Abstract
Methamphetamine (METH) is a psychostimulant with high abuse potential and severe neurotoxicity. Recent studies in animal models have indicated that METH can impair the blood–brain barrier (BBB), suggesting that some of the neurotoxic effects resulting from METH abuse could be due to barrier disruption. We report here that while chronic exposure to METH disrupts barrier function of primary human brain microvascular endothelial cells (HBMECs) and human umbilical vein endothelial cells (HUVECs), an early pro-survival response is observed following acute exposure by induction of autophagic mechanisms. Acute METH exposure induces an early increase in Beclin1 and LC3 recruitment. This is mediated through inactivation of the protein kinase B (Akt)/mammalian target of rapamycin (mTOR)/p70S6K pathway, and upregulation of the ERK1/2. Blockade of Kappa opioid receptor (KOR), and treatment with autophagic inhibitors accelerated METH-induced apoptosis, suggesting that the early autophagic response is a survival mechanism for endothelial cells and is mediated through the kappa opioid receptor. Our studies indicate that kappa opioid receptor can be therapeutically exploited for attenuating METH-induced BBB dysfunction.
Keywords: methamphetamine, autophagy, cell death, Kappa opioid receptor
Methamphetamine (METH) is a central nervous system (CNS) stimulant that is similar in structure to amphetamine. The escalating abuse of this drug in recent years and its long-lasting neurotoxic effects have placed an increased burden upon health care costs. Previous studies have shown that METH-induced autophagy in neurons is associated with the formation of ubiquitin-positive aggregates and multilamellar bodies.1, 2 In addition to its direct effects on neurons, blood–brain barrier (BBB) dysfunction could also contribute to METH-induced neurotoxicity.3 The BBB is a specialized structure formed by brain endothelial cells that are tightly interconnected to form a boundary between the CNS and periphery .4 Recent findings suggest that METH impairment of glucose transporter protein-1(GLUT1) at the brain endothelium may affect energy-associated disruption of tight junction assembly and loss of BBB integrity.5 Furthermore, METH can act directly on cultured primary rat brain microvascular endothelial cells to compromise their barrier properties inducing eNOS activation and enhanced transcytosis.3
The primary goal of the current study was to investigate whether METH treatment induces autophagy in endothelial cells. Macroautophagy (hereafter referred to as autophagy) is an intracellular lysosomal (vacuolar) degradation process that is characterized by the formation of a cytosolic double-membrane vesicle, the autophagosome. During autophagy, cytoplasmic proteins, organelles or other materials are surrounded by phagophores, which expand and close to form double-membrane autophagosomes. These autophagosomes fuse with lysosomes (or vacuoles) to form autolysosomes, in which the cytoplasmic cargos are degraded by lysosomal enzymes. The primary role of autophagy is to ensure cell survival under stress conditions.6 Most assays for autophagy modulators use the autophagy marker protein microtubule-associated protein 1 light chain 3 (LC3) as a readout for autophagic activity. LC3 is a mammalian orthologue of the yeast ATG 8 protein, a ubiquitin-like protein that becomes lipidated and tightly associated with the autophagosomal membranes.7 Lysosomal-associated membrane protein1 (LAMP1) is among the most abundant lysosomal membrane protein. It is a heavily glycosylated protein creating a sugar coat or glycocalyx on the inner side of the lysosomal membrane which aids to the protection of the membrane from the hydrolytic enzymes and degradation. LAMP1 is expressed mainly in the endosome-lysosomal membrane of cells which is involved in maintaining lysosomal membrane integrity and phagolysosome formation.8, 9
We show that METH exposure-induced autophagy early on and progressively changed into apoptosis in a time-dependent manner. The METH-induced autophagy was specific and initiated even in the presence of nutrient-rich medium. Inhibition of METH-induced autophagy accelerated apoptosis of endothelial cells. METH-induced autophagy was mediated through activation of Kappa opioid receptor (KOR) because KOR antagonist blocked METH-induced autophagy. These studies suggest that METH induces autophagy as a pro-survival response in endothelial cells, which transits into apoptosis upon continuous exposure to the drug.
Results
METH inhibits endothelial cell viability and disrupts cell–cell barrier integrity
We initially determined if METH treatment reduced cell viability. The blood concentrations of METH in drug abusers are in the low micromolar range, with a median concentration of 1.25 μM.10 Therefore, human umbilical vein endothelial cells (HUVECs) were exposed to 1 μM METH for 24, 48 or 72 h and cell viability determined using a MTT assay. Although, no obvious change in viability was observed at 24 and 48 h with low concentration (1 μM) of METH, significant decrease in viability was observed at 72 h compared with control (Figure 1a).
Figure 1.
METH induces autophagy in HUVECs. (a) Cell viability of HUVECs after 24, 48 and 72 h of METH treatment (MTT assay). Cell viability was expressed as a ratio of the absorbance between treated and untreated cells. The P-value for overall comparison between vehicle and METH was <0.05 and significant difference was seen at the dose of 1 μM METH after 72 h. Three independent experiments were performed. (b–c) METH-induced decrease in endothelial resistance. HBMECs and HUVECs resistance values (normalized) were monitored for up to 72 h, with measurements being collected every few seconds by the ECIS device. HUVECs were treated with 1 μM METH in the presence or absence of 50 μM zVAD-fmk. (d) Treatment of HUVECs with METH (1 μM) and Rapamycin (100 ng/ml) for 24 h shows a significant increase in the LC3 punctate staining pattern (green) when compared with untreated cells. Nuclei were labeled with DAPI. Rapamycin-treated HUVECs samples were used as a positive control. Panel ii shows granular/vesicular structures indicative of autophagosome formation. (e) The conversion of soluble LC3-I to lipid-bound LC3-II which is associated with the formation of autophagosomes. HUVECs were treated with METH (1 μM) for 3, 6 and 24 h in the presence of pepstatin A and E-64d. Cell lysates were separated on 12.5% SDS-PAGE and analyzed by western blot using anti-LC3 antibody (Cell Signaling). (f) Data in bar graphs represent the ratio of lipidated LC3-II to a non-lipidated LC3-I. *,**P<0.05. Shown are representative results from one of three independent experiments. (g) HUVECs were transiently transfected with GFP-LAMP1 and cultured for 24 h. Cells were then treated with METH for 24 and 48 h. Images were visualized by fluorescence microscope
Next, we evaluated whether 1 μM METH affected endothelial cell barrier integrity. Human brain microvascular endothelial cells (HBMECs) and HUVECs were cultured on ECIS array electrodes, and transendothelial electrical resistance was monitored continuously over the course of 72 h. As shown in Figures 1b and c, no significant decrease in average transendothelial electrical resistance measurements was observed with 1 μM METH at 24 h but at 72 h, a 50% decrease in resistance was observed compared with control.
Inhibition of apoptosis using 50 μM zVAD-fmk, a pan-caspase inhibitor-attenuated METH-induced decrease in electrical resistance at 72 h supporting our hypothesis that METH exposure reduced electrical resistance at late time point due to cell death and autophagy maybe responsible for the early protective effects.
METH induces LC3 recruitment and vesicular compartmentalization of LAMP1
To further determine if acute METH treatment induces an autophagic pro-survival response, we monitored different autophagy markers: LC3 vesicular recruitment and LAMP1. To analyze autophagosome formation, we used anti-LC3 immunofluorescence in a cell-based detection system with HUVECs. In 1 μM METH-treated cells, LC3 protein increased in total abundance and was aggregated in vesicular structures (Figure 1d). LC3 (ATG 8) is first cleaved by ATG4 to generate LC3-I and then lipidated to produce LC3-II. LC3-II is incorporated into the inner and outer surfaces of autophagosomes, and as such, remains associated with the autophagosome throughout the pathway. The lipidated (LC3-II) form can be detected as a faster-migrating band in western blots.11, 12 The amount of LC3-II in the presence and absence of saturating levels of inhibitors, which can be used to examine the transit of LC3-II through the autophagic pathway; if autophagy flux is occurring, the levels of LC3-II will be higher in the presence of the inhibitors. Lysosomal degradation can be prevented through the use of protease inhibitors (e.g., pepstatin A and E-64d).13 Our data demonstrate that the LC3-I/LC3-II conversion occurred rapidly, within 6 h, after METH treatment and was still detectable at 24 h post treatment in the presence of the protease inhibitors pepstatin A and E-64d (Figures 1e and f).
We also examined LAMP1 which is an integral glycoprotein residing in late endosomes and lysosomes.14 HUVECs were transfected with lysosomal-associated membrane protein 1-green fluorescent protein (LAMP1-GFP) and then treated with vehicle or METH. In Figure 1g we show increased LAMP1-GFP-labeled vesicles in METH-treated HUVECs after 24 and 48 h, indicating induction of an autophagic response following 1 μM METH treatment.
METH effects on endothelial cells are mediated via Beclin1/Bcl-2 interactions
Beclin1, a novel Bcl-2 homology 3 (BH3) domain-only protein, has an important part in both autophagosome formation and autolysosome fusion. Beclin1 and the class III phosphatidylinositide 3-kinase (PI3K) Vps34 form two distinct complexes. In complex I, which contributes to an autophagosome formation, Atg14L bridges Beclin1 and the Vps34–p150 complex. In complex II, which regulates the vacuolar protein sorting pathway, UV radiation resistance-associated gene product bridges Beclin1 and the Vps34–p150 complex. Bcl-2 family proteins negatively regulate the kinase activity of the Beclin1–Vps34 complex.15, 16 Thus, accumulation of Beclin1 protein can be used as an indicator of autophagy induction. To further confirm METH-induced autophagy, we measured Beclin1 protein level using confocal microscopy. Our results showed an upregulation of Beclin1 in METH-treated HUVECs (Figure 2a).
Figure 2.
METH-induced autophagy is mediated via Beclin1/Bcl-2 interactions. (a) Confocal microscopy images of HUVECs showing endogenous Beclin1 protein expression (green) at 24 and 48 h following METH treatment. Nuclei were labeled with DAPI. (b) Representative western blot analysis for Beclin1, Bcl-2 and α-tubulin in METH-treated HUVECs. Total cell lysates from METH-treated HUVECs were collected at different time points and subjected to western blot analysis. (c) Densitometric analysis of Beclin1 and Bcl-2 levels. Values were normalized against α-tubulin and presented as a ratio, compared with the basal level (0 h of treatment). Data represent the means of three independent blots. *P<0.05. (d) Co-immunoprecipitation of Beclin1 and Bcl-2 in HUVECs. Cell lysates from METH-treated HUVECs were collected at 24 h. The lysates (100 μg per sample) were incubated with anti-Beclin1 antibodies covalently coupled to protein A/G Plus–Sepharose. Immunoprecipitates were analyzed for Beclin1 and Bcl-2 by immunoblotting. (e) Densitometric analysis of Beclin1 and Bcl-2 levels. *P<0.05. Shown are representative results from one of three independent experiments
Previous studies have shown that formation of a Beclin1/Bcl-2 complex may function as a rheostat that regulates autophagic capacity and ensures that autophagy levels remain within a homeostatic range.17 To determine whether METH treatment affected Beclin1 and Bcl-2 expression, total cellular levels of each protein were analyzed by western blots (Figure 2b). Beclin1 levels increased around sixfold, whereas Bcl-2 levels decreased over the period of METH treatment (Figure 2c). The expression of Beclin1 was induced at 24 and 48 h of exposure, indicating induction of autophagy in METH-treated cells. To address the possibility that METH-modulated Beclin1 expression and the interaction with Bcl-2, Beclin1/Bcl-2 complexes were immunoprecipitated with antibodies against Beclin1. The western blots in Figures 2d and e showed that in control cells Bcl-2 co-immunoprecipitated with Beclin1 which decreased following 24 h of METH treatment. Together, these results suggested that METH-induced autophagy in endothelial cells was mediated by upregulating Beclin1 expression and disrupting the balance between Beclin1 and Bcl-2.
Expression of Beclin1 is through the KOR
KOR, is highly expressed on HUVECs.18 A growing body of evidence indicate that KOR may be involved in the modulation of abuse-related effects of CNS stimulants.19, 20 Stimulation of KOR decreases acute amphetamine-induced behavior and mRNA expression of neuropeptides in the rat striatum.21 To determine if KOR has a role in METH-induced autophagy, we examined the effects of the KOR antagonist nor-Binaltorphiminedihydrochloride (nor-BNI) pretreatment on inhibition of METH-induced Beclin1 expression. nor-BNI in a concentration-dependent manner inhibited METH-induced Beclin1 mRNA expression at the indicated concentrations of 1, 10 and 20 μM nor-BNI. A total of 20 μM nor-BNI significantly inhibited METH-induced Beclin1 mRNA expression compared with METH treatment alone (Figure 3a). Subsequently, 20 μM nor-BNI was used in the following experiments. Pretreatment with nor-BNI inhibited Beclin1 protein expression (Figures 3b and c). Immunofluoresence experiments further illustrated the downregulation of Beclin1 induced by METH in nor-BNI pretreated HUVECs (Figure 3d).
Figure 3.
METH-induced Beclin1 expression is mediated through the kappa opioid receptor (KOR). (a) HUVECs were treated with 1 μM, 10 μM and 20 μM nor-BNI 1 h before METH treatment for 24 h. Beclin1 mRNA expression was examined by real-time PCR analysis. (b) HUVECs were treated with nor-BNI (20 μM) 1 h before METH treatment for 24 h. Cell lysates were separated on 10% SDS-PAGE and level of Beclin1 was determined by western blot. (c) Densitometric analysis of Beclin1 expression. Values were normalized against α-tubulin and presented as fold increase compared with the basal level (0 h of treatment). *P<0.05. Shown are representative results from one of three independent experiments. (d) Confocal microscopy images of HUVECs showed endogenous Beclin1 protein expression (green). Nuclei were labeled with DAPI. HUVECs were treated with nor-BNI (20 μM) 1 h before METH treatment for 24 h. (e) HUVECs were treated with nor-BNI (20 μM) 1 h before METH treatment for 24 h. Cells were then analyzed by TEM. Images were obtained at × 10 000 (i) and × 40 000 (ii) magnification using a JEOL 1200EX transmission electron microscope (JEOL). Scale bar: 1 μm. Black arrow indicates autophagosomes. White arrow shows partially enclosed autophagosome. (f) HUVECs were transiently transfected with either scramble siRNA or KOR siRNA. KOR mRNA expression was analyzed by real-time RT-PCR. (g) Beclin1 mRNA expression was analyzed by real-time RT-PCR when HUVECs were treated with METH for 24 h after siRNA transfection
To further evaluate METH effects on endothelial cells at the ultrastructural level, transmission electron microscopy (TEM) was performed on cells treated with METH. Vehicle treated HUVECs exhibited the normal ultrastructural morphology of cytoplasm, organelles and nuclei (Figure 3e). In contrast, exposure to 1 μM METH for 24 h revealed double-membrane vacuolar structures (Figure 3eii, Black arrow). Noticeably, pre-incubation with 20 μM nor-BNI 1 h before METH treatment did not cause significant appearance of double-membrane vacuolar structures, nor did treatment alter the integrity of mitochondria (Figure 3eii, White arrow), implicating the role of KOR in METH-induced autophagy.
To confirm the specific role of the KOR antagonist nor-BNI in regulating Beclin1 expression, HUVECs were transfected with scramble siRNA or KOR siRNA. KOR and Beclin1 mRNA expression were detected by real-time PCR analysis. As shown in Figure 3f, KOR siRNA dramatically downregulated KOR mRNA level compared with scramble siRNA-transfected cells. We further examined Beclin1 mRNA expression in response to knockdown of KOR in the presence of METH treatment. As shown in Figure 3g, METH-induced Beclin1 mRNA expression was significantly decreased upon knockdown of KOR.
METH activates the ERK1/2 pathway and inhibits the Akt/mTOR pathway
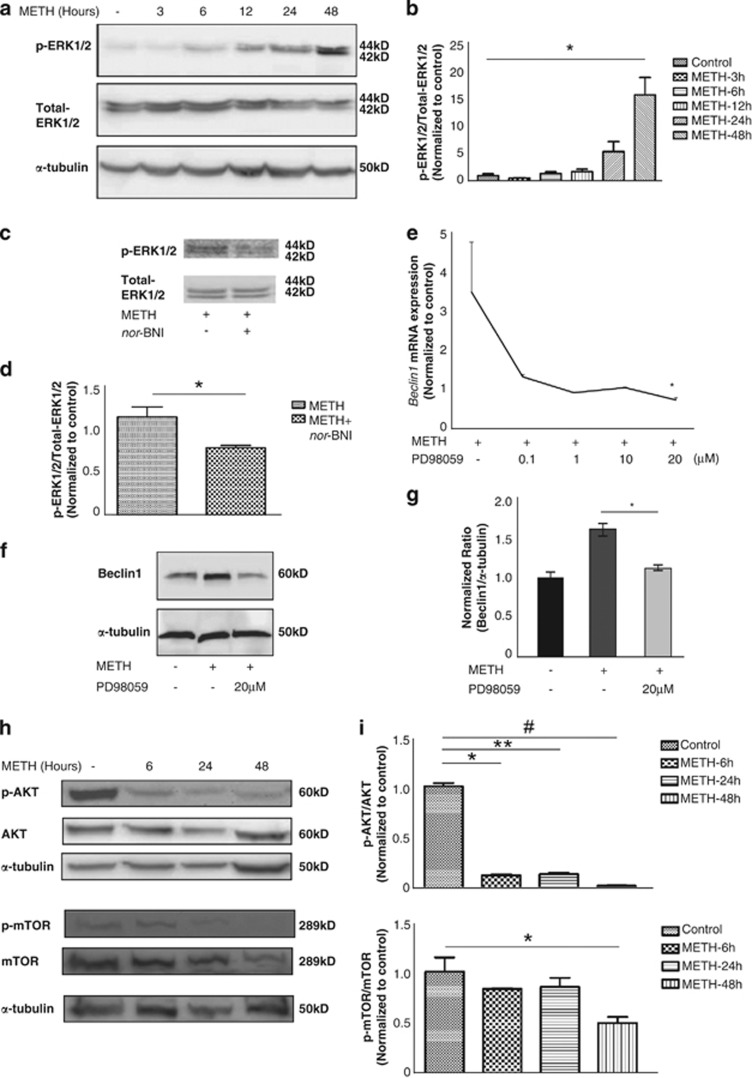
Autophagy is a multi-step process and is highly regulated through the action of various kinases, phosphatases and guanosine triphosphatases(GTPases).22, 23, 24 The following two pathways are known to be important in the regulation of autophagy: (i) The class I phosphatidylinositol 3-phosphate kinase (PI3K)/Akt/mammalian target of Rapamycin (mTOR)/p70 ribosomal protein S6 kinase (p70S6K) signaling pathway; and (ii) the Ras/Raf-1/mitogen-activated protein kinase 1/2 (MEK1/2)/extracellular signal-regulated kinase 1/2 (ERK1/2) pathway.25 Therefore, we examined the upstream mediators that are involved in METH-induced autophagy using western blotting. METH treatment showed a sustained increase in the level of phosphorylated ERK1/2, the active form, visible 6 h post treatment (Figures 4a and b). METH-induced ERK phosphorylation was inhibited when cells were pretreated with the KOR antagonist nor-BNI establishing the role of KOR in METH-induced autophagy (Figures 4c and d).
Figure 4.
METH-induced autophagy involves Akt/mTOR and the ERK pathways in endothelial cells. (a) Cells exposed to METH for 3, 6, 12, 24 and 48 h showed an increase in ERK1/2 phosphorylation after 6 h of treatment. (b) Densitometric analysis of p-ERK1/2 level. Values were normalized against α-tubulin and presented as a ratio compared with the basal level (0 h of treatment). Data represent the means of three independent blots. *P<0.05. (c) HUVECs were treated with nor-BNI (20 μM) 1 h before METH treatment for 24 h. nor-BNI decreased METH-induced ERK1/2 phosphorylation.(d) Data in bar graphs represent the ratio of p-ERK1/2 to total ERK1/2. *P<0.05. (e) Cells were treated with METH for 24 h in the presence of 0.1 μM, 1 μM, 10 μM and 20 μM PD98059 (MEK inhibitor) separately, followed by real-time PCR for detection of Beclin1 mRNA expression. *P<0.05. Three independent experiments were performed. (f) Cells were treated with METH for 24 h in the presence or absence of 20 μM PD98059 (MEK1 inhibitor), followed by western blot analysis for detection of Beclin1 protein expression. (g) Densitometric analysis of Beclin1 protein levels. *P<0.05. Shown are representative results from one of three independent experiments. (h) Cells exposed to METH for 6, 24 and 48 h showed a decrease in the levels of phosphorylated Akt and mTOR. (i) Densitometric analysis of p-AKT and p-mTOR protein levels. *,**,#P<0.05. Shown are representative results from one of three independent experiments
We then asked if in response to autophagic stimuli Beclin1 is upregulated through ERK signaling. In total, 0.1 μM, 1 μM, 10 μM and 20 μM PD98059 in a concentration-dependent manner inhibited METH-induced Beclin1 mRNA expression. A total of 20 μM PD98059 significantly inhibited METH-induced Beclin1 mRNA expression compared with METH treatment alone (Figure 4e). Twenty micrometer PD98059 was used in the following experiments. Twenty micrometer PD98059 also significantly inhibited METH-induced Beclin1 protein expression (Figures 4f and g). These data suggest that upregulation of Beclin1 is ERK-dependent. In contrast, treatment with METH showed a downregulation of phosphorylated Akt and phosphorylated mTOR, 6 h post treatment (Figures 4h and i). These results showed that METH induced autophagy in HUVECs by inhibiting the Akt/mTOR pathway and upregulating the ERK1/2 pathway.
Inhibiting METH-induced autophagy triggers apoptotic cell death
Under pathological conditions, autophagy is considered to have a pro-survival role.24 Therefore, we asked whether suppression of METH-induced autophagy may accelerate cell death. Therefore, two well-characterized inhibitors of autophagy, 3-methyladenine (3-MA) and chloroquine, were used in parallel in this study. 3-MA and chloroquine have different structures, different mechanisms of action and act at early and late stages of autophagy progression, respectively. 3-MA is a class III phosphatidylinositol 3-kinase (PI3K) inhibitor that prevents autophagy at the earliest stage of autophagosome formation. Chloroquine is a weak base that concentrates in acidic vesicles such as lysosomes and disrupts vesicular acidification compromising their degradative and recycling capacity.26 First, we used 3-MA to examine whether inhibition of autophagy can enhance METH-induced apoptosis. HUVECs were either pretreated with or without 3-MA before the addition of 1 μM METH for 72 h. Cells were stained with Annexin V (green) and analyzed by fluorescence microscopy. 3-MA markedly increased apoptosis induction by METH at 72 h, as measured by Annexin V labeling. 3-MA alone caused minimal apoptosis at 72 h (Figures 5a and b). These findings suggest that inhibition of autophagy can accelerate apoptosis. Second, 100 μM chloroquine also significantly increased METH-induced caspase-3 activity (Figure 5c). These findings suggest that although 3-MA and chloroquine have different mechanisms of action, they both can accelerate METH-induced apoptosis by inhibiting autophagy.
Figure 5.
METH-induced apoptosis is accelerated by inhibiting autophagy. (a) Cells were either pretreated with or without autophagy inhibitor 3-MA before the addition of METH for 72 h. Cells were stained using an Annexin (green) V-FLUOS assay kit (Roche Diagnostics), and nuclei (blue) with DAPI and analyzed by fluorescence microscope. Images were recorded on a Nikon ECLIPSE Ti inverted fluorescence confocal microscope at × 400 magnification.(b) Quantitative analysis of apoptotic cells by Annexin V staining. The vertical axis indicated the ratio of the Annexin V–positive cells relative to total cells (DAPI-positive nuclei). *P<0.05. Shown are representative results from one of three independent experiments.(c) Cells were with or without autophagy inhibitor chloroquine in the presence of METH. Caspase activity was measured with Caspase-Glo assay systems (Promega). Results were expressed as percentage of the treated cells to the untreated control cells. (d) HUVECs were treated with nor-BNI (20 μM) 1 h before METH treatment for 24 h. Apoptosis was analyzed with Annexin V-PE staining. (e) Caspase activity was measured with Caspase-Glo assay systems (Promega). Results were expressed as percentage of the treated cells to the untreated control cells. Treatment with METH for 24 h showed a significant increase in Caspase-8 and Caspase-3/7 activation in HUVECs as compared with the control. Compared with only METH treatment, METH-induced Caspase-3/7 activation significantly elevated when pretreated with 20 μM PD98059 (MEK inhibitor). Three independent experiments were performed. *,**,#P<0.05 (t-test). (f) HUVECs were transiently transfected with either scramble siRNA or Beclin1 siRNA. Beclin1 mRNA expression was analyzed by real-time RT-PCR. (g) Caspase activity was measured with Caspase-Glo assay systems (Promega). Results were expressed as percentage of the treated cells to negative control cells
We have shown that KOR has a role in METH-induced autophagy (Figure 3). To determine whether inhibiting METH-induced autophagy triggers apoptosis, we also examined effects of the KOR antagonist pretreatment on METH- induced apoptotic cell death. Pretreatment with nor-BNI resulted in an increase in the number of apoptotic cells (12.90%) compared with METH treatment (8.14%) and control (1.32%) (Figure 5d). To further determine the mechanism underlying METH-induced apoptosis in the context of an autophagic block, caspase activity was measured in METH-treated HUVECs using Caspase-Glo 8, Caspase-Glo 3/7 and Caspase-Glo 9 assay systems in the presence or absence of MEK inhibitor PD98059. Although METH treatment alone resulted in modest induction of caspase-3/7, the induction was dramatically increased when cells were pretreated with the MEK inhibitor PD98059 (Figure 5e). We also investigated the effect of knockdown of Beclin1 on the caspase-3 activity of HUVECs. HUVECs were transfected with Beclin1 siRNA. Beclin1 mRNA expression was detected by real-time PCR analysis. The activity of caspase-3 was also measured in HUVECs transfected with Beclin1 knockdown. The results showed that Beclin1 siRNA transfection significantly decreased the mRNA expression of Beclin1 in the transfected HUVECs compared with scramble siRNA transfection (Figure 5f). METH significantly increased caspase-3 activity in the HUVECs with Beclin1 knockdown compared with METH treatment alone (Figure 5g). In conclusion, METH promoted cell apoptosis in the presence of Beclin1 knockdown in HUVECs.
These results further supported our hypothesis that acute treatment with METH triggered a pro-survival mechanism, and inhibition of signaling pathways leading to autophagy resulted in accelerated apoptosis.
Discussion
Our study demonstrated that METH disrupted HBMECs and HUVECs barrier function in a time-dependent manner. Low-dose METH at early time points induced the conversion of LC3-I into LC3-II and increased LAMP1 in endothelial cells. Interestingly, METH upregulated Beclin1 expression through KOR and ERK1/2 pathways. Furthermore, METH-induced autophagy in endothelial cells was triggered by inactivation of the AKT/mTOR/p70S6K pathway and activation of the ERK1/2 pathway (Figure 6). METH-induced autophagy was regulated by the Bcl-2/Beclin1 complex. Changes in Bcl-2/Beclin1 complex have been observed in endothelial cell autophagy when treated with anti-angiogenic inhibitors.27 METH also promoted caspase activity and suppression of autophagy accelerated METH-induced apoptosis.
Figure 6.
Schematic diagram showing the signaling pathways leading to METH-induced autophagy. METH-induced Beclin1 expression is through kappa opioid receptor. METH inhibits the downstream targets Akt and mTOR and activates the Raf/mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) cascade, resulting in induction of autophagy. METH downregulates Bcl-2, which releases Beclin1 from its asscociation with Bcl-2 thereby promoting autophagy. MEK Inhibitor PD98059 downregulates Beclin1 expression and promotes apoptosis
METH is an illicit recreational drug known to have effects on multiple organ systems. Exposure to METH leads to neurotoxic effects, which include damage to dopaminergic and serotonergic terminals, dopamine release, endoplasmic reticulum stress, activation of mitochondrial death cascades, glutamate release and hyperthermia.28 In an attempt to better clarify the molecular and cellular mechanisms responsible for neurotoxicity, most studies have focused on oxidative stress, activation of transcription factors, DNA damage, excitotoxicity, microglial activation and various apoptotic pathways.29, 30 More recent studies suggested that METH-induced neurotoxicity may also result from its ability to induce impairment of BBB function. The BBB has a predominant role in actively transporting nutrients to the brain and filtering harmful compounds from the brain back to the bloodstream. This process guarantees the homeostasis of the brain microenvironment, necessary for neuronal activity and proper functioning of the CNS.31 METH induces a transient increase in the permeability of the BBB in the hippocampus comparing with the frontal cortex and striatum, which can be explained by alterations in the tight junction proteins and matrix metalloproteinase-9 (MMP-9).32 However, the direct effects of low-dose METH in modulating BBB breakdown are not fully understood.
Most human studies have assessed only the effects of high-dose, long-term METH abuse (e.g., greater than 1000 mg/day) in individuals meeting DSM-IV (Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition) criteria for METH dependence. For the majority of METH abusers, however, patterns of METH exposure that consist of lower doses remain less well understood. The blood concentration of METH in drug abusers are in the low micromolar range, with median and mean concentrations of 1.3 μM (0.19 mg/l) and 2 μM (0.3 mg/l), respectively.10 Endothelial cell barrier dysfunction was also observed at 1 μM of METH, suggesting that the BBB is compromised under typical abuse conditions.3 Interestingly, low-dose METH (1 μM) exposure, in two dopaminergic neuronal-derived cell lines (rat pheochromocitoma PC12 and human neuroblastoma SH-SY5Y) resulted in induction of autophagy and suppression of autophagy precipitated neuronal cell death.1 In addition, another study showed that METH triggers an autophagic sequestering process in the dopaminergic SK-N-SH cell line by inducing the expression of LC3-II in a dose-dependent manner.33 However, no studies to date have investigated the effects of METH at doses seen in METH addicts in BBB endothelial cells.
We therefore investigated the functional role of autophagy in low-dose METH on endothelial cells. LC3 and Beclin1 are two key proteins for autophagy function. In this study, we first demonstrate that METH induced higher levels of LC3-II. LC3-II formation is regarded as a reliable biochemical evidence of autophagy as the amount of LC3-II usually correlates well with the number of autophagosomes.34 In our study, Beclin1 expression was also increased in a time-dependent manner. Beclin1 has a role in the initiation step and is essential for the formation of the autophagosome. The anti-apoptotic protein Bcl-2 has a negative regulatory role in autophagy by blocking an essential protein in the signaling pathway, Beclin1.15 METH induces autophagy in the human neuroblastoma SK-N-SH cell line by inhibiting the dissociation of Bcl-2/Beclin1 complex and its upstream pathway that leads to cell death.35 Our data revealed that METH downregulated Bcl-2, which released Beclin1 from its asscociation with Bcl-2 thereby promoting autophagy (Figure 6). Beclin1 interacts with Bcl-2, suggesting cross-talk between two potential programmed cell death pathways. It has been reported that activation of MEK/ERK downstream of AMPK leads to an increase in Beclin1 expression.36 To examine if the activation of the ERK pathway is directly related to the increase of Beclin1, we found that MEK1 inhibitor PD98059 prevented METH-induced Beclin1 mRNA and protein expressions in the endothelial cells (Figures 4e and f).
Although several signaling pathways regulate autophagy, signaling from the cytoplasm to the autophagy machinery is controlled in a negative manner through serine/threonine kinase, a mTOR. Protein kinase B or Akt (PKB/Akt), a positive regulator of mTOR, suppresses the formation of autophagosomes and inhibits autophagy.34 In addition, the Raf-1/MEK1/extracellular signal-related kinase (ERK) 1/2 pathway is known to be involved in regulating autophagy.37 A recent study shows that inhibition of the pathway by a MAPK kinase (MAPKK) MEK1/2 inhibitor (PD98059), suppresses starvation-induced autophagy in the human colon cancer HT-29 cell line.38 Our study clearly showed that METH inhibited the Akt/mTOR/p70S6K pathway and activated the ERK signaling pathway, resulting in the induction of autophagy in endothelial cells (Figures 4 and 6).
Autophagy has been shown to engage in a complex interplay with apoptosis in both physiological and pathological settings. Autophagy can serve as a cell survival pathway in some cellular settings by suppressing apoptosis, and in other settings, it can lead to programmed cell death, either in collaboration with apoptosis or as a back-up mechanism when the former is defective.39 It has been reported that suppression of autophagy precipitated neuronal cell death following low doses of METH.1 To evaluate the protective or the detrimental role of autophagy by METH in endothelial cells, we determined cell death (Figure 5a) in HUVECs for up to 72 h. Our results showed an increase in cell death in the presence of 3-MA, an inhibitor of class III PI3k activity widely used to inhibit autophagy. KOR antagonist and MEK1 inhibitor also accelerated METH-induced apoptosis, suggesting that the early autophagic response is a survival mechanism for endothelial cells and were mediated through the KOR (Figure 6). These results help to rule out that autophagy represents a detrimental mechanism. More importantly, our data emphasize that autophagy is an early response to METH-induced stress and may have a protective role in cell death.
In conclusion, our data indicate that autophagy is an early response to the METH-induced stress and may have a protective role during early exposure to the drug. However, chronic exposure to METH allows for progression through autophagic flux and commits cells to apoptotic death. These results help to explain how chronic exposure of METH can contribute to endothelial injury. These findings clarify the link between autophagy and apoptosis signaling induced by METH abuse.
Materials and Methods
Materials
METH, MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide), zVAD-fmk, pepstatin A, E-64d, 3-methyladenine (3-MA), chloroquine and α-tubulin antibody were purchased from Sigma (St. Louis, MO, USA). DyLight TM488-conjugated donkey anti-rabbit IgG was from Jackson Immuno Research Laboratories, Inc. (West Grove, PA, USA). SuperFect Transfection Reagent was from QIAGEN (Valencia, CA, USA). Protein A/G Plus-Agarose, Bcl-2 and Beclin1 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). AKT, p-AKT, mTOR, p-mTOR, ERK1/2, p-ERK1/2 and LC3 antibodies were from Cell Signaling (Danvers, MA, USA). KOR antagonist nor-Binaltorphiminedihydrochloride (nor-BNI) was from Tocris Bioscience (Ellisville, MO, USA). MEK Inhibitor PD98059 was from Cell Signaling (Danvers, MA, USA).
Cell culture
Human umbilical vein cells (HUVECs) were grown in tissue-culture flasks pre-coated with 0.2% gelatin (Sigma, St. Louis, MO, USA) and maintained in complete endothelial growth medium (EGM; Clonetics, San Diego, CA, USA). Primary HBMECs were cultured in RPMI 1640 containing 10% heat-inactivated fetal bovine serum, 10% Nu-Serum, 2 mM glutamine, 1 mM pyruvate, penicillin (100 U/ml), streptomycin (100 μg/ml), essential amino acids and vitamins (Life Technologies, Grand Island, NY, USA). All cell-culture dishes were coated with rat-tail collagen type I (BD Biosciences, Bedford, MA, USA).
Resistance measurements with electric cell-substrate impedance sensing (ECIS)
Electric cell-substrate impedance sensing (ECIS) electrode arrays (8W10E+) were obtained from Applied BioPhysics (Troy, NY, USA). Each array slide consists of eight individual wells treated for cell culture. Cells were harvested from 100-mm dishes and plated at 3 × 105 cells/500 μl in each well of ECIS arrays to obtain a confluent layer of cells covering the electrode surface. Complete single-layer electrode coverage was confirmed microscopically before treatment. To determine whether METH impaired endothelial cell barrier function, we used the ECIS to examine the effects of METH on endothelial cell monolayer resistances. HBMECs and HUVECs were treated with METH 1 μM in the absence or presence of zVAD-fmk, and changes in the resistance were continuously monitored for up to 72 h.
Western blot
Cell lysates were prepared in RIPA buffer (Sigma) and stored at −80 °C. Total protein concentration was determined by Bradford method. Whole-cell extracts (20 μg) were electrophoresed on a sodium dodecyl sulfate/polyacrylamide gel (SDS-PAGE), and electrotransferred to a nitrocellulose membranes (Amersham Biosciences, Pittsburgh, PA, USA). Blots were blocked by Tris-buffered saline solution containing 5% dry milk. Primary and secondary antibody-HRP conjugates were prepared in blocking buffer. Bands were visualized by the enhanced chemiluminescence (ECL) detection system (Amersham Biosciences). To verify the quality of loading, the blots were reprobed with anti-α-tubulin antibody (Santa Cruz Biotechnology).
Confocal and immunofluorescence microscopy
Immunofluorescence staining was carried out using antibodies against LC3 (Cell Signaling) and Beclin1 (Santa Cruz Biotechnology). DyLight TM488-conjugated donkey anti-rabbit IgG (Jackson Immuno Research Laboratories Inc.) was used as the secondary antibody. HUVECs were transiently transfected with LAMP1-GFP by using Lipofectamine 2000 (Life Technologies). LAMP1-GFP construct is a generous gift from Dr Sundaram Ramakrishnan (University of Minnesota, Minneapolis, MN, USA).
Rhodaminephalloidin or DAPI (Life Technologies) was used to visualize F-actin and the nucleus, respectively. Fields were chosen randomly from various regions to ensure objectivity of sampling. Digital images at × 400 magnification were acquired using a Nikon confocal microscope.
Transmission electron microscopy
Transmission Electron Microscopy (TEM) of HUVECs was carried out as previously described40 on cellular pellets obtained after centrifugation at 1000 × g for 5 min. Cell pellets were fixed with 2.5% glutaraldehyde for 40 min. After three washes with 0.1 M sodium cacodylate, samples were post-fixed with 1% osmium tetroxide (OsO4) for 30 min. Following three rinses, samples were dehydrated in a graded series of ethanol. Samples were subsequently embedded in Epon 812 resin. Thin sections (65 nm) were cut by using a Leica UC6 ultramicrotome and stained with uranyl acetate and lead citrate, then examined using a JEOL 1200EX electron microscope (JEOL, Peabody, MA, USA).
Caspase activation
Caspase-8, Caspase-3/7 and Caspase-9 activities were analyzed using Caspase-Glo luminescent-based assays according to the manufacturer's instructions (Promega Corporation, Madison, WI, USA). Cells (1 × 104) were seeded into 96-well white opaque plates and a corresponding optically clear 96-well plate, then allowed to adhere overnight. The next day, MEK1 inhibitor PD98059 (20 μM) was added 2 h before the cells were treated with METH for 24 h. At the end of the incubation time, 100 μl of the appropriate Caspase-Glo reagent was added to each well containing 100 μl of blank, negative control or treated cells in culture medium. Plates were gently mixed and incubated for 1 h at room temperature. The luminescence was then read in a FLUOstar OPTIMA Microplate Reader (BMG LABTECH, Cary, NC, USA). Caspase activity was normalized to vehicle treatment.
Transfection with siRNA
ON-TARGET plus SMART Pool Human Beclin1 siRNA (L-010552-00) and human KOR siRNA (L-005684-00) were used as per the manufacturer's instructions. Transfections were carried out as previously described.41 Scrambled siRNA was used as a negative control. The reduction in Beclin1 and KOR mRNA expression were estimated by real-time PCR analysis. Caspase-3/7 activity was analyzed using Caspase-Glo luminescent-based assays according to the manufacturer's instructions (Promega Corporation).
Real-time RT-PCR
Total RNA isolated from HUVECs was subjected to two-step reverse transcription-PCR (RT-PCR). Real-time SYBR green PCR analysis was performed for Beclin1 (sense, 5′-GCTGAGAGACTGGATCAGGA-3′ antisense, 5′-ATTGTGCCAAACTGTCCACT-3′) and human KOR(sense, 5′-GAAACAAGCCCTGGTGAAAT-3′ antisense, 5′-TGAGCCAAGACTGTGCTACC-3′). Relative quantification was performed using the ABI Prism 7500 real-time PCR system (Applied Biosystems, Grand Island, NY, USA). Beclin1 transcript levels were normalized to β-actin (sense, 5′-GGACTTCGAGCAAGAGATGG-3′ antisense, 5′-AGCACTGTGTTG GCGTACAG-3′) transcript levels from the same preparations of cDNA.
Assessment of apoptosis
Apoptosis was determined by Annexin V staining after METH 72 h treatment. Cells were stained using an Annexin V (green) -FLUOS assay kit (Roche Diagnostics, Indianapolis,IN, USA), and nuclei (blue) with DAPI and analyzed by fluorescence microscopy. Images were recorded with a Nikon confocal microscope at × 400 magnification. In some experiments, Annexin V-PE Apoptosis Detection Kit I (BD Biosciences, San Diego, CA, USA) was used to detect apoptosis.
Statistical analysis
Values are expressed as mean ±S.E. Significant difference between the control and experimental group was analyzed by unpaired Student's t-test and P<0.05 was considered as statistically significant.
Acknowledgments
This work was supported by National Institutes of Health grants RO1 DA 12104, RO1 DA 022935, RO1 DA031202, K05DA033881, P50 DA 011806 and 1R01DA034582 (SR). We are grateful to Ms Fang Zhou for the assistance of TEM. TEM was carried out in the Characterization Facility, University of Minnesota and a member of the NSF-funded Materials Research Facilities Network (www.mrfn.org) via the MRSEC program.
Glossary
- BBB
blood–brain barrier
- CNS
central nervous system
- LC3
microtubule-associated protein 1 light chain 3
- LC3-II
microtubule-associated protein 1 light chain 3 lipidated form
- LAMP1
lysosomal-associated membrane protein 1
- 3-MA
3-methyladenine
- TEM
transmission electron microscopy
- siRNA
small interfering RNA
The authors declare no conflict of interest.
Footnotes
Edited by GM Fimia
References
- Castino R, Lazzeri G, Lenzi P, Bellio N, Follo C, Ferrucci M, et al. Suppression of autophagy precipitates neuronal cell death following low doses of methamphetamine. J Neurochem. 2008;106:1426–1439. doi: 10.1111/j.1471-4159.2008.05488.x. [DOI] [PubMed] [Google Scholar]
- Lin M, Chandramani-Shivalingappa P, Jin H, Ghosh A, Anantharam V, Ali S, et al. Methamphetamine-induced neurotoxicity linked to ubiquitin-proteasome system dysfunction and autophagy-related changes that can be modulated by protein kinase C delta in dopaminergic neuronal cells. Neuroscience. 2012;210:308–332. doi: 10.1016/j.neuroscience.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins T, Burgoyne T, Kenny BA, Hudson N, Futter CE, Ambrósio AF, et al. Methamphetamine-induced nitric oxide promotes vesicular transport in blood-brain barrier endothelial cells. Neuropharmacology. 2013;65:74–82. doi: 10.1016/j.neuropharm.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott NJ, Patabendige AaK, DEM Dolman, Yusof SR, Begley DJ. Structure and function of the blood-brain barrier. Neurobiol Dis. 2010;37:13–25. doi: 10.1016/j.nbd.2009.07.030. [DOI] [PubMed] [Google Scholar]
- Abdul Muneer PM, Alikunju S, Szlachetka AM, Murrin LC, Haorah J. Impairment of brain endothelial glucose transporter by methamphetamine causes blood-brain barrier dysfunction. Mol Neurodegener. 2011;6:23. doi: 10.1186/1750-1326-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Klionsky DJ. The regulation of autophagy-unanswered questions. J Cell Sci. 2011;124 (Pt 2:161–170. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TE, Johansen T. Following autophagy step by step. BMC Biol. 2011;9:39. doi: 10.1186/1741-7007-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen SS, Aaberg-Jessen C, Christensen KG, Kristensen B. Expression of the lysosomal-associated membrane protein-1 (LAMP-1) in astrocytomas. Int J Clin Exp Pathol. 2013;6:1294–1305. [PMC free article] [PubMed] [Google Scholar]
- Pereira EM, do Monte SJ, do Nascimento FF, de Castro JA, Sousa JL, Filho HC, et al. Lysosome-associated protein 1 (LAMP-1) and Lysosome-associated protein 2 (LAMP-2) in a larger family carrier of Fabry disease. Gene. 2014;536:118–122. doi: 10.1016/j.gene.2013.11.063. [DOI] [PubMed] [Google Scholar]
- Melega W, Cho A, Harvey D, Laćan G. Methamphetamine blood concentrations in human abusers: application to pharmacokinetic modeling. Synapse. 2007;61:216–220. doi: 10.1002/syn.20365. [DOI] [PubMed] [Google Scholar]
- Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier HJ, Britton P. Involvement of autophagy in coronavirus replication. Viruses. 2012;4:3440–3451. doi: 10.3390/v4123440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols MS, van Meel E, Oorschot V, ten Brink C, Fukuda M, Swetha MG, et al. hVps41 and VAMP7 function in direct TGN to late endosome transport of lysosomal membrane proteins. Nat Commun. 2013;4:1361. doi: 10.1038/ncomms2360. [DOI] [PubMed] [Google Scholar]
- Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27 (Suppl 1:S137–S148. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima Y, Kyoi S, Zhai P, Liu T, Li H, Ivessa A, et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med. 2013;19:1478–1488. doi: 10.1038/nm.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Yamamizu K, Furuta S, Katayama S, Narita M, Kuzumaki N, Imai S, et al. The κ opioid system regulates endothelial cell differentiation and pathfinding in vascular development. Blood. 2011;118:775–785. doi: 10.1182/blood-2010-09-306001. [DOI] [PubMed] [Google Scholar]
- Mello NK, Negus SS. Interactions between kappa opioid agonists and cocaine. Preclinical studies. Ann NY Acad Sci. 2000;909:104–132. doi: 10.1111/j.1749-6632.2000.tb06678.x. [DOI] [PubMed] [Google Scholar]
- Shippenberg TS, Chefer VI, Zapata A, Heidbreder CA. Modulation of the behavioral and neurochemical effects of psychostimulants by kappa-opioid receptor systems. Ann NY Acad Sci. 2001;937:50–73. doi: 10.1111/j.1749-6632.2001.tb03558.x. [DOI] [PubMed] [Google Scholar]
- Tzaferis JA, McGinty JF. Kappa opioid receptor stimulation decreases amphetamine-induced behavior and neuropeptide mRNA expression in the striatum. Brain Res Mol Brain Res. 2001;93:27–35. doi: 10.1016/s0169-328x(01)00178-4. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ, Arbor A. Development by self-digestion: molecular mechanisms and biological functions of autophagy review. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Meijer AJ, Codogno P. Regulation and role of autophagy in mammalian cells. Int J Biochem Cell Biol. 2004;36:2445–2462. doi: 10.1016/j.biocel.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Saiki S, Sasazawa Y, Imamichi Y, Kawajiri S, Fujimaki T, Tanida I, et al. Caffeine induces apoptosis by enhancement of autophagy via PI3K/Akt/mTOR/p70S6K inhibition. Autophagy. 2011;7:176–187. doi: 10.4161/auto.7.2.14074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliyev BK, Menshikov M. Differential effects of the autophagy inhibitors 3-methyladenine and chloroquine on spontaneous and TNF-α-induced neutrophil apoptosis. Apoptosis. 2012;17:1050–1065. doi: 10.1007/s10495-012-0738-x. [DOI] [PubMed] [Google Scholar]
- Nguyen TMB, Subramanian IV, Kelekar A, Ramakrishnan S. Kringle 5 of human plasminogen, an angiogenesis inhibitor, induces both autophagy and apoptotic death in endothelial cells. Blood. 2007;109:4793–4802. doi: 10.1182/blood-2006-11-059352. [DOI] [PubMed] [Google Scholar]
- Kaushal N, Matsumoto RR. Role of sigma receptors in methamphetamine-induced neurotoxicity. Curr Neuropharmacol. 2011;9:54–57. doi: 10.2174/157015911795016930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet JL, Krasnova IN. Molecular bases of methamphetamine-induced neurodegeneration. Int Rev Neurobiol. 2009;88:101–119. doi: 10.1016/S0074-7742(09)88005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinton MS, Yamamoto BK. Causes and consequences of methamphetamine and MDMA toxicity. AAPS J. 2006;8:E337–E347. doi: 10.1007/BF02854904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Wedel-Parlow M, Schrot S, Lemmen J, Treeratanapiboon L, Wegener J, Galla HJ. Neutrophils cross the BBB primarily on transcellular pathways: an in vitro study. Brain Res. 2011;1367:62–76. doi: 10.1016/j.brainres.2010.09.076. [DOI] [PubMed] [Google Scholar]
- Martins T, Baptista S, Gonçalves J, Leal E, Milhazes N, Borges F, et al. Methamphetamine transiently increases the blood-brain barrier permeability in the hippocampus: role of tight junction proteins and matrix metalloproteinase-9. Brain Res. 2011;1411:28–40. doi: 10.1016/j.brainres.2011.07.013. [DOI] [PubMed] [Google Scholar]
- Kongsuphol P, Mukda S, Nopparat C, Villarroel A, Govitrapong P. Melatonin attenuates methamphetamine-induced deactivation of the mammalian target of rapamycin signaling to induce autophagy in SK-N-SH cells. J Pineal Res. 2009;46:199–206. doi: 10.1111/j.1600-079X.2008.00648.x. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nopparat C, Porter JE, Ebadi M, Govitrapong P. The mechanism for the neuroprotective effect of melatonin against methamphetamine-induced autophagy. J Pineal Res. 2010;49:382–389. doi: 10.1111/j.1600-079X.2010.00805.x. [DOI] [PubMed] [Google Scholar]
- Sridharan S, Jain K, Basu A. Regulation of autophagy by kinases. Cancers. 2011;3:2630–2654. doi: 10.3390/cancers3022630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki H, Takada Y, Kondo S, Sawaya R, Aggarwal BB, Kondo Y. Evidence that curcumin suppresses the growth of malignant gliomas in vitro and in vivo through induction of autophagy: role of Akt and extracellular signal-regulated kinase signaling pathways. Mol Pharmacol. 2007;72:29–39. doi: 10.1124/mol.106.033167. [DOI] [PubMed] [Google Scholar]
- Ogier-Denis E, Pattingre S, El Benna J, Codogno P. Erk1/2-dependent phosphorylation of Galpha-interacting protein stimulates its GTPase accelerating activity and autophagy in human colon cancer cells. J Biol Chem. 2000;275:39090–39095. doi: 10.1074/jbc.M006198200. [DOI] [PubMed] [Google Scholar]
- Eisenberg-Lerner A, Bialik S, Simon H-U, Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16:966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- Smith DM, Patel S, Raffoul F, Haller E, Mills GB, Nanjundan M. Arsenic trioxide induces a beclin1-independent autophagic pathway via modulation of SnoN/SkiL expression in ovarian carcinoma cells. Cell Death Differ. 2010;17:1867–1881. doi: 10.1038/cdd.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujumdar N, Mackenzie TN, Dudeja V, Chugh R, Antonoff MB, Borja-Cacho D, et al. Triptolide induces cell death in pancreatic cancer cells by apoptotic and autophagic pathways. Gastroenterology. 2010;139:598–608. doi: 10.1053/j.gastro.2010.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]