Abstract
Socially learned behaviours leading to genetic population structure have rarely been described outside humans. Here, we provide evidence of fine-scale genetic structure that has probably arisen based on socially transmitted behaviours in bottlenose dolphins (Tursiops sp.) in western Shark Bay, Western Australia. We argue that vertical social transmission in different habitats has led to significant geographical genetic structure of mitochondrial DNA (mtDNA) haplotypes. Dolphins with mtDNA haplotypes E or F are found predominantly in deep (more than 10 m) channel habitat, while dolphins with a third haplotype (H) are found predominantly in shallow habitat (less than 10 m), indicating a strong haplotype–habitat correlation. Some dolphins in the deep habitat engage in a foraging strategy using tools. These ‘sponging’ dolphins are members of one matriline, carrying haplotype E. This pattern is consistent with what had been demonstrated previously at another research site in Shark Bay, where vertical social transmission of sponging had been shown using multiple lines of evidence. Using an individual-based model, we found support that in western Shark Bay, socially transmitted specializations may have led to the observed genetic structure. The reported genetic structure appears to present an example of cultural hitchhiking of mtDNA haplotypes on socially transmitted foraging strategies, suggesting that, as in humans, genetic structure can be shaped through cultural transmission.
Keywords: bottlenose dolphin, cultural hitchhiking, genetic structure, Tursiops sp., social learning
1. Introduction
Darwinian selection acts on phenotypes, which are manifested through both genetically and non-genetically inherited traits. Both inheritance mechanisms may be adaptive and, thus, of evolutionary consequence [1]. Vertical social transmission (i.e. learning from a biological parent) closely follows genetic inheritance patterns. For example, the diets and/or foraging strategies of offspring have been shown to resemble that of their mother in a wide range of mammalian taxa (e.g. orangutans, Pongo pygmaeus wurmbii; sea otters, Enhydra lutris nereis; and bottlenose dolphins, Tursiops sp.) [2–4]. All three species share prolonged maternal dependence and there is overlap between nursing and offspring-foraging during development, providing opportunities for social learning [2,3,5].
Social transmission can affect the evolutionary outcomes of genetic transmission and vice versa. Social transmission may change selection pressure on genes (gene-culture coevolutionary theory [6]), as for example documented for the spread of lactose tolerance in adult humans [7] and proposed for hundreds of human genes [8]. Further, individual learning capacity or the exposure to a socially learned trait can correlate with specific genetic marker systems [6]. For instance, in vertical social transmission, patterns of transmission of the socially learned trait and uniparentally inherited genetic units, such as mitochondrial DNA (mtDNA) and the Y chromosome, may be closely correlated. If the socially learned trait increases its bearer's fitness, population frequencies of these correlated genetic units will increase, even if there is no other active selection on genetic units. This phenomenon is called ‘cultural hitchhiking’ [9] and was posed as a possible explanation for the low genetic diversity in human mtDNA and Y chromosome sequences [10] and the low mtDNA diversity in matrilineal whales [9]. However, stochastic modelling has shown that a reduction of genetic diversity is not a necessary consequence of parallel transmission of genes and socially learned phenotypes [10,11].
Cultural hitchhiking might occur with little or no fitness differences if there is fine-scale geographical population structure that relates to the cultural trait, as we discuss below. There are two general types of geographical population structure: continuous clines or sharp boundaries [12]. Genetic and geographical distances often correlate in natural populations (i.e. isolation-by-distance, e.g. [13]). On the other hand, geographical features resulting in habitat fragmentation or behavioural patterns limiting movement and genetic exchange may lead to discontinuous genetic structuring of populations. There is ample evidence for structuring by geographical boundaries [14]. However, there is limited evidence for geographical structuring of genetic variation due to behaviour and even less evidence due to cultural behaviour. Possible mechanisms include assortative mating and microhabitat specialization. For example, two species of cichlid fish (Amphilophus xiloaensis and Amphilophus sagittae) preferentially mate with a partner of the same colour morph, which has led to genetic differentiation between morphs [15]. Moreover, in killer whales (Orcinus orca), socially transmitted foraging specializations within an ecosystem were proposed to have led to sympatric ecotypes in the absence of physical barriers [16,17].
To date, studies describing the influence of social transmission or culture on genetic structure or selection have focused on genetic variation between populations [8]. Here, we provide an example of how social transmission may drive genetic structure within a single dolphin population.
Thirteen foraging strategies have been described for bottlenose dolphins in the eastern gulf of Shark Bay (ESB), Western Australia [3]. Individual females have been observed engaging in one to seven of these strategies [3], some of which are only observed in specific habitats. The most prominent foraging strategies observed only in shallow water include beach hunting, bottom grubbing and kerplunking [18,19]. Foraging strategies observed in deep water include ‘sponging’ [20–23]. The question of whether sponging is a socially transmitted behaviour has been studied extensively in Shark Bay in the past [3,20,21,24–28]. These studies used different approaches (i.e. genetics, individual follows, survey data, network-based approaches, individual-based modelling or combinations thereof) to infer social transmission of tool use in this population.
Sponging dolphins (‘spongers’ hereafter) carry conical marine sponges on their rostrums, most likely to protect them while foraging for prey hiding in the substrate [21,27]. The behaviour is almost exclusively vertically transmitted from mothers to their offspring through social learning [3,20,24]. With one exception, spongers in ESB share a maternally inherited mtDNA haplotype [24]. Spongers in ESB are also biparentally more closely related than expected by chance [24]. Haplotype sharing between sponging individuals is expected in cases of strong vertical social transmission [24], even though a genetic basis for sponging has been ruled out [24,25,28]. Sponging has also been documented in the western gulf of Shark Bay (WSB) [23,25,29]. The ESB and WSB study sites are approximately 120 km apart by sea, divided by a prominent peninsula, and there is no direct evidence of dispersal between the sites [30]. In both gulfs, sponging is almost exclusively limited to deep channels where sponges occur [22,23]. However, not all individuals inhabiting deep channels use sponges to forage. Therefore, a purely ecological explanation for sponging would not account for the heterogeneity of foraging strategies observed in these deep channels [22,24].
This study focuses on the fine-scale population genetic structure of bottlenose dolphins in WSB. We investigated the relationship between maternal relatedness, habitat and behaviour. Our study significantly extends previous studies in that it explores the possibility of social transmission shaping genetic structure in an animal population. This is a very important advance because, to our knowledge, this study provides the first example of the potential impact of social learning on within-population structure in non-human animals (as opposed to its influence on between-population structure [15–17]). Briefly, we observed striking spatial homogeneity in mtDNA in contrast to other parts of the bay, where haplotype distribution was much more heterogeneous [30]. We aimed to test the hypothesis that vertical social transmission of a habitat-dependent trait can lead to fine-scale genetic structure. To do this, we modelled the possible process using an individual-based model based on empirical data. We used this model to investigate whether the observed homogeneous mtDNA haplotype structure could be a result of a random process or, alternatively, would require that transmission patterns of a socially learned trait and uniparentally inherited genetic units correlate, as is already known in this case [3,20,21,24–28]. The model showed that the observed geographical structuring of mtDNA haplotypes is only possible if genetic and social transmissions are correlated.
2. Material and methods
(a). Study site and data collection
Shark Bay is located on the west coast of Australia, 850 km north of Perth. It consists of an eastern and western gulf, divided by Peron Peninsula. Our main study area is off the township of Useless Loop in the western gulf and consists of approximately 260 km2, about five times the size of the average home range of an adult ESB bottlenose dolphin [31]. We will refer to our main study area as WSB. WSB is characterized by channels (more than 10 m deep) dividing shallow water areas that are closer to land (figure 1) [23].
Figure 1.
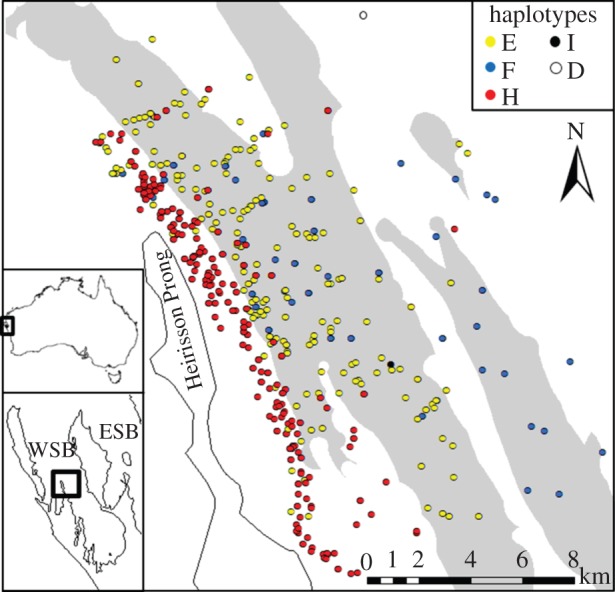
Segregation of dolphin haplotypes by habitats in WSB. Survey locations of dolphins with known mtDNA haplotypes are indicated. Survey colours represent haplotypes of dolphins. Each sighting of a sampled dolphin was plotted, thus individuals can appear multiple times. The sightings include all types of behaviour, including foraging, travelling, resting, socializing and unknown. White areas represent shallow (less than 10 m) and grey areas represent deep (more than 10 m) water. Insert illustrates the study area within Shark Bay; ESB indicates the location of the eastern gulf of Shark Bay.
We carried out systematic photo-identification and behavioural surveys on bottlenose dolphins from a 5.4 m boat with an outboard motor. Dolphin group composition (10 m chain rule [32]), water depth, GPS and predominant dolphin activity (forage, rest, travel, social or unknown) were recorded. For our analyses, we did not discriminate between these activities. Dolphins were individually identified by natural markings and the shape of their dorsal fin [33]. Biopsy sampling [34] was conducted under a Licence to use and/or supply Animals for Scientific Purposes from the Western Australian Department of Environment and Conservation, and ethics approval was obtained from the University of New South Wales (08/33B), Murdoch University and the University of Zurich.
A sponger was defined as a dolphin that was seen sponging at least twice [20]. We were able to use water depth as a proxy for habitat because Tyne et al. [23] reported that sponges only occur in water deeper than 10 m, while seagrass occurs in shallow water (less than 10 m) in WSB.
(b). Analyses
The genetic sampling and laboratory methods are detailed in the electronic supplementary material. We ran a nested ANOVA in PASW Statistics 18 to test for significance of association between water depth distribution of dolphins with different mtDNA haplotypes. Because multiple sightings of the same individuals were included, individual dolphin identification (ID) was used as a random factor. To exclude the possibility that the observed mtDNA haplotype distribution might simply be an artefact of significant autosomal population structure, we tested whether the observed segregation of mtDNA haplotypes and depth is reflected in biparentally inherited microsatellite markers. Such autosomal structure would have to be very strong, causing a severe restriction of gene flow, if it was to secondarily produce the very clear mitochondrial geographical structuring. To investigate this, we ran a STRUCTURE analysis (burn-in length of 105 and 106 Markov chain Monte Carlo steps) using mtDNA haplotype as Locprior [35–37]. The use of a Locprior model significantly increases the chance to detect population structure even in weakly structured populations [37].
We analysed whether spongers shared an mtDNA haplotype more often than would be expected by chance and whether they were biparentally more related than would be expected by chance. Individuals (spongers and non-spongers) were included in these analyses when they were sampled in an area specified by a 95% kernel utilization distribution of locations where sponging had been observed by any animal [24] (electronic supplementary material, figure S1). The kernel was calculated in ArcMap 9.2 (ESRI) using the extension Home Range Tools [38]. Pairwise comparisons between the observed mtDNA haplotype distribution within spongers were compared to pairwise comparisons of 10 000 times the number of sampled spongers randomly drawn. The haplotype frequencies used as a basis for randomization reflected the frequencies calculated for the WSB dolphins. The randomizations were carried out in a ‘macro’ written in Microsoft Excel.
To test whether spongers are biparentally more related than expected by chance, we calculated the average pairwise relatedness among spongers and compared it to the average pairwise relatedness of the population. We calculated the Queller & Goodnight [39] estimator of pairwise relatedness (R) in SPAGEDi [40]. In order to assess statistical significance, we programmed a randomization test in MATLAB R2010a (MathWorks, Natick, MA, USA). Biparental genotypes of all sampled individuals of the population were randomized and the average pairwise relatedness among 22 or 15 (for WSB and ESB, respectively, representing the number of biopsy sampled spongers) randomly drawn genotypes were calculated. In the randomization, the observed sex ratio of spongers was retained.
(c). Individual-based model
We used simulations to test the hypothesis that vertical social transmission of a habitat-dependent trait can lead to fine-scale genetic structure using an individual-based model based on empirical data. The model is described in detail in the electronic supplementary material. We included simulations of a null model with no fitness benefits for specialist, as well as one with fitness benefits. Most simulations were run with 5% fitness benefits for specialists, even though empirical data suggest that fitness advantages of spongers may be larger than that, albeit being non-significant [20].
3. Results
(a). Mitochondrial DNA haplotypes and habitat
We found a strong and significant association between dolphin mtDNA haplotypes and water depth in WSB. Dolphins with mtDNA haplotypes E or F were predominantly found in deep water (more than 10 m deep) and dolphins with haplotype H in shallow water (less than 10 m, figures 1 and 2). The geographical segregation of mtDNA haplotypes in WSB was not reflected in biparentally inherited genetic markers: STRUCTURE plots showed no indication of genetic structure (electronic supplementary material, figure S2). The depth distribution for both adult males and females with one of the three common haplotypes E, F and H differed significantly (tables 1 and 2), as revealed by a nested ANOVA and a subsequent Tukey's HSD post hoc test, based on 90 dolphins (59 females and 31 males) and 756 sightings (mean number of sightings ± s.e./individual: 8.4 ± 0.68, range = 1–33). Three other haplotypes (B, D and I) were rare (frequency less than 0.03).
Figure 2.
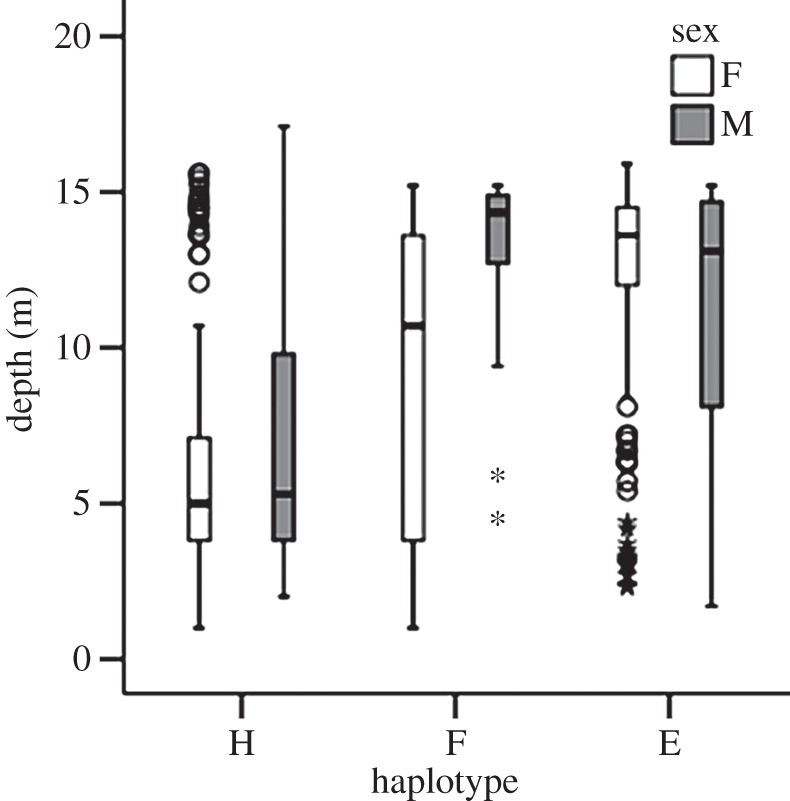
Depth preference of bottlenose dolphins with the three common haplotypes: H, F and E (sponger haplotype). Boxes contain 50% of data points. Medians are indicated by black horizontal lines within boxes. Whiskers delimit the lower and upper quartiles, respectively. Circles and asterisks represent outliers that are more than 1.5 and 3 times the box length away from either end of the box, respectively.
Table 1.
Depth preference of WSB dolphins. Nested ANOVA comparing depth spectra of dolphins with different mtDNA haplotypes. Dolphin ID was used as a random factor nested within haplotypes.
| type III sum of squares | d.f. | mean square | F | significance | |
|---|---|---|---|---|---|
| intercept | |||||
| hypothesis | 19 563.33 | 1 | 19 563.33 | 561.81 | <0.001 |
| error | 3744.74 | 107.54 | 34.82 | ||
| haplotype | |||||
| hypothesis | 1320.41 | 2 | 660.21 | 18.31 | <0.001 |
| error | 3824.89 | 106.09 | 36.05 | ||
| ID (haplotype) | |||||
| hypothesis | 6815.46 | 87 | 78.34 | 13.34 | <0.001 |
| error | 3912.11 | 666 | 5.87 | ||
Table 2.
Differences in depth preference for dolphins with different haplotypes (WSB). Tukey HSD post hoc test for nested ANOVA. hap, haplotype; depth diff., mean depth difference.
| hap. 1 | hap. 2 | depth diff. (m) | s.e. | significance | 95% CI depth (m) |
|
|---|---|---|---|---|---|---|
| lower | upper | |||||
| E | H | 5.7a | 0.19 | <0.001 | 5.2 | 6.1 |
| E | F | 1.5a | 0.28 | <0.001 | 0.9 | 2.2 |
| F | H | 4.1a | 0.28 | <0.001 | 3.5 | 4.8 |
aIndicates significance.
(b). Genetic relatedness of spongers
Forty spongers have been identified in WSB, which represents about a quarter (25.9%) of dolphins identified in deep water [29]. All 22 biopsied spongers in WSB share the same mtDNA haplotype E, which is significantly different (p < 0.001) from what would be expected by chance, based on the observed mtDNA haplotype frequencies in the population. The sponger haplotype E in WSB is different from the haplotype found among ESB spongers (haplotype H [24]). All 22 sampled spongers were biparentally more closely related than the population average (Rspongers = 0.0259, Rpopulation = −0.0104, Npopulation = 108), although this difference was not statistically significant (p = 0.092). We also re-ran the pairwise relatedness analyses for ESB [24], including additionally genotyped microsatellite loci and individuals. Our result confirms the previous publication [24]: spongers in ESB were found to be biparentally more closely related than the population average and than expected by chance (Rspongers = 0.0712, Rpopulation = −0.0044, Nspongers = 15, Npopulation = 238, p = 0.006).
(c). Individual-based model
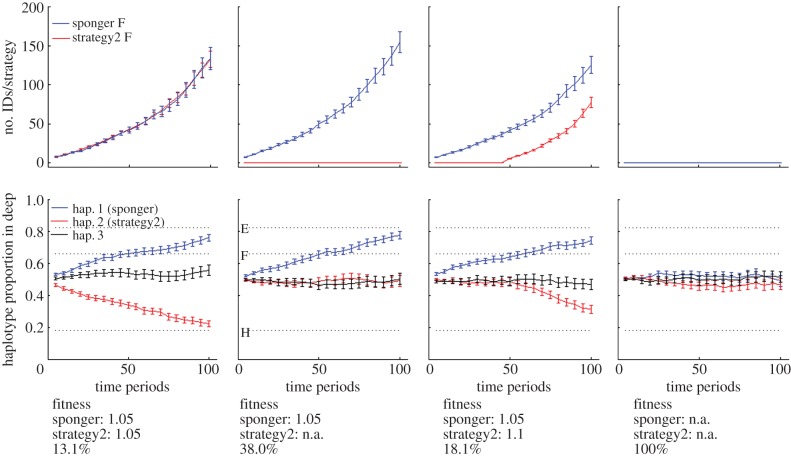
Simulations showed that fine-scale genetic structure based on mtDNA haplotypes can be driven by vertically, socially transmitted habitat-dependent traits (figure 3; electronic supplementary material, figure S3). The geographical segregation of haplotypes occurred even if the learning fidelity was not 100% (electronic supplementary material, figure S3), but in the absence of vertically, socially transmitted specializations, no geographical segregation of mtDNA haplotypes was observed (figure 3, fourth column).
Figure 3.
mtDNA haplotype segregation by habitat in an individual-based model. Three mtDNA haplotypes (hap.1, 2 and 3) and two habitat specializations (sponging and strategy2) were present. All spongers had hap.1 and all strategy2 individuals had hap.2. Top row: number of different females per strategy (no. IDs/strategy). Bottom row: proportion of individuals with a particular mtDNA haplotype in deep water relative to all individuals with this particular mtDNA haplotype. Proportions were calculated for every haplotype separately. Error bars represent 1 s.e. Dashed lines indicate the observed haplotype proportion in deep water for the three mtDNA haplotypes E, F and H in WSB. Fitness benefits for specialists are shown below graphs. Because random cultural drift [41] is a strong force counteracting the establishment of new innovations, we indicated the likelihood (%) of at least one specialist/strategy to persist for 100 time periods; this is shown below the fitness benefits. Learning fidelities equalled 1 for daughters born to specialists for the simulations shown here. Simulation results with various other learning fidelities and fitness benefits are shown in the electronic supplementary material, figure S3. In the first column, individuals of both strategies had the same fitness benefits. In the second column, only one strategy (sponging) was present, in the third column, strategy2 was innovated 50 time periods after sponging and had a higher fitness, and in the fourth column, there were no specializations present.
4. Discussion
Through a combination of empirical data and individual-based modelling, we were able to infer that the vertical cultural transmission of a foraging behaviour involving tools has led to the clear separation of mtDNA haplotypes within our study area. The observed geographical distribution of dolphins with different mtDNA haplotypes might represent a case of cultural hitchhiking. To our knowledge, this is one of the first studies to provide evidence for fine-scale geographical genetic structure driven by socially transmitted behaviour within a single wild animal population.
In what circumstances would we predict correlations between habitats and mtDNA haplotypes? We propose that four prerequisites must be met. First, a population must exhibit vertically socially transmitted, habitat-dependent skills, such as foraging or predator avoidance strategies. Second, philopatry must keep haplotypes localized, although if one sex disperses, the pattern could still be found in the philopatric sex. Third, habitat differentiation needs to be on a scale that is larger than an individual's home range. Fourth, in order for habitat specializations to drive genetic differentiation over small distances, these specializations must be stable over an individual's life time [42] and also present in following generations. The clear separation of matrilines and habitats we present here suggests that habitat specializations have been in place for many more generations than, for instance, the four generations of spongers that have been observed in ESB so far.
The segregation of mtDNA haplotypes by habitat appears to be driven by vertically transmitted foraging specializations (and/or other behaviours leading to habitat specialization). The absence of fine-scale genetic structure based on biparentally inherited markers shows that the correlation of maternally inherited mtDNA haplotype and water depth cannot be explained solely by geographical separation. Of course, it is possible that there is undetected autosomal structure, but it is most unlikely that this would be strong enough to secondarily cause the extremely sharp mtDNA patterning that we observed. That any autosomal structuring within our study area must be only weak is supported by two points. First, our use of Locprior ensures detection of relatively weak structure [37]. Second, the relatively weak autosomal genetic differentiation between ESB and WSB was documented using a much smaller number of markers [30] than used in this study.
Different depth preferences exhibited by dolphins with different mtDNA haplotypes coincide with different habitats. Seagrass meadows are characteristic of shallow water, and conical sponges only grow in water deeper than 10 m in WSB [23]. Shallow and deep habitats intergrade and are not divided by a barrier or distance that could prevent dolphins from moving between deep and shallow water. The absence of genetic structure based on biparental genetic markers indicates that there is no mating barrier between deep and shallow habitat and may be explained by the dispersal patterns of Shark Bay dolphins. Both sexes are philopatric, with males expanding their natal range [30,31]. In the WSB study site, we observed the dolphins foraging, travelling, resting and socializing. Therefore, the geographical segregation of dolphins with different mtDNA haplotypes indicates that dolphins stay in their natal habitat for all those activities (we do not know about mating). Male bottlenose dolphins in Shark Bay cooperate with one another in pairs and trios to consort females [43]. It appears that allied males can direct a female outside her regular home range. Some sightings of females with haplotype H in deep water or haplotype E in shallow water might be explained by the coercion of males. It is possible that these groupings involved consorting of females by males; however, we did not confirm the consortships by the standards of ESB [43]. In ESB, males have been observed consorting females never seen in the study site before and well-studied females altered their depth-use during consortships [44].
Although our study site is not likely to encompass the entire home ranges of some individuals [45] included in this study, it is about five times the size of an average adult ESB dolphin home range [31]. Accordingly, we expect our identification of each individual's habitat usage to be suitably representative. In addition, the number of resightings per individual (up to 33) suggests that the study site covers a great portion of the home ranges of at least some animals.
Our individual-based simulations emphasize that social transmission of habitat-dependent specializations can lead to fine-scale genetic structure (figure 3, first, second and third columns); whereas this fine-scale structure is unlikely to be seen if there is no social transmission (figure 3, fourth column). Social transmission of foraging strategies in WSB appears likely, given the mtDNA haplotype sharing among WSB spongers, as occurs in ESB [24]. Our finding that spongers in WSB are not more closely related than expected by chance provides further evidence that sponging is transmitted socially, rather than genetically, and in a vertical fashion. Furthermore, our simulations indicate that two specializations are necessary, one for the deep and one for the shallow habitat, in order to obtain the observed segregation of two mtDNA haplotypes by habitat. It is conceivable that traits other than sponging are vertically socially transmitted, such that matrilines H and F also exhibit habitat specialization. Without any habitat specialization, no mtDNA haplotype segregation is observed (figure 3, fourth column).
Although all spongers sampled in WSB belong to one matriline, it is a different matriline from that of spongers in ESB [24]. The presence of different sponging matrilines in each gulf can be explained most parsimoniously by the occurrence of at least two independent innovations, one in each gulf. Alternatively, there could have been horizontal transmission between matrilines. However, this would have to be extremely rare to fit the patterns that we observe.
What we have inferred for sponging may be just one example of a more widespread phenomenon of interactions between culture and geographical structure of genetic variation. Genetic differentiation on relatively small scales has been reported for many bottlenose dolphin populations (both Tursiops aduncus or Tursiops truncatus) in all oceans. This structuring was attributed largely to habitat specialization and was found based on maternal and biparentally inherited markers [46–50]. However, most of these studies (except [46]) gave no indication of what the specializations might be, or how they could have driven the genetic structure. Furthermore, the geographical scales at which genetic differentiation have been documented (tens to hundreds of km) in previous studies are much greater than those reported here: deep and shallow habitats in WSB are separated by just tens of metres. The largest delphinid, the killer whale, may present a similar case. Sympatric ecotypes were shown to differ genetically and in their foraging behaviour [16,51]. The genetic differentiation between ecotypes based on microsatellites suggests that this separation may be more stringent and/or has been in place for longer than that in WSB.
A correlation between habitat and genetic structure has also been found in tool-using New Caledonian crows (Corvus moneduloides) [52]. Rutz et al. [52] found biparental and maternal (e.g. mtDNA haplotypes) genetic structure among three habitats (dry forest, farmland and beachside habitat, less than 10 km apart), in which different tool use has been observed. Restricted dispersal may lead to localized occurrence of particular tool use [52]. In female mountain gorillas (Gorilla beringei beringei) and western gorillas (Gorilla gorilla diehli), Guschanski et al. [53] reported a correlation between microsatellite-based genetic and geographical structure (i.e. altitude). This correlation did not hold for males. In gorillas, both sexes disperse, but females usually join a neighbouring group. Hence, female dispersal distance is limited and often occurs within habitat types. The authors concluded that dispersal usually results in females staying within the habitat into which they were born, probably owing to food preferences [53]. Unfortunately, data on maternally inherited markers was not presented. The correlation between habitat and mitochondrial genetic structure in bottlenose dolphins reported here holds for both adult females and males.
If vertical social transmission influences the fine-scale genetic structure of a population (e.g. within WSB), it may have consequences for the genetic structure at larger scales (e.g. between the two gulfs of Shark Bay). As WSB matrilines seem to stay in their natal habitats, it is likely that they have behaviourally adapted to those habitats. If male and female dolphins in Shark Bay are philopatric because of learnt foraging strategies, genetic differentiation would occur—as observed—over small geographical distances. Limited dispersal for both sexes could potentially cause inbreeding. Indeed, elevated levels of inbreeding were measured in ESB, but the costs may not be high enough to outweigh the benefits of philopatry [54]. Philopatry bears benefits of habitat familiarity and the potential for kin cooperation [55]. This study highlights that cultural forces can shape genetic population structure outside humans and on a small geographical scale.
Acknowledgements
We are extremely grateful to Shark Bay Resources Inc. and the Useless Loop community for their ongoing and extensive logistical support. We thank our field crew, including Kathrin Bacher, Deirdre McElligott, Sina Kreicker, Dominik Noll and Richard Mardens. Thanks to Celine Frere, Michael Hassell and three anonymous reviewers for helpful comments on the manuscript.
Data accessibility
Genotypes used in this study have been archived in the Dryad Digital Repository (doi:10.5061/dryad.k5b1j).
Funding statement
This work was supported by the Sea World Research and Rescue Foundation, National Geographic Society, Winifred Violet Scott Foundation, Claraz-Schenkung, A.-H. Schultz Stiftung and Julius-Klaus Stiftung.
References
- 1.Bonduriansky R, Day T. 2009. Nongenetic inheritance and its evolutionary implications. Annu. Rev. Ecol. Evol. Syst. 40, 103–125 (doi:10.1146/annurev.ecolsys.39.110707.173441) [Google Scholar]
- 2.Jaeggi AV, Dunkel LP, Van Noordwijk MA, Wich SA, Sura AAL, Van Schaik CP. 2010. Social learning of diet and foraging skills by wild immature Bornean orangutans: implications for culture. Am. J. Primatol. 72, 62–71 (doi:10.1002/ajp.20752) [DOI] [PubMed] [Google Scholar]
- 3.Mann J, Sargeant BL. 2003. Like mother, like calf: the ontogeny of foraging traditions in wild Indian Ocean bottlenose dolphins (Tursiops sp.). In The biology of traditions: models and evidence (eds Fragaszy D, Perry S.), pp. 236–266 Cambridge, UK: Cambridge University Press [Google Scholar]
- 4.Estes J, Riedman M, Staedler M, Tinker M, Lyon B. 2003. Individual variation in prey selection by sea otters: patterns, causes and implications. J. Anim. Ecol. 72, 144–155 (doi:10.1046/j.1365-2656.2003.00690.x) [Google Scholar]
- 5.Staedler MM, Tinker MT, Estes J. 2009. Individual variation in maternal care and provisioning in the southern sea otter (Enhydra lutris nereis): causes and consequences of diet specialization in a top predator. In 18th Biennial Conf. on the Biology of Marine Mammals, Quebec City, Canada, 12–16 October 2009, pp. 242–243. Society for Marine Mammology [Google Scholar]
- 6.Feldman MW, Laland KN. 1996. Gene-culture coevolutionary theory. Trends Ecol. Evol. 11, 453–457 (doi:10.1016/0169-5347(96)10052-5) [DOI] [PubMed] [Google Scholar]
- 7.McCracken RD. 1971. Lactase deficiency: an example of dietary evolution. Curr. Anthropol. 12, 479–517 (doi:10.1086/201234) [Google Scholar]
- 8.Laland KN, Odling-Smee J, Myles S. 2010. How culture shaped the human genome: bringing genetics and the human sciences together. Nat. Rev. Genet. 11, 137–148 (doi:10.1038/nrg2734) [DOI] [PubMed] [Google Scholar]
- 9.Whitehead H. 1998. Cultural selection and genetic diversity in matrilineal whales. Science 282, 1708–1711 (doi:10.1126/science.282.5394.1708) [DOI] [PubMed] [Google Scholar]
- 10.Whitehead H, Richerson PJ, Boyd R. 2002. Cultural selection and genetic diversity in humans. Selection 3, 115–125 (doi:10.1556/Select.3.2002.1.9) [Google Scholar]
- 11.Whitehead H. 2005. Genetic diversity in the matrilineal whales: models of cultural hitchhiking and group-specific non-heritable demographic variation. Mar. Mamm. Sci. 21, 58–79 (doi:10.1111/j.1748-7692.2005.tb01208.x) [Google Scholar]
- 12.Manel S, Schwartz MK, Luikart G, Taberlet P. 2003. Landscape genetics: combining landscape ecology and population genetics. Trends Ecol. Evol. 18, 189–197 (doi:10.1016/S0169-5347(03)00008-9) [Google Scholar]
- 13.Pogson GH, Taggart CT, Mesa KA, Boutilier RG. 2001. Isolation by distance in the Atlantic cod, Gadus morhua, at large and small geographic scales. Evolution 55, 131–146 (doi:10.1111/j.0014-3820.2001.tb01279.x) [DOI] [PubMed] [Google Scholar]
- 14.Storfer A, Murphy MA, Spear SF, Holderegger R, Waits LP. 2010. Landscape genetics: where are we now? Mol. Ecol. 19, 3496–3514 (doi:10.1111/j.1365-294X.2010.04691.x) [DOI] [PubMed] [Google Scholar]
- 15.Elmer KR, Lehtonen TK, Meyer A. 2009. Color assortative mating contributes to sympatric divergence of neotropical cichlid fish. Evolution 63, 2750–2757 (doi:10.1111/j.1558-5646.2009.00736.x) [DOI] [PubMed] [Google Scholar]
- 16.Morin PA, et al. 2010. Complete mitochondrial genome phylogeographic analysis of killer whales (Orcinus orca) indicates multiple species. Genome Res. 20, 908–916 (doi:10.1101/gr.102954.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoelzel AR, Dover GA. 1991. Genetic differentiation between sympatric Killer whale populations. Heredity 66, 191–195 (doi:10.1038/hdy.1991.24) [Google Scholar]
- 18.Connor RC, Heithaus MR, Berggren P, Miksis JL. 2000. ‘Kerplunking’: surface fluke-splashes during shallow-water bottom foraging by bottlenose dolphins. Mar. Mamm. Sci. 16, 646–653 (doi:10.1111/j.1748-7692.2000.tb00959.x) [Google Scholar]
- 19.Sargeant BL, Mann J, Berggren P, Krützen M. 2005. Specialization and development of beach hunting, a rare foraging behavior, by wild bottlenose dolphins (Tursiops sp.). Can. J. Zool. 83, 1400–1410 (doi:10.1139/z05-136) [Google Scholar]
- 20.Mann J, Sargeant BL, Watson-Capps J, Gibson QA, Heithaus MR, Connor RC, Patterson E. 2008. Why do dolphins carry sponges? PLoS ONE 3, e3868 (doi:10.1371/journal.pone.0003868) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smolker R, Richards A, Connor R, Mann J, Berggren P. 1997. Sponge carrying by dolphins (Delphinidae, Tursiops sp.): a foraging specialization involving tool use? Ethology 103, 454–465 (doi:10.1111/j.1439-0310.1997.tb00160.x) [Google Scholar]
- 22.Sargeant BL, Wirsing AJ, Heithaus MR, Mann J. 2007. Can environmental heterogeneity explain individual foraging variation in wild bottlenose dolphins (Tursiops sp.)? Behav. Ecol. Sociobiol. 61, 679–688 (doi:10.1007/s00265-006-0296-8) [Google Scholar]
- 23.Tyne J, Loneragan N, Kopps AM, Allen SJ, Krützen M, Bejder L. 2012. Ecological characteristics contribute to sponge distribution and tool use in bottlenose dolphins (Tursiops sp.). Mar. Ecol. Prog. Ser. 444, 143–153 (doi:10.3354/meps09410) [Google Scholar]
- 24.Krützen M, Mann J, Heithaus MR, Connor RC, Bejder L, Sherwin WB. 2005. Cultural transmission of tool use in bottlenose dolphins. Proc. Natl Acad. Sci. USA 102, 8939–8943 (doi:10.1073/pnas.0500232102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bacher K, Allen S, Lindholm A, Bejder L, Krützen M. 2010. Genes or culture: are mitochondrial genes associated with tool use in bottlenose dolphins (Tursiops sp.)? Behav. Genet. 40, 706–714 (doi:10.1007/s10519-010-9375-8) [DOI] [PubMed] [Google Scholar]
- 26.Mann J, Stanton MA, Patterson EM, Bienenstock EJ, Singh LO. 2012. Social networks reveal cultural behaviour in tool-using dolphins. Nat. Commun. 3, 980 (doi:10.1038/ncomms1983) [DOI] [PubMed] [Google Scholar]
- 27.Patterson EM, Mann J. 2011. The ecological conditions that favor tool use and innovation in wild bottlenose dolphins (Tursiops sp.). PLoS ONE 6, e22243 (doi:10.1371/journal.pone.0022243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopps AM, Sherwin WB. 2012. Modelling the emergence and stability of a vertically transmitted cultural trait in bottlenose dolphins. Anim. Behav. 84, 1347–1362 (doi:10.1016/j.anbehav.2012.08.029) [Google Scholar]
- 29.Kopps AM, Krützen M, Allen SJ, Bacher K, Sherwin WB. In press. Characterizing the socially transmitted foraging tactic ‘sponging’ by bottlenose dolphins (Tursiops sp.) in the western gulf of Shark Bay, Western Australia. Mar. Mamm. Sci. (doi:10.1111/mms.12089) [Google Scholar]
- 30.Krützen M, Sherwin WB, Berggren P, Gales N. 2004. Population structure in an inshore cetacean revealed by microsatellite and mtDNA analysis: bottlenose dolphins (Tursiops sp.) in Shark Bay, Western Australia. Marine Mammal Science 20, 28–47 (doi:10.1111/j.1748-7692.2004.tb01139.x) [Google Scholar]
- 31.Tsai Y-JJ, Mann J. 2012. Dispersal, philopatry, and the role of fission-fusion dynamics in bottlenose dolphins. Mar. Mamm. Sci. 29, 261–279 (doi:10.1111/j.1748-7692.2011.00559.x) [Google Scholar]
- 32.Smolker RA, Richards AF, Connor RC, Pepper JW. 1992. Sex-differences in patterns of association among Indian-Ocean bottle-nosed dolphins. Behaviour 103, 38–69 (doi:10.1163/156853992X00101) [Google Scholar]
- 33.Würsig B, Würsig M. 1977. The photographic determination of group size, composition, and stability of coastal porpoises (Tursiops truncatus). Science 198, 755–756 (doi:10.1126/science.198.4318.755) [Google Scholar]
- 34.Krützen M, Barre LM, Möller LM, Heithaus MR, Simms C, Sherwin WB. 2002. A biopsy system for small cetaceans: darting success and wound healing in Tursiops SPP. Mar. Mamm. Sci. 18, 863–878 (doi:10.1111/j.1748-7692.2002.tb01078.x) [Google Scholar]
- 35.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hubisz MJ, Falush D, Stephens M, Pritchard JK. 2009. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 9, 1322–1332 (doi:10.1111/j.1755-0998.2009.02591.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodgers AR, Carr AP, Smith L, Kie JG. 2005. HRT: home range tools for ArcGIS. Thunder Bay, Ontario, Canada: Centre for Northern Forest Ecosystem Research, Ontario Ministry of Natural Resources; (http://www.blueskytelemetry.com/downloads.asp) [Google Scholar]
- 39.Queller DC, Goodnight KF. 1989. Estimating relatedness using genetic markers. Evolution 43, 258–275 (doi:10.2307/2409206) [DOI] [PubMed] [Google Scholar]
- 40.Hardy OJ, Vekemans X. 2002. SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2, 618–620 (doi:10.1046/j.1471-8286.2002.00305.x) [Google Scholar]
- 41.Koerper HC, Stickel EG. 1980. Cultural drift: a primary process of culture change. J. Anthropol. Res. 36, 463–469 [Google Scholar]
- 42.Knudsen R, Primicerio R, Amundsen P-A, Klemetsen A. 2010. Temporal stability of individual feeding specialization may promote speciation. J. Anim. Ecol. 79, 161–168 (doi:10.1111/j.1365-2656.2009.01625.x) [DOI] [PubMed] [Google Scholar]
- 43.Connor RC, Smolker RA, Richards AF. 1992. Two levels of alliance formation among male bottle-nosed dolphins (Tursiops Sp). Proc. Natl Acad. Sci. USA 89, 987–990 (doi:10.1073/pnas.89.3.987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watson J. 2005. Female mating behavior in the context of sexual coercion and female ranging behavior of bottlenose dolphins (Tursiops sp.) in Shark Bay, Western Australia. Washington, DC: Georgetown University [Google Scholar]
- 45.Nicholson K, Bejder L, Allen SJ, Krützen M, Pollock KH. 2012. Abundance, survival and temporary emigration of bottlenose dolphins (Tursiops sp.) off Useless Loop in the western gulf of Shark Bay, Western Australia. Mar. Freshw. Res. 63, 1059–1068 (doi:10.1071/MF12210) [Google Scholar]
- 46.Hoelzel AR, Potter CW, Best PB. 1998. Genetic differentiation between parapatric ‘nearshore’ and ‘offshore’ populations of the bottlenose dolphin. Proc. R. Soc. Lond. B 265, 1177–1183 (doi:10.1098/rspb.1998.0416) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Natoli A, Birkun A, Aguilar A, Lopez A, Hoelzel AR. 2005. Habitat structure and the dispersal of male and female bottlenose dolphins (Tursiops truncatus). Proc. R. Soc. B 272, 1217–1226 (doi:10.1098/rspb.2005.3076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sellas AB, Wells RS, Rosel PE. 2005. Mitochondrial and nuclear DNA analyses reveal fine scale geographic structure in bottlenose dolphins (Tursiops truncatus) in the Gulf of Mexico. Conserv. Genet. 6, 715–728 (doi:10.1007/s10592-005-9031-7) [Google Scholar]
- 49.Parsons KM, Durban JW, Claridge DE, Herzing DL, Balcomb KC, Noble LR. 2006. Population genetic structure of costal bottlenose dolphins (Tursiops truncatus) in the northern Bahamas. Mar. Mamm. Sci. 22, 276–298 (doi:10.1111/j.1748-7692.2006.00019.x) [Google Scholar]
- 50.Möller LM, Wiszniewski J, Allen SJ, Beheregaray LB. 2007. Habitat type promotes rapid and extremely localised genetic differentiation in dolphins. Mar. Freshw. Res. 58, 640–648 (doi:10.1071/MF06218) [Google Scholar]
- 51.Hoelzel AR, Hey J, Dahlheim ME, Nicholson C, Burkanov V, Black N. 2007. Evolution of population structure in a highly social top predator, the killer whale. Mol. Biol. Evol. 24, 1407–1415 (doi:10.1093/molbev/msm063) [DOI] [PubMed] [Google Scholar]
- 52.Rutz C, Ryder T, Fleischer R. 2012. Restricted gene flow and fine-scale population structuring in tool using New Caledonian crows. Naturwissenschaften 99, 313–320 (doi:10.1007/s00114-012-0904-6) [DOI] [PubMed] [Google Scholar]
- 53.Guschanski K, Caillaud D, Robbins MM, Vigilant L. 2008. Females shape the genetic structure of a gorilla population. Curr. Biol. 18, 1809–1814 (doi:10.1016/j.cub.2008.10.031) [DOI] [PubMed] [Google Scholar]
- 54.Frère CH, Krützen M, Kopps AM, Ward P, Mann J, Sherwin WB. 2010. Inbreeding tolerance and fitness costs in wild bottlenose dolphins. Proc. R. Soc. B 277, 2667–2673 (doi:10.1098/rspb.2010.0039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Handley LJ, Perrin N. 2007. Advances in our understanding of mammalian sex-biased dispersal. Mol. Ecol. 16, 1559–1578 (doi:10.1111/j.1365-294X.2006.03152.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genotypes used in this study have been archived in the Dryad Digital Repository (doi:10.5061/dryad.k5b1j).