Abstract
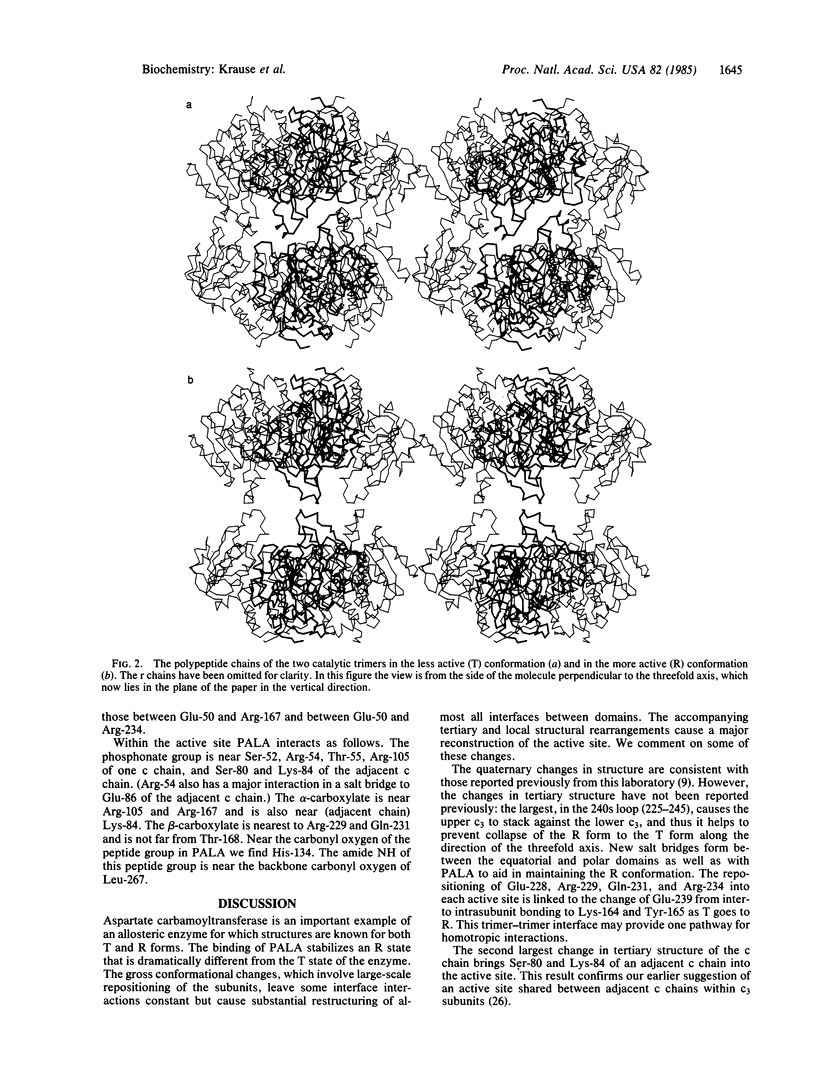
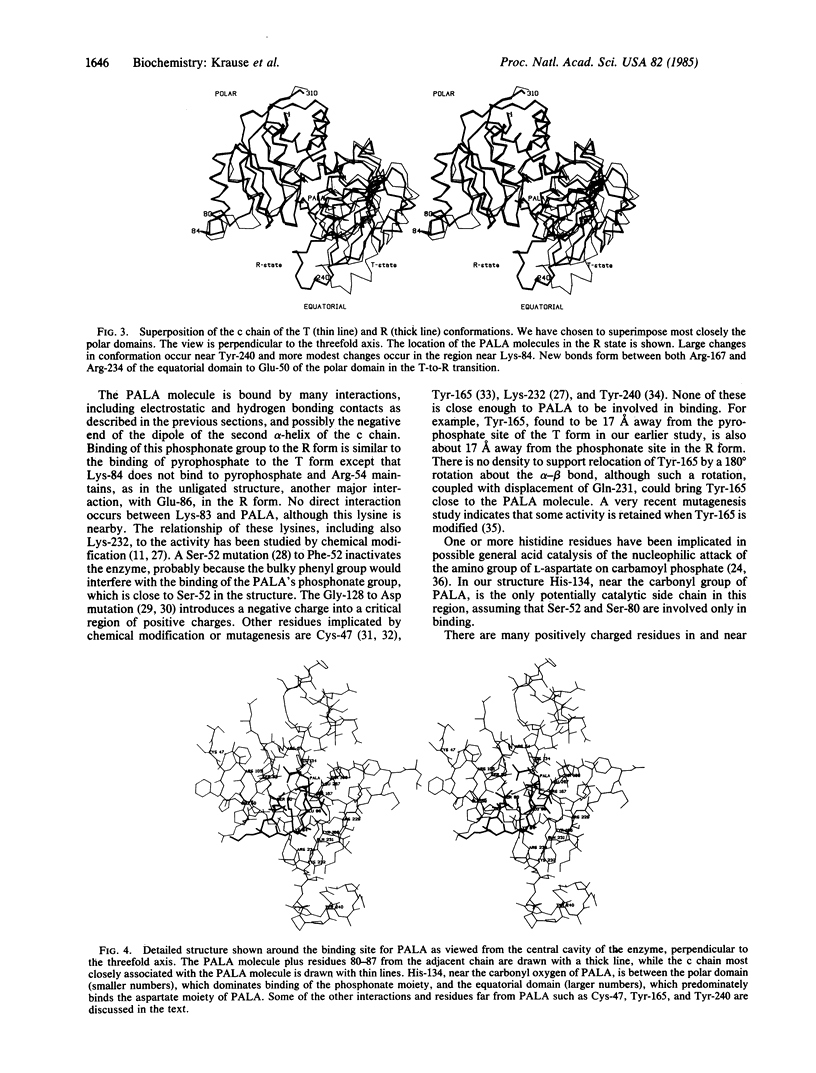
In an x-ray diffraction study by the isomorphous replacement method, the structure of the complex of aspartate carbamoyltransferase (EC 2.1.3.2) bound to the bisubstrate analogue N-(phosphonacetyl)-L-aspartate has been solved to 2.9-A resolution (R = 0.24). The large quaternary structural changes previously deduced by molecular replacement methods have been confirmed: the two catalytic trimers (c3) move apart by 12 A and mutually reorient by 10 degrees, and the regulatory dimers (r2) reorient each about its twofold axis by about 15 degrees. In this, the T-to-R transition, new polar interactions develop between equatorial domains of c chains and the Zn domain of r chains. Within the c chain the two domains, one binding the phosphonate moiety (polar) and the other binding the aspartate moiety (equatorial) of the inhibitor N-(phosphonacetyl)-L-aspartate, move closer together. The Lys-84 loop makes a large relocation so that this residue and Ser-80 bind to the inhibitor of an adjacent catalytic chain within c3. A very large change in tertiary structure brings the 230-245 loop nearer the active site, allowing Arg-229 and Gln-231 to bind to the inhibitor. His-134 is close to the carbonyl group of the inhibitor, and Ser-52 is adjacent to its phosphonate group. However, no evidence exists in the literature for phosphorylation of serine in the mechanism. Residues studied by other methods, including Cys-47, Tyr-165, Lys-232, and Tyr-240, are too far from the inhibitor to have a direct interaction.
Full text
PDF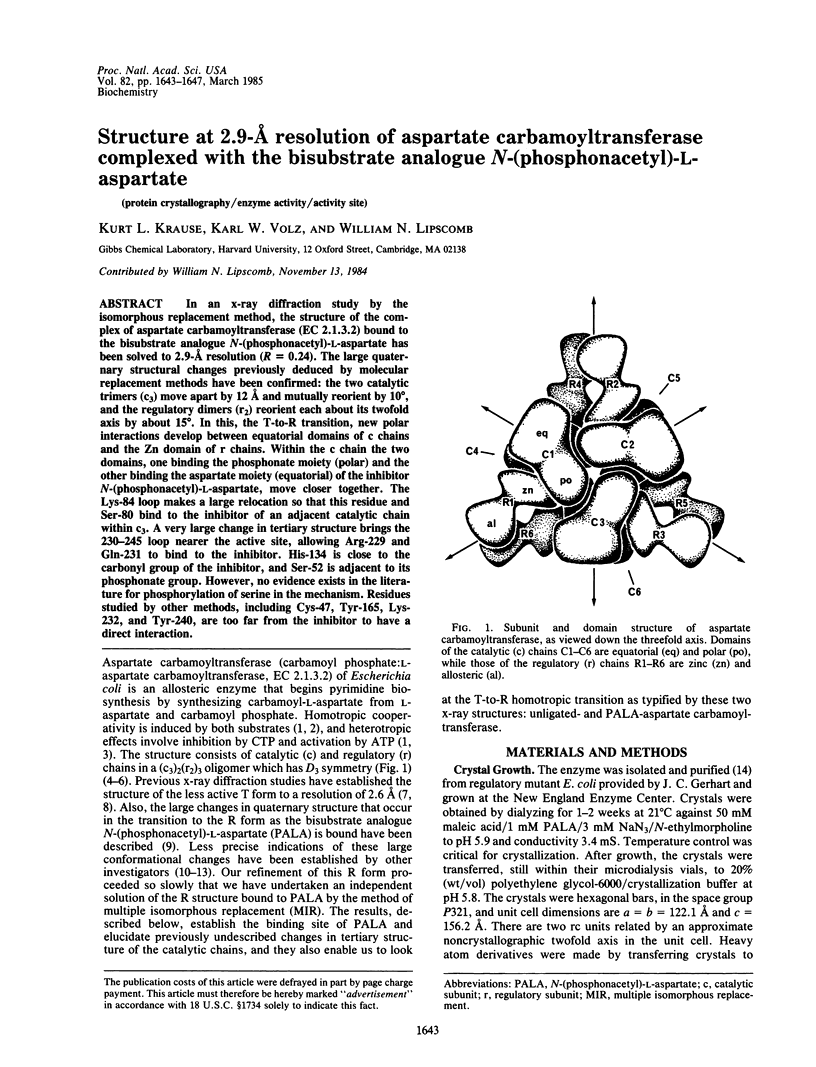


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bethell M. R., Smith K. E., White J. S., Jones M. E. Carbamyl phosphate: an allosteric substrate for aspartate transcarbamylase of Escherichia coli. Proc Natl Acad Sci U S A. 1968 Aug;60(4):1442–1449. doi: 10.1073/pnas.60.4.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. D., Stark G. R. Aspartate transcarbamylase. Interaction with the transition state analogue N-(phosphonacetyl)-L-aspartate. J Biol Chem. 1971 Nov;246(21):6599–6605. [PubMed] [Google Scholar]
- Evans D. R., Lipscomb W. N. The modification of the catalytic chain sulfhydryl group of aspartate transcarbamylase with mercurinitrophenols. J Biol Chem. 1979 Nov 10;254(21):10679–10685. [PubMed] [Google Scholar]
- GERHART J. C., PARDEE A. B. ASPARTATE TRANSCARBAMYLASE, AN ENZYME DESIGNED FOR FEEDBACK INHIBITION. Fed Proc. 1964 May-Jun;23:727–735. [PubMed] [Google Scholar]
- GERHART J. C., PARDEE A. B. The enzymology of control by feedback inhibition. J Biol Chem. 1962 Mar;237:891–896. [PubMed] [Google Scholar]
- Gerhart J. C., Holoubek H. The purification of aspartate transcarbamylase of Escherichia coli and separation of its protein subunits. J Biol Chem. 1967 Jun 25;242(12):2886–2892. [PubMed] [Google Scholar]
- Greenwell P., Jewett S. L., Stark G. R. Aspartate transcarbamylase from Escherichia coli. The use of pyridoxal 5'-phosphate as a probe in the active site. J Biol Chem. 1973 Sep 10;248(17):5994–6001. [PubMed] [Google Scholar]
- Hampson R. K., Taylor M. K., Olson M. S. Regulation of the glycine cleavage system in the isolated perfused rat liver. J Biol Chem. 1984 Jan 25;259(2):1180–1185. [PubMed] [Google Scholar]
- Heyde E. A unifying concept for the active site region in aspartate transcarbamylase. Biochim Biophys Acta. 1976 Nov 8;452(1):81–88. doi: 10.1016/0005-2744(76)90059-0. [DOI] [PubMed] [Google Scholar]
- Honzatko R. B., Crawford J. L., Monaco H. L., Ladner J. E., Ewards B. F., Evans D. R., Warren S. G., Wiley D. C., Ladner R. C., Lipscomb W. N. Crystal and molecular structures of native and CTP-liganded aspartate carbamoyltransferase from Escherichia coli. J Mol Biol. 1982 Sep 15;160(2):219–263. doi: 10.1016/0022-2836(82)90175-9. [DOI] [PubMed] [Google Scholar]
- Honzatko R. B., Lipscomb W. N. Interactions of phosphate ligands with Escherichia coli aspartate carbamoyltransferase in the crystalline state. J Mol Biol. 1982 Sep 15;160(2):265–286. doi: 10.1016/0022-2836(82)90176-0. [DOI] [PubMed] [Google Scholar]
- Howlett G. J., Schachman H. K. Allosteric regulation of aspartate transcarbamoylase. Changes in the sedimentation coefficient promoted by the bisubstrate analogue N-(phosphonacetyl)-L-aspartate. Biochemistry. 1977 Nov 15;16(23):5077–5083. doi: 10.1021/bi00642a021. [DOI] [PubMed] [Google Scholar]
- Ke H. M., Honzatko R. B., Lipscomb W. N. Structure of unligated aspartate carbamoyltransferase of Escherichia coli at 2.6-A resolution. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4037–4040. doi: 10.1073/pnas.81.13.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempe T. D., Stark G. R. Pyridoxal 5'-phosphate, a fluorescent probe in the active site of aspartate transcarbamylase. J Biol Chem. 1975 Sep 10;250(17):6861–6869. [PubMed] [Google Scholar]
- Ladner J. E., Kitchell J. P., Honzatko R. B., Ke H. M., Volz K. W., Kalb A. J., Ladner R. C., Lipscomb W. N. Gross quaternary changes in aspartate carbamoyltransferase are induced by the binding of N-(phosphonacetyl)-L-aspartate: A 3.5-A resolution study. Proc Natl Acad Sci U S A. 1982 May;79(10):3125–3128. doi: 10.1073/pnas.79.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landfear S. M., Evans D. R., Lipscomb W. N. Elimination of cooperativity in aspartate transcarbamylase by nitration of a single tyrosine residue. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2654–2658. doi: 10.1073/pnas.75.6.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritzen A. M., Landfear S. M., Lipscomb W. N. Inactivation of the catalytic subunit of aspartate transcarbamylase by nitration with tetranitromethane. J Biol Chem. 1980 Jan 25;255(2):602–607. [PubMed] [Google Scholar]
- Lauritzen A. M., Lipscomb W. N. Modification of three active site lysine residues in the catalytic subunit of aspartate transcarbamylase by D- and L-bromosuccinate. J Biol Chem. 1982 Feb 10;257(3):1312–1319. [PubMed] [Google Scholar]
- Monaco H. L., Crawford J. L., Lipscomb W. N. Three-dimensional structures of aspartate carbamoyltransferase from Escherichia coli and of its complex with cytidine triphosphate. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5276–5280. doi: 10.1073/pnas.75.11.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody M. F., Vachette P., Foote A. M. Changes in the x-ray solution scattering of aspartate transcarbamylase following the allosteric transition. J Mol Biol. 1979 Oct 9;133(4):517–532. doi: 10.1016/0022-2836(79)90405-4. [DOI] [PubMed] [Google Scholar]
- Muirhead H., Cox J. M., Mazzarella L., Perutz M. F. Structure and function of haemoglobin. 3. A three-dimensional fourier synthesis of human deoxyhaemoglobin at 5.5 Angstrom resolution. J Mol Biol. 1967 Aug 28;28(1):117–156. doi: 10.1016/s0022-2836(67)80082-2. [DOI] [PubMed] [Google Scholar]
- Pastra-Landis S. C., Evans D. R., Lipscomb W. N. The effect of pH on the cooperative behavior of aspartate transcarbamylase from Escherichia coli. J Biol Chem. 1978 Jul 10;253(13):4624–4630. [PubMed] [Google Scholar]
- Schachman H. K., Pauza C. D., Navre M., Karels M. J., Wu L., Yang Y. R. Location of amino acid alterations in mutants of aspartate transcarbamoylase: Structural aspects of interallelic complementation. Proc Natl Acad Sci U S A. 1984 Jan;81(1):115–119. doi: 10.1073/pnas.81.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanaman T. C., Stark G. R. A study of the sulfhydryl groups of the catalytic subunit of Escherichia coli aspartate transcarbamylase. The use of enzyme--5-thio-2-nitrobenzoate mixed disulfides as intermediates in modifying enzyme sulfhydryl groups. J Biol Chem. 1970 Jul 25;245(14):3565–3573. [PubMed] [Google Scholar]
- Wall K. A., Flatgaard J. E., Schachman H. K., Gibbons I. Purification and characterization of a mutant aspartate transcarbamoylase lacking enzyme activity. J Biol Chem. 1979 Dec 10;254(23):11910–11916. [PubMed] [Google Scholar]
- Wall K. A., Schachman H. K. Primary structure and properties of an inactive mutant aspartate transcarbamoylase. J Biol Chem. 1979 Dec 10;254(23):11917–11926. [PubMed] [Google Scholar]
- Weber K. New structural model of E. coli aspartate transcarbamylase and the amino-acid sequence of the regulatory polypeptide chain. Nature. 1968 Jun 22;218(5147):1116–1119. doi: 10.1038/2181116a0. [DOI] [PubMed] [Google Scholar]
- Wiley D. C., Evans D. R., Warren S. G., McMurray C. H., Edwards B. F., Franks W. A., Lipscomb W. N. The 5.5 Angstrom resolution structure of the regulatory enzyme, asparate transcarbamylase. Cold Spring Harb Symp Quant Biol. 1972;36:285–290. doi: 10.1101/sqb.1972.036.01.038. [DOI] [PubMed] [Google Scholar]
- Wiley D. C., Lipscomb W. N. Crystallographic determination of symmetry of aspartate transcarbamylase. Nature. 1968 Jun 22;218(5147):1119–1121. doi: 10.1038/2181119a0. [DOI] [PubMed] [Google Scholar]



