Abstract
Inhibition of human mitochondrial peptide deformylase (HsPDF) depolarizes the mitochondrial membrane, reduces mitochondrial protein translation and causes apoptosis in Burkitt's lymphoma. We showed that HsPDF mRNA and protein levels were overexpressed in cancer cells and primary acute myeloid leukemia samples. Myc regulates mitochondria and metabolism; we also demonstrated c-myc regulated the expression of HsPDF, likely indirectly. Inhibition of HsPDF by actinonin blocked mitochondrial protein translation and caused apoptotic death of myc-positive Burkitt's lymphoma, but not myc-negative B cells. Inhibition of mitochondrial translation by chloramphenicol or tetracycline, structurally different inhibitors of the mitochondrial ribosome, which is upstream of deformylase activity, followed by treatment with actinonin, resulted in reversal of the biochemical events and abrogation of the apoptosis induced by actinonin. This reversal was specific to inhibitors of HsPDF. Inhibition of HsPDF resulted in a mitochondrial unfolded protein response (increased transcription factors CHOP and CEB/P and the mitochondrial protease Lon), which may be a mechanism mediating cell death. Therefore, HsPDF may be a therapeutic target for these hematopoietic cancers, acting via a new mechanism.
Keywords: umbilical cord blood derived mesenchymal stem cells, galectin-1, extracellular matrix proteins, motility
In the mitochondria, like in bacteria, a formyl group is added to the N terminus of the mitochondrially encoded proteins.1 This formyl group is removed by the peptide deformylase (PDF) in the majority of bacterial proteins. These essential activities are required for viability and proliferation of bacteria.2 Inhibition of the bacterial PDF causes stasis of bacterial cells,3 and PDF is currently an emerging target for antibiotic research.4, 5 Our lab has characterized the functions of human mitochondrial protein peptide deformylase (HsPDF).6, 7, 8, 9 Mammalian PDFs are capable of removing the formyl group of N-terminally formylated peptides, which are derived from the proteins encoded by the mitochondrial DNA.9, 10 Inhibiting HsPDF in cancer cells results in inhibition of mitochondrial translation.7 Furthermore, genetic and pharmacologic inhibition of HsPDF also selectively reduces proliferation of cancer, but not normal cells.8 HsPDF expression in healthy and malignant cells has not been studied, nor has the mechanism of its selective effect on cancer cells. c-Myc (myc) is a transcription factor that has a central role in carcinogenesis, cancer cell metabolism, proliferation and cell size.11, 12 Myc is also a vital factor in the oncogenesis of acute myeloid leukemia (AML) and a hallmark of Burkitt's lymphoma. Furthermore, Myc has been shown to regulate mitochondrial biogenesis through direct transcription of its targets, some of which promote transcription of the mitochondrial DNA and control mitochondrial DNA replication13 in Burkitt's lymphoma models. Furthermore, increases in c-Myc expression correspond to increases in mitochondrial function, oxygen consumption and mitochondrial mass.14 Differential expression of c-Myc between normal and cancer cells and the control of mitochondrial function by this transcriptional regulator led us to investigate the relationship between HsPDF and c-Myc in Burkitt's lymphoma cells. Here we show that HsPDF protein and mRNA were overexpressed in most cancer cell lines and primary myeloid leukemias, independent of mitochondrial mass and that HsPDF was regulated by c-Myc, likely indirectly. The inhibition of HsPDF by the peptidomimetic actinonin disrupted mitochondrial translation in c-Myc-overexpressing cells, resulting in c-Myc selective apoptotic death. In addition, the newly discovered mitochondrial unfolded protein response (mtUPR) was induced by inhibition of HsPDF and is therefore one possible mechanism contributing to cell death. Therefore, HsPDF offers a mechanistically different therapeutic target for these hematopoietic cancers.
Results
HsPDF is overexpressed in cancer cell lines and primary myeloid leukemias
Although inhibition of HsPDF has been reported to result in decreased proliferation of cancer cells in vitro and in xenograft mouse models without toxicity, the differences in HsPDF protein expression between cancer and normal cells in not known. Therefore, we created a monoclonal antibody to HsPDF in order to quantify HsPDF protein levels in multiple cell lines by use of flow cytometry, confirmed with three independent antibody clones, and then proceeded to do our experiments with one representative clone, 20D10.
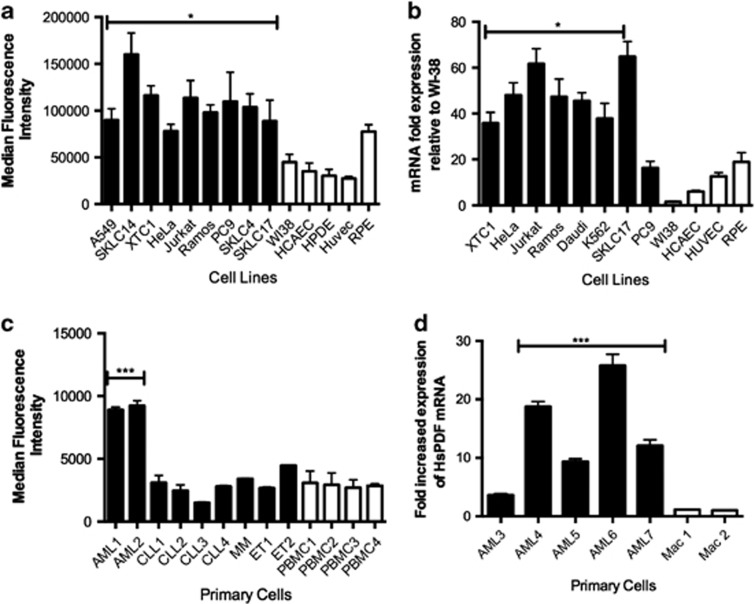
Nine human cancer cell lines (shown in black) and five human normal cell lines (shown in white) of different origins were assayed by flow cytometry (Figure 1a). Three of the normal cell types were primary (human coronary arterial endothelial cell, WI38 and human umbilical vein endothelial cells), while two of them were immortalized (retinal pigmented epithelium and human pancreatic duct epithelial cells). The normal cell lines (median fluorescence intensity=38 000) had lower HsPDF expression than the nine cancer cell lines (median fluorescence intensity=69 000).
Figure 1.
HsPDF is overexpressed in human cancers. (a) HsPDF protein levels in cancer (black) and normal (white) cell lines as measured by flow cytometry. Median fluorescence intensity for 10 000 events was counted for each sample in triplicate (shown as mean±S.E.M.). Average sample median fluorescence intensity was significantly higher in cancer cells than the median fluorescence intensity in normal cells (*P<0.05) (b) HsPDF mRNA levels in cancer (black) and normal (white) cell lines as measured by qRT-PCR. Samples were normalized to B-actin and then compared with WI38, the lowest value. Mean mRNA fold HsPDF expression was significantly higher in cancer cells relative to normal cells (**P<0.01). (c) HsPDF protein levels in AML, CLL, multiple myeloma (MM), essential thrombocythemia (ET) and peripheral blood mononuclear cells (PBMCs). Median HsPDF AML fluorescence intensity=8800; median CLL fluorescence intensity=2400; median ET fluorescence intensity=3500; median PBMC fluorescence intensity=2100. (d) HsPDF mRNA relative to B-actin and then normalized to macrophages. Median AML message was 12-fold higher than macrophages (***P<0.001).
The expression of HsPDF at the mRNA level was also significantly higher in cancer cells compared with normal cell line WI38 as the baseline level (median mRNA relative to WI38=45.5-fold) relative to normal cells (median mRNA relative to WI38=6.02 fold; Figure 1b).
HsPDF protein in primary leukemia cells in primary blood samples of patients with chronic lymphocytic leukemia (CLL) and AML were measured in comparison with peripheral blood mononuclear cells and macrophages from normal controls. The AML cells, but not CLL cells, showed that expression of the HsPDF protein (median fluorescence intensity=8900) was twice as high as that of monocytes (median fluorescence intensity=2300) (Figure 1c). Quantitative RT-PCR consistently showed that four of five AML samples had substantially elevated HsPDF mRNA expression relative to normal, confirming the protein data (Figure 1d). These data were further confirmed using immunoprecipitation and in-cell western assays with a subset of the cancer and normal cell lines as well (Supplementary Figures S1a and b).
The overexpression of the HsPDF could be a reflection of increases in mitochondria, so we estimated mitochondrial mass using the mitochondrial DNA content across the same panel of cells. The ratio of cytochrome c oxidase subunit II, a mitochondrial DNA-encoded protein to RNAseP, a single copy-number nuclear-encoded protein was measured in isolated total DNA of cancer and normal cells. Mitochondrial content between both populations was comparable (Supplementary Figure S1c).
To exclude the possibility that the increased levels of HsPDF in cancer cell lines might be due to a generalized overexpression of mitochondrial proteins, we profiled the mRNA expression of another cytoplasmically translated mitochondrial protein, manganese superoxide 2 (MnSOD). The expression of MnSOD was also similar between cancer and normal cell lines (Supplementary Figure S1d).
We also determined that there was no difference in the localization of the HsPDF protein in cancer and normal cells by examining the colocalization of mitotracker red and HsPDF by use of a secondary antibody to mouse anti-HsPDF conjugated to alexa-488. The micrographs showed that the HsPDF in cancer and normal cells localized to the mitochondria (Supplementary Figure S1e).
HsPDF is regulated by c-Myc
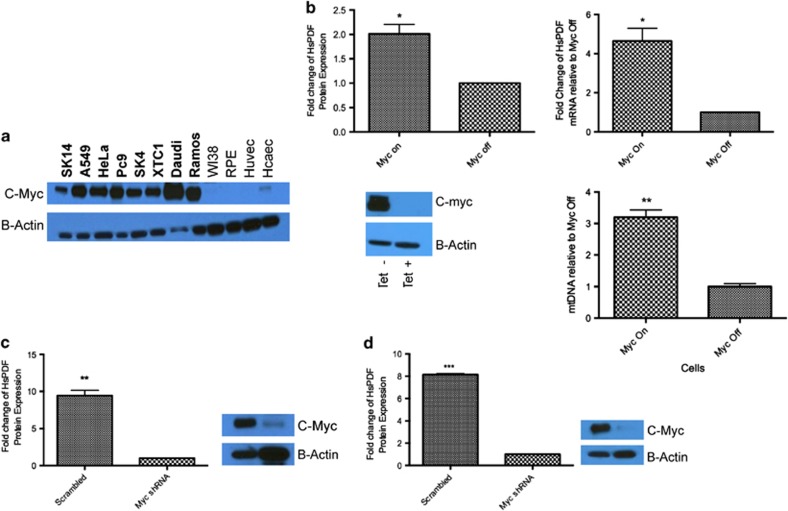
We investigated whether c-Myc also regulated HsPDF, by examining the expression of c-Myc in the cell lines overexpressing HsPDF, based on Figure 1a, by western blot. The cancer cells that overexpressed HsPDF (Figure 1a) also overexpressed c-Myc (Figure 2a). Next, we examined the HsPDF expression in the P493 cell line,13 which has tetracyline-off-mediated overexpression of c-Myc. When tetracycline was added, the reduction in myc expression resulted in a twofold reduction in HsPDF protein levels and an eightfold reduction in HsPDF mRNA (Figure 2b).
Figure 2.
HsPDF is regulated by c-Myc. (a) c-Myc is overexpressed in all cancer cell lines that also overexpress HsPDF. Normal cell lines did not have detectable levels of c-Myc. B-actin was used as a protein loading control. (b) Clockwise from top left: HsPDF protein expression doubles with c-Myc overexpression in P493 cells; HsPDF mRNA goes up eightfold with c-Myc overexpression; mitochondrial mass increase threefold as measured by mitochondrial DNA (ratio of complex III subunit 1/18sRNA). Western blot showing expression of c-Myc in the absence and presence of 1 μg/ml tetracycline. (c) HsPDF protein expression is abrogated in Ramos Burkitt's lymphoma cells when c-Myc is knocked down by small hairpin RNA (shRNA; left), western blot showing knockdown of c-Myc reduced HsPDF relative to B-actin (right). (d) HsPDF protein expression is abrogated when c-Myc is knocked down in Daudi Burkitt's lymphoma cells (right); western blot showing knockdown of c-Myc relative to B-actin. *P<0.05; **P<0.01; ***P<0.001
The Burkitt's lymphoma cells (containing t(8;14)(q24;q32)), Daudi and Ramos, were used as models for c-myc-overexpressing cancers. Both the Daudi and Ramos cells were killed by actinonin in a dose-dependent manner with IC50s of 6 and 16 μM, respectively (Supplementary Figure S1f).
To confirm the association of c-Myc with HsPDF levels, we transduced the two Burkitt's lymphoma cell lines, Ramos and Daudi, with c-Myc small hairpin RNA carrying lentiviral particles. The knockdown of c-Myc in the cells (Figures 2c and d) resulted in elimination of HsPDF protein expression (Figures 2c and d). These sets of data provided strong evidence that c-Myc regulates the expression of HsPDF, although it may not be directly.
Inhibition of HsPDF selectively targets c-Myc-positive tumors
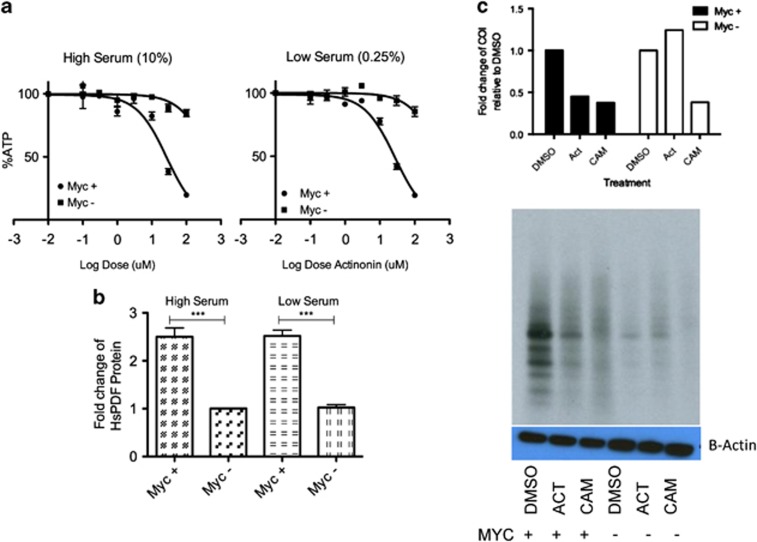
Treatment of c-myc-overexpressing P493 cells with actinonin resulted in dose-dependent killing of the cells (Figure 3a, left) and cells overexpressing c-Myc have a faster doubling time than the cells that lack c-myc. This raised the possibility that actinonin may be more cytotoxic to proliferating cells. To answer this question, we reduced the serum levels in the culture media on the myc-overexpressing P493 cells to 0.25%, a level at which the cells do not proliferate, but still remain viable. Trypan blue staining showed that these cells were viable and not proliferating for at least 24 h after moving to the low serum condition (not shown). The nonproliferating cells were also killed by actinonin with similar kinetics to the proliferating cells (Figure 3a, right). The data implied that the cytotoxicity of actinonin was associated with c-Myc overexpression and not the rate of cell proliferation. We also confirmed that the protein expression of HsPDF by flow cytometry remained constant during the serum deprivation to ensure the cytotoxicity of actinonin was not due to loss of protein expression (Figure 3b).
Figure 3.
Actinonin is selectively cytotoxic to c-Myc-positive cells. (a) Actinonin selectively kills c-Myc-positive P493 cells whether cells are proliferating (10% serum; IC50s Myc-positive=26 μM, Myc-negative≥100 μM, left panel) or not (0.25% serum; IC50s Myc-positive=27 μM, Myc-negative≥100 μM, right panel). (b) HsPDF levels do not change with serum deprivation or changes in the rate of proliferation (LS, low serum; HS, high serum). (c) Actinonin (ACT, 20 μM) selectively inhibits mitochondrial translation in c-Myc-positive cells (black bars), but not c-Myc-negative cells (white bars). Gel shows raw data and loading control. CAM (100 μM), a positive control, inhibits mitochondrial translation in both c-Myc-positive cells and c-Myc-negative cells. ***P<0.001
To further confirm whether the cytotoxicity from actinonin was directly related to the functional activity of HsPDF, we assayed mitochondrial protein translation with actinonin in the presence and absence of c-myc. HsPDF is required for mitochondrial protein translation,7 and so we used this as a surrogate for HsPDF function in live cells. In the presence of overexpressed c-myc, actinonin caused inhibition of mitochondrial protein translation relative to the vehicle control (Figure 3c). Chloramphenicol (CAM), an inhibitor of the large subunit of the mitochondrial ribosome was used as a positive control. In the absence of c-Myc, mitochondrial translation occurred at a lower level, but it was not inhibited by actinonin. In contrast, CAM still inhibited mitochondrial protein translation, showing the specificity of c-Myc in regulation of HsPDF and its function.
Apoptosis induced by actinonin originates in the mitochondria and is HsPDF specific
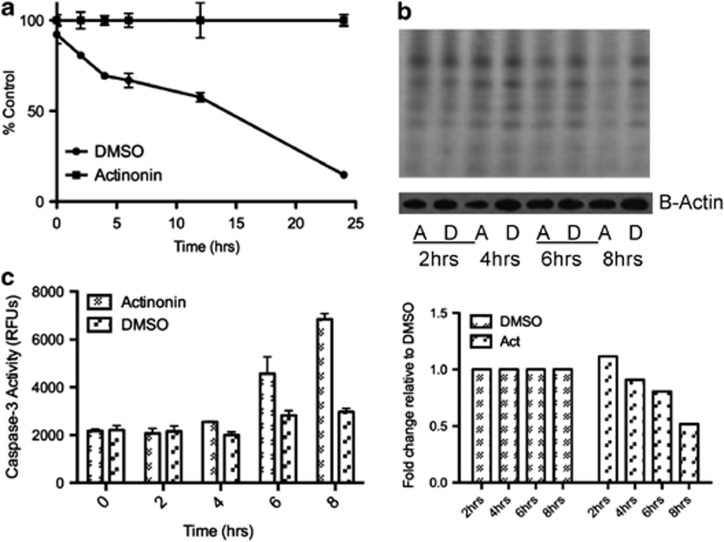
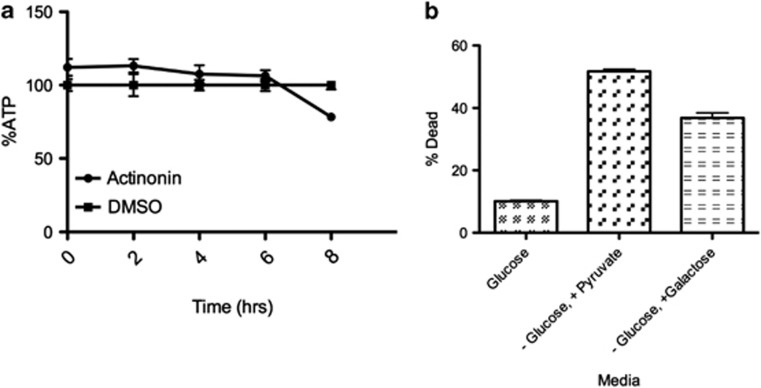
To prove that the mechanism of actinonin-induced apoptosis was occurring through the inhibition of HsPDF and not possible off targets of this drug, we performed a series of confirmatory experiments. First, we investigated whether the damage was initiated in the mitochondria by use of a temporal study of the events that occurred after treatment with actinonin. The earliest measurable event, which occurred at 2 h, was the depolarization of the mitochondrial membrane (Figure 4a). This depolarization occurs likely because of the inhibition of mitochondrial protein translation, which, owing to limitations in the dynamic range of the assay can be visualized earliest at 4–6 h (Figure 4b). Eventually, caspases-3/7 get activated at 6–8 h and apoptosis occurs (Figure 4c).
Figure 4.
Actinonin-mediated apoptosis begins in the mitochondria. (a) Actinonin (15 μM) causes mitochondrial membrane depolarization in Ramos cells as measured by JC1 dye beginning by 2 h. (b) Inhibition of mitochondrial protein translation begins at 4 h as shown by incorporation of 35S (top) and densitometric measurement (bottom) of the same (c) Caspase-3/7 activation begins at 6 h
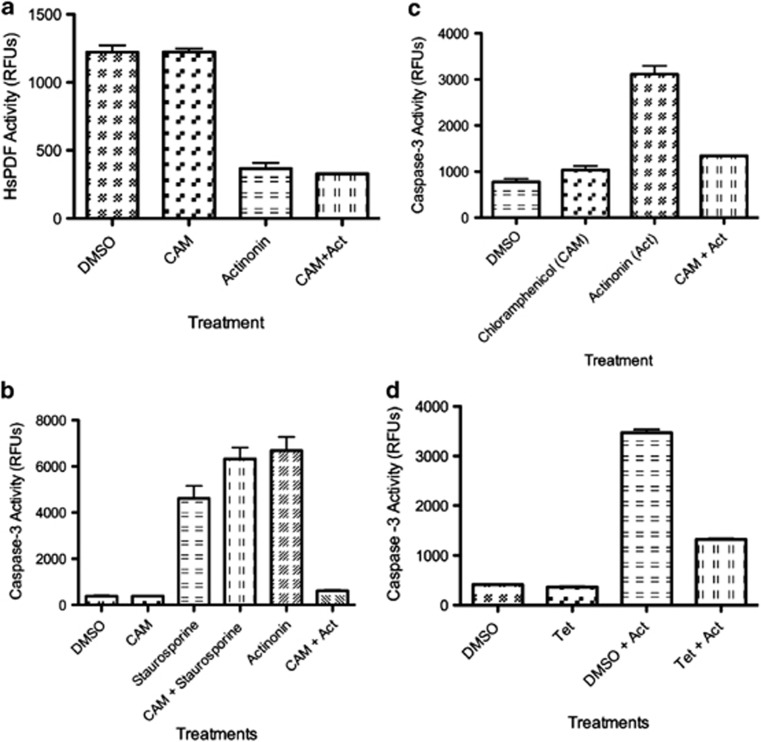
Actinonin possibly could inhibit other metalloproteases via its hydroxamic acid moiety, such as matrix metalloproteases, and possibly histone deacetylases. To show that the cytotoxicity mediated by actinonin on the Daudi and Ramos cells was directly related to the inhibition of HsPDF and not via another off-target enzyme, we used an assay that has been established to determine HsPDF specificity.7, 15 In brief, this assay involves the pretreatment of cells by CAM before addition of actinonin. CAM stops mitochondrial ribosome translation of mitochondrial DNA-encoded proteins. HsPDF acts downstream of the ribosome, and inhibiting translation with CAM would deprive HsPDF of its mitochondrially translated nascent chain substrates. If the deprivation of substrates for HsPDF eliminated actinonins cytotoxic activity, then the mechanism of apoptosis mediated by actinonin would have to be via HsPDF and not an off-target of actinonin. CAM does not inhibit HsPDF function itself, nor does it directly interact with actinonin in vivo (Figure 5a). Pretreatment of Ramos cells with CAM prevented actinonin-mediated apoptosis, but not the positive control staurosporine-mediated apoptosis (Figure 5b). This was confirmed in Daudi cells (Figure 5c). CAM or tetracycline alone did not activate caspase-3. Further, we confirmed this result using another structurally different inhibitor of mitochondrial translation, tetracycline, which also prevented actinonin-mediated apoptosis (Figure 5d).
Figure 5.
Actinonin causes apoptosis through specific inhibition of HsPDF. (a) Recombinant HsPDF activity is inhibited by actinonin (100 nM) treatment for 2 h and is not prevented with pretreatment of CAM (100 μM). DMSO=vehicle control, Act=100 nM actinonin, CAM=100 μM CAM. (b) Actinonin (15 μM) causes caspase-3 activation that is prevented with pretreatment of CAM (100 μM) for 2 h in Ramos Burkitt's lymphoma cells. DMSO=vehicle control, CAM=100 μM CAM, staurosporine=2 μM, CAM+staurosporine=100 μM CAM 2 h before addition of 2 μM staurosporine. All caspase-3 activation assays were conducted 14 h after the addition of CAM or 12 h after the addition of actinonin or staurosporine. (c) Actinonin causes caspase-3 activation in Daudi Burkitt's lymphoma cells that is prevented with pretreatment with CAM (100 μM). (d) Actinonin (15 μM) causes caspase-3 activation that is prevented by pretreatment with tetracycline (10 μM) for 2 h. Caspase-3 activation assays were conducted at 12 h after addition of actinonin
To further investigate contributing roles of other possible off-target enzymes, we treated Daudi and Ramos cells with inhibitors of histone deacetylases, trichostatin A and SAHA, both hydroxamic acid peptidomimetics. CAM did not protect against the drug-induced apoptosis, showing that these other non-HsPDF-based mechanisms were not involved (Supplementary Figure S2a). Furthermore, the pan matrix metalloprotease inhibitor, NNGH, killed these cells through a non-caspase-3-mediated apoptosis pathway (Supplementary Figures S2b and c). We also used two structurally similar aminopeptidase N (APN) inhibitors, Chroma 2863 and Chroma 6768. Of these, only Chroma 2863 killed the cells; however, this death was neither apoptotic nor protected by CAM (Supplementary Figures S2d and e).
It was also clear that CAM protected actinonin-mediated mitochondrial membrane depolarization, but not staurosporine-mediated depolarization, implying a unique protective effect (Supplementary Figure S2f).
Next, we treated the cells with bestatin, another inhibitor of APN. Although the majority of reports suggest that Daudi and Ramos cells do not express APN on the cell surface, there are reports that suggest that intracellular APN may exist in these cells, which can be inhibited by bestatin. We measured the dose response effects of bestatin in these cells (Supplementary Figure S2g), and there did not appear to be any inhibition of adenosine triphosphate (ATP) levels up to doses of 100 μM, whereas actinonin caused cytotoxicity at doses of 6 μM in Daudi and 16 μM in Ramos. Further, we tested the activity of a structurally similar molecule to actinonin, actinonamide (AA), which lacks the hydroxamic acid element (Supplementary Figure S2b) and thus does not inhibit HsPDF. AA also showed no inhibition of viability at doses up to 80 μM. This showed that the death was not through one of the known or possible off-targets of actinonin. This series of experiments conclusively showed that actinonin-mediated apoptosis was occurring through the inhibition of HsPDF.
Inhibition of HsPDF results in a mtUPR
We have previously shown that HsPDF enables the synthesis of mitochondrial encoded proteins. Here we also show that inhibition of HsPDF leads to apoptosis. Therefore, we asked whether this death was due to impaired mitochondrial ATP production. Cancer cells, however, may rely on aerobic glycolysis to produce ATP as proposed by Warburg.17 If these leukemic cells depend on aerobic glycolysis for energy, then the toxic effect of disrupting mitochondrial ATP production should be negligible, provided glucose is present. Therefore, we treated Daudi and Ramos cells with media lacking glucose and containing pyruvate, which would force the cells to respire through the mitochondria. In the presence of pyruvate and absence of glucose, the Ramos and Daudi cells die within 10 h (Figures 6a and b). To confirm that this cytotoxicity was not due to the absence of glycosylation of proteins, we also added galactose to the media with pyruvate. Cells still died, suggesting that energy in these cells is predominantly glycolytic and the mechanism of HsPDF-mediated death was unrelated to mitochondrial ATP production.
Figure 6.
Burkitt's lymphoma cells depend on glycolysis for energy production. (a) ATP levels in the actinonin-treated Ramos cells do not decrease until 8 h as measured by ATPLite kit (Perkin Elmer, Santa Clara, CA, USA). (b) Cells die in 12 h when they are transferred to media without glucose, but with pyruvate (56% dead) or galactose (40% dead). Cell death was measured by propidium iodide
Prokaryotic deformylases are associated with the chaperone trigger factor and a methionine aminopeptidase. In the human mitochondria, a methionine aminopeptidase 1D (MAP1D) has been described, but there is no known trigger factor chaperone. This prompted us to hypothesize that HsPDF may provide a function outside cleaving the formyl group on the nascent chain. Thus, a lack of HsPDF function would result in poorly folded or unprocessed proteins capable of stimulating a mtUPR. Hence, we looked for a mtUPR after treatment with actinonin, and whether this unfolded protein response was abrogated by treatment with CAM.
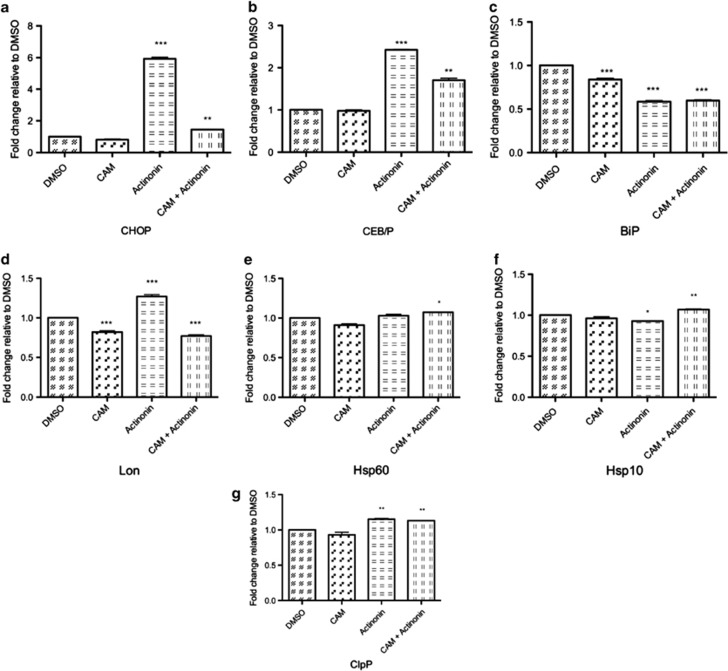
When Ramos cells were treated with actinonin, CHOP and CEB/P, two proteins that are characteristic of the mitochondrial unfolded response, increased at 4 h (Figures 7a and b). This increase was prevented by pretreatment with CAM. At the same time, there was no upregulation in BiP, a protein associated with an endoplasmic reticulum (ER)-specific response, nor were BiP levels protected by CAM (Figure 7c). The mitochondrial protease Lon, which is responsible for degrading damaged proteins, was also upregulated after treatment with actinonin (Figure 7d). However, there was no increase in other proteins involved in the mtUPR such as the mitochondrial chaperones Hsp10 and Hsp60, and a minor, but significant, increase in the protease ClpP (Figures 7e–g).
Figure 7.
Inhibition of HsPDF induces mtUPR. mRNA for protein markers of mtUPR was measured by qPCR, normalized to B-actin and then represented relative to DMSO vehicle control in Ramos Cells. Actinonin=15 μM, CAM=100 μM, CAM was added to pretreated cells for 2 h before the addition of actinonin. All measurements were taken at 4 h post treatment (6 h post pretreatment). (a) CHOP message was upregulated sixfold relative to vehicle control and reduced by pretreatment of CAM. (b) CEB/P was upregulated 2.5-fold relative to vehicle control and is reduced by pretreatment with CAM. (c) BiP, a marker of ER stress is reduced after treatment with actinonin and unchanged by CAM pretreatment. (d) Mitochondrial protease Lon is upregulated on treatment with actinonin, and reduced by pretreatment with CAM. There is no change after addition of actinonin or pretreatment with CAM in chaperones Hsp60 (e), Hsp10 (f) and the protease ClpP (g). *P<0.05; **P<0.01; ***P<0.001
Discussion
The role of mitochondria in cancer has recently been under intense investigation.16 Furthermore, recent studies have drawn insight into new roles for the mitochondria, which have been validated as potential cancer targets.18
Previously, we have shown pharmacologically and genetically that inhibition of HsPDF can lead to a marked decline in proliferation in many cancer cell lines and apoptosis in leukemia cell lines,19 (Supplementary Figure S3). Furthermore, an evolutionary binding partner and functionally linked protein of HsPDF, MAP1D, is also overexpressed in colon cancers.20 However, there has never been an in-depth study to investigate the relationship between cancer and the value of HsPDF as a cancer-specific target. Here, we found that (1) HsPDF expression in cancer was upregulated in both cancer cell lines and primary myeloid leukemia samples, (2) HsPDF expression was regulated by the oncogene c-Myc, which is known to control components of mitochondrial biogenesis and mitochondrial translation, (3) inhibition of HsPDF resulted in an unfolded protein stress response, and (4) apoptosis in the c-Myc-overexpressing Burkitt's lymphoma cell lines, but not c-Myc-negative lines.
There have been several studies tying c-Myc to mitochondrial biogenesis.13, 21, 22 An analysis of the promoter sequence of HsPDF, which has recently been cloned suggests, however, that it is unlikely that this regulatory relationship is direct.23 There are no E-boxes on the currently cloned promoter region of the HsPDF protein; however, it is possible that there may be a distal promoter region. In addition, a likely candidate protein regulating HsPDF expression is the protein MZF1, which is a direct target of c-Myc, which binds to the cloned promoter region of HsPDF.23
Actinonin selectively inhibited and killed c-Myc-positive Burkitt's lymphoma cells, regardless of proliferation rate, implying that HsPDF was a c-Myc-specific cancer therapeutic target. The specificity of actinonin for HsPDF in these lymphoma cells was evidenced by several distinct sets of data, both kinetic and pharmacologic (Supplementary Figures S2–5). First, the kinetics of actinonin-induced apoptosis suggested that the apoptotic death began with mitochondrial membrane depolarization, followed by mitochondrial translation inhibition, eventually leading to caspase activation. Second, we used specific assays in which we deprived HsPDF of its putative mitochondrial electron transport chain mitochondrially translated substrates, by CAM or tetracyline. Following CAM or tetracycline treatments, there was protection against apoptosis induced by actinonin.7, 15 Third, CAM and tetracycline did not prevent the cytotoxicity of other small molecules that inhibit possible or published off targets of HsPDF. CAM or tetracycline do not inhibit HsPDF activity themselves. These data confirmed that the actinonin-mediated apoptosis was due to the specific inhibition of HsPDF function.
Kinetic measurements of ATP levels during the apoptotic process argued against a loss of energy as the cause. Our data showed that Burkitt's lymphoma was dependent on glycolysis for survival and that the mitochondrial contribution to ATP production was minor and unable to independently sustain the cellular energy needs. Thus, the death is not caused by actinonin's disruption of electron transport complex function, but by a distinct other mechanism.
There have been reports suggesting that the deformylation of mitochondrial DNA-encoded proteins is not significant in bovine liver,10 but others suggest that the lack of formylation of the mitochondrial proteins results in the pathology of Leigh syndrome.24 A recent study has suggested that HsPDF inhibition results in antiproliferative activity by causing HsPDF to block the ribosomal exit site. This causes an accumulation of amino acids in the P site of the ribosome, which results in stress and retrograde signaling, which eventually leads to a inhibition of proliferation.15 Our results confirm this thesis, and we propose a mechanism by which inhibition of HsPDF could lead to cell death.
One of the main binding partners of bacterial PDF is a trigger factor, a chaperone. There is no homolog of a trigger factor in mammals or higher eukaryotes. One could therefore speculate that HsPDF serves a role as a chaperone or in some other protein-processing capacity to ensure that properly folded proteins function correctly and are delivered to their appropriate location. Indeed, inhibition of PDF has been shown to cause ribosomal degradation and an accumulation of amino acids in the P site of the ribosome.15 We have also shown that HsPDF inhibition resulted in a mtUPR. Furthermore, other components of the mitochondrial translation apparatus such as EFTu have been shown to have chaperone activities.25 HsPDF does not appear to behave like a traditional stress chaperone, as we did not see an increase in the expression of HsPDF when the cells were stressed either by heat or reactive oxygen species. However, we hypothesize it is involved in handing over the nascent chains to downstream assembly chaperones like Oxa1.
The mtUPR has recently been implicated as a mechanism for death in cancer cells.26 The known markers of this phenomenon are the upregulation of the stress response transcription factors CHOP and CEB/P.27 However, these transcription factors are also upregulated under conditions of ER stress. To confirm that the effects seen were due to the inhibition of HsPDF, and not secondary to apoptosis in general or off-targets of actinonin, we pretreated the cells with CAM and examined the mRNA expression at 4 h, before caspase-3 was activated. In this time frame, CAM did not promote a stress response of its own. CHOP and CEB/P were both increased, but BiP was not, in actinonin-treated cells, and both these effects were reversed by CAM, suggesting a role for an unfolded protein response in response to PDF inactivation. We saw a modest increase in the expression of the protease Lon, which is known to degrade mitochondrially unfolded proteins. This effect was also reversed by CAM. Although Lon has not yet been implicated in the mtUPR, the model systems used have always studied mitochondrial proteins that are translated in the cytoplasm and exported to the mitochondria rather than the electron transport chain proteins encoded by the mtDNA. Thus, it is possible that Lon may have a role in degrading either the unfolded proteins that are translated by the mitochondrial genome.
On the basis of recent studies, it is also possible that these stress proteins were upregulated as a result of accumulation of tRNAs in the P site or blocked ribosomes, both of which could result in stressing the mitochondria.15 Furthermore, it is possible that Lon is involved in the eventual degradation of the mitochondrial ribosome. However, these data suggest a mitochondrial stress response following HsPDF inhibition. As the mtUPR response can lead to death in mammalian cells,26 one can speculate that this stress is related to cell death in our system.
In conclusion, we have discovered that the protein HsPDF is upregulated in cancer, and that HsPDF is regulated by the oncogene product c-Myc. Furthermore, the inhibition of this protein resulted in selective cell death of c-Myc-overexpressing leukemias and lymphomas, regardless of their proliferation rate. This death was apoptotic and began with the depolarization of the mitochondrial membrane, followed by an inhibition of translation eventually resulting in caspase-3/7 activation. This apoptosis was HsPDF specific, and was not due to other targets of the inhibitor, actinonin. Actinonin-mediated apoptosis was prevented by depriving HsPDF of substrates. Finally, the inhibition of HsPDF resulted in a mitochondrial stress response, which may be contributing to the apoptosis of these cancer cells. Therefore, inhibition of HsPDF is potentially a mechanistically different approach to therapy of these hematopoietic cancers.
Materials and Methods
Production of HsPDF monoclonal antibody
Monoclonal antibodies were produced in mice after immunization with 500 μg of HsPDF-MBP along with the adjuvant titermax every 3 weeks for 15 weeks. For details please refer to the Supplementary Methods.
HsPDF expression immunoassays
Cells (0.25–0.75 × 106) were fixed using 4% paraformaldehyde in media (RPMI or DMEM) for 30 min at room temperature. After two washes with PBS for 5 min each, they were resuspended in blocking buffer (10% goat serum, 2% BSA, 100 mM glycine, 0.2% Triton-X) for an hour on a shaker. Antibody (1 μg/ml) was added to each sample for an hour, mouse IgG (Invitrogen (Life Technologies, Grand Island, NY, USA) for flow cytometry) was added as a negative control and samples were incubated overnight at 4 °C. The following morning samples were washed three times in wash buffer (PBS+0.5% BSA+0.2% Triton-X) for 10 min each and incubated with secondary antibody (goat anti-mouse-alexa-488, Invitrogen). Samples were washed and read by flow cytometry. Flow cytometry was performed using an Accuri C6 flow cytometer (Accuri, Ann Arbor, MI, USA). Ten thousand events were acquired in list mode for each treatment group and analyzed with FlowJo 8.7.3 software (Treestar, Ashland, OR, USA). HsPDF expression was measured as a median fluorescence emission at 529 nm (FL1). IgG control and 4T1 (mouse cell line) cell line fluorescence intensity values were used as negative controls and were subtracted from readings. All measurements were performed in triplicate and are expressed as the mean±S.E.M.
Human samples were obtained under approved IRB protocols and animal experiments were conducted under approved IACUC protocols.
Acknowledgments
We thank the members of the Scheinberg Laboratory, Giovanni Manfredi and Cole Haynes for thoughtful discussions. This work was supported by grants from the National Institutes of Health (RO1CA55349), the Experimental Therapeutics Center and the Cancer Research Fund.
Glossary
- AA
actinonamide
- AML
acute myeloid leukemia
- APN
aminopeptidase N
- ATP
adenosine triphosphate
- CAM
chloramphenicol
- CLL
chronic lymphocytic leukemia
- ER
endoplasmic reticulum
- HsPDF
human mitochondrial peptide deformylase
- MAP1D
methionine aminopeptidase 1D
- MnSOD
manganese superoxide dismutase
- mtUPR
mitochondrial unfolded protein response
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Raschellà
Supplementary Material
References
- Bianchetti R, Lucchini G, Crosti P, Tortora P. Dependence of mitochondrial protein synthesis initiation on formylation of the initiator methionyl-tRNAf. J Biol Chem. 1977;252:2519–2523. [PubMed] [Google Scholar]
- Mazel D, Pochet S, Marlière P. Genetic characterization of polypeptide deformylase, a distinctive enzyme of eubacterial translation. EMBO J. 1994;13:914–923. doi: 10.1002/j.1460-2075.1994.tb06335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Khuller GK, Sharma S. Peptide deformylase – a promising therapeutic target for tuberculosis and antibacterial drug discovery. Expert Opin Ther Targets. 2009;13:753–765. doi: 10.1517/14728220903005590. [DOI] [PubMed] [Google Scholar]
- East SP, Ayscough A, Toogood-Johnson I, Taylor S, Thomas W. Peptidomimetic inhibitors of bacterial peptide deformylase. Bioorg Med Chem Lett. 2011;21:4032–4035. doi: 10.1016/j.bmcl.2011.04.132. [DOI] [PubMed] [Google Scholar]
- Goemaere E, Melet A, Larue V, Lieutaud A, Alves de Sousa R, Chevalier J, et al. New peptide deformylase inhibitors and cooperative interaction: a combination to improve antibacterial activity. J Antimicrob Chemother. 2012;67:1392–1400. doi: 10.1093/jac/dks058. [DOI] [PubMed] [Google Scholar]
- Lee MD, Antczak C, Li Y, Sirotnak FM, Bornmann WG, Scheinberg DA. A new human peptide deformylase inhibitable by actinonin. Biochem Biophys Res Commun. 2003;312:309–315. doi: 10.1016/j.bbrc.2003.10.123. [DOI] [PubMed] [Google Scholar]
- Escobar-Alvarez S, Gardner J, Sheth A, Manfredi G, Yang G, Ouerfelli O, et al. Inhibition of human peptide deformylase disrupts mitochondrial function. Mol Cell Biol. 2010;30:5099–5109. doi: 10.1128/MCB.00469-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MD, She Y, Soskis MJ, Borella CP, Gardner JR, Hayes PA, et al. Human mitochondrial peptide deformylase, a new anticancer target of actinonin-based antibiotics. J Clin Invest. 2004;114:1107–1116. doi: 10.1172/JCI22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Alvarez S, Goldgur Y, Yang G, Ouerfelli O, Li Y, Scheinberg DA. Structure and activity of human mitochondrial peptide deformylase, a novel cancer target. J Mol Biol. 2009;387:1211–1228. doi: 10.1016/j.jmb.2009.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Carroll J, Altman MC, Fearnley IM. Chapter 6 Mass spectrometric characterization of the thirteen subunits of bovine respiratory complexes that are encoded in mitochondrial DNA. Meth Enzymol. 2009;456:111–131. doi: 10.1016/S0076-6879(08)04406-6. [DOI] [PubMed] [Google Scholar]
- Berta MA, Baker CM, Cottle DL, Watt FM. Dose and context dependent effects of Myc on epidermal stem cell proliferation and differentiation. EMBO Mol Med. 2010;2:16–25. doi: 10.1002/emmm.200900047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, et al. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- Richter U, Lahtinen T, Marttinen P, Myöhänen M, Greco D, Cannino G, et al. A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation. Curr Biol. 2013;23:535–541. doi: 10.1016/j.cub.2013.02.019. [DOI] [PubMed] [Google Scholar]
- Scatena R. Mitochondria and cancer: a growing role in apoptosis, cancer cell metabolism and dedifferentiation. Adv Exp Med Biol. 2012;942:287–308. doi: 10.1007/978-94-007-2869-1_13. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Neuzil J, Dong L-F, Rohlena J, Truksa J, Ralph SJ.Classification of mitocans, anti-cancer drugs acting on mitochondria. Mitochondrion [Internet]. 28 July 2012 (cited 19 August 2012); Available from: http://www.ncbi.nlm.nih.gov/pubmed/22846431 . [DOI] [PubMed]
- Grujic M, Zavasnik-Bergant T, Pejler G, Renko M. Actinonin induces apoptosis in U937 leukemia cells. Cancer Lett. 2005;223:211–218. doi: 10.1016/j.canlet.2004.10.048. [DOI] [PubMed] [Google Scholar]
- Leszczyniecka M, Bhatia U, Cueto M, Nirmala NR, Towbin H, Vattay A, et al. MAP1D, a novel methionine aminopeptidase family member is overexpressed in colon cancer. Oncogene. 2006;25:3471–3478. doi: 10.1038/sj.onc.1209383. [DOI] [PubMed] [Google Scholar]
- Dang CV, Li F, Lee LA. Could MYC induction of mitochondrial biogenesis be linked to ROS production and genomic instability. Cell Cycle. 2005;4:1465–1466. doi: 10.4161/cc.4.11.2121. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee J, Iyer VR. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One. 2008;3:e1798. doi: 10.1371/journal.pone.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Castro I, Costa LT, da, Amorim A, Azevedo L. Transcriptional regulation of the human mitochondrial peptide deformylase (PDF) Biochem Biophys Res Commun. 2012;421:825–831. doi: 10.1016/j.bbrc.2012.04.097. [DOI] [PubMed] [Google Scholar]
- Tucker EJ, Hershman SG, Köhrer C, Belcher-Timme CA, Patel J, Goldberger OA, et al. Mutations in MTFMT underlie a human disorder of formylation causing impaired mitochondrial translation. Cell Metab. 2011;14:428–434. doi: 10.1016/j.cmet.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Ueda T, Taguchi H, Takeuchi N. Chaperone properties of mammalian mitochondrial translation elongation factor Tu. J Biol Chem. 2007;282:4076–4084. doi: 10.1074/jbc.M608187200. [DOI] [PubMed] [Google Scholar]
- Siegelin MD, Dohi T, Raskett CM, Orlowski GM, Powers CM, Gilbert CA, et al. Exploiting the mitochondrial unfolded protein response for cancer therapy in mice and human cells. J Clin Invest. 2011;121:1349–1360. doi: 10.1172/JCI44855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.