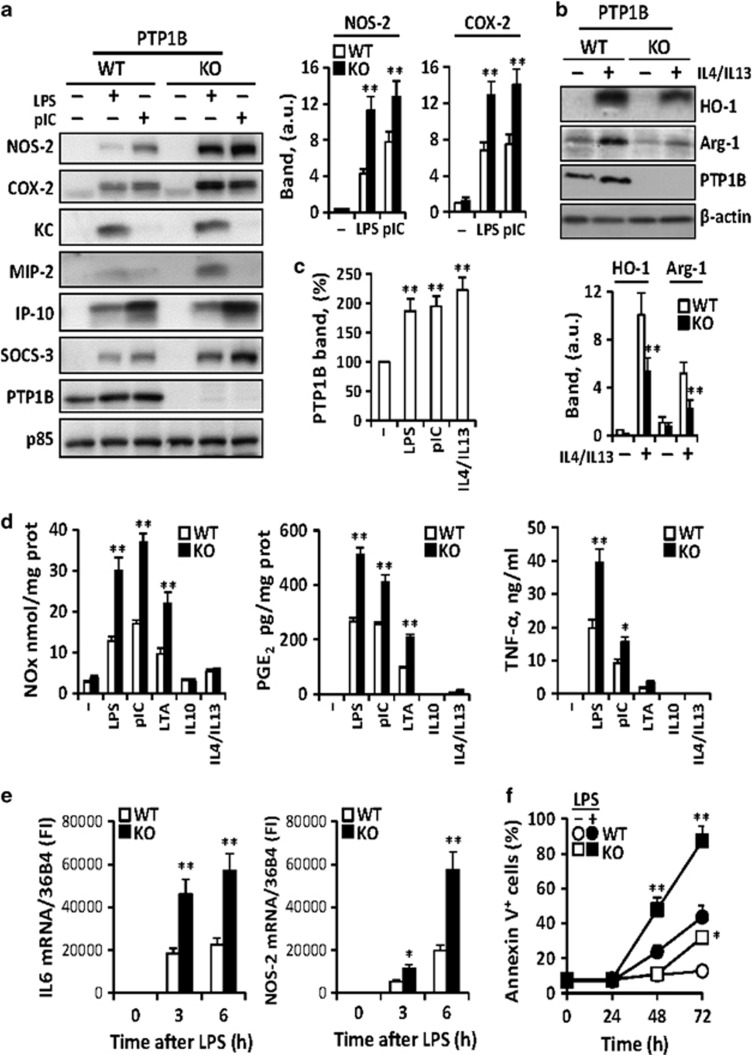
Peritoneal macrophages from wild-type (WT) and PTP1B-deficient mice were activated with lipopolysaccharide (LPS) and polyriboinosinic:polyribocytidylic acid (polyI:C) (M1 stimuli) and the expression of genes characteristic of the innate immune response was determined. PTP1B deficiency results in an enhancement in the levels of these M1 representative genes (Figure 1a). In the same way, challenge with combinations of IL-4 and IL-13 (M2 cytokines) decreased heme oxygenase 1 (HO-1) and arginase-1 (Arg-1) expression reflecting attenuation of M2 responses in PTP1B knockout (KO) macrophages (Figure 1b). Interestingly, PTP1B levels were upregulated upon M1 or M2 challenge (Figure 1c). The accumulation of nitric oxide (NO), prostaglandin E2 (PGE2) and TNF-α in the culture medium (Figure 1d), and the time course of the rise in the mRNA levels of IL-6 and nitric oxide synthase-2 (NOS-2) (Figure 1e) were potentiated in PTP1B KO macrophages upon M1 activation. Moreover, a significant loss of viability was observed in PTP1B-deficient cells after 48–72 h of culture (Figure 1f).
Figure 1.
Effect of PTP1B deficiency on the polarization of macrophages. Peritoneal macrophages from wild-type (WT) or PTP1B-deficient (KO) mice were isolated after thioglycollate eliciting. Cells were stimulated with 200 ng/ml of LPS or 25 μg/ml polyI:C (M1 stimuli) and the levels of the indicated proteins were determined at 24 h (a). The response to M2 stimuli was analyzed after challenge for 24 h with a mixture of 20 ng/ml each IL-4+IL-13 (b). The levels of PTP1B in M1 and M2 polarized macrophages were determined (c). The accumulation of NO, PGE2 and TNF-α in the incubation medium was determined after 24 h of activation with 200 ng/ml LPS, 25 μg/ml polyI:C, 5 μg/ml LTA, 20 ng/ml IL-10 or 20 ng/ml of IL-4+IL-13 (d). The time course of mRNA levels of IL-6 and NOS-2 after activation with 200 ng/ml LPS (e) and of cells undergoing apoptosis by annexin V binding criteria (f) were determined. Results show the mean±S.D. of three independent experiments. *P<0.05; **P<0.01 versus the same condition in the WT cells or non-treated cells (c). (a–c) The band intensities of NOS-2, COX-2 (a), HO-1, Arg-1 (b) and PTP1B (c) after normalization using p85 and β-actin, respectively, are shown
Early pro-inflammatory signaling in PTP1B-deficient peritoneal macrophages
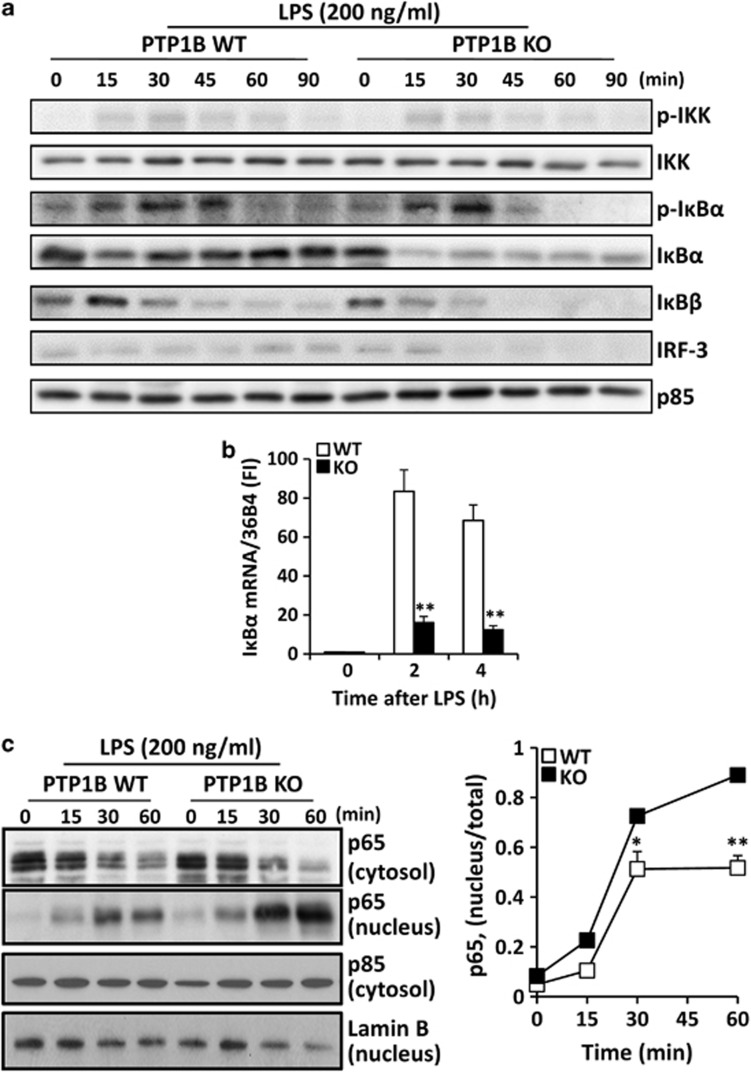
To evaluate the impact of PTP1B deficiency on the early response of macrophages to LPS, the nuclear factor-κB (NF-κB) pathway was analyzed. The degradation of IκBα was more efficient in PTP1B KO versus the WT counterparts, at the time that the late recovery by resynthesis was impaired (Figure 2a). This was also confirmed when the kinetics of the IκBα mRNA levels was determined (Figure 2b). Similar results were observed for IκBβ. Regarding the interferon (IFN) regulated factor 3 (IRF-3) pathway, our data show a lesser upregulation in PTP1B KO macrophages, confirming previous work.18 Finally, the absence of PTP1B resulted in a higher translocation of p65 from the cytosol to the nucleus (Figure 2c).
Figure 2.
PTP1B deficiency enhances IκBα degradation and impairs IκBα recovery. Macrophages from WT and PTP1B KO mice were stimulated with 200 ng/ml of LPS for the indicated times and the NF-κB and IRF3 pathways were analyzed (a). The upregulation of IκBα mRNA expression as a sensor of resetting of NF-κB was determined by qPCR (b). The nuclear translocation of p65 was determined by western blotting using p85 and lamin B as markers of the corresponding cytosolic and nuclear fractions, respectively (c). Results show the mean±S.D. of four independent experiments (b) or a representative blot out of four (a, c). *P<0.05; **P<0.01 versus the same condition in the WT cells
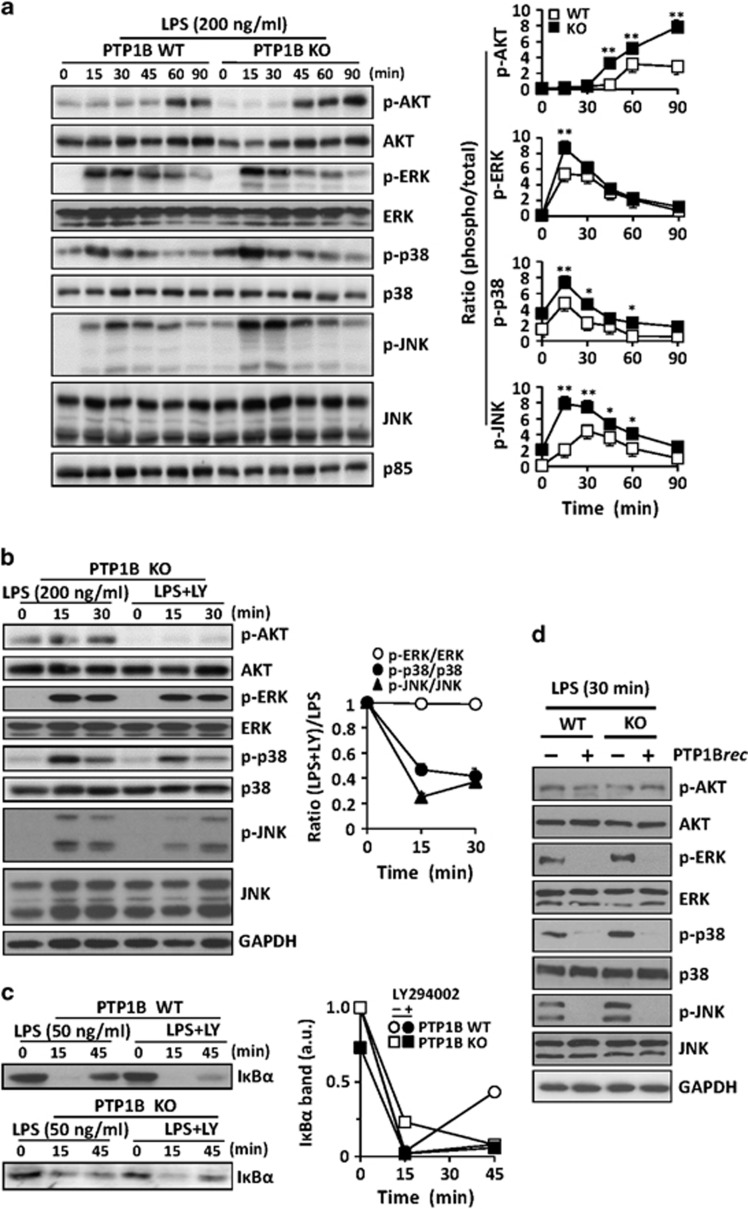
The phosphorylation state of protein kinase B (AKT) and the mitogen-activated protein kinases (MAPKs), two well-known targets activated after LPS challenge, was investigated. As Figure 3a shows, AKT phosphorylation was sustained in cells lacking PTP1B. The same response was evidenced in the MAPKs extracellular signal-regulated kinase (ERK), p38 and c-Jun N-terminal kinase (JNK). Since PI3K has a role in the attenuation of the response to LPS in macrophages,19, 20 cells lacking PTP1B were incubated with the PI3K inhibitor LY294002 and then activated with LPS. As Figure 3b shows, AKT phosphorylation was suppressed whereas the phosphorylation of p38 and JNK was significantly attenuated. However, the levels of phospho-ERK remained unchanged regardless the presence of LY294002. Since PI3K inhibition results in enhanced IκBα degradation in response to LPS,20, 21 the contribution of this pathway to the phenotype observed in PTP1B-deficient cells was investigated.19, 21, 22, 23 Macrophages challenged with low doses of LPS exhibited a similar response in terms of IκBα degradation with negligible recovery at 45 min after PI3K inhibition regardless the presence of PTP1B (Figure 3c). In the same regard, treatment with recombinant PTP1B of extracts from PTP1B WT or KO macrophages activated for 30 min with LPS resulted in a robust dephosphorylation of all MAPKs but not of AKT (Figure 3d).
Figure 3.
PTP1B deficiency enhances AKT and MAPK activation. Macrophages from WT and PTP1B KO mice were stimulated with 200 ng/ml of LPS and the phosphorylation of AKT, ERK, p38 and JNK was determined at the indicated times (a). To evaluate the effect of the PI3K pathway in the modulation of AKT and MAPK activities, macrophages from PTP1B KO mice were treated for 10 min before LPS activation with 10 μM LY29004. At the indicated times, the phosphorylation state of these proteins was determined (b). In parallel to the precedent assays, the levels of IκBα were determined using a lower LPS concentration (50 ng/ml) as stimulus (c). The effect of recombinant PTP1B on the dephosphorylation of extracts from WT and PTP1B macrophages activated for 30 min with 200 ng/ml of LPS was assayed in vitro after 30 min of incubation at 30 °C with recombinant PTP1B (d). Results show the mean±S.D. of three independent experiments, or a representative dephosphorylation experiment (d). *P<0.05; **P<0.01 versus the same condition in the WT cells
Mini-array and microarray analysis of pro-inflammatory responses in macrophages from PTP1B-deficient mice
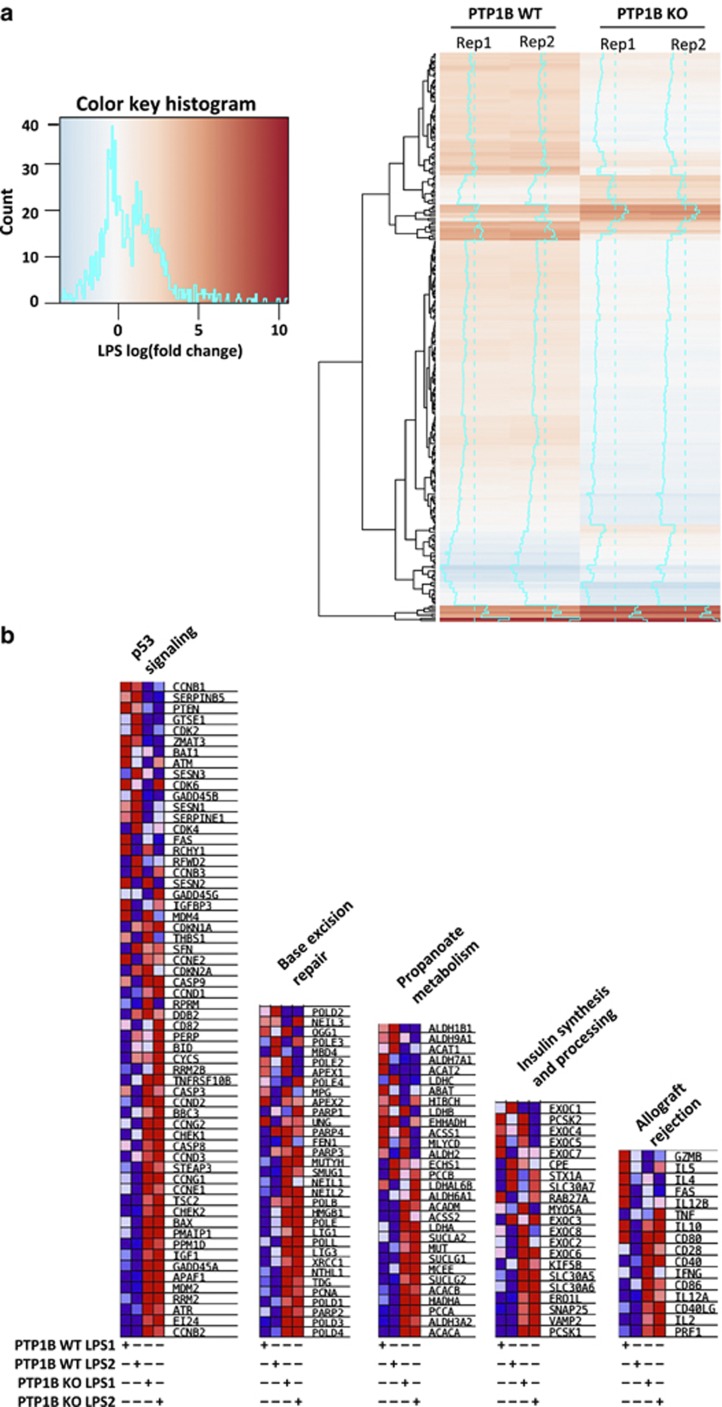
Because PTP1B-deficient macrophages have an enhanced response to pro-inflammatory stimuli a mini-array analysis of chemokine and receptors was performed. As Supplementary Figure S1 summarizes the levels of Ccl2, Ccl5, Ccl7 and Ccl9, chemokines that recruit inflammatory cells, were significantly elevated in PTP1B KO macrophages. To better understand these results, we performed microarray gene expression analysis in PTP1B WT or KO macrophages. In all, 276 probes were differentially expressed (DE, false discovery rate (FDR)<0.05; Figure 4a) in WT versus PTP1B KO macrophages stimulated with LPS. Among the DE genes, we identified chemokines (IL-6, CCL2 and CCL17), which showed significantly higher expression in PTP1B KO cells, in agreement with qPCR and mini-array results. Additionally, chemokine (C-X-C motif) ligand 5 (CXCL5) also showed higher expression in PTP1B-deficient macrophages. However, an important fraction of DE genes tend to show lower upregulation after 24 h of LPS challenge in PTP1B KO cells (Figure 4a, heatmap of only DE genes). Interestingly, Toll-like receptor 4 (TLR4) was downregulated at 24 h in PTP1B KO macrophages, suggesting a complex scenario where deregulation of the inflammatory response in PTP1B-deficient macrophages is occurring at different levels of the regulatory network. To get biological insights about the pathways altered, we performed gene set enrichment analysis (GSEA) using Kyoto encyclopedia of genes and genomes (KEGG) and REACTOME pathways (Figure 4b). Four KEGG pathways were significantly upregulated (FDR<0.25) in PTP1B KO macrophages (‘p53 signaling', ‘Base excision repair', ‘Propanoate metabolism' and ‘Allograft rejection') and one REACTOME pathway (‘Insulin Synthesis and Processing'). The alterations in insulin synthesis reflect the known role of PTP1B in insulin signaling,1, 2, 3, 5, 7, 12, 24 while the DE in propanoate metabolism genes suggests a possible novel role for PTP1B in this pathway. In fact, propionic acid has been associated with lipogenesis and glucose uptake and has an anti-inflammatory effect on human macrophages.25, 26 The so-called ‘Allograft rejection' pathway included PTP1B upregulated inflammatory genes such as perforin 1, IL-2, CD40LG and IL-12A. Interestingly, the alteration in p53 and base-excision pathways could be directly related with the higher sensitivity of PTP1B-deficient mice to high doses of irradiation. Notably, PTP1B KO macrophages showed upregulation of many genes activated after DNA damage (i.e., PCNA, ATR, CHEK2, GADD45A and BAX) and this exacerbated response could explain the apoptosis of these cells at 48 h (Figure 1e).
Figure 4.
Gene expression profiling of WT and PTP1B-deficient macrophages treated with LPS. Macrophages were activated with 200 ng/ml of LPS for 24 h and the RNA was extracted, analyzed for quality in a Bioanalyzer and submitted to microarray hybridization (Agilent). Heatmap showing the microarray probes with differential response (FDR<0.05) to LPS between WT and PTP1B KO macrophages (a). Gene set enrichment analysis (GSEA) of KEGG and REACTOME pathways in macrophages from PTP1B KO mice after pro-inflammatory stimulation (b). The genes in significantly enriched pathways (FDR<0.25) are shown and the color code corresponds to the standardized gene expression per row (not LPS fold change as in a). Red indicates higher expression and blue lower expression
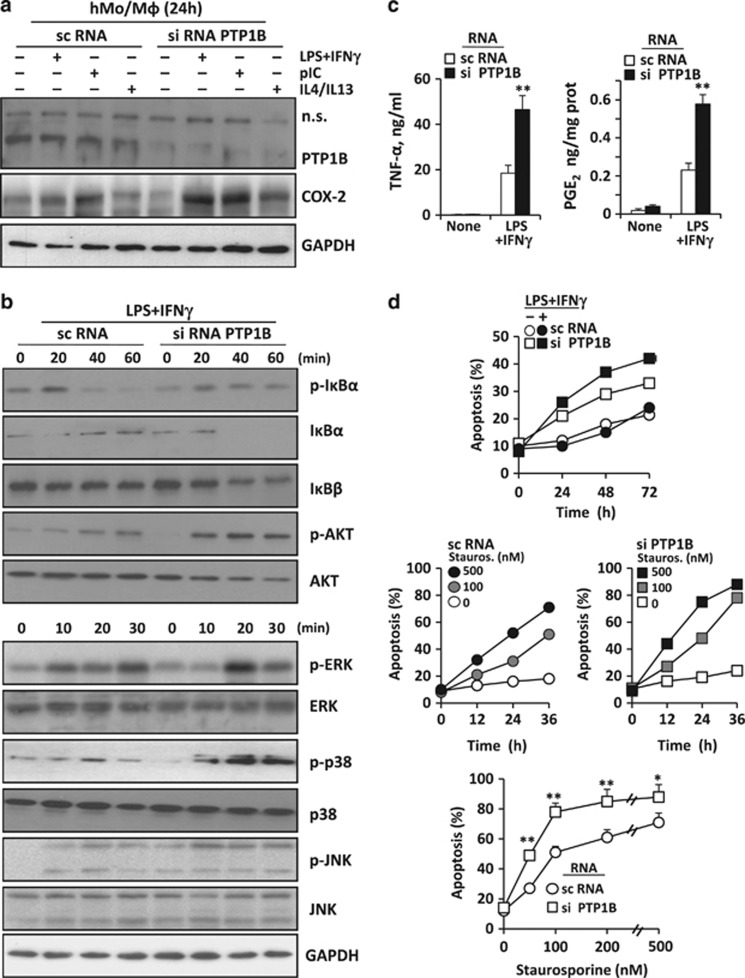
Effect of silencing human PTP1B on the response of human monocytes/macrophages to pro-inflammatory stimulation
PTP1B siRNA efficiently decreased the levels of PTP1B at the time that moderately increased the expression of cyclooxygenase-2 (COX-2) after pro-inflammatory challenge (Figure 5a). In addition to this, the response to LPS+IFN-γ resulted in a decrease in IκBα (without recovery as in the murine counterparts) and accelerated IκBβ degradation (Figure 5b). The phosphorylation of AKT and MAPKs was notably enhanced after silencing PTP1B (Figure 5b). Also, TNF-α and PGE2 levels measured in the culture medium increased in cells treated with PTP1B siRNA (Figure 5c). This enhanced response to pro-inflammatory challenge in cells with reduced PTP1B levels resulted in a higher susceptibility to apoptosis in response to LPS and staurosporine (Figure 5d). Notably, the bright-field morphology of murine and human macrophages lacking PTP1B was very similar, exhibiting a more fusiform shape than the corresponding WT cells (Supplementary Figure S2A). Finally, inhibition of PTP activity with peroxovanadate (POV) showed a similar profile to that observed after PTP1B silencing, including the lack in recovery of IκBα, a higher translocation of p65 to the nucleus (Supplementary Figure S2B), and a loss in viability, a process that was partially inhibited by the caspase inhibitor z-VAD (Supplementary Figure S2C).
Figure 5.
Effect of PTP1B silencing on the response of human macrophages. Human monocytes from buffy coats were differentiated into macrophages and treated with a mixture of sc- or siRNA oligonucleotides to silence PTP1B. After incubation for 24 h with 200 ng/ml of LPS plus 20 ng/ml of human IFN-γ, 25 μg/ml of polyI:C or 20 ng/ml of each human IL-4+IL-13, the levels of PTP1B and COX2 were determined (a). The time course of the phosphorylation and total levels of proteins related to NF-κB, AKT and MAPKs were determined (b). The release of TNF-α and PGE2 to the culture medium was determined with specific kits (c). The induction of apoptosis, measured as the annexin V-positive population, was determined in both resting and LPS+IFN-γ activated cells (d). In addition to this, to determine the sensitivity of cells silenced for PTP1B to apoptosis in response to pro-inflammatory dependent stimuli the time course and the sensitivity at 36 h to different concentrations of staurosporine (as an stressor to promote mitochondrial-dependent apoptosis) were determined (d). Results show a representative experiment or the mean±S.D. of three independent experiments. *P<0.05; **P<0.01 versus the same condition in cells treated with scRNA
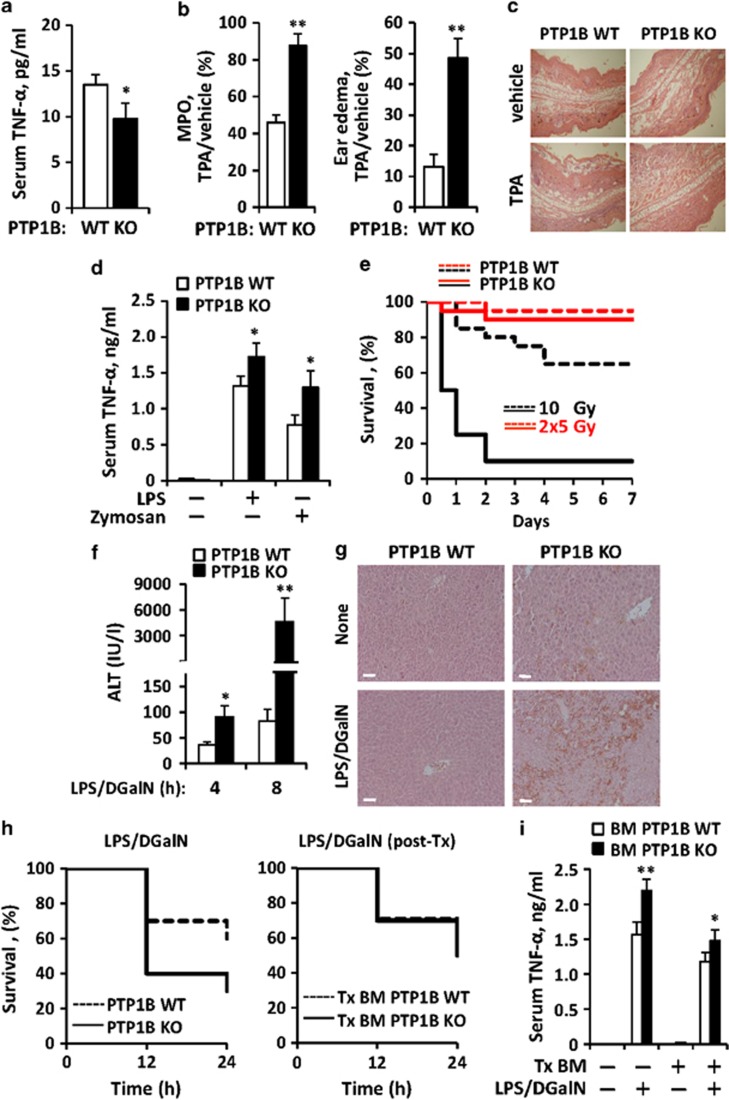
In vivo effects of pro-inflammatory activation in PTP1B-deficient mice
To determine the in vivo contribution of PTP1B to pro-inflammatory responses, the basal levels of TNF-α in serum (Figure 6a), the development of ear edema (Figures 6b and c), the rise in TNF-α levels in serum after administration of LPS or zymosan (Figure 6d), the sensitivity to irradiation (Figure 6e) and the response to LPS/D-galactosamine (D-GalN) challenge (Figures 6f–i) were analyzed. The basal levels of TNF-α in serum were significantly lower in PTP1B KO mice than in the WT counterparts. However, in the ear pro-inflammatory model induced after tetradecanoylphorbol acetate (TPA) application, PTP1B KO mice exhibited a higher accumulation of neutrophils, as measured by the myeloperoxidase (MPO) activity criteria and edema (Figures 6b and c). Also, the TNF-α levels measured in serum after i.p. challenge with LPS or zymosan were significantly higher in the PTP1B KO counterparts. Interestingly, these animals were very sensitive to irradiation and at least 90% died in <2 days when treated with 10 Gy, linking this response to the activation of the p53 and excision base repair pathways observed in the microarray (Figure 4b). Opposite to this situation 70% of the WT mice were alive. When the same quantity of irradiation was administered in two doses in a 4-h interval, >90% of the animals survived regardless the absence of PTP1B.
Figure 6.
In vivo effect of PTP1B deficiency on the response of mice to different pro-inflammatory challenges. WT or PTP1B KO male mice matched in age were used in these experiments. The serum levels of TNF-α of untreated animals (n=12) were determined (a). The ear MPO activity and edema (n=10 animals per condition) after topic administration of TPA was calculated as ratio versus the ear treated with vehicle (b). Fixed ears were stained with eosin/hematoxylin to show the structure and with (sagittal sections; c). The serum levels of TNF-α of untreated animals or i.p. injected LPS (1 mg/kg body weight) or zymosan (30 mg/kg body weight) were determined at 1 h (n=7 animals per condition) (d). The sensitivity to irradiation after a unique dose of 10 Gy or two doses of 5 Gy administered in an interval of 4 h (n=15 per group) was evaluated as survival after 7 days post transplantation (Tx) of the corresponding bone marrow; P=0.0005 versus the WT at 10 Gy (e). The liver injury, determined by the measurement of the ALT activity in the serum (f) and structure (eosin/hematoxylin staining at 4 h; white bar=100 μm; g), and the survival of animals after i.p. administration of a single dose of LPS/D-GalN (n=12 animals per condition) were determined; P=0.074 versus the WT (h, left). Alternatively, a series of WT animals underwent irradiation (10 Gy) as depicted in (e), and were subsequently submitted to bone marrow transplantation from WT or PTP1B KO mice. After restitution of the immune system (12 weeks), these animals were submitted to LPS/D-GalN challenge and survival was determined (h, right). The TNF-α levels after 1 h of treatment with LPS/D-GalN were determined in serum from animals of (i). Results show the mean±S.D. of the indicated number of animals and/or the Kaplan–Meier representation of survival (e and h). *P<0.05; **P<0.01 versus the same condition in the WT animals or samples
PTP1B KO mice challenged with LPS/D-GalN exhibited a higher liver injury than the WT counterparts as assessed by the rapid rise in serum alanine aminotransferase (ALT) activity (Figure 6f) and the loss of liver integrity (Figure 6g). These PTP1B KO mice exhibited a higher rate of death upon LPS/D-GalN administration (Figure 6h, left), that was abrogated when the experiments were repeated in irradiated WT animals reconstituted with bone marrow from PTP1B KO donor mice (Figure 6h, right), suggesting that resident liver macrophages (Kupffer cells?), or even hepatocytes, have a key role in the liver-induced animal death. Interestingly, the TNF-α levels in the serum of these animals at 1 h reflected a minimal, but statistically significant increase when the circulating monocytes were from PTP1B KO mice (Figure 6i). In addition to this approach, mice were treated with clodronate to remove resident macrophages. Under these conditions, reconstitution of WT-irradiated mice with the bone marrow from PTP1B KO animals failed to show differences in survival after LPS/D-GalN challenge; however, the levels of TNF-α in serum after LPS/D-GalN were significantly lower than those obtained in the above reported experimental models (avoiding clodronate treatment), which might explain the absence of differences in survival between both groups (Supplementary Figure S3A and B). Regarding the quality of the macrophage replenishment, RT-PCR analysis ensured that the liver population after transplantation was from the PTP1B KO donors (Supplementary Figure S3C) and, in the absence of transplantation, all F4/80-positive liver population was removed after 48 h of clodronate treatment (Supplementary Figure S3D).
Discussion
Macrophages respond very efficiently and regulate several signaling pathways in a strict temporal pattern in which resolution is a key component.27, 28 Among the systems that modulate this response, protein phosphatases exert a significant role.29 Several reports suggested that PTP1B is an important modulator of the activation process of macrophages restricting the time of pro-inflammatory signaling in response to TLR and to both type I and type II IFN signaling.14, 18, 30, 31, 32 Also, PTP1B could be increased or activated by inflammatory factors such as TNF-α.33 Intriguingly, these observations contrast with results obtained in myeloid-cell (LysM)-PTP1B KO mice, despite to show enhanced NOS-2 expression and accumulation of nitrite in the medium upon LPS challenge,34 and in a murine model of experimental colitis.35 Opposite to these results, other studies have identified an essential role for PTP1B in obesity-induced inflammation, in regulating leukocyte recruitment during allergic inflammation and in macrophage development.30, 32, 36 At the same time, evidence from the area of diabetes has suggested that inhibition of PTP1B might be envisaged as a way to prolong insulin action.3, 5, 12, 36 Keeping in mind these ideas, we have investigated whether the deficiency of PTP1B in murine and human macrophages affects the regulation of the response of these cells to pro-inflammatory and anti-inflammatory stimuli. Our data show that peritoneal macrophages from PTP1B KO mice presented an exacerbated inflammatory response versus the WT, with increased production of M1 polarization markers. Moreover, this enhanced activation in PTP1B KO macrophages was not restricted to TLR4 signaling but also occurred in response to TLR2 (lipoteichoic acid, LTA) and TLR3 (polyI:C) challenge, confirming preliminary data in the Raw 264.7 macrophage cell line.18 Conversely, in the absence of PTP1B, macrophages show an impaired response to IL-4 plus IL-13 resulting in attenuated M2 polarization that suggests a dual role for PTP1B in the regulation of M1/M2 commitment. It is well known that induction of target genes upon TLR stimulation requires a combination of downstream signaling events, including transcriptional activation via NF-κB factors and activation of the PI3K/AKT and MAPK pathways.37, 38, 39 The enhanced signaling of these three pathways in PTP1B-deficient macrophages suggests that PTP1B acts as a common negative regulator of NF-κB, AKT and MAPK activities. In this regard, the more efficient degradation of IκBα together with its delayed resynthesis in PTP1B KO macrophages resulted in a persistent and augmented p65 translocation to the nucleus and, therefore, of NF-κB DNA binding activity consistently with the observed exacerbated inflammatory responses.
Several studies have shown that the PI3K/AKT pathway exerts a key role in the regulation of the innate immune responses under pro-inflammatory conditions and against different pathogens.20, 21, 22, 23, 39 In agreement with previous reports performed in hepatocytes,7, 36 in this study we have shown that PTP1B deficiency induces an increased and persistent AKT phosphorylation in human and mouse macrophages. This AKT activation was dependent on PI3K activity as far as it was suppressed by the broad PI3K inhibitor LY294002. Moreover, in the presence of this inhibitor the levels of phospho-p38 and phospho-JNK were significantly reduced whereas the phosphorylation of ERK remained unaffected, indicating a complex regulatory network at early steps of the pro-inflammatory signaling, and orchestrated by the PI3K/AKT pathway. In this regard, the inhibition of PI3K/AKT resulted in an almost indistinguishable overactivation of the NF-κB pathway with a delayed resetting of IκB levels, regardless the presence or absence of PTP1B (see Figure 3c). PTP1B dephosphorylates downstream substrates through its phosphatase activity and we found that PTPT1B deficiency significantly increased the phosphorylation of MAPKs and AKT; however, the treatment of extracts from WT or PTP1B KO macrophages with recombinant PTP1B lead to a significant dephosphorylation of all MAPKs but not of AKT. These findings suggest that MAPKs are direct substrates of PTP1B and, therefore, PTP1B negatively regulates LPS responses in macrophages through its phosphatase activity.
Another relevant aspect in PTP1B biology is its involvement in the endocytosis of IFN-α/β receptor chain 1 in human but not in murine cells31 and therefore, restricting antiviral therapies. However, in terms of regulation of inflammatory responses both human and murine macrophages behave similarly. Indeed, we observed that silencing PTP1B in human macrophages significantly reduced cell viability, counteracting in this way the potential beneficial aspects of the use of PTP1B inhibitors.40, 41, 42 Moreover, this loss of viability due to reduced PTP1B activity, even under basal conditions, was also reproduced by broad and transient PTP inhibitors, such as POV, indicating that the activity of these PTPs is required at specific moments of cell activation to maintain cell viability. Therefore, caution should be taken in the development of strategies of inhibition of PTP1B activity, at least under circumstances in which maintenance of cell viability is a requisite (i.e., phagocytosis). In addition to this, the mini-array data obtained after short periods of pro-inflammatory activation confirmed the enhanced pro-inflammatory responses expected from targets downregulated by this PTP. However, when we performed a microarray analysis at 24 h after LPS challenge we realized that the main differences in gene expression were not related to pro-inflammatory genes, but to very specific pathways: the p53 and DNA excision/repair pathways and the propanoate metabolism pathway. Interestingly, the link between the p53 pathway and the DNA excision/repair pathway fits with the observed loss of viability of PTP1B-deficient cells. Moreover, when we irradiated the animals to perform bone marrow transplantation, we were surprised by the extreme sensitivity of the PTP1B KO mice, compatible with an enhanced DNA injury. This role of PTP1B in protecting mice against irradiation injury is a relevant issue of the present work and we reasoned that perhaps strategies to improve transient PTP1B activity in the course of irradiation will have beneficial effects. Moreover, the levels of PTP1B are consistently increased after pro-inflammatory challenge in macrophages, perhaps as a way to improve cell viability under conditions of intense synthesis of reactive oxygen and/or nitrogen species that may damage the DNA.
Regarding the in vivo experiments in PTP1B-deficient animals, according to our results and those from other groups,30 we observed an enhanced ear edema in these animals, compatible with an increased leukocyte trafficking, as deduced by the higher levels of MPO in the ear samples. In addition to this, we evaluated the effects of PTP1B deficiency in the LPS/D-GalN model that evidences TNF-α toxicity in the liver. In this way, under identical conditions, animal death (mainly by liver hemorrhage) was significantly more prominent in the PTP1B KO mice, together with minimally, but statistically significant higher serum levels of TNF-α in PTP1B KO mice. However, when we performed bone marrow transplantation and reintroduced the PTP1B KO myeloid system into the WT animals, the rates of survival were identical between both groups, suggesting that perhaps the main contribution to the animal death upon LPS/D-GalN challenge was not the circulating monocyte/macrophages, but the tissue resident population of the liver. Using clodronate administration as a strategy to deplete tissue macrophages and to replenish the organs with the myeloid cells coming from the transplanted bone marrow,43 we analyzed the effect of reintroduction of myeloid cells from PTP1B KO mice into irradiated WT counterparts and then performed the LPS/D-GalN challenge; however, again it was impossible to distinguish between the two groups. One possible explanation is that the levels of TNF-α reached in both models were very similar and lower than those measured in the absence of clodronate depletion. Alternatively, the fact that our PTP1B KO mice lack of this enzyme in all tissues, including the liver, cannot exclude a contribution of the hepatocytes as a result of a paracrine effect to the increased sensitivity to LPS/D-GalN observed in the intact PTP1B KO model. Finally and in an intriguing way, it has been reported that PTP1B deficiency confers resistance against apoptosis induced by pro- and anti-inflammatory stimuli such as transforming growth factor β (TGF-β), TNF-α or resveratrol in hepatocytes and brown adipocytes.8 Our observation that even under basal conditions the viability of human and murine macrophages is reduced in the absence of PTP1B suggests the existence of alternative pathways linking cell viability to its presence in immune cells. In fact, a cross-talk between different groups of PTP members has been described as a regulatory network to ensure sufficient immunity against pathogen challenge and in preventing adverse autoimmune responses.29 In conclusion, our data show a previously unrecognized genetic damage in myeloid cell upon pro-inflammatory activation, and suggest that the broad use of PTP1B inhibitors, although with potential benefits over the insulin signaling pathway, might exert undesirable effects in response to stressors of the immune system including the fine tuning of the pro-inflammatory and pro-resolution balance.
Materials and Methods
Materials
Common reagents including z-VAD.FMK were from Sigma-Aldrich (St Louis, MO, USA) or Roche (Darmstadt, Germany). Murine or human cytokines were obtained from PeproTech (London, UK). Antibodies were from Ambion (Austin, TX, USA), Santa Cruz Biotech (Santa Cruz, CA, USA) or Cell Signaling (Danvers, MA, USA). Reagents for electrophoresis were from Bio-Rad (Hercules, CA, USA) and Sigma-Aldrich. Tissue culture dishes were from Falcon (Lincoln Park, NJ, USA), and serum and culture media were from Invitrogen and Gibco (Life Technologies/Thermo Fisher, Madrid, Spain).
Animal care and preparation of macrophages
Male and female PTP1B heterozygous (HET) mice, maintained on a mixed genetic background (C57BL/6 × 129sv), were intercrossed to yield the three genotypes of mice (WT, HET and KO). In this study, we used 8- to 12-week-old WT and PTP1B KO male mice housed under 12 h light/dark cycle and food and water was provided ad libitum. Animals were cared for according to a protocol approved by the Ethical Committee of our institution (following directive 2010/63/EU of the European Parliament). PTP1B-deficient mice were used as previously described.36 Elicited peritoneal macrophages were obtained from male mice 4 days after intraperitoneal (i.p.) administration of 2.5 ml of 3% thioglycollate broth essentially as described previously.44 Cells were seeded in 6-multiwell plates at a density of 4 × 106 cells/plate and cultured in RPMI-1640 medium supplemented with 10% heat-inactivated fetal calf serum (FCS) and antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin) at 37 °C in a humidified atmosphere with 5% CO2. After 2 h of incubation, non-adherent cells were removed. Adherent cells were maintained in these culture conditions and used within 2 days after plating. After overnight serum reduction, cells were challenged with LPS, polyI:C and LTA (as M1 stimuli) or with IL-4, IL-13 and IL-10 (as M2 stimuli).
Isolation of human monocytes
PBMCs were isolated from enriched buffy coats of healthy donors by separation on Ficoll-Hypaque Plus (GE Healthcare, Madrid, Spain) following the manufacturers' protocol. After washing the upper layer, CD14+ cells were obtained using immunomagnetic isolation (Miltenyi Biotec, Bergisch, Germany). Cells were maintained for 4 h at 1 × 106 cells/ml in DMEM supplemented with antibiotics (100 IU/ml penicillin and 100 μg/ml streptomycin). After this period, the supernatant was removed and adherent cells were cultured in the same medium supplemented with 10% heat-inactivated FCS. Cells were maintained overnight with this medium and differentiated into macrophages with hM-CSF (20 ng/ml; PeproTech) for 7 days.
Silencing of PTP1B in human monocyte/macrophages
Human differentiated macrophages were incubated with Opti-Mem (Gibco) supplemented with 10% FCS and after 4 h in this medium were treated with 50 nM scrambled (sc)- or siRNAs specific for human PTP1B (Dharmacon, Madrid, Spain). Transfection was accomplished with lipofectamine RNAiMAX (Invitrogen) following the instructions of the supplier. After 12 h, the dishes were supplemented with 2 vol of complete medium containing 10% FCS and antibiotics and maintained for an additional 24-h period. Cell viability was ensured after this incubation and the plates were washed twice with PBS and incubated with fresh medium containing 2% FCS. Cells were used 12 h after this step.
Preparation and treatment of murine or human macrophages with POV
A fresh solution of POV was used to mimic broad PTP inhibition. Briefly, POV was prepared by incubating 1 mM vanadate in PBS with 1 mM H2O2 for 10 min at 30 °C.45 The mixture was treated with purified recombinant catalase (200 μg/ml) to remove the residual H2O2 and was used in the next 10 min by adding the solution to the culture medium. As a control, cells received a solution prepared in parallel, but lacking vanadate (this preparation was referred to as vehicle in the same set of experiments).
Determination of ALT activity and NO, TNF-α and PGE2 levels
The ALT activity was determined in serum samples using a Reflotron plus device (Roche). The amount of nitrate and nitrite in the culture medium was measured after reduction of nitrate to nitrite, and quantification of nitrite using a previous protocol.44 The accumulation of TNF-α or PGE2 in the culture medium was measured using specific immunoassay kits (GE Healthcare). The levels of TNF-α in serum were measured under similar conditions following the instructions of the immunoassay kit supplier.
Measurement of cell viability
Macrophage viability was determined by flow cytometry as follows: cells were seeded in TPP dishes that favor its detachment with cold PBS. After centrifugation at 4 °C for 5 min at 1000 × g, cells were resuspended in 10 mM HEPES; pH 7.4, 140 mM NaCl, 2.5 mM CaCl2 and labeled with annexin V–FITC (Invitrogen) solution and/or propidium iodide (PI) (10 μg/ml) for 15 min at RT in the dark and then analyzed in a BD-Canto flow cytometer (BD Biosciences, Madrid, Spain). PI is impermeable to living but stains necrotic and apoptotic dying cells with impaired membrane integrity in contrast to annexin V, which stains early apoptotic cells.
Preparation of total protein cell extracts
Macrophages were homogenized in a buffer containing 10 mM Tris-HCl, pH 7.5; 1 mM MgCl2, 1 mM EGTA, 10% glycerol, 0.5% CHAPS, 1 mM β-mercaptoethanol and 0.1 mM PMSF and a protease and phosphatase inhibitor cocktail (Sigma-Aldrich). The extracts were vortexed for 30 min at 4 °C and after centrifuging for 20 min at 13 000 × g, the supernatants were stored at −20 °C. Protein levels were determined using Bradford reagent (Bio-Rad).
Western blotting
Protein extracts were boiled in loading buffer (250 mM Tris-HCl; pH 6.8, 2% sodium dodecyl sulfate (SDS), 10% glycerol and 2% β-mercaptoethanol) and 30 μg of protein were subjected to 8–10% SDS polyacrylamide gel electrophoresis (SDS-PAGE) electrophoresis gels. Proteins were transferred onto polyvinylidene fluoride (PVDF) membranes (GE Healthcare). Membranes were incubated for 1 h with low-fat milk powder (5%) in PBS containing 0.1% Tween-20. Blots were incubated for 2 h or overnight at 4 °C with primary antibodies against NOS-2, COX-2, KC, MIP2, IP10, SOCS3, PTP1B, HO-1, Arg-1, IRF-3, p65, and the specific anti-phospho- or total AKT, ERK, p38 and JNK at the dilutions recommended by the suppliers. The blots were developed with ECL Advance protocol (GE Healthcare) and different exposure times were performed for each blot with a Charged Coupling Device camera in a luminescent image analyzer (Molecular Imager; Bio-Rad) to ensure the linearity of the band intensities. Blots were normalized for lane charge using antibodies against p85-PI3K, GAPDH or β-actin. Values of densitometry were determined using the Quantity One software (Bio-Rad).
RNA isolation and RT-PCR analysis
RNA was extracted with TRIzol Reagent (Invitrogen) and reverse transcribed using Transcriptor First Strand cDNA Synthesis Kit for RT-PCR following the indications of the manufacturer (Roche). Real-time PCR was conducted with SYBR Green Master (Roche) on a MyiQ Real-Time PCR System (Bio-Rad). Primer sequences for mouse IL-6, NOS-2, IκBα and PTP1B are available on request. Validation of amplification efficiency was performed for each pair of primers.44 PCR thermocycling parameters were 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. Each sample was run in duplicate and was normalized versus 36B4. The fold induction (FI) was determined in a ΔΔCt-based fold-change calculation.
Mini-array and microarray analysis
Differential expression at 0 and 3 h was tested using the ‘Chemokine and Receptors' 96-well PCR mini-array (Qiagen, Madrid, Spain; ref. PAMM-022Z) and RNA samples of peritoneal macrophages from WT and PTP1B KO mice challenged with LPS. RNA extraction and expression analysis was performed according to the instructions of the supplier (Qiagen). In addition to this, normalized expression data in macrophages after 24 h of stimulation were obtained in our core facility, using a mouse microarray platform (Agilent-014868 Whole Mouse Genome Microarray 4 × 44K G4122F). Quality of the samples was ensured in a Bioanalyzer 2100. Processing, normalization and differential expression were performed using Limma Bioconductor package.46 Enrichment of gene sets of interest from KEGG and REACTOME was accomplished using the GSEA software as described by Mootha et al.47
Bone marrow transplantation and clodronate treatment
Eight- to nine-week-old C57BL/6 × 129sv WT male mice were irradiated with a dose of 1000 rads (10 Gy) by a cesium γ-source. One-week prior and 2-week post bone marrow transplantation, 100 mg/l neomycin and 10 mg/l polymyxin B sulfate (Sigma-Aldrich) were added to the acidified water. The bone marrow was collected from the femurs and tibias of 8- to 9-week-old donor male PTP1B WT or KO mice (C57BL/6 × 129sv) by flushing femurs with phosphate-buffered saline. Each recipient mouse was injected with about 5–6 × 106 bone marrow cells by retroorbital injection 4 h after irradiation. In another set of animals, 2 weeks after irradiation, mice were injected 200 μl of clodronate- or PBS-loaded liposomes (www.clodronateliposomes.com) by retroorbital injection.43 Twelve weeks after transplantation, genomic DNA was harvested from whole blood and liver for use in PCR-based genotyping of WT and PTP1B-deficient alleles.36 In addition to this, liver sections were prepared and stained with F4/80-FITC and laminin to ensure the depletion of Kupffer and resident macrophages.
In vivo evaluation of sensitivity to LPS/D-GalN treatment
To evaluate the in vivo response to inflammatory challenge, PTP1B WT and KO animals were injected (i.p.) a low dose of LPS (2 μg/kg) in combination with D-GalN (800 mg/kg) and animal survival was evaluated. In addition to this, WT mice transplanted with bone marrow from PTP1B KO or WT donors with or without treatment with clodronate liposomes to decrease resident macrophage populations were also used for applying the LPS/D-GalN protocol. Serum from these animals was prepared from blood obtained by retroorbital puncture. Liver sections were prepared and stained with eosin/hematoxylin to determine the macroscopic integrity of the tissue upon LPS/D-GalN administration.
Measurement of ear edema and MPO activity
TPA (2.5 μg) dissolved in 20 μl acetone was applied in 10 μl volumes to both the inner and the outer surfaces of the left ear of mice (20–25 g), and the same procedure was repeated in the right ear with the vehicle. Animals were killed after 4 h, and equal-sized samples of both ears were punched out and weighed. Edema was measured as the weight difference between the right and left ear samples. These ear samples were homogenized in 500 μl of ice-old PBS, snap frozen and thawed four times, subsequently submitted to sonication (twice for 10 s) and centrifuged at 10 000 × g for 15 min at 4 °C. MPO activity was measured in the supernatants as a marker for neutrophil infiltration. Aliquots of supernatants (50 μl) were assayed in a reaction mixture that contained 110 μl PBS, 20 μl of 0.22 M NaH2PO4 (pH 5.4), 20 μl of 0.026% (v/v) H2O2, and 20 μl of 18 mM tetramethylbenzidine in 8% (v/v) aqueous dimethylformamide. After 10 min of reaction at 37 °C, 30 μl sodium acetate (1.5 M; pH 3) was added, and the absorbance at 620 nm was read in a microtiter plate reader. The activity was expressed as mU/mg protein.48, 49 In addition to this, ears were fixed with paraformaldehyde and stained with eosin/hematoxylin to visualize their structure and extension of the edema.
Analysis of PTP1B-mediated dephosphorylation
Macrophages from PTP1B KO mice were stimulated with LPS (200 ng/ml) for 30 min and immediately homogenized in buffer containing 20 mM imidazole-HCl, 2 mM EGTA and 2 mM EDTA (pH 7.0) supplemented with protease inhibitors (10 mg/ml leupeptin, 10 mg/ml aprotinin and 1 mM PMSF). Samples were sonicated 3 × 15 s at 1.5 mA and lysates were clarified by centrifugation at 12 000 × g for 10 min. After protein content determination, 60 μl of cell lysates (1 mg/ml) was incubated with 10 units of recombinant PTP1B (Upstate, Merck-Millipore, Darmstadt, Germany) for 30 min at 37 °C. The reaction was stopped by adding Laemmli's buffer and samples were analyzed by western blotting.
Statistical analysis
The values in graphs correspond to the means±S.D. The statistical significance was determined with Student's t test for unpaired observation. Data were analyzed by the SPSS for Windows statistical package, v21 (IBM, Madrid, Spain).
Acknowledgments
This work was supported by grants BFU2011-24760 and SAF2012-33283 from MINECO, S2010/BMD-2378 and S2010/BMD-2423 from Comunidad de Madrid and FIS-RIC RD06/0014/0025 and EFSD and Amylin Paul Langerhans Grant. Ciberdem and Ciberehd are funded by the Instituto de Salud Carlos III.
Glossary
- ALT
alanine aminotransferase
- AKT
protein kinase B
- Arg-1
arginase-1
- CXCL
chemokine (C-X-C motif) ligand
- COX-2
cyclooxygenase-2
- DE
differentially expressed
- D-GalN
D-galactosamine
- ERK
extracellular signal-regulated kinase
- F4/80
EGF-like module-containing mucin-like hormone receptor-like 1
- FDRs
false discovery rates
- GSEA
gene set enrichment analysis
- HO-1
heme oxygenase 1
- IFN
interferon
- IL
interleukin
- i.p.
intraperitoneal
- IRF-3
IFN regulated factor 3
- JAK/STAT
janus kinase/signal transducer and activator of transcription
- JNK
c-Jun N-terminal kinase
- KEGG
Kyoto encyclopedia of genes and genomes
- LPS
lipopolysaccharide
- KO
knockout
- LTA
lipoteichoic acid
- MAPK
mitogen-activated protein kinase
- M-CSF
macrophage colony-stimulating factor
- MPO
myeloperoxidase
- NF-κB
nuclear factor κB
- NO
nitric oxide
- NOS-2
nitric oxide synthase-2
- PGE2
prostaglandin E2
- PI3K
phosphatidylinositol 3-kinase
- polyI:C
polyriboinosinic:polyribocytidylic acid
- PVDF
polyvinylidene fluoride
- q(RT)-PCR
quantitative real-time (RT) PCR
- POV
peroxovanadate
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TGF-β
transforming growth factor β
- TLRs
Toll-like receptors
- TNF-α
tumor necrosis factor-α
- TPA
tetradecanoylphorbol acetate
- TYK
non-receptor tyrosine-protein kinase
- WT
wild type
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by T Brunner
Supplementary Material
References
- Haj FG, Markova B, Klaman LD, Bohmer FD, Neel BG. Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatase-1B. J Biol Chem. 2003;278:739–744. doi: 10.1074/jbc.M210194200. [DOI] [PubMed] [Google Scholar]
- Salmeen A, Andersen JN, Myers MP, Tonks NK, Barford D. Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Mol Cell. 2000;6:1401–1412. doi: 10.1016/s1097-2765(00)00137-4. [DOI] [PubMed] [Google Scholar]
- Seely BL, Staubs PA, Reichart DR, Berhanu P, Milarski KL, Saltiel AR, et al. Protein tyrosine phosphatase 1B interacts with the activated insulin receptor. Diabetes. 1996;45:1379–1385. doi: 10.2337/diab.45.10.1379. [DOI] [PubMed] [Google Scholar]
- Elchebly M, Payette P, Michaliszyn E, Cromlish W, Collins S, Loy AL, et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science. 1999;283:1544–1548. doi: 10.1126/science.283.5407.1544. [DOI] [PubMed] [Google Scholar]
- Haj FG, Zabolotny JM, Kim YB, Kahn BB, Neel BG. Liver-specific protein-tyrosine phosphatase 1B (PTP1B) re-expression alters glucose homeostasis of PTP1B-/-mice. J Biol Chem. 2005;280:15038–15046. doi: 10.1074/jbc.M413240200. [DOI] [PubMed] [Google Scholar]
- Klaman LD, Boss O, Peroni OD, Kim JK, Martino JL, Zabolotny JM, et al. Increased energy expenditure, decreased adiposity, and tissue-specific insulin sensitivity in protein-tyrosine phosphatase 1B-deficient mice. Mol Cell Biol. 2000;20:5479–5489. doi: 10.1128/mcb.20.15.5479-5489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rodriguez A, Mas-Gutierrez JA, Mirasierra M, Fernandez-Perez A, Lee YJ, KO HJ, et al. Essential role of protein tyrosine phosphatase 1B in obesity-induced inflammation and peripheral insulin resistance during aging. Aging Cell. 2012;11:284–296. doi: 10.1111/j.1474-9726.2011.00786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rodriguez A, Escribano O, Alba J, Rondinone CM, Benito M, Valverde AM. Levels of protein tyrosine phosphatase 1B determine susceptibility to apoptosis in serum-deprived hepatocytes. J Cell Physiol. 2007;212:76–88. doi: 10.1002/jcp.21004. [DOI] [PubMed] [Google Scholar]
- Adachi M, Sekiya M, Arimura Y, Takekawa M, Itoh F, Hinoda Y, et al. Protein-tyrosine phosphatase expression in pre-B cell NALM-6. Cancer Res. 1992;52:737–740. [PubMed] [Google Scholar]
- Yi T, Cleveland JL, Ihle JN. Identification of novel protein tyrosine phosphatases of hematopoietic cells by polymerase chain reaction amplification. Blood. 1991;78:2222–2228. [PubMed] [Google Scholar]
- Myers MP, Andersen JN, Cheng A, Tremblay ML, Horvath CM, Parisien JP, et al. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J Biol Chem. 2001;276:47771–47774. doi: 10.1074/jbc.C100583200. [DOI] [PubMed] [Google Scholar]
- Aoki N, Matsuda T. A cytosolic protein-tyrosine phosphatase PTP1B specifically dephosphorylates and deactivates prolactin-activated STAT5a and STAT5b. J Biol Chem. 2000;275:39718–39726. doi: 10.1074/jbc.M005615200. [DOI] [PubMed] [Google Scholar]
- Lu X, Malumbres R, Shields B, Jiang X, Sarosiek KA, Natkunam Y, et al. PTP1B is a negative regulator of interleukin 4-induced STAT6 signaling. Blood. 2008;112:4098–4108. doi: 10.1182/blood-2008-03-148726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen KM, Dube N, Bourdeau A, Lapp WS, Tremblay ML. Protein tyrosine phosphatase 1B negatively regulates macrophage development through CSF-1 signaling. Proc Natl Acad Sci USA. 2006;103:2776–2781. doi: 10.1073/pnas.0508563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorst ST, Krueger A, Muerkoster S, Fas SC, Golks A, Gruetzner U, et al. Suramin inhibits death receptor-induced apoptosis in vitro and fulminant apoptotic liver damage in mice. Nat Med. 2004;10:602–609. doi: 10.1038/nm1049. [DOI] [PubMed] [Google Scholar]
- Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Garlanda C, Locati M. Macrophage diversity and polarization in atherosclerosis: a question of balance. Arterioscler Thromb Vasc Biol. 2009;29:1419–1423. doi: 10.1161/ATVBAHA.108.180497. [DOI] [PubMed] [Google Scholar]
- Xu H, An H, Hou J, Han C, Wang P, Yu Y, et al. Phosphatase PTP1B negatively regulates MyD88- and TRIF-dependent proinflammatory cytokine and type I interferon production in TLR-triggered macrophages. Mol Immunol. 2008;45:3545–3552. doi: 10.1016/j.molimm.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Laird MH, Rhee SH, Perkins DJ, Medvedev AE, Piao W, Fenton MJ, et al. TLR4/MyD88/PI3K interactions regulate TLR4 signaling. J Leukoc Biol. 2009;85:966–977. doi: 10.1189/jlb.1208763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Guerra MJ, Castrillo A, Martin-Sanz P, Bosca L. Negative regulation by phosphatidylinositol 3-kinase of inducible nitric oxide synthase expression in macrophages. J Immunol. 1999;162:6184–6190. [PubMed] [Google Scholar]
- Guha M, Mackman N. The phosphatidylinositol 3-kinase-Akt pathway limits lipopolysaccharide activation of signaling pathways and expression of inflammatory mediators in human monocytic cells. J Biol Chem. 2002;277:32124–32132. doi: 10.1074/jbc.M203298200. [DOI] [PubMed] [Google Scholar]
- Fukao T, Koyasu S. PI3K and negative regulation of TLR signaling. Trends Immunol. 2003;24:358–363. doi: 10.1016/s1471-4906(03)00139-x. [DOI] [PubMed] [Google Scholar]
- Ruse M, Knaus UG. New players in TLR-mediated innate immunity: PI3K and small Rho GTPases. Immunol Res. 2006;34:33–48. doi: 10.1385/IR:34:1:33. [DOI] [PubMed] [Google Scholar]
- Nieto-Vazquez I, Fernandez-Veledo S, de Alvaro C, Rondinone CM, Valverde AM, Lorenzo M. Protein-tyrosine phosphatase 1B-deficient myocytes show increased insulin sensitivity and protection against tumor necrosis factor-alpha-induced insulin resistance. Diabetes. 2007;56:404–413. doi: 10.2337/db06-0989. [DOI] [PubMed] [Google Scholar]
- Al-Lahham S, Roelofsen H, Rezaee F, Weening D, Hoek A, Vonk R, et al. Propionic acid affects immune status and metabolism in adipose tissue from overweight subjects. Eur J Clin Invest. 2012;42:357–364. doi: 10.1111/j.1365-2362.2011.02590.x. [DOI] [PubMed] [Google Scholar]
- Al-Lahham SH, Roelofsen H, Priebe M, Weening D, Dijkstra M, Hoek A, et al. Regulation of adipokine production in human adipose tissue by propionic acid. Eur J Clin Invest. 2010;40:401–407. doi: 10.1111/j.1365-2362.2010.02278.x. [DOI] [PubMed] [Google Scholar]
- Gordon S, Pasare C, Medzhitov R. The macrophage: past, present and future. Toll-like receptors: linking innate and adaptive immunity. Eur J Immunol. 2007;37 (Suppl 1:S9–S17. doi: 10.1002/eji.200737638. [DOI] [PubMed] [Google Scholar]
- Mantovani A. Molecular pathways linking inflammation and cancer. Curr Mol Med. 2010;10:369–373. doi: 10.2174/156652410791316968. [DOI] [PubMed] [Google Scholar]
- Rhee I, Veillette A. Protein tyrosine phosphatases in lymphocyte activation and autoimmunity. Nat Immunol. 2012;13:439–447. doi: 10.1038/ni.2246. [DOI] [PubMed] [Google Scholar]
- Berdnikovs S, Pavlov VI, Abdala-Valencia H, McCary CA, Klumpp DJ, Tremblay ML, et al. PTP1B deficiency exacerbates inflammation and accelerates leukocyte trafficking in vivo. J Immunol. 2012;188:874–884. doi: 10.4049/jimmunol.1004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone CJ, Zheng H, Bhattacharya S, Lewis JR, Reiter AM, Henthorn P, et al. Protein tyrosine phosphatase 1B is a key regulator of IFNAR1 endocytosis and a target for antiviral therapies. Proc Natl Acad Sci USA. 2012;109:19226–19231. doi: 10.1073/pnas.1211491109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen KM, Bourdeau A, Doody KM, Tremblay ML. Protein tyrosine phosphatases PTP-1B and TC-PTP play nonredundant roles in macrophage development and IFN-gamma signaling. Proc Natl Acad Sci USA. 2009;106:9368–9372. doi: 10.1073/pnas.0812109106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabolotny JM, Kim YB, Welsh LA, Kershaw EE, Neel BG, Kahn BB. Protein-tyrosine phosphatase 1B expression is induced by inflammation in vivo. J Biol Chem. 2008;283:14230–14241. doi: 10.1074/jbc.M800061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant L, Shearer K, Czopek A, Lees E, Owen C, Agouni A, et al. Myeloid-cell protein tyrosine phosphatase-1B deficiency in mice protects against high-fat diet and lipopolysaccharide induced inflammation, hyperinsulinemia and endotoxemia through an IL10 STAT3 dependent mechanism. Diabetes. 2013. [DOI] [PubMed]
- Zhang J, Wang B, Zhang W, Wei Y, Bian Z, Zhang CY, et al. Protein tyrosine phosphatase 1B deficiency ameliorates murine experimental colitis via the expansion of myeloid-derived suppressor cells. PLoS ONE. 2013;8:e70828. doi: 10.1371/journal.pone.0070828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rodriguez A, Clampit JE, Escribano O, Benito M, Rondinone CM, Valverde AM. Developmental switch from prolonged insulin action to increased insulin sensitivity in protein tyrosine phosphatase 1B-deficient hepatocytes. Endocrinology. 2007;148:594–608. doi: 10.1210/en.2006-0644. [DOI] [PubMed] [Google Scholar]
- Rao KM. MAP kinase activation in macrophages. J Leukoc Biol. 2001;69:3–10. [PubMed] [Google Scholar]
- Traves PG, de Atauri P, Marin S, Pimentel-Santillana M, Rodriguez-Prados JC, Marín de Mas I, et al. Relevance of the MEK/ERK signaling pathway in the metabolism of activated macrophages: a metabolomic approach. J Immunol. 2012;188:1402–1410. doi: 10.4049/jimmunol.1101781. [DOI] [PubMed] [Google Scholar]
- Hazeki K, Nigorikawa K, Hazeki O. Role of phosphoinositide 3-kinase in innate immunity. Biol Pharm Bull. 2007;30:1617–1623. doi: 10.1248/bpb.30.1617. [DOI] [PubMed] [Google Scholar]
- He R, Yu Z, He Y, Zeng LF, Xu J, Wu L, et al. Double click reaction for the acquisition of a highly potent and selective mPTPB inhibitor. ChemMedChem. 2010;5:2051–2056. doi: 10.1002/cmdc.201000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang ZX, Zhang ZY. Targeting PTPs with small molecule inhibitors in cancer treatment. Cancer Metastasis Rev. 2008;27:263–272. doi: 10.1007/s10555-008-9113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Zhou B, Yang H, He Y, Jiang ZX, Kumar S, et al. Aryl vinyl sulfonates and sulfones as active site-directed and mechanism-based probes for protein tyrosine phosphatases. J Am Chem Soc. 2008;130:8251–8260. doi: 10.1021/ja711125p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J, Trinidad NJ, Moayeri M, Kintzer AF, Wang SB, van Rooijen N, et al. Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature. 2012;490:107–111. doi: 10.1038/nature11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Prados JC, Traves PG, Cuenca J, Rico D, Aragones J, Martín-Sanz P, et al. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol. 2010;185:605–614. doi: 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- Diaz-Guerra MJ, Castrillo A, Martin-Sanz P, Bosca L. Negative regulation by protein tyrosine phosphatase of IFN-gamma-dependent expression of inducible nitric oxide synthase. J Immunol. 1999;162:6776–6783. [PubMed] [Google Scholar]
- Smyth GK.Limma: linear models for microarray dataIn: Bioinformatics and Computational Biology Solutions using R and Bioconductor Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, (eds)Springer: New York; 2005 [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Ota H, Sasagawa S, Sakatani T, Fujikura T. Assay method for myeloperoxidase in human polymorphonuclear leukocytes. Anal Biochem. 1983;132:345–352. doi: 10.1016/0003-2697(83)90019-2. [DOI] [PubMed] [Google Scholar]
- Gross S, Gammon ST, Moss BL, Rauch D, Harding J, Heinecke JW, et al. Bioluminescence imaging of myeloperoxidase activity in vivo. Nat Med. 2009;15:455–461. doi: 10.1038/nm.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.