Abstract
Apoptotic cell death is an integral part of cell turnover in many tissues, and proper corpse clearance is vital to maintaining tissue homeostasis in all multicellular organisms. Even in tissues with high cellular turnover, apoptotic cells are rarely seen because of efficient clearance mechanisms in healthy individuals. In Caenorhabditis elegans, two parallel and partly redundant conserved pathways act in cell corpse engulfment. The pathway for cytoskeletal rearrangement requires the small GTPase CED-10 Rac1 acting for an efficient surround of the dead cell. The CED-10 Rac pathway is also required for the proper migration of the distal tip cells (DTCs) during the development of the C. elegans gonad. Parkin, the mammalian homolog of the C. elegans PDR-1, interacts with Rac1 in aged human brain and it is also implicated with actin dynamics and cytoskeletal rearrangements in Parkinsons's disease, suggesting that it might act on engulfment. Our genetic and biochemical studies indicate that PDR-1 inhibits apoptotic cell engulfment and DTC migration by ubiquitylating CED-10 for degradation.
Keywords: Parkin, Rac1, C. elegans, engulfment, proteasomal degradation
Multicellular organisms execute the majority of unwanted cell populations in a regulated manner via the process of apoptosis.1, 2 Remarkably, even in tissues with high cellular turnover, apoptotic cells are rarely seen in situ, which is thought to be owing to efficient clearance mechanisms.
Early studies in the nematode Caenorhabditis elegans (C. elegans) suggested that apoptotic cells are recognized and cleared before they are ‘fully dead'.3, 4
Two evolutionary conserved and partially redundant signaling pathways act together in the clearance of cell corpses during development and in the germline in C. elegans: (a) CED-2, CED-5 and CED-12 and (b) CED-1, CED-6 and CED-7, both converging in the activation of the small GTPase CED-10 (Rac1 in mammals), which, eventually, promote cytoskeletal reorganization and cell corpse engulfment.5, 6 In addition, two independent signaling pathways have been described very recently: the one acting through the ABI-1 tyrosine kinase and the other being regulated by the SLI-1 ligase.7, 8 However, the redundancy of these new pathways to the canonical engulfment branches still cannot be discarded.
CED-10/Rac1 is a member of the Rho family GTPases. GTP-bound Rac has an evolutionary conserved positive effect on engulfment, and Rac activation at sites of apoptotic cell recognition subsequently leads to cytoskeletal rearrangement.9, 10 Once Rac1 is activated, it is ubiquitylated for targeting to proteasomal degradation.11, 12 The specific E3 ubiquitin ligase regulating Rac1 degradation in the scenario of the engulfment remains elusive.
E3 ubiquitin ligases constitute the third step of the ubiquitination process and have an obligatory role in catalyzing the conjugation of ubiquitin to a lysine residue in the target proteins. Rac1 activity is regulated by the proteasomal machinery in other scenarios, and the E3 ligases HACE1, c-IAP and XIAP are some examples.13, 14, 15 HACE-1 has been involved in the mechanism of pathogen infection and c-IAP and XIAP regulate carcinogenesis.16, 17 In the IAPs context, the downregulation of the E3 ligases leads to an increase of Rac1 levels and activity in cells, promoting cell elongation and migration.
Parkin encodes a protein with E3 ligase activity whose loss of function is considered causative of autosomal recessive-juvenile parkinsonism. Human Parkin (hParkin) consists of four domains: an N-terminal ubiquitin-like domain (UBL), and two RING-finger domains flanking a cysteine-rich in-between domain. The UBL domain has been implicated in proteasome binding,18 substrate recognition19 and regulation of Parkin stability.20 A number of substrates of Parkin have been identified in various experimental systems.21, 22, 23, 24 Some of the identified mutations in the gene encoding for Parkin have been shown to impair its E3 ubiquitin ligase activity for several substrates.25, 26
Parkin function has been associated with multiple cellular processes, among them is the stability of cytoskeletal components. Thus, Parkin is associated with actin filaments in neuronal and non-neuronal cells.27
Protein degradation by ubiquitin-mediated targeting to the proteasomal machinery has a crucial role in cell signaling, notably in pathways that control actin cytoskeleton dynamics.11, 28 We hypothesized that PDR-1/hParkin might act in engulfment pathways. In addition, we asked whether it did so by interacting with the C. elegans CED-10.
We now present evidence that PDR-1 inhibits apoptotic cell engulfment. Interestingly, we find that PDR-1 regulates the turnover of CED-10 by polyubiquitylation. This work provides the first link between cellular processes of ubiquitylation/proteasomal degradation and the ability to clear apoptotic cells efficiently in the nematode. Further studies are needed to elucidate the exact mechanism working in humans.
Results
Pdr-1 mutations decreased the number of cell corpses of engulfment mutants
Searching for Parkin interactors involved in cytoskeletal rearrangements, we detected a positive interaction between Parkin and Rac1 in aged human brains (Supplementary Figure S1). The nematode C. elegans was then used as the model to study the regulation of ced-10 (human Rac1, hRac1) by pdr-1 (hParkin) because of the simplicity of C. elegans as an animal model to study genetic interactions, and because mechanisms controlling the engulfment in C. elegans are conserved in metazoa.4 To test first whether pdr-1 had a role in engulfment, the number of unengulfed cell corpses was counted in the heads of first larval stage (L1) animals, harboring mutations in pdr-1 and ced-10. The number of unengulfed corpses varies with the strength of the engulfment defect and defines a quantitative assay of engulfment abnormalities.29 In the ced-10 gene, the presence of the n1993 lesion is a G-to-T transition that results in a change of valine 190 to glycine.30 The mutation ced-10(n3246) is a G-to-A transition resulting in a change of glycine 60 of CED-10 to arginine (G60R).31 This mutation results in altered function (gain-of-function or dominant-negative phenotypes described in Reddien and Horvitz31 and Shakir et al.30). Because of the increased defects in the engulfment, this allele is considered to be strong, in comparison with ced-10(n1993) allele that is considered to be weak. Both these alleles are not null and they still maintain some residual activity.30, 31
Two presumptive null alleles of pdr-1 were used in this study, lg103 and tm395. The allele lg103 carries in-frame deletion encoding the internally truncated PDR-1 protein Δaa24–247, resulting in the loss of the UBL and the complete unique Parkin domain and the following RING domain.22 PDR-1 activity is being abolished as a consequence of this loss-of-function mutation.22, 32 The allele tm395 is an out-of-frame deletion that results in an earlier stop codon so that its capacity of association with the proteasome is not modified.22 Mutant alleles lg103 and tm395 were backcrossed with N2 wild-type 7 at least twice respectively.22
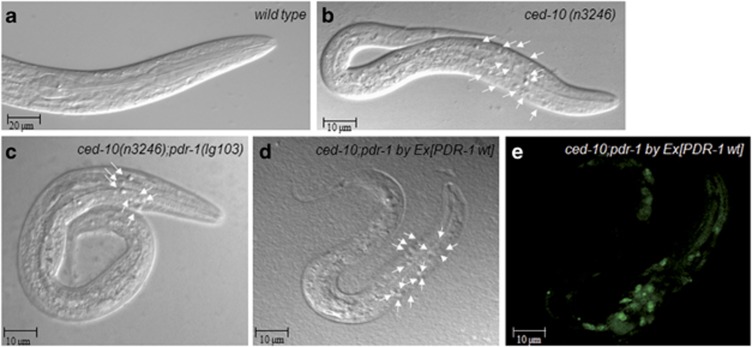
Pdr-1 effect on engulfment was first tested in a ced-10/hRac1 background. Neither wild-type nor pdr-1 mutation alone had any obvious effect on engulfment. However, mutations of pdr-1 suppressed both ced-10 alleles, as the number of unengulfed corpses were decreased in the heads of the two ced-10;pdr-1 double mutants (Table 1 and Figures 1a–c).
Table 1. pdr-1 mutations suppress the engulfment defects of engulfment ced-10 gene mutations.
| Genotype | Number of cell corpses±s.d. | n | P-value |
|---|---|---|---|
| N2 | 0±1 | 30 | — |
| pdr-1(tm395) | 0 | 30 | — |
| pdr-1(lg103) | 0±1 | 30 | — |
| ced-10(n3246) | 21.4±0.6 | 67 | — |
| ced-10(n1993) | 15.41±1.054 | 30 | — |
| ced-10(n1993);pdr-1(tm395) | 6.1±0.3 | 48 | <0.0001 to ced-10(n1993) |
| ced-10(n1993);pdr-1(lg103) | 7.4±0.4 | 30 | <0.0001 to ced-10(n1993) |
| ced-10(n3246);pdr-1(lg103) | 8.9±0.3 | 121 | <0.0001 to ced-10(n3246) |
| ced-10(n3246);pdr-1(tm395) | 10.5±0.7 | 35 | <0.0001 to ced-10(n3246) |
| pdr-1(lg103);byEx[PDR-1 wild type;sel-12::gfp] | 0±1 | 30 | — |
| ced-10(n3246);pdr-1(lg103);byEx[PDR-1 wild type;sel-12::gfp] | 19.48±0.5 | 63 | 0.0117 to ced-10(n3246) |
| ced-5(n1812)a | 25.07±0.8 | 30 | — |
| ced-5(n1812);pdr-1(lg103) | 19.05±0.7 | 31 | <0.0001 to ced-10(n3246) |
| ced-1(e1375)b | 16.7±0.5 | 31 | — |
| ced-1(e1375);pdr-1(lg103) | 9.9±0.6 | 51 | <0.0001 to ced-10(n1993) |
pdr-1 accelerates the engulfment process. First larval stage (L1) animals were anesthetized and viewed using DIC microscopy. Cell corpses were counted in the head of L1s. The P-value refers to comparisons between the strain with the engulfment mutation alone, ced-10(n3246), ced-10(n1993), ced-5(n1812), ced-1(n1375), and the strains with the engulfment mutation with either the pdr-1(tm395) or pdr-1(lg103) alleles or the rescue strain ced-10(n3246);pdr-1(lg103);byEx[PDR-1 wt::sel-12-gfp]. All data are means±s.d. (statistics: paired t-test; two-tailed, unequal variances)
Figure 1.
Loss of pdr-1 accelerates the engulfment machinery in ced-10 mutants. (a–e) DIC microscopy images of freshly hatched L1 larvae of the indicated genotypes. Arrowheads indicate persistent cell corpses. Scale bar, 20 and 10 μm as indicated. (d and e) Expression of PDR-1 wild type rescues the ced-10(n3246);pdr-1(lg103) phenotype. Alleles used were as follows: ced-10(n3246), pdr-1(lg103), ced-10(n3246);pdr-1(lg103) and the rescue strain ced-10(n3246);pdr-1(lg103); byEx[PDR-1 wt::sel-12-gfp]
The suppression of the ced-10 stronger allele (such as that generated by the (n3246) mutation) by pdr-1 mutations is consistent with a general inhibition of the engulfment by pdr-1. We could not use the ced-10(t1875) null allele in these studies owing to its embryonic lethality phenotype, which prevented scoring larval corpses.33
In addition to ced-10, the loss-of-function ced-5(n1812) and ced-1(e1735) alleles were also partially suppressed by pdr-1(lg103) (Table 1). Dyn-1 mutants died at the embryonic stage and were not analyzed. As the main purpose was to investigate the interaction between Parkin and Rac1, the other engulfment mutant alleles were not tested.
The fact that loss of pdr-1 function suppresses the engulfment defects caused by ced-1, ced-5 and ced-10 mutations suggested that PDR-1 may act in parallel or downstream to both engulfment pathways. Because of our previous results obtained in human brains (Supplementary Figure S1), the regulation of ced-10 by pdr-1 cannot be excluded.
Like CED-10,34 PDR-1 protein is ubiquitously expressed. Expression starts in embryogenesis and is maintained throughout development until adulthood.22 Both CED-10 and PDR-1 act in the same subcellular compartments.22, 35 To determine whether pdr-1 was responsible for the engulfment increase detected in ced-10;pdr-1 worms, we used the rescue strain, pdr-1(lg103);byEx429[K08E3]sel-12::gfp,22 as described in the Methodology section. The cosmid K08E3 is a genomic subclone containing the complete pdr-1 operon.22 PDR-1 wild type in double mutants ced-10;pdr-1 rescued the suppressor activity of pdr-1 (Table 1 and Figures 1d and e). This indicates a solid rescue of mutant pdr-1 suppressor activity and identifies PDR-1 as a new inhibitor in the engulfment of apoptotic cells.
Pdr-1 is not involved in the execution of apoptosis
A reduction in persistent cell corpse numbers can be achieved not only through a restoration of engulfment activity but also through reduced apoptosis or delayed initiation of cell death.36 To discard any role of pdr-1 in the apoptosis execution, pdr-1 influence in the generation of apoptotic cells was subsequently tested. Whereas 11–13 cells remained alive in the anterior pharynx of ced-3 loss-of-function (lf) mutants (allele ced-3(n717)),36, 37, 38 no surviving extra cells were observed in pdr-1 mutants alone (Table 2). In each ced-3(lf) and ced-3(lf);pdr-1 mutant animals from our experiment, some undead cells were not found as referenced also by the detailed protocol described in Schwartz.38 Even so the number of extra cells was not altered when ced-3 (lf) worms were crossed with pdr-1 mutants (Table 3). The same effect was obtained under conditions limiting caspase activity through ced-3(n2438).36, 37 Therefore, taking into account the genetic interaction of pdr-1 with the engulfment machinery described above, pdr-1 selectively affects the engulfment of apoptotic cells.
Table 2. pdr-1 is not involved in the execution of apoptosis.
| Genotype | Extra cells±s.d. | n | P-value |
|---|---|---|---|
| N2 | 0.1±0.2 | 15 | — |
| pdr-1(lg103) | 0.1±0.2 | 15 | — |
| ced-3(n717) | 8±0.4 | 15 | — |
| ced-3(n717);pdr-1(lg103) | 8±0.6 | 35 | 0.711 to ced-3(n717) |
| ced-3(n2438) | 2.3±0.1 | 13 | — |
| ced-3(n2438);pdr-1(lg103) | 1.9±0.2 | 35 | 0.463 to ced-3(n2438) |
pdr-1 does not affect the execution of apoptosis. Third larval stage (L3) animals were anesthetized and viewed using DIC microscopy. The number of extra cell nuclei in the anterior pharynx (pro- and metacorpus) was counted. Alleles used were as follows: ced-3(n717), ced-3(n2438) and pdr-1(lg103). All data are means±s.d. (statistics: paired t-test; two-tailed, unequal variances)
Table 3. ced-10 activity in the engulfment of cell corpses depends on the proteasomal activity.
| Genotype | Treatment | Number of cell corpses±s.d. | n | P-value |
|---|---|---|---|---|
| wild type | MG-132 | 0 | 15 | — |
| ced-10(n3246) | DMSO | 16.2±3.9 | 20 | |
| ced-10(n3246) | MG-132 | 5.7±2.3 | 25 | P<0.001 |
Loss of pdr-1 accelerates the engulfment of dying cells
Apoptotic cell corpses are visible from the time they are generated until they are engulfed and degraded by neighboring cells. The kinetics of the engulfment was investigated by four-dimensional (4D) Nomarski time-lapse video microscopy (Leica DM 6000, Madrid, Spain) to follow the first 13 programmed cell deaths in the AB cell lineage3 (Figure 2 and Supplementary Table S2 and Figure S2).
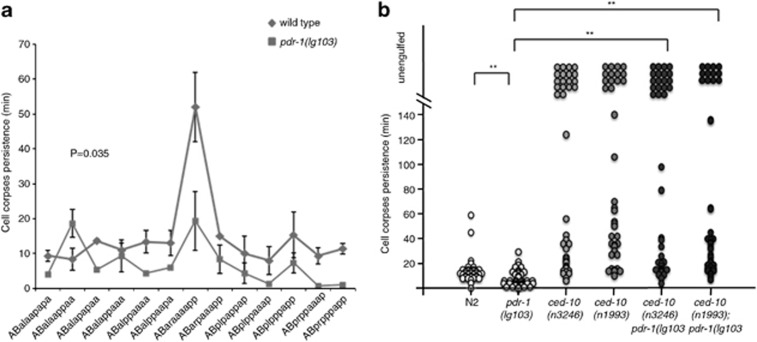
Figure 2.
Loss of pdr-1 increases engulfment kinetics. The first 13 apoptotic cell deaths in the AB lineage were followed by 4D microscopy and the cell corpse persistence was measured. Cells indicated as not engulfed either floated off the tissue into the egg-shell cavity or their lineage could not be followed anymore owing to the beginning of muscle contraction at the 1.5-fold stage (320 min after fertilization or approximately 120 min after the onset of cell death). Analysis was performed in three independent individuals in each genetic background (n=3 × 13 cells). Alleles used were as follows: ced-10(n3246), ced-10(n1993), pdr-1(lg103) and pdr-1(tm395). (a) The name of each analyzed cell was represented by the X axis and statistical analysis was carried out using Wilcoxon's rank-sum test. (b) Each circle represents a single cell and the statistical analysis was carried out using Kolmogorov–Simirnov test (**P<0.01)
Surprisingly, alterations were found in both the kinetics of the engulfment time and the onset of apoptosis in pdr-1 allele alone (Figure 2 and Supplementary Figure S2A, respectively). As demonstrated by Supplementary Figure S2B, the first mutant embryo development occurred mostly synchronized with the wild type, but the cell cycle was progressively delayed (Supplementary Figure 2B). Both cell lineages, respectively, are evolving unsynchronized with the increasing embryonic developmental time. Therefore, the delay in the onset of apoptosis cannot be ascribed to a function of pdr-1 in the initiation of apoptosis per se, but may correspond to a general defect in cell cycle progression affecting the developing embryo. Strikingly, the mean engulfing time was shorter in pdr-1 compared with wild-type worms (Figure 2 and Supplementary Table S2). Given the general delay in the developing embryos, a delay in the engulfment resulting in accumulated cell corpses at L1 stage could then also be expected. Moreover, it is worth noting the lack of differences in cell corpse persistence between ced-10 mutants alone in comparison with ced-10;pdr-1 double mutant animals (Figure 2b and Supplementary Table S2), albeit a clear reduction in cell corpse number (Figure 1 and Table 1). While in the 4D recordings only the first 13 cell deaths are being analyzed (from this developing time the embryo is moving faster and the engulfing time is impossible to follow appropriately39), the counting of cell corpses at L1 stage represents the final process and reflects the global efficiency of the engulfment, including all the unphagocyted cell corpses. Therefore, the increase in the rate of engulfment observed in the 4D recordings reinforces and lends further support to the initial enhancement in the engulfment observed in pdr-1 animals.
Pdr-1 has cell-autonomic functions in the engulfing cells
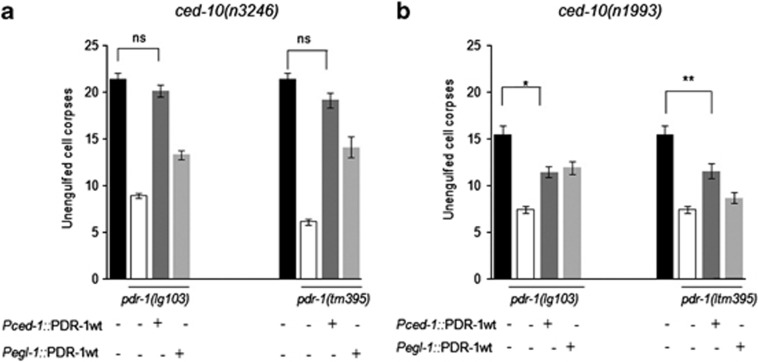
To corroborate the hypothesis of pdr-1 acting in the engulfing cell, PDR-1::GFP was expressed in a ‘tissue-specific' manner by two different cell-type-specific promoters.40, 41 Overexpression of PDR-1 controlled by the CED-1 promoter (Pced-1::pdr-1::gfp), which drives gene expression specifically in engulfing cells,6 fully rescued the reduced cell corpses observed in ced-10(n3246);pdr-1(lg103) mutants. This rescuing activity was not observed when PDR-1 expression was controlled by the egl-1 promoter (Pegl-1::pdr-1::gfp), which drives gene expression specifically in dying cells42 (Figure 3a and Supplementary Table S1). However, the rescue data on the ced-10 weak allele (n1993) were not fully accomplished when pdr-1(lg103) was the allele improving the engulfment (Figure 2b and Supplementary Table S1). As already reported, the use of hypomorphic alleles in the study of epistasis and double mutant analysis is problematic in the nematode.43 Still, a partial rescue was obtained when this weak allele of ced-10 was in combination with pdr-1(tm395). These results demonstrate that PDR-1 functions in engulfing rather than in dying cells, but a minor function of pdr-1 in the apoptotic cell cannot be excluded completely.
Figure 3.
Overexpression of pdr-1 in the engulfing cell reverses the effect of pdr-1 mutations. (a) The rescue of pdr-1::gfp in the engulfing cell (driven by Pced-1) was partially or completely achieved in the double mutant strains ced-10(3246);pdr-1(lg103) and ced-10(n3246);pdr-1(tm395). (b) The rescue of the transgenic strain pdr-1::gfp in the engulfing as well as in the apoptotic cells (driven by Pegl-1) was partially accomplished in the ced-10(n1993);pdr-1(lg103) double mutant. The rescue of pdr-1::gfp in the engulfing cell was also partially achieved in the ced-10(n1993);pdr-1(tm395) double mutant strain. This P-value refers to comparisons between the strains with the wild-type pdr-1 under the two different promoters (Pced-1 or Pegl-1) with the double mutant strains in which pdr-1 decreases the number of cell corpses. All data are means±s.d. (Statistics: paired t-test; two-tailed, unequal variances) (**P<0.01 and *P<0.05). Between 30 and 50 animals were scored per genotype
PDR-1 suppresses the distal tip cell migration defect associated with CED-10
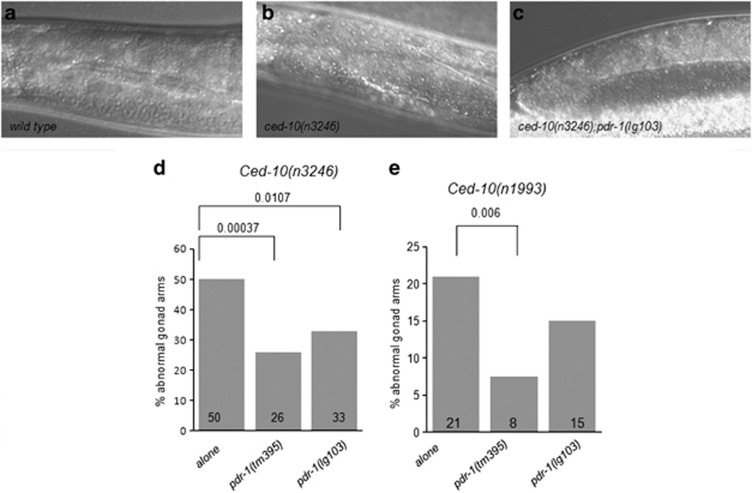
The two distal tip cells (DTCs) migrate during development from the center of the animal outward and then back again, meeting approximately in the center of the animal. As they move, the gonads form behind them, resulting in two U-shaped gonads.44 In ced-10 pathway mutants, the gonads often have extra turns or arms caused by abnormal DTC migration (Wu et al.45 and Figures 4b and c). To elucidate whether this pathway was also affected by pdr-1, DTC migration defects were analyzed in pdr-1 mutants alone and in combination with ced-10. Animals containing only any of the mutations of pdr-1 presented here showed the normal U-shaped gonadal arms. However, mutations of pdr-1 decreased the percentage of gonadal morphology defects in the two ced-10 alleles tested (Figure 4). In the context of DTC, the effect exerted by both alleles of pdr-1 on the most severe mutation ced-10(n3246) robustly confirms the genetic role of the modulation of the DTC migration phenotype by pdr-1.
Figure 4.
Pdr-1 mutation suppresses DTC migration defects in both hypomorphic ced-10 mutants. The gonads of ced-10(n3246) or ced-10(n1993) animals at the fourth larval stage (L4) with or without mutations in pdr-1 were scored for morphology using DIC microscopy (d, left and right, respectively). (a) Representative picture showing the normal U-shaped gonad in wild type, pdr-1 and a percentage of ced-10;pdr-1 animals. (b and c) Representative DIC pictures showing the abnormalities scored and analyzed as in (d). (d and e) Bar graph representing the scoring of DTC abnormalities in the two ced-10 backgrounds, ced-10(n3246) and ced-10(n1993), respectively. More than 50 gonad arms were scored for all genotypes. P-values derived using Fisher's exact test are shown
The amount of CED-10 is increased in the absence of PDR-1
The results presented above strongly suggest PDR-1 as a negative modulator of the engulfment.
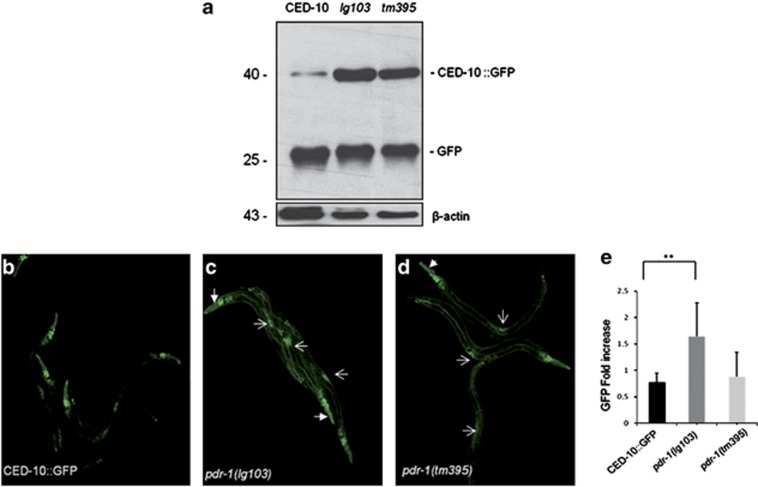
Both ced-10 alleles analyzed still maintain some remaining activity.30, 31 An increase in the pool of CED-10 active protein will also accelerate the engulfment machinery, resulting in less cell corpses in L1 animals. The decrease in the cell corpses observed in ced-10;pdr-1 double mutants might be the consequence of CED-10 accumulation because of PDR-1 miss function. This hypothesis was analyzed biochemically by immunoblot. The genetic strain ced-10(n3417);CED-10::GFP (described in the Supplementary Methodology section) was crossed with both pdr-1 mutants used in the engulfment assays. Worm lysates were prepared as described previously,46 and the amount of GFP was detected by chemiluminiscence. A band of 46 kDa was observed in all lanes, corresponding to CED-10::GFP (Figure 5a). CED-10::GFP was increased in the worms containing both alleles of pdr-1 (Figure 5a, second and third lane of the western blot analysis). The 25 kDa band observed in all samples corresponds to the molecular weight of the GFP protein.
Figure 5.
Parkin miss function affects the amount of CED-10. (a) The strain ced-10(n3417) harboring the integrated array CED-10::GFP was crossed individually with the pdr-1 mutant strains, and the amount of GFP correlated with the amount of CED-10 detected by immunoblotting. Twenty micrograms of protein was electrophoresed and blotted against GFP (upper panel). The amount of CED-10::GFP was increased in both pdr-1 mutant strains analyzed. Antibody anti-β-actin was used as a loading control (bottom panel). The same results were obtained in three independent experiments. (b–d) Confocal fluorescent micrographs of CED-10 translational GFP reporter expression pattern and quantification of total CED-10::GFP in L4 animals. The most evident fluorescent organs are labeled by white arrows, vulva by light arrow and head by thick arrow. Scale bars in (b–d)=40 μm. (e) Values are means±s.e.m. (n=10–15 worms imaged in three independent experiments). **P<0.01 in the strain containing the allele pdr-1(lg103) compared with the control strain. P-value in the strain containing the allele pdr-1(tm395)=0.78948142
These results strongly show a direct contribution of PDR-1/Parkin to the modulation of the CED-10/Rac1 turnover. Moreover, the increased accumulation of CED-10 induced by the strong loss of function allele lg103 clearly indicates that the elimination of PDR-1 E3 ubiquitin ligase activity (the deletion removes the UBL) effectively increased the amount of CED-10.
Similar results were obtained when the amount of CED-10::GFP was analyzed from confocal images and quantified (Figures 5b–e and Supplementary Figure S4). The lack of pdr-1 did not affect the subcellular expression of CED-10::GFP (Supplementary Figures S3 and S4). Total fluorescence was significantly increased in the worms containing the allele pdr-1(lg103), in comparison with the strain ced-10(n3417);CED-10::GFP, thus corroborating the results obtained by western blot analysis. Results were statistically significant only for the lg103 allele of pdr-1 (P<0.01) (Figure 5e and Supplementary Figure S3).
CED-10 is targeted by PDR-1 for proteasomal degradation
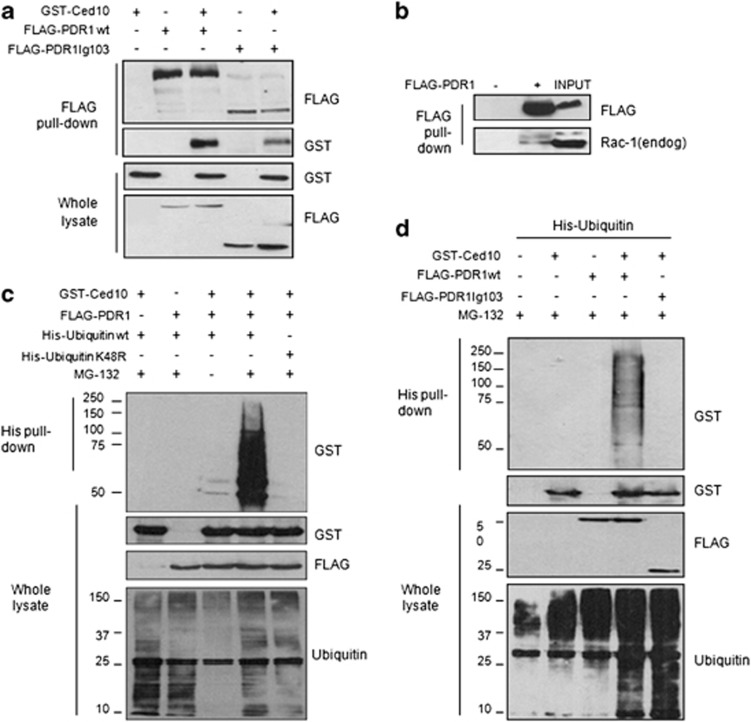
We show for the first the interaction between Rac1 and Parkin, the human orthologs of CED-10 and PDR-1, respectively, in aged human brain (Supplementary Figure S1). The genetic interaction between pdr-1 and ced-10 plus the biochemical studies performed in worms led us to test whether the two nematode proteins can also physically interact. The unavailability of specific C. elegans antibodies against PDR-1 and CED-10 for immunoprecipitation and for immunohistochemical approaches47 hampered analysis of the interaction between the endogenous proteins in the nematode. In consequence, interaction between the nematode proteins was tested in cells. Co-immunoprecipitation experiments were performed in human embryonic kidney 293T (HEK293T) cells transiently transfected with the nematode-tagged proteins FLAG-PDR1 and GST-CED-10. Figure 6a (first panel, lanes 3 and 5, respectively) shows that CED-10 co-precipitate with the two tested forms of PDR-1, the wild-type and the mutant.
Figure 6.
PDR-1 loss-of-function fails to ubiquitylate CED-10. (a) HEK293T cells were transfected with the indicated expression plasmids, and 500 μg of lysates were used to purify overexpressed FLAG-PDR1 using a FLAG-agarose resin. Immune complexes were immunoblotted for CED-10 and PDR-1 using anti-GST and anti-FLAG antibodies, respectively. Similar results were obtained in three independent experiments. (b) One microgram of HEK293T cells overexpressing FLAG-PDR1 was immunoprecipitated with anti-FLAG-agarose beads. Immune complexes were immunoblotted for human endogenous Rac1 and PDR-1 using anti-Rac1 and anti-FLAG antibodies. (c and d) Cells overexpressing the indicated plasmids were treated with 20 μM MG-132 for 18 h. His-tagged ubiquitin was purified using using Ni2+-NTA-agarose beads and the ubiquitylated forms of CED-10 were visualized by immunoblotting using an anti-GST antibody
As it was previously described for pdr-1 mutants, they retain the capability of interaction with their protein partners,32 suggesting that the PDR-1-CED-10 interaction was not affected by PDR-1 mutations.
Importantly, overexpressed PDR-1 interacted with endogenous human Rac1, the human ortholog of CED-10 (Figure 6b), suggesting a common mechanism of regulation in metazoan.
As PDR-1-CED-10 interaction was not affected by PDR-1 mutations, we next investigated whether the ubiquitylation capacity of PDR-1 was compromised by PDR-1 mutation. First, it was analyzed whether PDR-1 induced the ubiquitylation of CED-10. To test this, HEK293T cells were transiently transfected with the constructs encoding FLAG-PDR-1 and GST-CED-10 and/or His-ubiquitin, and further treated either with or without the proteasome inhibitor MG-132. Levels of ubiquitylation were determined by pulling down His-ubiquitin with Ni2+-NTA-agarose beads and immunoblotting for GST-CED-10 protein (Figure 6c). Slowly migrating species of CED-10 corresponding to the ubiquitylated forms were detected only in cells treated with MG-132. No CED-10 immunostaining was observed when ubiquitin-K48R was used in pull-down assays, indicating that PDR-1 ubiquitylated CED-10 through K48 ubiquitin linkages (Figure 6c, lanes 4 and 5, respectively). As expected, the E3 ligase-null mutant of PDR-1 (lg103) failed to induce CED-10 ubiquitylation in the presence of MG-132 in comparison with the non-mutated PDR-1 (Figure 6d, lanes 4 and 5, respectively).
The proteasomal machinery regulates the engulfment pathway
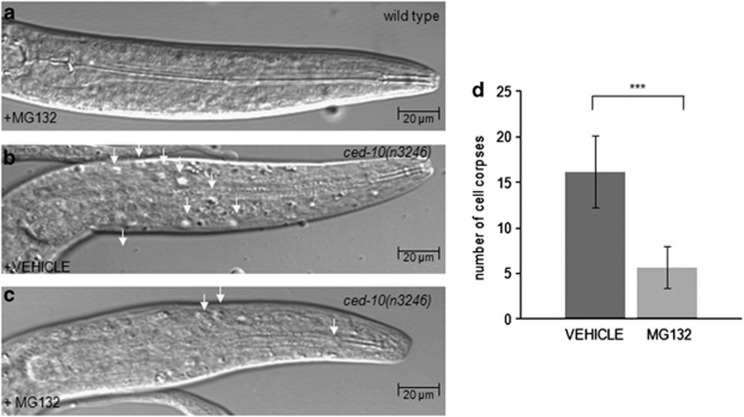
The modulation of CED-10 turnover by PDR-1 strongly suggested that the proteasomal machinery was a mechanism underlying the regulation of CED-10 activity during the engulfment. To further corroborate this hypothesis in vivo, ced-10 mutant animals were treated by the proteasomal inhibitor MG-132 and the amount of cell corpses were counted as described in the head of L1 larvae (Figure 7). Indeed, the amount of cell corpses was decreased in ced-10 mutant animals after MG-132 treatment, thus confirming the proteasomal regulation of the CED-10 activity during the engulfment.
Figure 7.
The activity of ced-10 during engulfment is dependent on proteasomal activity. (a–c) DIC microscopy images of synchronized L1 larvae of the indicated genotypes. Arrowheads indicate persistent cell corpses. Scale bar, 20 μm. (a) The proteasomal inhibitor MG-132 is not affecting the number of cell corpses in wild-type animals. (b and c) MG-132 treatment decreased the amount of cell corpses in ced-10(n3246) animals, (c) compared with (b), in which the worms were incubated only with the solvent DMSO 0.2% as a control. All data are means±s.d. (Statistics: paired t-test; two-tailed, unequal variances). Between 15 and 25 animals were scored per genotype. (d) Values are means±s.d. ***P<0.001 in ced-10 mutant animals treated with MG-132 compared with the ced-10 animals incubated only with the DMSO 0.2% as a vehicle for the treatment
Discussion
Our findings support that PDR-1 negatively regulates two mechanisms in which CED-10 has a pivotal role: the engulfment of apoptotic cells and the migration of DTCs during gonadogenesis. Ectopic expression experiments indicate that PDR-1 action in engulfing cells is partly dependent on its UBL-binding domain. PDR-1 ubiquitin ligase function is crucial to modulate the CED-10 ubiquitylation and turnover. Consistent with these findings, the general proteasomal inhibition also ameliorates the engulfment phenotype in ced-10 mutants, thus confirming for the first time a proteasomal modulation of the engulfment machinery.
Proteasomal regulation of CED-10/Rac1 in the engulfment
Protein degradation represents the most reliable way to dampen the amplitude and duration of signal-transduction pathways.48 It has been previously shown in vitro that Rac1, after activation, suffers ubiquitylation and proteasomal degradation.11, 12 Ubiquitin-mediated degradation of Rac1 (and RhoA) is specifically impaired in different immortalized and cancer cell lines, suggesting that several enzymes specifically target Rho protein members to ubiquitin-mediated proteasomal degradation.49 In fact, it has been recently described that Rac1 activity is regulated by the proteasomal machinery in other scenarios, and, as described in the introduction, the E3 ligases HACE1, c-IAP and XIAP are some examples.13, 14, 15 Our MG-132 experiments clearly demonstrate that the activity of CED-10 during the engulfment is regulated by the proteasomal machinery as well. Concretely from the proteasomal machinery ligases, PDR-1 interacts with human Rac1and ubiquitylates CED-10. The ameliorated phenotypes of ced-10;pdr-1 mutants might be the consequence of an increased CED-10 stability owing to the lack of degradation by loss-of-function PDR-1. The use of hypomorphic but severe alleles with remaining activity, such as ced-10(n3246), is crucial to investigate activity modulators. Obviously, this aim cannot be achieved by using null alleles such as ced-10(n3417) or ced-10(t1875) in which the whole activity is absent.
In conclusion, our results add Parkin/PDR-1 to the ligases regulating Rac1/CED10, and propose Parkin/PDR-1 as the one regulating cellular engulfment. Both proteins Parkin/PDR-1 and Rac1/CED10 act in the same cellular compartment,22, 35 and according to our scenario, PDR-1 miss function would increase CED-10 stability during the development of the worm, thus improving the GTPase activity, and lastly resulting in an increase in the rate of engulfment.
Although the high degree of homology between PDR-1 and CED-10 and their human counterparts has been well established so far, further work must be performed to extrapolate these results to humans.
PDR-1/Parkin as a suppressor of the engulfment machinery
Negative regulators of the engulfment machinery in C. elegans are swan-1,50 mtm-1,40 abl-1,7 pgrn-1,51 srgp-139 and the recently reported E3 ligase sli-1,8 in which the E3 ligase activity is dispensable to modulate engulfment, contrary to that detected by pdr-1. Among them, only SRGP-1 (Slit-Robo Gap-1)39 and PGRN-151 show effects on the kinetics of engulfment as PDR-1 does.
The multitude of regulatory molecules acting on CED-10 (Rac1) emphasizes the key position of this GTPases in the engulfment. The CED-5–CED-12 complex acting as a GEF (by promoting the active, GTP-bound state)52 and SRGP-1 acting as a GAP (by promoting the GDP-bound state and inactivation)39 are the main regulators described so far acting in engulfment machinery. Our results add a further complexity in this process. We propose PDR-1 as the E3 ligase responsible for the maintenance of the right ubiquitylation state, to guarantee the stabilization of CED-10 and its efficient regulation by the corresponding GEFs and GAPs. However, it is likely that additional regulators exist that act at other points within these pathways.
All living tissues have some mechanism(s) in place to handle corpse clearance, and most cell types posses the ability to phagocyte apoptotic cells, underlining the relevance of this process in metazoan cells. Engulfment of apoptotic debris induces anti-inflammatory responses that result in a quiet death, free from inflammation and cytotoxicity to neighboring cells.53 In contrast, materials associated with Alzheimer's disease or Parkinson's disease appear to induce a proinflammatory glial response that is neurotoxic (summarized in Sokolowski and Mandell54). In this scenario, engulfment might be associated with the clearance of neuronal debris and increased generation of reactive oxygen species as a consequence. Besides, a correct and efficient clearance will avoid an exaggerated immune response after cell death. Further work is required to study which are the consequences of the Rac1/Parkin interaction in human disease and whether this interaction is relevant for the development of the juvenile variant Parkinson's disease, characterized by Parkin mutations. Finally, future therapeutic drugs designed to modulate efficiently the activity of Rac1 might contribute to the amelioration of diseases in which Rac1 activity and cell clearance are clearly involved, such as cancer and autoimmunity.
Materials and Methods
The complete methodology is extensively described in the Supplementary Methodology section.
General methods and strains
C. elegans strains were maintained at 20 °C on nematode growth medium plates seeded with Escherichia coli strain OP50 as described previously by Brenner.55 N2 (Bristol) was used as the wild-type strain. The mutations used in this study are listed by chromosomes as follows: linkage group LGI – ced-1(e1375); LGIII – pdr-1(lg103), pdr-1(tm395); LGIV – ced-3(n717), (n2438), ced-5(n1812), ced-10(n1993), (n3246). Extrachromosomal array – pdr-1(lg103);byEx429[K08E3]sel-12::gfp.22 Integrated array – ced-10(n3417)::CED-10::GFP (gift from Dr. Erik Lundquist, University of Kansas, Lawrence, KS, USA; strain LE999).
Molecular biology
Recombinant DNA, polymerase chain reaction and cloning procedures were performed using standard protocols.56
Quantification of engulfment defects
Unengulfed apoptotic cell corpses were visualized directly in the heads of young larvae as refractile discs using Nomarski differential interference contrast (DIC) microscopy (Leica DM 6000 microscope).57, 58 Statistical analysis was performed using GraphPadPrism Version 4 software (GraphPAd Software, La Jolla, CA, USA). P-values for pairwise comparisons were calculated using the Student's t-test.
Quantification of cell-death defects
For quantification of cell-death defects in the anterior pharynx, animals in the third larval stage (L3) were anesthetized and viewed with DIC microscopy as described.38
Rescue experiments
Ced-10(n3246);pdr-1(lg103) hermaphrodites were crossed with males of the already generated rescue strain pdr-1(lg103);byEx434[pBY1500], in which pBY1500 contains the complete operon of pdr-1 named K08E3 and sel-12::gfp has been used as a comarker.22 To facilitate the understanding of the rescue experiments, the cosmid name has been substituted by ‘PDR-1 wild type' and the comarker sel-12::gfp has been omitted. L1 green worms from the F3 generation were scored for cell corpses as described previously.
Cell autonomous rescue
To address the question of the role of pdr-1 in the engulfing and/or the dying cell, we used two already well-characterized promoters of ced-1 and egl-1, respectively.6, 42 CED-1 is a cell surface phagocytic receptor that recognizes cell corpses,6 and EGL-1 localizes specifically in apoptotic cells.42 The corresponding translational fusions were generated as described previously.22 Pced-1::gfp and Pegl-1::gfp were kindly provided by Dr. Z Zhou (Baylor College of Medicine, Houston, TX, USA) and Dr. B Conradt (Ludwig-Maximilians-Universität München (LMU), Munich, Germany), respectively.
Video recording
4D microscopy was carried out using standard live-animal mounting techniques on a Leica DM 6000 microscope fitted with Nomarski optics. The use of Nomarski optics allows identification of key time points during apoptosis without using any dye or fluorescent marker that might alter the process. Embryonic cell death was determined as described.59
C. elegans western blot analysis
To elucidate the dependence on the amount of CED-10 by PDR-1, we took advantage of the strain LE999 in which ced-10(n3417) IV harbors the transgene CED-10::GFP in an array (strain kindly provided by Dr. E Lundquist).35 Ced-10(n3417) lacks exons 2 and 3 (from 666 bp 3′ of the ced-10 ATG, within intron 2, to 1637 bp 3′ of the ced-10 ATG, within intron 3) of the ced-10 locus, which is predicted to result in the expression of only the first 35 amino acids of CED-10.35 The pdr-1 mutant strains pdr-1(lg103) and pdr-1(tm395) were crossed individually with the strain LE999. Worm pellets from the corresponding genotypes were processed for western blot analysis as described.46 The expanded method is detailed in the Supplementary Methodology section.
Confocal analysis of the CED-10::GFP reporter
In vivo analysis of the CED-10 reporter was investigated by analyzing the strain LE999 described above. To quantify GFP expression, L4 stage animals were mounted on 5%, 20 mM sodium azide agar pads and confocal images were collected from 10–15 animals. We then measured the pixels produced by the fluorescence in the whole body of animals and determined the fluorescence level relative to worm area, using the ImageJ software (http://imagej.nih.gov/ij/plugins/index.html). The final value, by means of internal density, was obtained by subtracting the background of individual images by using the pluggin ‘background subtraction from ROI' included in the software. P-values for pairwise comparisons were calculated using the Student's t-test.
Cell culture, transient transfection, immunoprecipitation and immunoblotting
HEK293T cells were cultured in 10-cm diameter dishes at 37 °C in Dulbecco's modified Eagle's medium (Sigma, Barcelona, Spain), containing 10% fetal bovine serum and antibiotics. Cells were lysed in ice-cold lysis buffer (50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 150 mM NaCl, 0.5% Nonidet P-40 and protease inhibitor cocktail), centrifuged at 12 000 × g for 15 min at 4 °C and supernatants were then stored at −20 °C until use. Immunoprecipitation experiments were performed as described previously.60
Ubiquitination in vivo assay
The purification of His-ubiquitinated conjugates was performed following the method described by Erazo et al.61
Proteasome inhibitor treatment
To test in vivo whether the CED-10 activity during the engulfment was regulated by the proteasomal machinery, the worms were treated with the commonly used proteasome inhibitor MG-132 (Calbiochem, Barcelona, Spain). The MG-132 experiments were performed as described previously, with some modifications.62, 63 Briefly, L4 stage larvae of the corresponding genotypes were grown on E. coli (strain OP50)-seeded NGM plates containing fresh MG-132 50 μM with 0.2% DMSO as a solvent control both in the agar media and in the OP50-like bacteria. After 2 days, the mothers were washed off the plates and bleached, and the hatched L1 progeny was incubated in M9+MG-132 50 μM with 0.2% DMSO as a solvent control overnight at 20 °C. To see the difference between MG-132-treated and -non-treated worms, animals were mounted on agar pads and the quantification of the engulfment defects, by means of the amount of cell corpses, was performed as described above. Ced-10(n3246) worms treated with 0.2% DMSO were used as a control, to discard the effects of the solvent per se in the quantification of the cell corpses. P-values for pairwise comparisons were calculated using the Student's t-test.
Acknowledgments
We thank Nathalie Launay (of Aurora Pujol's Neurometabolic Disease Lab, IDIBELL, Spain) for comments and discussion and Tom Yohannan for editorial assistance. Some strains were supplied by the Caenorhabditis Genetic Center (CGC). The genetic strain LE999 was a kind gift from Dr. Eric Lundquist. The genetic strains ced-10(n3246), ced-10(n1993) were kindly given by Dr. Hengartner's lab. The genetic constructs Pced-1::gfp and Pegl-1::gfp were gifts from Drs. Zheng Zou and Barbara Conradt, respectively. The constructs PDR-1-FLAG and PDR-1(LG103)-FLAG were kindly provided by Dr. Wolfdieter Springer. This work was supported in part by The Spanish Ministry of Science and Competitivity grants PC0009/003 and PI1100968 (E Dalfo and I Ferrer respectively), Riojasalud Foundation and The Spanish Ministry of Science and Innovation Grant BFU2010-21794 (J Cabello and E Gómez), Grant DFG (Deutsche Forschungsgemeinschaft) SFB850 and SFB746 (R Baumeister) and PIF fellowship from Universitat Autònoma de Barcelona and Spanish Ministerio de Educación y Ciencia grant BFU2007-60268 (T Erazo and J Lizcano respectively).
Glossary
- ced
cell-death abnormality
- DTC
distal tip cell
- PD
Parkinson's disease
- UBL
ubiquitin-like domain
- DIC
differential interference contrast
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by D Bano
Supplementary Material
References
- Henson PM, Hume DA. Apoptotic cell removal in development and tissue homeostasis. Trends Immunol. 2006;27:244–250. doi: 10.1016/j.it.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell. 2010;140:619–630. doi: 10.1016/j.cell.2010.02.014. [DOI] [PubMed] [Google Scholar]
- Hoeppner DJ, Hengartner MO, Schnabel R. Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature. 2001;412:202–206. doi: 10.1038/35084103. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR. The engulfment process of programmed cell death in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2004;20:193–221. doi: 10.1146/annurev.cellbio.20.022003.114619. [DOI] [PubMed] [Google Scholar]
- Hasegawa E, Karashima T, Sumiyoshi E, Yamamoto M. C. elegans CPB-3 interacts with DAZ-1 and functions in multiple steps of germline development. Dev Biol. 2006;295:689–699. doi: 10.1016/j.ydbio.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- Hurwitz ME, Vanderzalm PJ, Bloom L, Goldman J, Garriga G, Horvitz HR. Abl kinase inhibits the engulfment of apoptotic [corrected] cells in Caenorhabditis elegans. PLoS Biol. 2009;7:e99. doi: 10.1371/journal.pbio.1000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C, Zhou S, Sawin E, Horvitz HR, Hurwitz ME. SLI-1 Cbl inhibits the engulfment of apoptotic cells in C. elegans through a ligase-independent function. PLoS Genet. 2012;8:e1003115. doi: 10.1371/journal.pgen.1003115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano F, Montcourrier P, Chavrier P. Membrane recruitment of Rac1 triggers phagocytosis. J Cell Sci. 2000;113 (Part 17:2955–2961. doi: 10.1242/jcs.113.17.2955. [DOI] [PubMed] [Google Scholar]
- Doye A, Mettouchi A, Bossis G, Clement R, Buisson-Touati C, Flatau G, et al. CNF1 exploits the ubiquitin–proteasome machinery to restrict Rho GTPase activation for bacterial host cell invasion. Cell. 2002;111:553–564. doi: 10.1016/s0092-8674(02)01132-7. [DOI] [PubMed] [Google Scholar]
- Lerm M, Pop M, Fritz G, Aktories K, Schmidt G. Proteasomal degradation of cytotoxic necrotizing factor 1-activated rac. Infect Immun. 2002;70:4053–4058. doi: 10.1128/IAI.70.8.4053-4058.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan BZ, Chapman JA, Reynolds SH. Proteasome inhibitor MG132 induces apoptosis and inhibits invasion of human malignant pleural mesothelioma cells. Transl Oncol. 2008;1:129–140. doi: 10.1593/tlo.08133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacon G, Mettouchi A, Lemichez E. The tumor suppressor HACE1 targets Rac1 to ubiquitin-mediated proteasomal degradation. Med Sci (Paris) 2012;28:39–41. doi: 10.1051/medsci/2012281014. [DOI] [PubMed] [Google Scholar]
- Nethe M, Hordijk PL. The role of ubiquitylation and degradation in RhoGTPase signalling. J Cell Sci. 2010;123 (Part 23:4011–4018. doi: 10.1242/jcs.078360. [DOI] [PubMed] [Google Scholar]
- Torrino S, Visvikis O, Doye A, Boyer L, Stefani C, Munro P, et al. The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Dev Cell. 2011;21:959–965. doi: 10.1016/j.devcel.2011.08.015. [DOI] [PubMed] [Google Scholar]
- Oberoi TK, Dogan T, Hocking JC, Scholz RP, Mooz J, Anderson CL, et al. IAPs regulate the plasticity of cell migration by directly targeting Rac1 for degradation. EMBO J. 2012;31:14–28. doi: 10.1038/emboj.2011.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata E, Yamaguchi Y, Kurimoto E, Kikuchi J, Yokoyama S, Yamada S, et al. Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain. EMBO Rep. 2003;4:301–306. doi: 10.1038/sj.embor.embor764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- Finney N, Walther F, Mantel PY, Stauffer D, Rovelli G, Dev KK. The cellular protein level of parkin is regulated by its ubiquitin-like domain. J Biol Chem. 2003;278:16054–16058. doi: 10.1074/jbc.C300051200. [DOI] [PubMed] [Google Scholar]
- Imai Y, Soda M, Hatakeyama S, Akagi T, Hashikawa T, Nakayama KI, et al. CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol Cell. 2002;10:55–67. doi: 10.1016/s1097-2765(02)00583-x. [DOI] [PubMed] [Google Scholar]
- Springer W, Hoppe T, Schmidt E, Baumeister RA. Caenorhabditis elegans Parkin mutant with altered solubility couples alpha-synuclein aggregation to proteotoxic stress. Hum Mol Genet. 2005;14:3407–3423. doi: 10.1093/hmg/ddi371. [DOI] [PubMed] [Google Scholar]
- Corti O, Brice A. Parkinson's disease: what have we learned from the genes responsible for familial forms. Med Sci (Paris) 2003;19:613–619. doi: 10.1051/medsci/2003195613. [DOI] [PubMed] [Google Scholar]
- Ko HS, von Coelln R, Sriram SR, Kim SW, Chung KK, Pletnikova O, et al. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J Neurosci. 2005;25:7968–7978. doi: 10.1523/JNEUROSCI.2172-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H, Schlossmacher MG, Hattori N, Frosch MP, Trockenbacher A, Schneider R, et al. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson's disease. Science. 2001;293:263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci USA. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh DP, Scoles DR, Ho TH, Del Bigio MR, Pulst SM. Parkin is associated with actin filaments in neuronal and nonneural cells. Ann Neurol. 2000;48:737–744. [PubMed] [Google Scholar]
- Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, et al. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- Horvitz HR, Sternberg PW. Multiple intercellular signalling systems control the development of the Caenorhabditis elegans vulva. Nature. 1991;351:535–541. doi: 10.1038/351535a0. [DOI] [PubMed] [Google Scholar]
- Shakir MA, Gill JS, Lundquist EA. Interactions of UNC-34 Enabled with Rac GTPases and the NIK kinase MIG-15 in Caenorhabditis elegans axon pathfinding and neuronal migration. Genetics. 2006;172:893–913. doi: 10.1534/genetics.105.046359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat Cell Biol. 2000;2:131–136. doi: 10.1038/35004000. [DOI] [PubMed] [Google Scholar]
- Nyamsuren O, Faggionato D, Loch W, Schulze E, Baumeister R. A mutation in CHN-1/CHIP suppresses muscle degeneration in Caenorhabditis elegans. Dev Biol. 2007;312:193–202. doi: 10.1016/j.ydbio.2007.09.033. [DOI] [PubMed] [Google Scholar]
- Patel FB, Bernadskaya YY, Chen E, Jobanputra A, Pooladi Z, Freeman KL, et al. The WAVE/SCAR complex promotes polarized cell movements and actin enrichment in epithelia during C. elegans embryogenesis. Dev Biol. 2008;324:297–309. doi: 10.1016/j.ydbio.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Chen S, Yap SF, Lim L. The Caenorhabditis elegans p21-activated kinase (CePAK) colocalizes with CeRac1 and CDC42Ce at hypodermal cell boundaries during embryo elongation. J Biol Chem. 1996;271:26362–26368. doi: 10.1074/jbc.271.42.26362. [DOI] [PubMed] [Google Scholar]
- Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, Bargmann CI. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development. 2001;128:4475–4488. doi: 10.1242/dev.128.22.4475. [DOI] [PubMed] [Google Scholar]
- Stanfield GM, Horvitz HR. The ced-8 gene controls the timing of programmed cell deaths in C. elegans. Mol Cell. 2000;5:423–433. doi: 10.1016/s1097-2765(00)80437-2. [DOI] [PubMed] [Google Scholar]
- Shaham S, Reddien PW, Davies B, Horvitz HR. Mutational analysis of the Caenorhabditis elegans cell-death gene ced-3. Genetics. 1999;153:1655–1671. doi: 10.1093/genetics/153.4.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz HT. A protocol describing pharynx counts and a review of other assays of apoptotic cell death in the nematode worm Caenorhabditis elegans. Nat Protoc. 2007;2:705–714. doi: 10.1038/nprot.2007.93. [DOI] [PubMed] [Google Scholar]
- Neukomm LJ, Frei AP, Cabello J, Kinchen JM, Zaidel-Bar R, Ma Z, et al. Loss of the RhoGAP SRGP-1 promotes the clearance of dead and injured cells in Caenorhabditis elegans. Nat Cell Biol. 2011;13:79–86. doi: 10.1038/ncb2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Lu Q, Zhao D, Li W, Mapes J, Xie Y, et al. Caenorhabditis elegans myotubularin MTM-1 negatively regulates the engulfment of apoptotic cells. PLoS Genet. 2009;5:e1000679. doi: 10.1371/journal.pgen.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Cameron S, Horvitz HR. Phagocytosis promotes programmed cell death in C. elegans. Nature. 2001;412:198–202. doi: 10.1038/35084096. [DOI] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell. 1999;98:317–327. doi: 10.1016/s0092-8674(00)81961-3. [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Cabello J, Klingele D, Wong K, Feichtinger R, Schnabel H, et al. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature. 2005;434:93–99. doi: 10.1038/nature03263. [DOI] [PubMed] [Google Scholar]
- Kimble JE, White JG. On the control of germ cell development in Caenorhabditis elegans. Dev Biol. 1981;81:208–219. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- Wu YC, Tsai MC, Cheng LC, Chou CJ, Weng NY. C. elegans CED-12 acts in the conserved crkII/DOCK180/Rac pathway to control cell migration and cell corpse engulfment. Dev Cell. 2001;1:491–502. doi: 10.1016/s1534-5807(01)00056-9. [DOI] [PubMed] [Google Scholar]
- Yang W, Li J, Hekimi S. A measurable increase in oxidative damage due to reduction in superoxide detoxification fails to shorten the life span of long-lived mitochondrial mutants of Caenorhabditis elegans. Genetics. 2007;177:2063–2074. doi: 10.1534/genetics.107.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Lim HH, Lim L. A new member of the ras superfamily, the rac1 homologue from Caenorhabditis elegans. Cloning and sequence analysis of cDNA, pattern of developmental expression, and biochemical characterization of the protein. J Biol Chem. 1993;268:320–324. [PubMed] [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Boyer L, Turchi L, Desnues B, Doye A, Ponzio G, Mege JL, et al. CNF1-induced ubiquitylation and proteasome destruction of activated RhoA is impaired in Smurf1−/− cells. Mol Biol Cell. 2006;17:2489–2497. doi: 10.1091/mbc.E05-09-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Lu J, Rovnak J, Quackenbush SL, Lundquist EA. SWAN-1, a Caenorhabditis elegans WD repeat protein of the AN11 family, is a negative regulator of Rac GTPase function. Genetics. 2006;174:1917–1932. doi: 10.1534/genetics.106.063115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao AW, Eisenhut RJ, Martens LH, Nakamura A, Huang A, Bagley JA, et al. A neurodegenerative disease mutation that accelerates the clearance of apoptotic cells. Proc Natl Acad Sci USA. 2011;108:4441–4446. doi: 10.1073/pnas.1100650108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wu YC, Fadok VA, Lee MC, Gengyo-Ando K, Cheng LC, et al. Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science. 2003;302:1563–1566. doi: 10.1126/science.1087641. [DOI] [PubMed] [Google Scholar]
- Elliott MR, Ravichandran KS. Clearance of apoptotic cells: implications in health and disease. J Cell Biol. 2010;189:1059–1070. doi: 10.1083/jcb.201004096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski JD, Mandell JW. Phagocytic clearance in neurodegeneration. Am J Pathol. 2011;178:1416–1428. doi: 10.1016/j.ajpath.2010.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW.Isolation of high-molecular-weight DNA from mammalian cells using proteinase K and phenol CSH Protoc 20062006pii:pdb.prot4036. [DOI] [PubMed] [Google Scholar]
- Sulston JE. Neuronal cell lineages in the nematode Caenorhabditis elegans. Cold Spring Harb Symp Quant Biol. 1983;48 (Part 2:443–452. doi: 10.1101/sqb.1983.048.01.049. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, White JG. Polyploid tissues in the nematode Caenorhabditis elegans. Dev Biol. 1985;107:128–133. doi: 10.1016/0012-1606(85)90381-1. [DOI] [PubMed] [Google Scholar]
- Nieto C, Almendinger J, Gysi S, Gomez-Orte E, Kaech A, Hengartner MO, et al. Ccz-1 mediates the digestion of apoptotic corpses in C. elegans. J Cell Sci. 2010;123 (Part 12:2001–2007. doi: 10.1242/jcs.062331. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Asiain A, Ruiz-Babot G, Romero W, Cubi R, Erazo T, Biondi RM, et al. Brain specific kinase-1 BRSK1/SAD-B associates with lipid rafts: modulation of kinase activity by lipid environment. Biochim Biophys Acta. 2011;1811:1124–1135. doi: 10.1016/j.bbalip.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Erazo T, Moreno A, Ruiz-Babot G, Rodriguez-Asiain A, Morrice NA, Espadamala J, et al. Canonical and kinase activity-independent mechanisms for extracellular signal-regulated kinase 5 (ERK5) nuclear translocation require dissociation of Hsp90 from the ERK5-Cdc37 complex. Mol Cell Biol. 2013;33:1671–1686. doi: 10.1128/MCB.01246-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Shim J, Bae J, Kim YJ, Lee J. Stabilization of RNT-1 protein, runt-related transcription factor (RUNX) protein homolog of Caenorhabditis elegans, by oxidative stress through mitogen-activated protein kinase pathway. J Biol Chem. 2012;287:10444–10452. doi: 10.1074/jbc.M111.314146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell KA, Tucci ML, Armagost J, Hodges TW, Chen J, Memon SB, et al. Investigating bacterial sources of toxicity as an environmental contributor to dopaminergic neurodegeneration. PLoS One. 2009;4:e7227. doi: 10.1371/journal.pone.0007227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.