Abstract
We describe a statistical framework for reconstructing the sequence of transmission events between observed cases of an endemic infectious disease using genetic, temporal and spatial information. Previous approaches to reconstructing transmission trees have assumed all infections in the study area originated from a single introduction and that a large fraction of cases were observed. There are as yet no approaches appropriate for endemic situations in which a disease is already well established in a host population and in which there may be multiple origins of infection, or that can enumerate unobserved infections missing from the sample. Our proposed framework addresses these shortcomings, enabling reconstruction of partially observed transmission trees and estimating the number of cases missing from the sample. Analyses of simulated datasets show the method to be accurate in identifying direct transmissions, while introductions and transmissions via one or more unsampled intermediate cases could be identified at high to moderate levels of case detection. When applied to partial genome sequences of rabies virus sampled from an endemic region of South Africa, our method reveals several distinct transmission cycles with little contact between them, and direct transmission over long distances suggesting significant anthropogenic influence in the movement of infected dogs.
Keywords: endemic disease, rabies virus, spatial epidemiology, transmission trees, surveillance
1. Introduction
Understanding the spatial aspects of disease transmission is increasingly recognized as an essential component of successful control strategies [1,2]. However, disease transmission is usually a highly elusive event and reconstructing ‘who-infected-whom’ in outbreaks of infectious disease remains a challenging problem. The advent of high volume and more affordable pathogen genome sequencing to complement conventional space-time incidence data promises a step-change in our ability to understand transmission at the population level. Yet, progress will only be made with advances in statistical methodology to accompany this ever increasing access to genetic and other data.
Two different but complementary approaches that use spatial, temporal and pathogen genetic information to reconstruct the dynamics of epidemics have been developed in recent years. The first approach is based on coalescent models that assume some form of population dynamic model to relate the demography of the pathogen to its evolution, while implementing a diffusion model to account for the movement of the pathogen over geographical space [3]. These models can be used to estimate various parameters of interest, such as the rate of spatial spread of the pathogen [4] and the rate of evolution over time [5]. This approach has the advantage that it is relatively robust to the intensity of epidemiological sampling, but because such models do not have an explicit epidemiological formulation, the inferences cannot easily be related to real epidemiological processes. The second approach is based on spatial epidemiological models of transmission and simple models of genetic drift and directly reconstructs the transmission tree reflecting ‘who-infected-whom’, thus explicitly recognizing the host population structure and the epidemiological processes that govern the interaction of host and pathogen. In this approach, an epidemiological model of disease progression in individuals is used to estimate probability distributions for possible dates of infection and the infectious period of all cases. When coupled with a model of spatial diffusion and a model of the accumulation of point mutations over time, the probability of any two cases A and B being causally related can be calculated based on the likelihood that case A was infectious and case B was infected during the same time window, the probability that the pathogen could have dispersed from the geographical location at which case A was observed to the location at which B was observed in the time between observations, and the probability that the pathogen genetic sequence from case A could have mutated to the sequence from case B in the time between observations. This approach enables inferences to be made about epidemiological processes [6], the transmission tree [6,7], the mechanism of transmission [8] and the rate of evolution ‘per transmission event’ [9]. More recently, the two approaches have been combined, using a coalescent model to account for the influence of intra-host population dynamics on the structure of pathogen genetic data while reconstructing the transmission tree, thus addressing an important source of inaccuracy at high sampling intensities [10]. However, current transmission tree-based methods cannot handle large numbers of missing infections, and therefore require a high proportion of infected hosts from the outbreak to be present within the sample.
In general, these techniques have been applied to epidemics, and to data that are assumed to arise from a single introduction to the region under study (thus making its structure monophyletic). When pathogens are sampled from infected hosts in an endemic context (i.e. where the pathogen is stably maintained in an area in the absence of introductions from outside of that area), the epidemiological situation is potentially more complex. In this context, the connection between cases applies at two scales (figure 1a). At the scale of the entire endemic region, all cases may be related in some way (through the global transmission tree), leading to genetic relatedness and spatial autocorrelation between sampled cases. However, in a given study region (even one that has been exhaustively sampled), only some cases will be directly related through chains of transmission, and many chains of transmission may exist that are only indirectly related to each other by virtue of sharing a common ancestor outside the sampled area. The sample of pathogens within the study area is therefore polyphyletic. The picture is further complicated because surveillance is unlikely to be exhaustive, and therefore the sampling will be incomplete. Undetected or unsampled cases will reduce the detectable correlation between cases that are nevertheless causally related. If we hope to use genetic data to understand the detailed transmission biology of endemic pathogens, the challenge will be to develop algorithms that can accommodate the polyphyletic nature of pathogen population structure, and account for and make inferences regarding the unobserved and unsampled infections.
Figure 1.

Modelling the transmission of endemic diseases. (a) All cases in the study region are in some way related both genetically and spatially because they form part of a larger epidemic that originated from a single progenitor. This, along with the fact that some cases go undetected, makes determining dependence among transmission chains difficult. Letters O represent sampled cases, while X represent unsampled cases. (b) Map of the KwaZulu Natal province of South Africa, showing the locations of the 176 cases used to infer the transmission tree (see also the electronic supplementary material, table S1). (c) Pathogens radiate both in terms of genetic diversity and in terms of the geographic space invaded. Triangles represent possible locations in the geographic–genetic space to which cases can move and evolve, with the grey triangle showing the radiation of the entire epidemic, which can also be viewed as the indirect radiation of the index case (represented by a black X) through its descendants. In the relatively short observation window, three types of relationships are apparent: direct transmissions (purple), introductions, which will be more closely related to the common ancestor of all sampled cases than to any other cases (red and yellow), and indirect transmissions via unsampled intermediary cases (green).
Here, we describe the extension of a spatial-genetic SEIR (susceptible/exposed/infectious/removed) model of transmission to accommodate the complexities inherent to polyphyletic and partially sampled outbreak data containing space, time and genetic information. In addition, we infer the infected host population size over the study period and region by developing a mark–recapture method applied to the virus lineages occurring in the transmission tree, thus providing upper and lower estimates of the number of undetected or unsampled cases. We test this technique by analysing various simulated scenarios, before applying it to endemic rabies virus in a province of South Africa (figure 1b), and show how it can be used to better understand the spatial epidemiology of the virus. Such knowledge is crucial for advancing the effectiveness of large-scale vaccination campaigns—some of which have been in place for decades, but have failed to eliminate the disease in question.
Rabies is a complex disease endemic to much of the developing world [11]. The mutation rates of RNA viruses are so high that population genetic and epidemiological processes occur on similar timescales, and spatial expansion and epidemiology leave a discernible fingerprint on the genetic structure of these viruses [12,13]. Rabies virus is typically transmitted by direct contact through biting [14]. However, the epidemiological dynamics of rabies are complicated by two factors. First, rabies has a highly variable incubation period [15,16] and second, rabies has a very large host range that includes all mammals, many of which would play no part in the onward transmission of the virus [14]. In southern Africa, two distinct genetic variants of rabies virus are known to circulate—one among members of the Canidae, including domestic dogs (Canis lupus familiaris), and the other among several members of the Herpestidae [17]. Nevertheless, the majority of infections in humans are associated with rabid domestic dogs [11,18], and it is in dogs that the disease must be controlled if the burden on humans is to be reduced [19].
2. Material and methods
(a). Data collection
In the KwaZulu Natal province of South Africa (KZN), suspected cases are primarily collected through a network of state and private veterinarians. Further cases are collected by travelling vaccination teams of a Bill and Melinda Gates Foundation-sponsored rabies elimination project active throughout KZN. All cases testing positive for rabies virus by the gold-standard fluorescent antibody test [20] between 1 March 2010 and 8 June 2011 were selected for analysis (n = 195; electronic supplementary material, table S1). Five cases were negative by polymerase chain reaction (PCR; see below) after multiple attempts and were excluded from further analysis. One sequence, from an unrecorded wildlife species, matched the herpestid variant of rabies virus by BLAST [21] and was also excluded. A further 13 cases lacked GPS coordinates and were therefore excluded from the transmission tree reconstruction.
(b). RT-PCR and sequencing
RNA was extracted from original brain material using TRIzol reagent (Invitrogen). Reverse transcription (RT)-PCR and sequencing were performed as described in the electronic supplementary material. Consensus sequences were aligned using the FFT-NS-i algorithm of MAFFT v. 6 [22]. Sequences were trimmed to equal length (760 nucleotides, encompassing the last 224 nucleotides of the glycoprotein gene, the G-L intergenic region and 118 nucleotides of the polymerase gene, based on the genome of the Pasteur rabies virus strain [23]). The overall mean distance between sequences in the alignment was calculated using MEGA v. 5 [24].
(c). Transmission tree reconstruction
The transmission trees linking cases were reconstructed using the trimmed alignment described above, which was realigned with MAFFT after exclusion of 13 cases lacking GPS coordinates (electronic supplementary material, table S1).
The core algorithm used here is a generalization of the algorithm of Morelli et al. [7] to allow its application to any directly transmitted disease. We start with an epidemiological model in which any susceptible host i becomes infected at time  . Following an incubation period Li, it becomes infectious for time-period Di before dying. Both Li and Di are random variables with informative prior distributions based on contact tracing data from Tanzania [15]. From this data, it is possible to calculate the probability of a transmission from any host j to any host i based on the probability of j being infectious at the time of i's infection, if we assume the known observation date occurred shortly after the end of the infectious period [7].
. Following an incubation period Li, it becomes infectious for time-period Di before dying. Both Li and Di are random variables with informative prior distributions based on contact tracing data from Tanzania [15]. From this data, it is possible to calculate the probability of a transmission from any host j to any host i based on the probability of j being infectious at the time of i's infection, if we assume the known observation date occurred shortly after the end of the infectious period [7].
However, this forms only part of the probability of transmission between hosts. The spatial component of the likelihood equation was modified to accommodate a wide variety of spatial transmission patterns by replacing the exponential transmission kernel used in [7] with the exponential-power spatial transmission kernel described by [25]. This kernel is often used in dispersal studies and can take a variety of shapes, making it well suited to a range of endemic situations where often little is known regarding spatial transmission patterns. We also replaced the simplified substitution model of [7] with the Kimura three-parameter model [26].
(d). Extension to polyphyletic transmission trees
In a partially sampled outbreak, any given infected host which was sampled might have been infected by: (i) another sampled host (through direct transmission), (ii) an unsampled host which had been infected directly or indirectly by a sampled infected host (termed ‘indirect transmission’ here) or (iii) an unsampled host which has no ancestors within the sample, i.e. transmission from an exogenous source (figure 1a,c). The model of [7] allows for only a single virus introduction (i.e. a single ‘exogenous’ transmission) followed by direct transmissions for the rest of the outbreak. We extended this model by allowing multiple unobserved cases to arise anywhere in both space and time within the set of inferred transmissions.
The likelihood equation of [7] models the spatial radiation and genetic evolution of cases over time to determine the likelihood of various parameters at any point in time and thus calculate the probability of different transmissions. In our model, this is equivalent to the approach taken for direct transmissions, where each sampled infected host species able to transmit the virus can be a source of infection. These are modelled by the probability distribution  defined over the geographical–genetic space and evolving in time (represented by coloured cones in figure 1c).
defined over the geographical–genetic space and evolving in time (represented by coloured cones in figure 1c).  is dependent on the infection time of the host (estimated as described above), its incubation duration (estimated), its removal or observation time (observed), a spatial dispersal kernel (estimated) and substitution rates for the sequence evolution (estimated).
is dependent on the infection time of the host (estimated as described above), its incubation duration (estimated), its removal or observation time (observed), a spatial dispersal kernel (estimated) and substitution rates for the sequence evolution (estimated).
Each sampled infected host which can spread the disease can also be an indirect source of observed infections after its removal, as a consequence of unsampled intermediate hosts: case A (sampled) infects B (unsampled) which infects C (unsampled) which infects D (sampled). As these unsampled cases extend the influence of case A in both geographical and genetic space, their effect can be modelled by allowing observed cases to continue moving and evolving after their death. This is represented by probability distribution 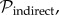 again defined over the geographical–genetic space and evolving in time and depending on the same parameters as
again defined over the geographical–genetic space and evolving in time and depending on the same parameters as  . The spatial influence contributed by unsampled cases is harder to determine. We considered two different specifications for the dispersal kernel governing indirect transmissions
. The spatial influence contributed by unsampled cases is harder to determine. We considered two different specifications for the dispersal kernel governing indirect transmissions 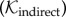 . In the first specification, we conservatively assume that
. In the first specification, we conservatively assume that  is the same as the spatial dispersal kernel used for the direct transmissions, thus allowing only movement over transmission distances observed for (single) direct transmissions. In this scenario, infections occurring after the death of the source host are attributed to unsampled intermediate hosts. However, this does not adequately accommodate a scenario encompassing multiple unsampled intermediate cases, where greater geographical distances between the indirectly connected cases would be possible. We therefore also considered a more liberal specification, where
is the same as the spatial dispersal kernel used for the direct transmissions, thus allowing only movement over transmission distances observed for (single) direct transmissions. In this scenario, infections occurring after the death of the source host are attributed to unsampled intermediate hosts. However, this does not adequately accommodate a scenario encompassing multiple unsampled intermediate cases, where greater geographical distances between the indirectly connected cases would be possible. We therefore also considered a more liberal specification, where 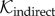 is a uniform distribution over the whole study region, thus allowing unsampled intermediate hosts to carry the virus to any location within the sampled region. These two scenarios form extremes between which the true process can reasonably be expected to occur.
is a uniform distribution over the whole study region, thus allowing unsampled intermediate hosts to carry the virus to any location within the sampled region. These two scenarios form extremes between which the true process can reasonably be expected to occur.
In a similar vein, the source of exogenous transmissions can be modelled as a probability distribution  defined over the geographical–genetic space and evolving in time (represented by the grey cone in figure 1c).
defined over the geographical–genetic space and evolving in time (represented by the grey cone in figure 1c). 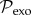 can be completely specified based on an ancestral virus sequence (determined a priori through ancestral state reconstruction, in our case using the FastML server under the generalized time reversible model [27]), a time for the ancestral sequence and the same substitution rates as above (both of which are co-estimated with the transmission tree). The ancestral sequence and the sampled infected hosts generate a mixture
can be completely specified based on an ancestral virus sequence (determined a priori through ancestral state reconstruction, in our case using the FastML server under the generalized time reversible model [27]), a time for the ancestral sequence and the same substitution rates as above (both of which are co-estimated with the transmission tree). The ancestral sequence and the sampled infected hosts generate a mixture  of spatio-temporal-genetic distributions (
of spatio-temporal-genetic distributions ( ,
,  and
and  ) from which the infection events are drawn. Estimating the source that infected a given host involves assessing in which component of the mixture model
) from which the infection events are drawn. Estimating the source that infected a given host involves assessing in which component of the mixture model  the infection of the host arose.
the infection of the host arose.
Conceptually, however, the source of both types of transmissions involving unobserved ancestors (indirect and exogenous) can be modelled in the same way—as being external to the sampled dataset, meaning the transmissions arise in  . Thus, to reduce complexity and computation time, we distinguished only between direct and ‘unsampled’ sources in the primary Markov chain Monte Carlo (MCMC) sampling procedure (only
. Thus, to reduce complexity and computation time, we distinguished only between direct and ‘unsampled’ sources in the primary Markov chain Monte Carlo (MCMC) sampling procedure (only  and
and  were used to define
were used to define  ), with a post-processing algorithm to distinguish between indirect and true exogenous transmissions. In the previously described monophyletic model [7], the posterior distributions of the incubation and infectious period durations can be deformed by indirect links between cases. We used narrow priors for the parameters governing these distributions, essentially forcing a decision between direct transmission or linkage to an exogenous source in the first step. To distinguish between exogenous and indirect transmissions, the post-processing analysis applies a Metropolis–Hastings update to the ‘unsampled’ transmission links determined by the MCMC algorithm, which involves comparing the probability that the transmission was really from an exogenous source (based on
), with a post-processing algorithm to distinguish between indirect and true exogenous transmissions. In the previously described monophyletic model [7], the posterior distributions of the incubation and infectious period durations can be deformed by indirect links between cases. We used narrow priors for the parameters governing these distributions, essentially forcing a decision between direct transmission or linkage to an exogenous source in the first step. To distinguish between exogenous and indirect transmissions, the post-processing analysis applies a Metropolis–Hastings update to the ‘unsampled’ transmission links determined by the MCMC algorithm, which involves comparing the probability that the transmission was really from an exogenous source (based on  as described above) with the probability that it was merely indirect (based on
as described above) with the probability that it was merely indirect (based on  ). This post-processing was applied under both the conservative and liberal specifications of the spatial transmission kernel
). This post-processing was applied under both the conservative and liberal specifications of the spatial transmission kernel  described above.
described above.
(e). Population size estimation
To determine the true number of cases represented by indirect links, we developed a mark–recapture technique applied to the virus lineages identified in the previous analysis. If we split the transmission tree dataset into two parts based on the sampling times of the hosts, any host sampled in the second time-period is considered as recaptured if it was directly or indirectly infected by a host observed in the first part of the dataset. Although the full transmission tree is not known, the previous analysis provides a sample of its posterior distribution. For each element of this sample, the number of recaptured virus lineages can be calculated, generating a posterior distribution of the number of recaptured virus lineages. With this distribution, one can determine the posterior distribution of the population size using a mark–recapture analysis, which takes into account uncertainty regarding changes in the population size from the first to the second time-period.
(f). Simulations
The accuracy of the method was assessed using 100 simulated datasets from each of six scenarios (i.e. 600 simulations in total). The first four scenarios were used to investigate overall accuracy and the effect of sampling rate on the reconstruction method with high (three-quarters of all cases), moderate (two-thirds of all cases), intermediate (one-half of all cases) and low (one-quarter of all cases) detection rates, respectively. A further two scenarios were used to test the sensitivity of the method to small and large misspecifications of epidemiological parameters. The simulation model was based on the probability distributions and specifications described above and in the electronic supplementary material, but contained a more realistic specification for the external source of infection. While the inference model assumes a single external source with a constant infection strength (constant in both space and time), the simulation model allows for multiple sources of novel lineages, occurring both inside and outside the sampling region, with infection strengths that are localized in time and space. The simulated epidemics were initiated from a single point in time and space outside the sampling period and region and allowed to progress until a set number of hosts had been infected. Only data from one-third of the region and time-period affected by the simulated epidemic were retained and subsampled with the detection rates above determining the probability of a case being retained.
A more formal description of the model, inference procedures and simulations described here can be found in the electronic supplementary material.
3. Results and discussion
Reconstructions of 600 simulated outbreaks reveal that the method described here accurately recovers most parameters regardless of sampling intensity or model misspecification (electronic supplementary material, table S2). As can be expected, reconstruction of transmission events is sensitive to the informative priors used for the incubation and infectious periods (electronic supplementary material, table S3). This limits the suitability of the approach to diseases where the epidemiology is reasonably well known. The reconstruction of direct transmissions remains fairly accurate regardless of sampling intensity (mean posterior probability of true transmission events more than 0.73; electronic supplementary material, table S3) and actually increases in accuracy when sampling intensity decreases. Reconstruction of transmissions involving unobserved cases is moderately accurate at high sampling intensities, but becomes increasingly unreliable when 50% or fewer of the cases in the sampling region are sampled. At these sampling intensities, the post-processing algorithm cannot accurately distinguish between indirect and exogenous connections, which in turn also leads to a significant underestimation of the total number of cases (electronic supplementary material, table S4). At high to moderate sampling intensities (three-quarter to two-thirds of all cases in the sampled area), however, the 95% posterior interval (PI) inferred for the total population size covers the true value in more than 97% of cases under both the conservative and liberal specifications of the model.
Between 1 March 2010 and 8 June 2011, 195 rabies virus-positive cases were detected in KZN. The majority of these cases occurred close to densely populated areas, often in the peri-urban townships surrounding cities and large towns (electronic supplementary material, figure S1). A 760 nucleotide fragment spanning the highly variable G-L intergenic region was sequenced from 190 of these samples (electronic supplementary material, table S1). Despite the small spatial and temporal scale, the overall mean distance between the 189 canid-associated rabies virus sequences generated was 8.42 nucleotides. However, many clusters of identical sequences exist, and the phylogenetic divergence was not sufficient to generate a well-resolved phylogeny (electronic supplementary material, figure S2).
The transmission trees linking cases were estimated using 176 canid-associated rabies cases for which detailed epidemiological data were available (electronic supplementary material, table S1). When considering only direct transmissions, there were several independent chains of transmission and many transmissions inferred to have taken place over long distances (figure 2 and electronic supplementary material, figure S3). The mean distance between the most probable directly connected cases was 14.9 km (0.025- and 0.975-quantiles: 0.0 and 56.1 km; electronic supplementary material, figure S3). This was despite the use of narrow prior distributions for the parameters governing the durations of infections, which would tend to minimize the distance between directly connected cases in favour of indirect or exogenous connections instead. Occasional long-distance transmissions in this region, particularly along the major highways that follow the KZN coast, have been identified before (based on phylogenetic patterns) and were ascribed to motorized transportation of dogs [28]. Road distances have also been shown to be a better predictor of rabies dissemination than absolute distances in northern Africa [29]. The long distances and short time-periods between cases in the transmission tree (electronic supplementary material, figures S3 and S4) provide further evidence for motorized transportation of infected dogs, but such transmissions were not restricted to any one area and instead appear to be a common feature of the epidemiology of rabies in this area. This might be owing to the high prevalence of circular human migration and migrant labour in many parts of KZN, with migrants visiting their rural households (and, it would seem, taking their dogs with them) on a regular basis [30].
Figure 2.
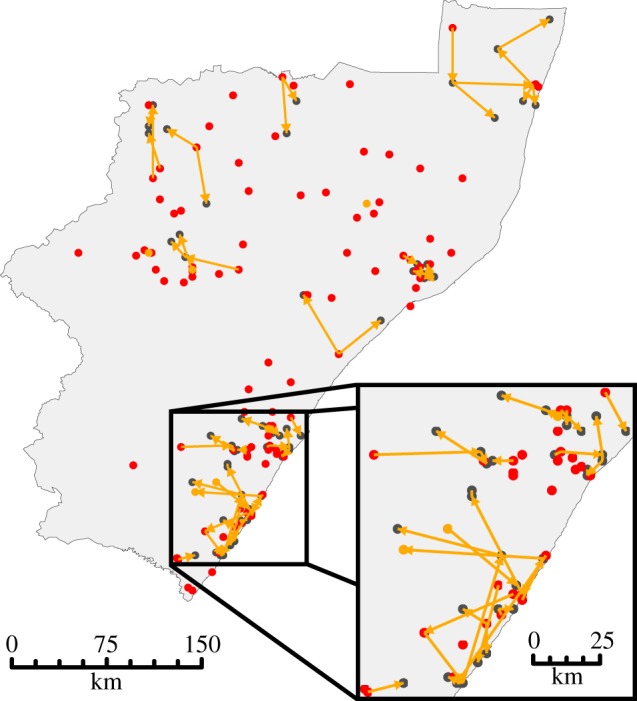
Transmission trees showing the direct pairwise transmissions with highest posterior probabilities. Transmission links between cases are represented by orange arrows. Red dots represent cases for which no direct ancestor was detected and black dots represent all other cases. The inset shows an enlarged view of connections in the southern coast of KZN, where the majority of cases were detected.
The majority of cases could not be linked through direct transmissions—69 (95% PI: 60–79) direct transmissions were identified, while unsampled sources were the most likely link for the remaining 107 (95% PI: 97–117) cases (electronic supplementary material, figure S5). The conservative specification of the post-processing algorithm identified a further 37 (95% PI: 27–47) indirect transmission links over the 15 month study period, while the liberal version of the algorithm identified 67 (95% PI: 57–78) indirect transmissions (figure 3). Sixteen cases were assigned different indirect ancestors by the two specifications, while a further 35 were interpreted as having an exogenous source by the conservative specification, but were assigned indirect ancestors by the liberal specification. There are no obvious similarities between cases assigned different ancestors by the two specifications, with no evidence of either phylogenetic clustering (assessed using Moran's I to measure autocorrelation to inverse phylogenetic distances between cases, p-value of 0.16 when the null hypothesis is no clustering) or spatial clustering (assessed using a spatial scan statistic with a null hypothesis that there is no more clustering among cases interpreted differently than among cases in general; p-value of 0.69 for the best supported cluster) [31–33]. The same was true for cases interpreted as having an exogenous source by one specification but not the other, with no evidence of either phylogenetic (p-value = 0.86) or spatial clustering (p-value = 0.08 for the best supported cluster).
Figure 3.
Pairwise transmissions with the highest posterior probabilities in each quarter of the sampled period, including indirect transmissions. Black dots represent all cases since the start of the sampling period, while red dots represent cases appearing in that quarter that have an exogenous source. Orange arrows represent direct transmission events. Blue arrows represent indirect transmissions inferred using the conservative (a) and liberal specification (b) of the post-processing algorithm. Note that detected cases (black dots) are displayed cumulatively. Q1–Q4: first to fourth quarter of the sampling period.
When considering both direct and indirect connections, there are many separate, unjoined transmission trees (electronic supplementary material, figure S6). For the most probable connections under both the conservative and liberal specifications of our algorithm, these transmission trees can be grouped into eight distinct spatial clusters. Transmission between different spatial clusters was rare—we detected only one such transmission with the conservative specification of the algorithm, and 10 such transmissions with the liberal specification. In addition, such transmissions do not appear to seed substantial additional numbers of cases, as only one instance of onward spread in the new cluster was detected under either specification, causing just one additional case in both instances. Interestingly, four of the inter-cluster transmissions identified under the liberal specification involved transmission from one cluster to another and then back to more-or-less the same location, before onward transmission in the original cluster, further supporting the hypothesis of migrants moving dogs back-and-forth between their urban and rural homes.
To gain a better understanding of the surveillance failures leading to the high number of indirect connections detected, we estimated the true number of cases occurring in the study area. This yielded a posterior median estimate of 389 cases (95% PI: 260–881) using the conservative specification of the post-processing algorithm, and 195 cases (95% PI: 182–298) using the liberal specification, over the 15 month study period (electronic supplementary material, figure S7). Our analyses of simulated datasets show that this mark–recapture approach is only accurate at fairly high sampling intensities, owing to difficulties in distinguishing between indirect and exogenous transmissions, and we note that the 95% PI of the number of recaptured lineages under the conservative specification is fairly wide (electronic supplementary material, figure S8). However, direct transmissions are accurately identified regardless of sampling intensity (electronic supplementary material, table S2), and in this dataset the conservative algorithm identified almost all infections involving unsampled individuals as exogenous transmissions, while the liberal algorithm identified most of these infections as indirect transmissions. Thus, the conservative algorithm minimized the number of recaptured lineages, while the liberal algorithm maximized it, which means the inferred population sizes can be interpreted as a lower and upper bound of the true value. As the herpestid-associated genetic variant of rabies virus is rare in KZN, the five cases which could not be sequenced were most likely representatives of the canid-associated variant. Thus, surveillance detected 194 cases of infection with the canid-associated variant, or between 49.87 and 99.49% of all canid-associated cases (based on the posterior medians of the conservative and liberal specifications, respectively). Such high detection rates are exceptional for rabies [34] and need further confirmation by contact tracing. However, surveillance effort (measured as the number of samples submitted per month) was fairly constant over the study period while incidence concurrently declined, suggesting that the ongoing intensive control programme is effectively driving rabies towards elimination, which could account for the low total number of cases inferred from this analysis. The areas where cases are still being missed can be deduced from our identification of indirect links (figure 3), providing a powerful tool for improving detection rates which would be particularly important if rabies is indeed close to being eliminated in this province.
4. Conclusion
To successfully control rabies and other endemic diseases in a changing landscape, a detailed understanding of its spatial epidemiology is required. The method described here allows for the detailed reconstruction of the transmission events of endemic infectious diseases, providing information that can be used both in designing more efficient control strategies and to measure and improve the quality of surveillance programmes. Importantly, key parameters could be recovered accurately regardless of sampling intensity.
The long distances characterizing many internal transmissions point to a significant anthropogenic influence on the epidemiology of rabies in KZN, the causes of which require further study. Despite these long-distance transmissions, clear spatial groupings could be discerned (electronic supplementary material, figure S6). In addition, the frequent long-distance transmissions cause most of these spatial clusters to consist of a relatively small core area and numerous surrounding cases (figure 3). Thus, identifying the connections of surrounding cases to specific clusters enables more directed vaccination, where targeting the smaller core areas would allow control of rabies over large areas. Identifying the spatial scale at which independent control strategies can be applied means it is possible to replace the thin spread of limited resources across the province with intense, focused campaigns that move across the province on an annual basis. Also crucial to the success of any disease elimination effort is effective surveillance. By identifying the true state of surveillance as well as the areas where cases are being missed from existing, routinely collected data, the method described here can be used as a starting point to investigate the causes of poor surveillance in specific parts of the region of concern.
By applying the methods described here to data from multiple years, important information will be revealed about how to iteratively improve surveillance and adapt rabies control strategies by identifying areas to be prioritized during annual vaccination campaigns. In addition, these methods can easily be adapted to other endemic diseases, and the high mutation rate of other RNA viruses makes them ideal candidates for this approach. Particularly encouraging is the fact that the small genome region sequenced here provided sufficient resolution for this analysis, making the generation of adequate data for large numbers of cases feasible even in resource-poor areas.
Acknowledgements
The authors wish to thank the staff of the Allerton Veterinary Laboratory of the KZN Department of Agriculture and Environmental Affairs for providing samples, and Roman Biek and Joseph Hughes for critical discussions. They also wish to thank two anonymous reviewers, whose contributions improved the manuscript.
Data accessibility
Funding statement
N.M. was financially supported by the NRF (grant no. 74606), the PRF (grant no. 11/85) and the University of Pretoria Postgraduate Study Abroad Programme. K.H. was supported by the Wellcome Trust (grant no. 095787/Z/11/Z). D.T.H. and S.T. were financially supported by the UK Medical Research Council (MRC, grant no. G0901135). S.S. was financially supported by the ANR (grant EMILE). The sequencing was financially supported by the MRC (grant no. G0901135). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Ferguson NM, Keeling MJ, Edmunds WJ, Gani R, Grenfell BT, Anderson RM, Leach S. 2003. Planning for smallpox outbreaks. Nature 425, 681–685 (doi:10.1038/nature02007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keeling MJ, Woolhouse MEJ, May RM, Davies G, Grenfell BT. 2003. Modelling vaccination strategies against foot-and-mouth disease. Nature 421, 136–142 (doi:10.1038/nature01343) [DOI] [PubMed] [Google Scholar]
- 3.Lemey P, Rambaut A, Welch JJ, Suchard MA. 2010. Phylogeography takes a relaxed random walk in continuous space and time. Mol. Biol. Evol. 27, 1877–1885 (doi:10.1093/molbev/msq067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pybus OG, et al. 2012. Unifying the spatial epidemiology and molecular evolution of emerging epidemics. Proc. Natl Acad. Sci. USA 109, 15 066–15 071 (doi:10.1073/pnas.1206598109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. 2002. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 161, 1307–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ypma RJF, Bataille AMA, Stegeman A, Koch G, Wallinga J, Van Ballegooijen WM. 2012. Unravelling transmission trees of infectious diseases by combining genetic and epidemiological data. Proc. R. Soc. B 279, 444–450 (doi:10.1098/rspb.2011.0913) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morelli MJ, Thébaud G, Chadœuf J, King DP, Haydon DT, Soubeyrand S. 2012. A Bayesian inference framework to reconstruct transmission trees using epidemiological and genetic data. PLoS Comput. Biol. 8, e1002768 (doi:10.1371/journal.pcbi.1002768) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ypma RJF, Jonges M, Bataille A, Stegeman A, Koch G, Van Boven M, Koopmans M, Van Ballegooijen WM, Wallinga J. 2013. Genetic data provide evidence for wind-mediated transmission of highly pathogenic avian influenza. J. Infect. Dis. 207, 730–735 (doi:10.1093/infdis/jis757) [DOI] [PubMed] [Google Scholar]
- 9.Cottam EM, Thébaud G, Wadsworth J, Gloster J, Mansley L, Paton DJ, King DP, Haydon DT. 2008. Integrating genetic and epidemiological data to determine transmission pathways of foot-and-mouth disease virus. Proc. R. Soc. B 275, 887–895 (doi:10.1098/rspb.2007.1442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ypma RJF, Van Ballegooijen WM, Wallinga J. 2013. Relating phylogenetic trees to transmission trees of infectious disease outbreaks. Genetics 195, 1055–1062 (doi:10.1534/genetics.113.154856) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization 2002. World survey of rabies number 35 for the year 1999. Geneva, Switzerland: World Health Organization [Google Scholar]
- 12.Grenfell BT, Pybus OG, Gog JR, Wood JLN, Daly JM, Mumford JA, Holmes EC. 2004. Unifying the epidemiological and evolutionary dynamics of pathogens. Science 303, 327–332 (doi:10.1126/science.1090727) [DOI] [PubMed] [Google Scholar]
- 13.Biek R, Henderson JC, Waller LA, Rupprecht CE, Real LA. 2007. A high-resolution genetic signature of demographic and spatial expansion in epizootic rabies virus. Proc. Natl Acad. Sci. USA 104, 7993–7998 (doi:10.1073/pnas.0700741104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rupprecht CE, Hanlon CA, Hemachudha T. 2002. Rabies re-examined. Lancet Infect. Dis. 2, 327–343 (doi:10.1016/S1473-3099(02)00287-6) [DOI] [PubMed] [Google Scholar]
- 15.Hampson K, Dushoff J, Cleaveland S, Haydon DT, Kaare M, Packer C, Dobson A. 2009. Transmission dynamics and prospects for the elimination of canine rabies. PLoS Biol. 7, e1000053 (doi:10.1371/journal.pbio.1000053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charlton KM, Nadin-Davis S, Casey GA, Wandeler AI. 1997. The long incubation period in rabies: delayed progression of infection in muscle at the site of exposure. Acta Neuropathol. 94, 73–77 (doi:10.1007/s004010050674) [DOI] [PubMed] [Google Scholar]
- 17.Nel LH, Sabeta CT, Von Teichman B, Jaftha JB, Rupprecht CE, Bingham J. 2005. Mongoose rabies in southern Africa: a re-evaluation based on molecular epidemiology. Virus Res. 109, 165–173 (doi:10.1016/j.virusres.2004.12.003) [DOI] [PubMed] [Google Scholar]
- 18.Cleaveland S, Kaare M, Knobel D, Laurenson MK. 2006. Canine vaccination: providing broader benefits for disease control. Vet. Microbiol. 117, 43–50 (doi:10.1016/j.vetmic.2006.04.009) [DOI] [PubMed] [Google Scholar]
- 19.Lembo T, et al. 2011. Renewed global partnerships and redesigned roadmaps for rabies prevention and control. Vet. Med. Int. 2011, 1–18 (doi:10.4061/2011/923149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean DJ, Abelseth MK, Atanasiu P. 1996. The fluorescent antibody test. In Laboratory techniques in rabies (eds Meslin FX, Kaplan MM, Koprowski H.), pp. 88–95, 4th edn Geneva, Switzerland: World Health Organization [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (doi:10.1016/S0022-2836(05)80360-2) [DOI] [PubMed] [Google Scholar]
- 22.Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinf. 9, 286–298 (doi:10.1093/bib/bbn013) [DOI] [PubMed] [Google Scholar]
- 23.Tordo N, Poch O, Ermine A, Keith G, Rougeon F. 1988. Completion of the rabies virus genome sequence determination: highly conserved domains among the L (polymerase) proteins of unsegmented negative-strand RNA viruses. Virology 165, 565–576 (doi:10.1016/0042-6822(88)90600-9) [DOI] [PubMed] [Google Scholar]
- 24.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (doi:10.1093/molbev/msr121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Austerlitz F, Dick CW, Dutech C, Klein EK, Oddou-Muratorio S, Smouse PE, Sork VL. 2004. Using genetic markers to estimate the pollen dispersal curve. Mol. Ecol. 13, 937–954 (doi:10.1111/j.1365-294X.2004.02100.x) [DOI] [PubMed] [Google Scholar]
- 26.Kimura M. 1981. Estimation of evolutionary distances between homologous nucleotide sequences. Proc. Natl Acad. Sci. USA 78, 454–458 (doi:10.1073/pnas.78.1.454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashkenazy H, Penn O, Doron-Faigenboim A, Cohen O, Cannarozzi G, Zomer O, Pupko T. 2012. FastML: a web server for probabilistic reconstruction of ancestral sequences. Nucleic Acids Res. 40, W580–W584 (doi:10.1093/nar/gks498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coetzee P, Nel LH. 2007. Emerging epidemic dog rabies in coastal South Africa: a molecular epidemiological analysis. Virus Res. 126, 186–195 (doi:10.1016/j.virusres.2007.02.020) [DOI] [PubMed] [Google Scholar]
- 29.Talbi C, et al. 2010. Phylodynamics and human-mediated dispersal of a zoonotic virus. PLoS Pathog. 6, e1001166 (doi:10.1371/journal.ppat.1001166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posel D, Marx C. 2013. Circular migration: a view from destination households in two urban informal settlements in South Africa. J. Dev. Stud. 49, 819–831 (doi:10.1080/00220388.2013.766717) [Google Scholar]
- 31.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (doi:10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 32.Gittleman JL, Kot M. 1990. Adaptation: statistics and a null model for estimating phylogenetic effects. Syst. Biol. 39, 227–241 (doi:10.2307/2992183) [Google Scholar]
- 33.Kulldorff M, Nagarwalla N. 1995. Spatial disease clusters: detection and inference. Stat. Med. 14, 799–810 (doi:10.1002/sim.4780140809) [DOI] [PubMed] [Google Scholar]
- 34.Townsend SE, Lembo T, Cleaveland S, Meslin FX, Miranda ME, Putra AAG, Haydon DT, Hampson K. 2013. Surveillance guidelines for disease elimination: a case study of canine rabies. Comp. Immunol. Microbiol. Infect. Dis. 36, 249–261 (doi:10.1016/j.cimid.2012.10.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



