Abstract
The chromosomal passenger complex (CPC) plays a pivotal role in controlling accurate chromosome segregation and cytokinesis during cell division. Aurora-B, one of the chromosomal passenger proteins, is important for the mitotic spindle assembly checkpoint (SAC). Previous reports noted that Aurora-C is predominantly expressed in male germ cells and has the same subcellular localization as Aurora-B. Increasing evidence indicates that Aurora-C is overexpressed in many somatic cancers, although its function is uncertain. Our previous study showed that the aberrant expression of Aurora-C increases the tumorigenicity of cancer cells. Here, we demonstrate that overexpressed Aurora-C displaces the centromeric localization of CPCs, including INCENP, survivin, and Aurora-B. When cells were treated with nocodazole to turn on SAC, both the Aurora-B protein stability and kinase activity were affected by overexpressed Aurora-C. As a result, the activation of spindle checkpoint protein, BubR1, and phosphorylation of histone H3 and MCAK were also eliminated in Aurora-C-overexpressing cells. Thus, our results suggest that aberrantly expressed Aurora-C in somatic cancer cells may impair SAC by displacing the centromeric localization of CPCs.
Keywords: Aurora-C, Aurora-B, spindle assembly checkpoint, chromosomal passenger proteins
The chromosomal passenger complex (CPC), which includes INCENP, survivin, borealin, and Aurora-B, moves from the centromere to the spindle midzone during cell division.1, 2 The CPC has multiple functions during cell division, including correcting errors in chromosome segregation, regulating the mitotic ckeckpoint and cytokinesis, and ultimately maintaining genomic stability.2 Activation of the CPC depends on Aurora-B activity. Aurora-B is the only protein within the CPC that has enzymatic activity. Interactions between Aurora-B and other CPC components are required for the localization and function of the CPC and are also necessary for the activation of Aurora-B.3, 4, 5 Aurora-B binds to the C-terminal IN box of INCENP, and subsequently phosphorylates the conserved threonine–serine–serine (TSS) domain near the C-terminus of INCENP that results in the activation of Aurora-B.4 Activated Aurora-B phosphorylates its substrates for their functions during cell division. To date, more than 50 proteins have been identified as Aurora-B substrates, including the CPC components, histone H3, and MCAK.2 Dysregulation of Aurora-B results in cytokinesis failure and polyploidy, and ultimately leads to chromosome instability, a hallmark of tumorigenesis.6
Aurora-C is the third member to be identified in the Aurora kinase family. It is highly similar to Aurora-B, both in the nucleotide sequence and subcellular localization.7, 8, 9 It was originally reported that Aurora-C is a testes-specific protein.9 However, increasing evidence indicates that Aurora-C is also overexpressed in numerous somatic cancer tissues and hematologic cancers.10, 11, 12, 13 Our previous report indicated that overexpressed Aurora-C can promote the tumorigenicity of cancer cells.14 The aberrant expression of Aurora-C is positively correlated with tumor progression in human cervical cancer and colorectal cancer. Aurora-C and Aurora-B are oppositely expressed in human colorectal cancer tissues. Overexpressed Aurora-C displaces the centromeric localization of Aurora-B in cancer cells.14 However, the mechanism by which Aurora-C disrupts the localization and decreases the expression of Aurora-B remains unclear.
In this study we found that overexpressed Aurora-C reduced the expression and displaced the centromeric localization of the CPC components independent of its kinase activity, thereby interfering with Aurora-B kinase activity. Aurora-B protein stability was maintained during spindle assembly checkpoint (SAC) activation, but the overexpression of Aurora-C resulted in increased degradation of the Aurora-B protein even after SAC activation. The centromeric substitution of Aurora-C resulted in an inability to turn on the mitotic checkpoint by checking the phosphorylation and subcellular localization of BubR1. Thus, overexpressed Aurora-C promotes tumorigenesis by increasing cell survival, migration, and invasiveness, and also by the avoidance of apoptosis.
Results
Overexpressed Aurora-C-GFP localizes at the centromeric region and reduces the expression levels of the CPC components
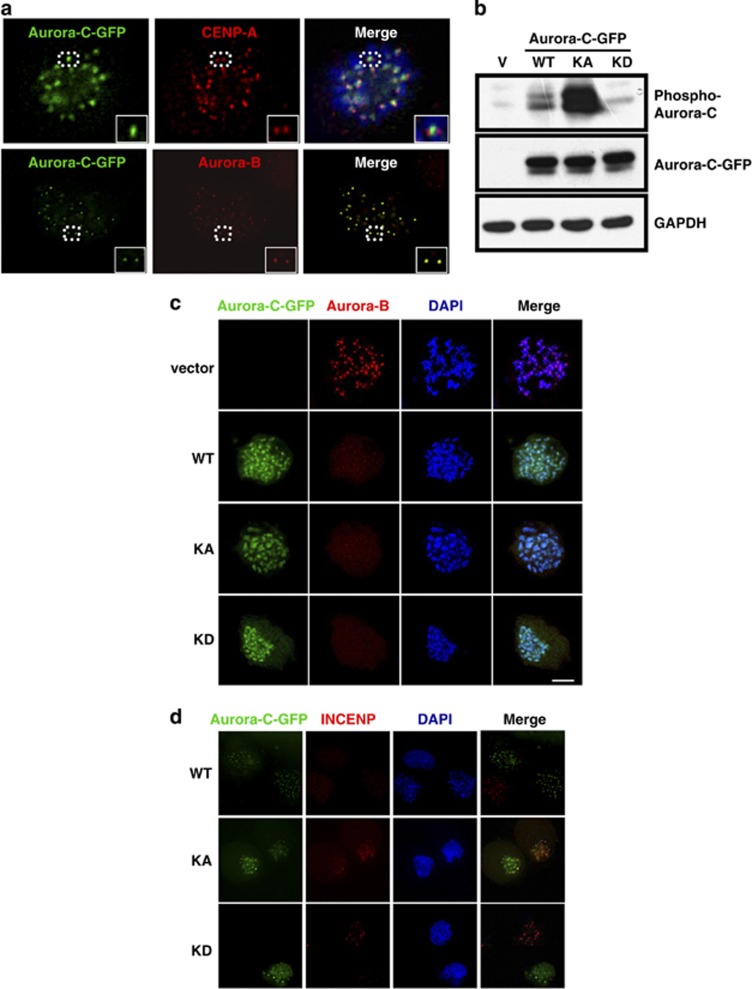
Several reports indicated that overexpressed Aurora-C is present in numerous somatic cancer cells,10, 11, 12, 13, 14 although the physiological role of overexpressed Aurora-C in tumorigenesis remains uncertain. To address this, the Aurora-C-GFP was ectopically expressed in HeLa cells and its subcellular localization subsequently analyzed by immunofluorescence (IF) assay. The results showed that some overexpressed Aurora-C-GFP, but not all, can localize to the interphase centrosomal region (data not shown). When cells entered mitosis, Aurora-C-GFP was located at the centromeric region and had the same subcellular localization as Aurora-B (Figure 1a). The centromeric localization of Aurora-C-GFP was independent of its kinase activity, as the overexpressed Aurora-C-GFP kinase hyperactive (KA) and kinase-dead (KD) mutants (Figure 1b, details in Materials and Methods) had the same subcellular localization as the Aurora-C-GFP wild type (WT) (Figure 1c). Overexpressed Aurora-C-GFP/WT, KA, and KD displaced the centromeric localization of the endogenous CPC proteins, Aurora-B, INCENP, and survivin (Figures 1c and d and Supplementary Figure 1).
Figure 1.
Aurora-C-GFP abolishes the centromeric localization of CPC components. (a) HeLa cells were transiently transfected with Aurora-C-GFP and stained with CENP-A (red, upper panel) or Aurora-B (red, lower panel). Insets show enlarged images of the dot circles. (b) HeLa cells transfected with the vector control (V), Aurora-C-GFP/WT, KA, or KD were collected for IB analysis to detect the expression and phosphorylation status of Aurora-C-GFP. GAPDH was used as a loading control. (c) Aurora-C-GFP/WT-, KA-, KD-, or vector control-transfected HeLa cells (green) were treated with nocodazole (45 ng/ml) for 16 h to synchronized cells at prometaphase and then stained with Aurora-B (red). DAPI is a specific DNA dye (blue). (d) Cells with Aurora-C-GFP/WT, KA, or KD expression were stained with INCENP (red) as described above. The centromeric localization of Aurora-B or INCENP was quite limited in Aurora-C-GFP-overexpressed cells
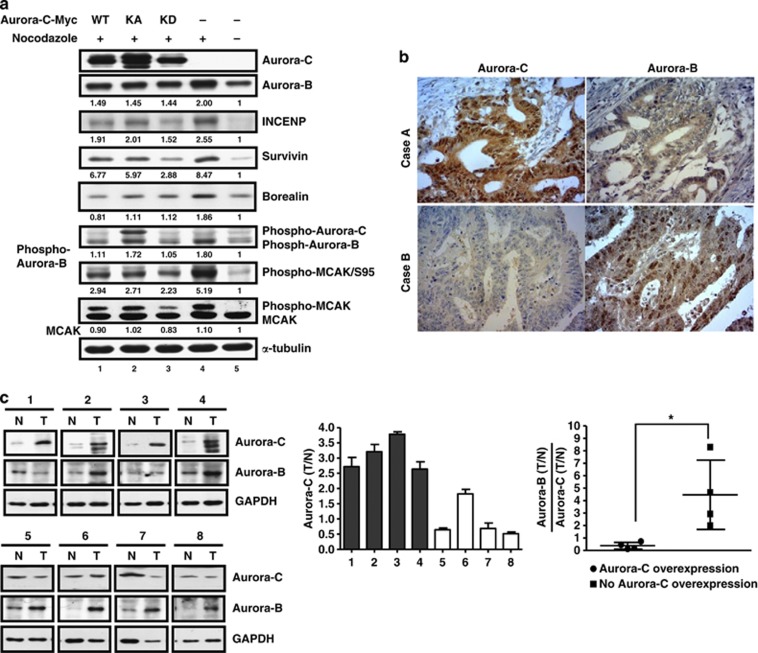
We hypothesize that the centromeric displacement of Aurora-B and CPC proteins by overexpressed Aurora-C could result from decreased expression levels of CPC proteins. To address this, we used immunoblot (IB) analysis to determine the expression levels of Aurora-B and CPC proteins in the presence of Aurora-C-GFP. When cells were treated with nocodazole, a microtubule disrupter, to turn on the SAC, the expressions of the CPC proteins increased (Figure 2a, compare lanes 4 and 5). However, in cells with overexpressed Aurora-C-GFP/WT, KA, or KD, the levels of Aurora-B and CPC proteins decreased (Figure 2a, lanes 1–3). The reduced expression of CPC proteins due to overexpressed Aurora-C-GFP was found to be kinase activity independent and showed a dose-dependent manner, whereas the vector control had no effect (Supplementary Figure 2). The expression levels of Aurora-B mRNA under the same conditions were confirmed using Q-PCR. These results showed that overexpressed Aurora-C did not alter the level of Aurora-B mRNA (Supplementary Figure 3). Importantly, the opposite expression patterns of Aurora-B and Aurora-C were confirmed in human clinical colorectal cancer tissues by immunohistochemistry (IHC) assay (Figure 2b)14 and IB analysis (Figure 2c).
Figure 2.
The overexpression of Aurora-C reduces the expression of chromosomal passenger components and Aurora-B kinase activity. (a) Cells with or without Aurora-C-Myc expression were treated with 45 ng/ml nocodazole for 4 h to turn on the mitotic checkpoint. The expression of the chromosomal passenger complex, INCENP, survivin, borealin, and Aurora-B, and the phosphorylation status of Aurora-B and MCAK were determined using IB analysis. α-Tubulin was used as a loading control. The relative expression levels of individual proteins are displayed as ratios. (b) Human colorectal cancer tissues were assessed for the expression of Aurora-B and Aurora-C using immunohistochemistry (IHC) analysis. Two representative cases are shown. (c) Inverse expression of Aurora-B and Aurora-C in human colorectal tissues. (Left) Eight representative specimens showed the reverse expression of Aurora-B and Aurora-C in human colorectal cancer tissues. Equal amounts of total lysates from paired human colorectal cancer tissue were used to check the expression levels of Aurora-C and Aurora-B by IB analysis. GAPDH was used as a loading control. (Middle) Cases 1–4 are specimens with Aurora-C overexpression; cases 5–8 are specimens without Aurora-C overexpression. The expression levels of Aurora-C in tumor tissues (T) were compared with normal tissues (N). (Right) The ratios between Aurora-B and Aurora-C in different individuals are shown. *P<0.05
Aurora-C-GFP interferes with the SAC by weakening Aurora-B kinase activity
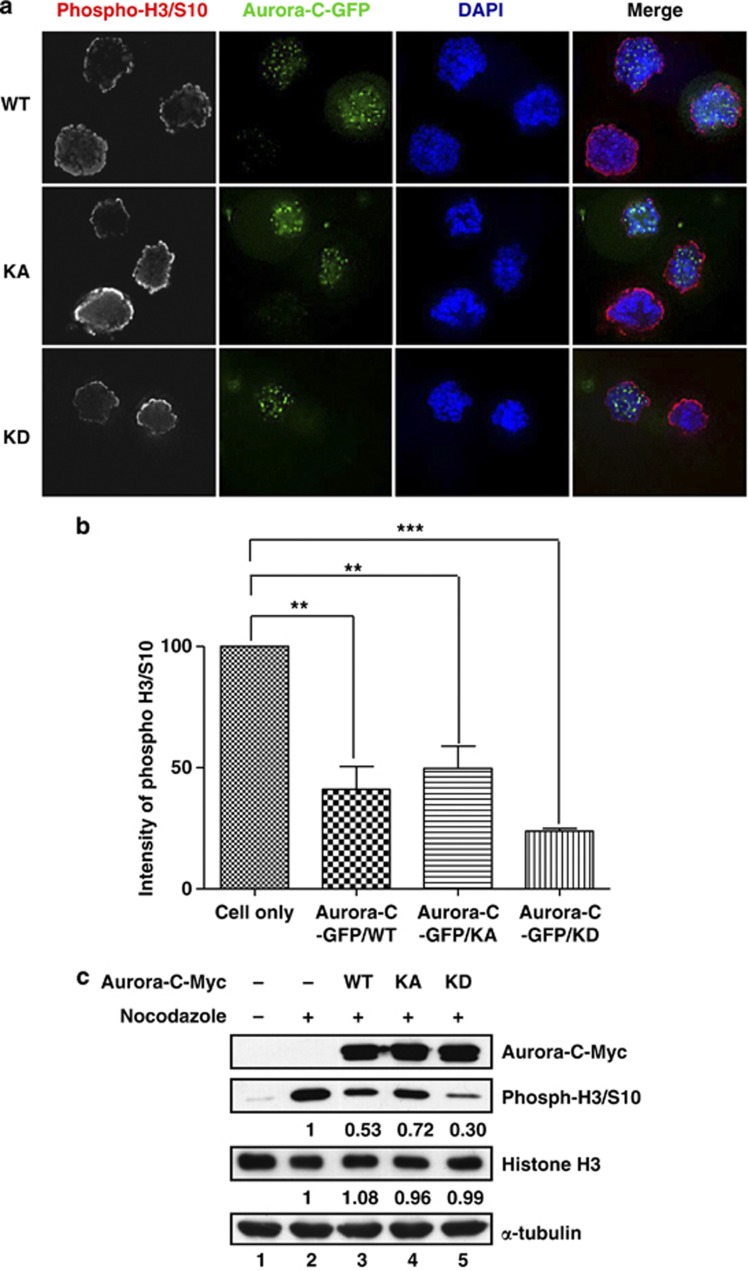
We note that overexpressed Aurora-C-GFP not only resulted in a decreased expression of Aurora-B, but also reduced its phosphorylation level (Figure 2a, compare lanes 1–3 and 4). To determine whether the reduced phosphorylation level resulted in lower Aurora-B kinase activity, we used IB analysis to evaluate the phosphorylation status of MCAK, a known Aurora-B substrate. As shown in Figure 2a, the phosphorylation levels of MCAK decreased because of overexpressed Aurora-C in a kinase-independent manner (Figure 2a, lanes 1–3) and a dose-dependent manner (Supplementary Figure 2). In addition, it is also known that phosphorylation of histone H3/Serine 10 (H3/S10) by Aurora-B is important for chromosome condensation during cell division.15 Using IF assay and IB analysis, the phosphorylation levels of H3/S10 were greatly reduced in Aurora-C-GFP/WT, KA, or KD overexpressed cells (Figure 3 and Supplementary Figure 4).
Figure 3.
Phosphorylation of histone H3/S10 is decreased in cells overexpressing Aurora-C. (a) HeLa cells were transfected with Aurora-C-GFP and then treated with nocodazole for 16 h to synchronize the cell cycle at prometaphase. The expression of phospho-histone H3/S10 (phosph-H3/S10) was determined using immunofluorescence (IF) assay. (b) Quantitative results from (a). (c) Cells overexpressing Aurora-C-Myc were treated with nocodazole as described above. Total lysates were prepared for IB analysis with the indicated antibodies. α-Tubulin was used as a loading control. The relative expression levels of individual proteins are displayed as ratios
Bub1, BubR1, and Bub3 are SAC proteins that are required for kinetochore–microtubule attachment.16 During prophase, SAC proteins are targeted to kinetochores to aid in proper kinetochore–microtubule interactions (Supplementary Figure 5).16 Aurora-B plays an important role in SAC activation by phosphorylating and targeting SAC proteins to the kinetochore region.17 Our results indicate that overexpressed Aurora-C-GFP decreases the expression level of Aurora-B as well as its kinase activity (Figures 2a and 3). Therefore, we evaluated the status of SAC in cells overexpressing Aurora-C-GFP. As shown in Figure 4a, the kinetochore localization of BubR1 was lost in cells with overexpressed Aurora-C-GFP independently of its kinase activity. In addition, nocodazole-induced phosphotylation of BubR1 was also reduced because of overexpressed Aurora-C-GFP (Figure 4b, compare lanes 3–5 with 2). However, cells having only vector expression did not affect the phosphorylation level of BubR1 (Figure 4c). These results suggest that overexpressed Aurora-C interferes with SAC by reducing the protein expression level and kinase activity of Aurora-B. As with the results described above, the status of Aurora-C kinase activity appeared not to affect its interference with SAC activity (Figure 4b).
Figure 4.
Aurora-C expression reduces BubR1 activation. (a) Aurora-C-GFP/WT-, KA-, or KD-transfected HeLa cells were treated with nocodazole for 4 h to turn on the spindle checkpoint. The kinetochore localization of BubR1 (red) was detected by IF analysis. DAPI is a DNA-specific dye. (b) Cells from (a) were used to examine BubR1 activation status using IB analysis. The expressions of Aurora-C-Myc/WT, KA, or KD are shown. GAPDH was used as a loading control. (c) HeLa cells transfected with vector control were treated with nocodazole and assessed for the phosphorylation status of BubR1 as described above. α-Tubulin was used as a loading control. The relative expression levels of individual proteins are displayed as ratios
Aurora-B and INCENP interaction is disturbed by overexpressed Aurora-C-GFP
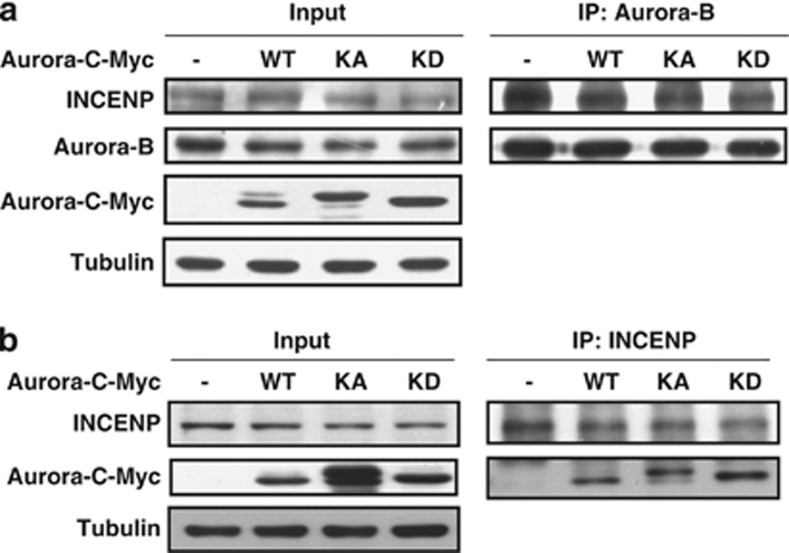
Our results described above suggest that overexpressed Aurora-C interferes with Aurora-B-mediated spindle checkpoint activation. To further investigate the underlying mechanism, cells with or without overexpressed Aurora-C were harvested to analyze the interactions between those CPC components. The immunoprecipitation (IP) assay showed that the overexpression of Aurora-C-Myc weakened the interaction between INCENP and Aurora-B (Figure 5a). According to a previous report, Aurora-C can form a complex with INCENP.8 Here, we confirm the interaction between overexpressed Aurora-C and INCENP using IP assay. The results show that Aurora-C-Myc forms a complex with INCENP (Figure 5b). This implies that, through its interaction with the CPC components, Aurora-C displaces the centromeric localization of Aurora-B and thereby decreases Aurora-B kinase activity and Aurora-B-mediated SAC activation.
Figure 5.
Aurora-C expression interferes with the interaction between Aurora-B and INCENP. (a) Aurora-C-Myc/WT-, KA-, or KD-transfected HeLa cells were lysed and immunoprecipitated (IP) using anti-Aurora-B antibody, followed by IB analysis using anti-INCENP antibody. (b) Cell lysates from cells overexpressing Aurora-C-Myc/WT, KA, or KD were prepared for IP using anti-INCENP antibody, followed by IB analysis using anti-Myc antibody
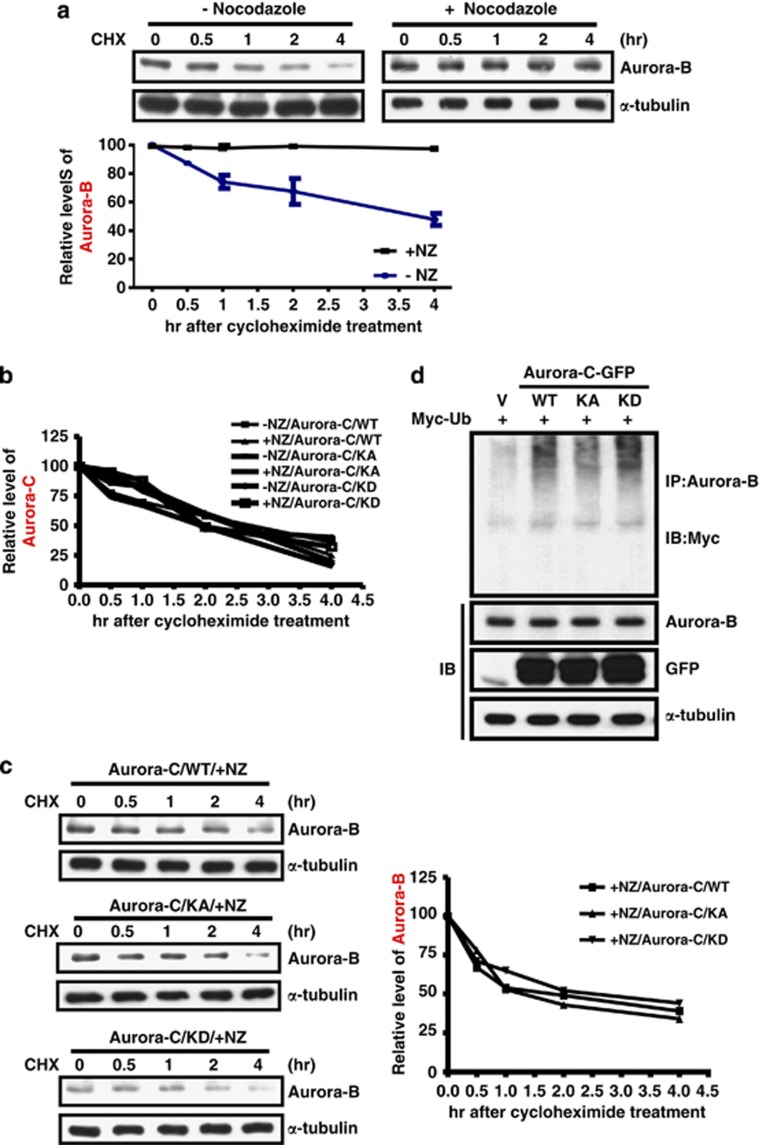
Nocodazole-induced Aurora-B protein stability is disrupted by overexpressed Aurora-C
Aurora-B is activated when SAC is turned on, and hence we evaluated the protein stability of Aurora-B under that condition. The data show that the half-life of Aurora-B is ∼4 h and that Aurora-B protein stability is maintained when cells were treated with nocodazole to turn on SAC (Figure 6a). This suggests that Aurora-B protein stability increases when SAC is activated that could be advantageous to its function of correcting aberrant kinetochore–microtubule attachment. The protein stability of Aurora-C did not change after nocodazole treatment (Figure 6b). In fact, its responses were to the opposite, implying that the role of Aurora-C during the SAC response may be different from Aurora-B. Because the expression level of Aurora-B was decreased due to overexpressed Aurora-C, we next evaluated the protein stability of Aurora-B in the presence of Aurora-C. Nocodazole-induced Aurora-B protein stability was eliminated in cells overexpressing Aurora-C independently of kinase activity (Figure 6c). In addition, the half-life of Aurora-B protein in cells overexpressing Aurora-C was lower than in control cells (∼4 h in Figure 6a and 2 h in Figure 6c). To further investigate the molecular mechanism of Aurora-C-induced degradation of Aurora-B protein, the expression level of Chd1, the activator of the anaphase-promoting complex/cyclosome (APC/C), was checked by IB analysis.18 The data show that overexpression of Aurora-C-GFP does not alter the level of Cdh1 (Supplementary Figure 6). The overexpression of Aurora-C-GFP increased the protein ubiquitination levels of Aurora-B, and this was independent of Aurora-C kinase activity (Figure 6d). These results are consistent with our previous results of overexpressed Aurora-C decreasing the expression of Aurora-B (Figure 2a).
Figure 6.
Aurora-B protein stability is enhanced upon SAC activation and is reduced in cells overexpressing Aurora-C. (a) HeLa cells with or without nocodazole treatment were further incubated with cyclohexamide (CHX) at different time points. Total lysates were prepared for IB analysis using anti-Aurora-B. The expression levels of Aurora-B were normalized by α-tubulin. The quantitative results of three independent experiments are shown below. (b) Cells expressing Aurora-C/WT, KA, or KD were treated as described above. Aurora-C expression levels were determined using IB analysis and normalized by α-tubulin. Quantitative results are shown. (c) The expression levels of Aurora-B in cells expressing Aurora-C/WT, KA, or KD expression were determined by IB analysis and normalized by α-tubulin. All cells were treated with nocodazole, followed by CHX as described above. Quantitative results are shown. (d) Cells were co-transfected with Aurora-C-GFP/WT, KA or KD, and Myc-Ubi. Equal amounts of total lysates were used for the IP assay using anti-Aurora-B antibody followed by IB analysis using anti-Myc antibody. The expressions of Aurora-B, Aurora-C-GFP/WT, KA, KD, and α-tubulin were determined using IB analysis
Overexpression of Aurora-C increases cell proliferation, migration, invasiveness, and survival
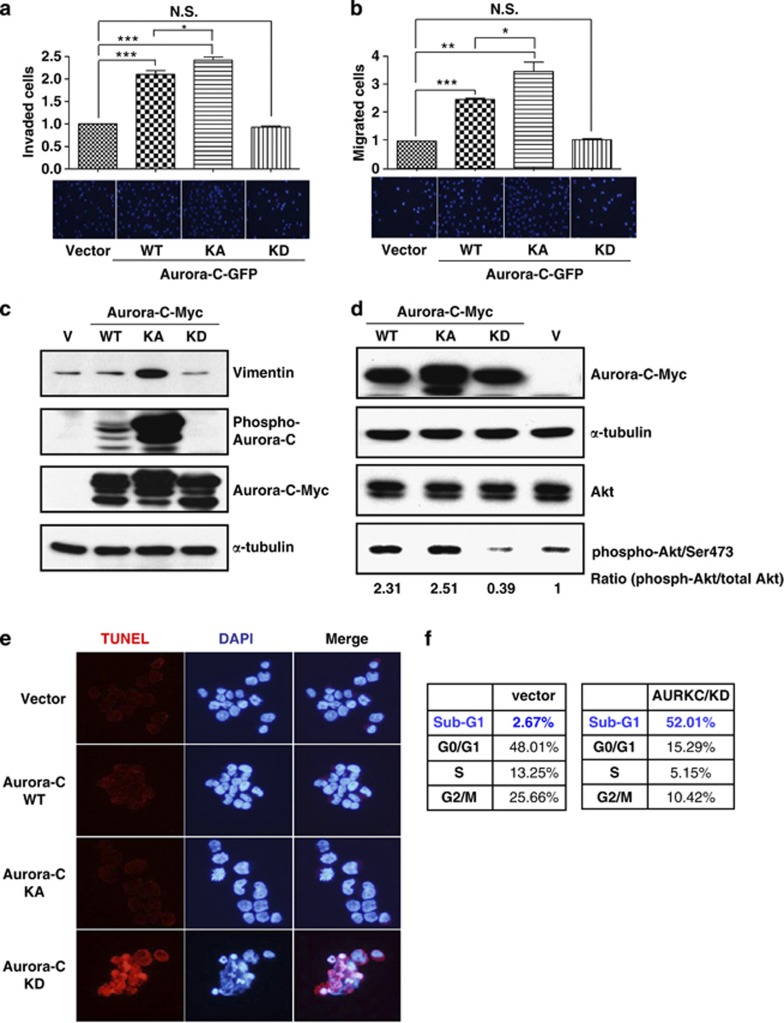
Our previous report showed that the augmented expression and increased kinase activity of Aurora-C can promote cell proliferation and migration abilities.14 To further confirm the role of Aurora-C in tumorigenesis, we used transwell migration and invasiveness assays. The results show that cells overexpressing Aurora-C-GFP/KA had greater migration and invasion capabilities than Aurora-C-GFP/WT-expressing cells, whereas Aurora-C-GFP/KD-transfected cells showed no significant differences when compared with vector-transfected cells (Figures 7a and b). In addition, IB analysis demonstrated that overexpressed Aurora-C/KA results in the predominant expression of vimentin, which plays an important role in promoting cancer cell metastasis,19 as compared with Aurora-C/WT and Aurora-C/KD (Figure 7c).
Figure 7.
Aurora-C-GFP enhances cell migration, invasiveness, and survival in a kinase-dependent manner. (a and b) Equal numbers of cells that expressed Aurora-C-GFP/WT, KA, or KD were seeded onto collagen-coated (a) or matrigel-coated (b) transwell inserts. Invasive or migrated cells were counted after 24 h of incubation and subsequent staining with DAPI. The quantitative results of three independent experiments are shown. *P<0.05; **P<0.01; ***P<0.001. NS, not significant. (c) HeLa cells expressing Aurora-C-Myc/WT, KA, or KD were used for IB. The expression levels of vimentin, Aurora-C, and phospho-Aurora-C are shown. Vector (V)-transfected cells were used as a background control. (d) Total lysates of HeLa cells with Aurora-C-Myc/WT, KA or KD expression were prepared to determine the expression levels of phospho-AKT (phosphor-Akt/Ser473), AKT, and Aurora-C-Myc using IB. α-Tubulin was used as a loading control. (e) HeLa cells that stably expressed Aurora-C-Myc/WT, KA, or KD were used for the TUNEL assay. Red signal indicates apoptotic cells. DAPI was used as a DNA-specific dye. Representative images are shown. (f) HeLa cells that stably expressed Aurora-C-Myc/KD or vector control were used to assess cell cycle distributions by flow cytometry. Quantitative results are shown
In our previous report, the expression of Aurora-C/KD did not promote tumor formation in a xenograft animal model.14 Furthermore, cell proliferation assay showed that cells with Aurora-C/KD expression have lower proliferative activity (Supplementary Figure 7). Here, we analyzed the activation status of Akt, a major regulator of cell proliferation and survival, in Aurora-C-transfected cells. The results show that Aurora-C-Myc/WT- or KA-transfected cells, but not Aurora-C/KD-transfected cells, had more activated Akt than vector-transfected cells (Figure 7d). TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) assay and flow cytometry also show that the expression of Aurora-C/KD results in an increased population of apoptotic cells (Figures 7e and f).
Discussion
Genomic instability is a hallmark of cancer development.6 One of the major devices for maintaining chromosome stability during cell division is the CPC. Loss or disruption of CPC functions results in aneuploidy, chromosome imbalances, and ultimately cancer development.20 When incorrect kinetochore–microtubule attachments occur during cell division, the SAC will be turned on to correct the error.21 In mammals, Aurora-B is activated to correct kinetochore–microtubule attachments when the SAC is turned on.22 Our results show that overexpressed Aurora-C can localize at the centromeric region and replace the localization and decrease the expression of CPC components. Importantly, using the kinase-active mutant of Aurora-C, we establish that the centromeric localization of Aurora-C interferes with the SAC by promoting the degradation of Aurora-B protein. Our results suggest that Aurora-C is not a compensatory partner of Aurora-B.
Several studies have reported that Aurora-C plays important roles in early development.23, 24, 25 They demonstrated that Aurora-C activity is essential for cell division during early mammalian development. The roles of Aurora-C in this stage are involved in CPC functions (i.e., SAC activation and cytokinesis), which are similar to those of Aurora-B. Although their observations seem to be opposite to ours, this may be because of different biological functions. Furthermore, other reports proposed that Aurora-B and Aurora-C play different roles in spermatogenesis and oogenesis.26, 27 In order to further evaluate whether overexpressed Aurora-C has opposite effects with overexpressed Aurora-B in somatic cancer cells, we checked the expression levels of CPC proteins (Supplementary Figure 8), phospho-H3/S10 (Supplementary Figure 9), and phospho-BubR1 (Supplementary Figure 10) in Aurora-B overexpressed cells. The results indicated that Aurora-B and Aurora-C have different effects (Supplementary Figures 8–10). Taken together, the results suggest that Aurora-B and Aurora-C in different biological situations may have different roles.
An increasing number of reports indicate that Aurora-C is aberrantly expressed in cancer tissues.7, 28 Two recent reports suggest that overexpressed Aurora-C promotes the tumorigenicity of somatic cancer cells.14, 29 However, its interaction and/or its correlation with Aurora-B, particularly for regulating the SAC, remain uncertain. Most previous studies used Aurora-C KD mutant to demonstrate the roles of Aurora-C in cancer cells and suggest that Aurora-C in somatic cancer cells has functions similar to those of Aurora-B.30, 31, 32 Our present results suggest that the overexpression of Aurora-C decreases the phosphorylation levels of MCAK and histone H3/S10, both of which are known substrates of Aurora-B (Figures 2a and 3). Furthermore, the overexpression of Aurora-C reduced the phosphorylation of BubR1, a marker for the activated SAC (Figure 4). All of these results were independent of Aurora-C kinase activity, as the same effects were found with Aurora-C/WT, KA, and KD for replacing the centromeric localization of Aurora-B. Importantly, Aurora-C/KA expression did not increase the phosphorylation levels of histone H3 and BubR1. These data again imply that Aurora-C has roles that are different from Aurora-B.
Our results demonstrate that the protein stability of Aurora-B is maintained when the SAC is turned on (Figure 6a). Nevertheless, the protein stability of Aurora-C did not change upon SAC activation (Figure 6b). This is additional evidence that the physiological role of Aurora-C is different from Aurora-B, in particular for regulating the SAC. Furthermore, overexpressed Aurora-C enhanced the degree of protein degradation in Aurora-B even in the presence of nocodazole (Figures 6c and d). This could explain the phenomena demonstrated in Figure 2a that overexpressed Aurora-C decreased the expression of Aurora-B as well as of other CPC components. Moreover, it is consistent with our previous observations in which the overexpression of Aurora-C was opposite to the expression of Aurora-B in human colorectal cancer tissues (Figures 2b and c).14 Based on these results, we propose that the centromeric replacement of Aurora-B by overexpressed Aurora-C in a kinase activity-independent manner may expose a specific recognition domain of Aurora-B to the ubiquitination machinery. Besides, our results indicate that after the centromeric localization of Aurora-B and INCENP is delocalized, overexpressed Aurora-C still maintains its centromeric localization (Figure 1). This may because of an interaction between Aurora-C and other centromeric proteins, such as CENP-A, HP1, and shugoshins.33, 34 However, the details of these molecular mechanisms require additional investigation.
Using a xenograft animal model, our previous report indicated that the overexpression of Aurora-C/KA strongly promoted tumorigenicity, whereas the overexpression of Aurora-C/KD induced almost no tumor formation.14, 29 In the present study, an increased phosphorylation level of Akt was found in cells expressing Aurora-C/KA or WT but not in cells expressing Aurora-C/KD (Figure 7d). Cells with Aurora-C/KD expression had an increased proportion of apoptotic cells (Figures 7e and f). An obvious augmented expression of vimentin was found in cells overexpressing Aurora-C/KA (Figure 7c). Whether the expression of Aurora-C plays a role in the epithelial–mesenchyal transition during carcinogenesis remains uncertain, and is currently under investigation.
Materials and Methods
Plasmid construction
Human Aurora-C cDNA was cloned from HeLa cells by RT-PCR, and then subcloned into pCDNA3.1-Myc/His (Invitrogen, Carlsbad, CA, USA) or pEGFP-N1 (Clontech, Mountain View, CA, USA) using EcoRI and BamHI sites, respectively. Primers used for Aurora-C cloning are: forward, 5′-CGGAATTCTAATGAGCTCCCCCAGAGCTG-3′ reverse, 5′-GTGGATCCTCGGAAGCCATCTGAGCACAG-3′. The Aurora-C mutants kinase-hyperactive (KA/T191D) and kinase-dead (KD/T202D) were created using a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). Primers used for Aurora-C/KA are: forward, 5′-GGCTGGTCTGTGCACGCCCCCTCCCTGAGGAGG-3′ reverse, 5′-CCTCCTCAGGGAGGGGGCGTGCACAGACCAGCC-3′. Primers used for Aurora-C/KD are: forward, 5′-GGCTGGTCTGTGCACGACCCCTCCCTGAGGAGG-3′ reverse, 5′-CCTCCTCAGGGAGGGGTCGTGCACAGACCAGCC-3′.
Cell culture and transfection
Human cervical adenocarcinoma HeLa cells were cultured in Dulbecco's modified Eagle's medium (Sigma, St. Louis, MO, USA) supplemented with 10% fetal bovine serum, 100 μg/ml streptomycin, and 100 U/ml penicillin. Cells were grown at 37°C under 5% CO2. Transfection was performed by Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. In general, cells grown in 60 mm dishes were transiently transfected with 4 μg of plasmid.
Total cell lysate preparation and IB analysis
For total cell lysates, cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA, and 2 mM EGTA) containing 1 mM sodium orthovanadate, 50 mM glycerol-2-phosphate, 100 × phosphatase inhibitor cocktail (Sigma), 1 μM okadaic acid, and 25 × protease inhibitor. For IB assay, 30 μg of total lysate was separated using 10% polyacrylamide gel electrophoresis-sodium dodecyl sulfate (PAGE-SDS) and analyzed for the specific antibodies indicated in figure legends. Anti-GAPDH, anti-INCENP, anti-phospho-histone H3/S10, anti-histone H3, and anti-CENP-A antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA); anti-Aurora-B and anti-α-tubulin antibodies were purchased from Sigma; anti-phospho-MCAK/S95 and anti-phospho-Aurora-B/T232 were purchased from Abcam (Cambridge, MA, USA); anti-MCAK was purchased from Bethyl (Montgomery, TX, USA); anti-BubR1 antibody was obtained from BD Biosciences (San Jose, CA, USA); anti-Myc antibody was purchased from Upstate (Lake Placid, NY, USA); anti-survivin antibody was purchased from Cell Signaling (Danvers, MA, USA); and anti-Aurora-C antibody was purchased from Zymed (San Francisco, CA, USA). Specific protein signals were detected using horseradish peroxidase-conjugated secondary antibodies and revealed by an enhanced chemiluminescence (ECL) Western blot system (PIERCE, Rockford, IL, USA).
IF assay
Cells were incubated with 0.06 M KCl for 30 min at room temperature, fixed with 3.7% formaldehyde, and permeabilized using 0.1% Tween-20/PBS for 10 min. Cells were probed with the specific antibodies indicated in figure legends and followed by incubation with Alexa 568 (Molecular Probes, Cincinnati, OH, USA). After mounting with ProLong Gold antifade reagent (Invitrogen) containing 4,6-diamidino-2-phenylindole (DAPI), images were acquired using a laser scanning confocal system (FV1000, Olympus, Center Valley, PA, USA).
IP assay
Cells were lysed with IP buffer (0.5% sodium deoxycholate, 0.5% Nonidet P-40, 25X protease inhibitor, 1 mM Na3VO4, 50 mM glycerol-2-phosphate, 100 × phosphatase inhibitor cocktail, and 1 μM okadaic acid) and incubated with anti-Aurora-B or anti-INCENP antibodies at 4°C for 2 h. Protein-A/G agarose beads were added to the mixture and incubated at 4°C for an additional 1 h. After washing the beads three times with immunoprecipitation washing buffer (PBS with 0.1% sodium deoxycholate, 0.1% Nonidet P-40), the immunoprecipitated complexes were resolved by immunoblot analysis.
Protein degradation assay
Cells were treated with or without 45 ng/ml of nocodazole (Sigma) for 4 h, followed by the addition of 200 μg/ml of cycloheximide, a de novo protein synthesis inhibitor, for the indicated time periods. Total cell lysates were prepared for immunoblot analysis.
TUNEL assay
TUNEL assay was performed with an In Situ Cell Death Detection kit according to the manufacturer's instructions (Roche, Basel, Switzerland). In brief, cells were fixed with 4% paraformaldehyde for 1 h at room temperature and then incubated with freshly prepared permeabilization solution (0.1% Triton X-100 in 0.1% sodium citrate) for 2 min at 4°C. After incubation with the TUNEL reaction mixture for 1 h in the dark, cells were washed with PBS, stained with DAPI, and mounted. Apoptotic cells were observed using an Olympus BX51 immunofluoresence microscope.
Flow cytometry
Cells were fixed using 80% cold ethanol for 2 h at −20°C and then incubated along with a staining solution (0.1% Triton X-100, 200 μg/ml of RNAse A, and 20 μg/ml of propidium iodide (PI)) for 30 min in the dark. Cell cycle populations were analyzed using a Cell Lab Quanta SC High Resolution Flow Cytometer (BECKMAN COULTER, Brea, CA, USA).
Transwell assay
Cells were seeded onto the transwell insert (pore size of 3.0 μm, Millipore, Bedford, MA, USA), which had been precoated with 5 μg/cm2 of collagen gel or 0.5 mg/ml of matrigel (BD Biosciences), and cultured in a serum-free medium. Transwell inserts were placed in a 24-well plate containing growth medium with 10% FBS. After 24 h of incubation, transwell filters were fixed with 3.7% formaldehyde and subsequently mounted with ProLong Gold antifade reagent with DAPI. Cells on the filters were counted using an Olympus BX51 immunofluorescence microscope.
Aurora-B ubiquitination assay
Cells co-transfected with Aurora-C and Myc-Ubiquitine were treated with 25 μM MG132 for 6 h and then lysed with IP buffer containing 10 mM N-ethylmaleimide (NEM). Equal amounts of total lysates were immunoprecipitated using anti-Aurora-B antibody followed by IB analysis using anti-Myc antibody.
Acknowledgments
We thank Mr. Buford Pruitt Jr. and Enago (www.enago.tw) for the English language review. This work was supported by grants NSC101-2325-B-006-001 and NSC102-2325-B-006-001 from the National Science Council; Project of Promoting Academic Excellence and Developing World Class Research Centers (C006); Infectious Disease and Signaling Research Center, National Cheng Kung University; and National Cheng Kung University Hospital (NCKUH-10102063, NCKUH-10202004).
Glossary
- APC/C
anaphase-promoting complex/cyclosome
- CPC
chromosomal passenger complex
- IB
immunoblot
- IF
immunofluorescence
- IHC
immunohistochemistry
- IP
immunoprecipitation
- KA
kinase hyperactive
- KD
kinase dead
- NEM
N-ethylmaleimide
- PAGE-SDS
polyacrylamide gel electrophoresis-sodium dodecyl sulfate
- SAC
spindle assembly checkpoint
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- WT
wild type
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Stephanou
Supplementary Material
References
- Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Waal MS, Hengeveld RC, van der Horst A, Lens SM. Cell division control by the Chromosomal Passenger Complex. Exp Cell Res. 2012;318:1407–1420. doi: 10.1016/j.yexcr.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330:235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Xu Z, Ogawa H, Vagnarelli P, Bergmann JH, Hudson DF, Ruchaud S, et al. INCENP-aurora B interactions modulate kinase activity and chromosome passenger complex localization. J Cell Biol. 2009;187:637–653. doi: 10.1083/jcb.200906053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Sasai K, Katayama H, Stenoien DL, Fujii S, Honda R, Kimura M, et al. Aurora-C kinase is a novel chromosomal passenger protein that can complement Aurora-B kinase function in mitotic cells. Cell MotilCytoskeleton. 2004;59:249–263. doi: 10.1002/cm.20039. [DOI] [PubMed] [Google Scholar]
- Tang CJ, Lin CY, Tang TK. Dynamic localization and functional implications of Aurora-C kinase during male mouse meiosis. Dev Biol. 2006;290:398–410. doi: 10.1016/j.ydbio.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Tseng TC, Chen SH, Hsu YP, Tang TK. Protein kinase profile of sperm and eggs: cloning and characterization of two novel testis-specific protein kinases (AIE1, AIE2) related to yeast and fly chromosome segregation regulators. DNA Cell Biol. 1998;17:823–833. doi: 10.1089/dna.1998.17.823. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Futamura M, Yoshimi N, Sano J, Katada M, Takagi Y, et al. Centrosomal kinases, HsAIRK1 and HsAIRK3, are overexpressed in primary colorectal cancers. Jpn J Cancer Res. 2000;91:1007–1014. doi: 10.1111/j.1349-7006.2000.tb00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulisse S, Delcros JG, Baldini E, Toller M, Curcio F, Giacomelli L, et al. Expression of Aurora kinases in human thyroid carcinoma cell lines and tissues. Int J Cancer. 2006;119:275–282. doi: 10.1002/ijc.21842. [DOI] [PubMed] [Google Scholar]
- Wiseman SM, Masoudi H, Niblock P, Turbin D, Rajput A, Hay J, et al. Anaplastic thyroid carcinoma: expression profile of targets for therapy offers new insights for disease treatment. Ann Surg Oncol. 2007;14:719–729. doi: 10.1245/s10434-006-9178-6. [DOI] [PubMed] [Google Scholar]
- Zekri A, Lesan V, Ghaffari SH, Tabrizi MH, Modarressi MH. Gene amplification and overexpression of Aurora-C in breast and prostate cancer cell lines. Oncol Res. 2012;20:241–250. doi: 10.3727/096504013x13589503482978. [DOI] [PubMed] [Google Scholar]
- Tsou JH, Chang KC, Chang-Liao PY, Yang ST, Lee CT, Chen YP, et al. Aberrantly expressed AURKC enhances the transformation and tumourigenicity of epithelial cells. J Pathol. 2011;225:243–254. doi: 10.1002/path.2934. [DOI] [PubMed] [Google Scholar]
- Giet R, Glover DM. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logarinho E, Bousbaa H. Kinetochore-microtubule interactions “in check” by Bub1, Bub3 and BubR1: the dual task of attaching and signalling. Cell Cycle. 2008;7:1763–1768. doi: 10.4161/cc.7.12.6180. [DOI] [PubMed] [Google Scholar]
- Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S, Fang G. Destruction box-dependent degradation of aurora B is mediated by the anaphase-promoting complex/cyclosome and Cdh1. Cancer Res. 2005;65:8730–8735. doi: 10.1158/0008-5472.CAN-05-1500. [DOI] [PubMed] [Google Scholar]
- Hu L, Lau SH, Tzang CH, Wen JM, Wang W, Xie D, et al. Association of Vimentin overexpression and hepatocellular carcinoma metastasis. Oncogene. 2004;23:298–302. doi: 10.1038/sj.onc.1206483. [DOI] [PubMed] [Google Scholar]
- Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- Liu D, Lampson MA. Regulation of kinetochore-microtubule attachments by Aurora B kinase. Biochem Soc Trans. 2009;37 (Pt 5:976–980. doi: 10.1042/BST0370976. [DOI] [PubMed] [Google Scholar]
- Avo Santos M, van de Werken C, de Vries M, Jahr H, Vromans MJ, Laven JS, et al. A role for Aurora C in the chromosomal passenger complex during human preimplantation embryo development. Hum Reprod. 2011;26:1868–1881. doi: 10.1093/humrep/der111. [DOI] [PubMed] [Google Scholar]
- Fernandez-Miranda G, Trakala M, Martin J, Escobar B, Gonzalez A, Ghyselinck NB, et al. Genetic disruption of aurora B uncovers an essential role for aurora C during early mammalian development. Development. 2011;138:2661–2672. doi: 10.1242/dev.066381. [DOI] [PubMed] [Google Scholar]
- Schindler K, Davydenko O, Fram B, Lampson MA, Schultz RM. Maternally recruited Aurora C kinase is more stable than Aurora B to support mouse oocyte maturation and early development. Proc Natl Acad Sci USA. 2012;109:E2215–E2222. doi: 10.1073/pnas.1120517109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmins S, Crosio C, Kotaja N, Hirayama J, Monaco L, Hoog C, et al. Differential functions of the Aurora-B and Aurora-C kinases in mammalian spermatogenesis. Mol Endocrinol. 2007;21:726–739. doi: 10.1210/me.2006-0332. [DOI] [PubMed] [Google Scholar]
- Sharif B, Na J, Lykke-Hartmann K, McLaughlin SH, Laue E, Glover DM, et al. The chromosome passenger complex is required for fidelity of chromosome transmission and cytokinesis in meiosis of mouse oocytes. J Cell Sci. 2010;123 (Pt 24:4292–4300. doi: 10.1242/jcs.067447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YS, Su LJ, Yu CT, Wong FH, Yeh HH, Chen SL, et al. Gene expression profiles of the aurora family kinases. Gene Expr. 2006;13:15–26. doi: 10.3727/000000006783991962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan J, Ezan F, Cremet JY, Fautrel A, Gilot D, Lambert M, et al. Overexpression of active Aurora-C kinase results in cell transformation and tumour formation. PLoS One. 2011;6:e26512. doi: 10.1371/journal.pone.0026512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HL, Tang CJ, Chen CY, Tang TK. Overexpression of an Aurora-C kinase-deficient mutant disrupts the Aurora-B/INCENP complex and induces polyploidy. J Biomed Sci. 2005;12:297–310. doi: 10.1007/s11373-005-0980-0. [DOI] [PubMed] [Google Scholar]
- Slattery SD, Mancini MA, Brinkley BR, Hall RM. Aurora-C kinase supports mitotic progression in the absence of Aurora-B. Cell Cycle. 2009;8:2984–2994. [PubMed] [Google Scholar]
- Yang KT, Li SK, Chang CC, Tang CJ, Lin YN, Lee SC, et al. Aurora-C kinase deficiency causes cytokinesis failure in meiosis I and production of large polyploid oocytes in mice. Mol Biol Cell. 2010;21:2371–2383. doi: 10.1091/mbc.E10-02-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattani A, Wolna M, Ploquin M, Helmhart W, Morrone S, Mayer B, et al. Sgol2 provides a regulatory platform that coordinates essential cell cycle processes during meiosis I in oocytes. Elife. 2013;2:e01133. doi: 10.7554/eLife.01133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery SD, Moore RV, Brinkley BR, Hall RM. Aurora-C and Aurora-B share phosphorylation and regulation of CENP-A and Borealin during mitosis. Cell Cycle. 2008;7:787–795. doi: 10.4161/cc.7.6.5563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.