Abstract
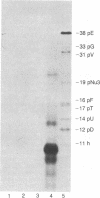
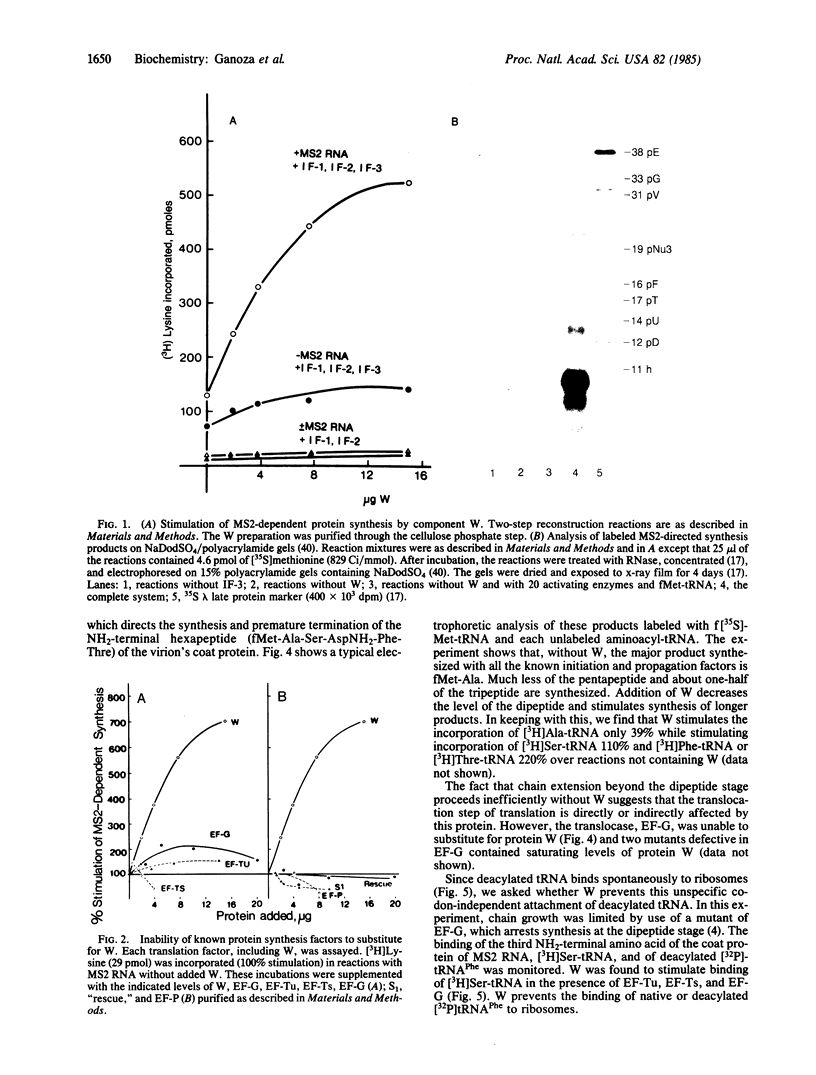
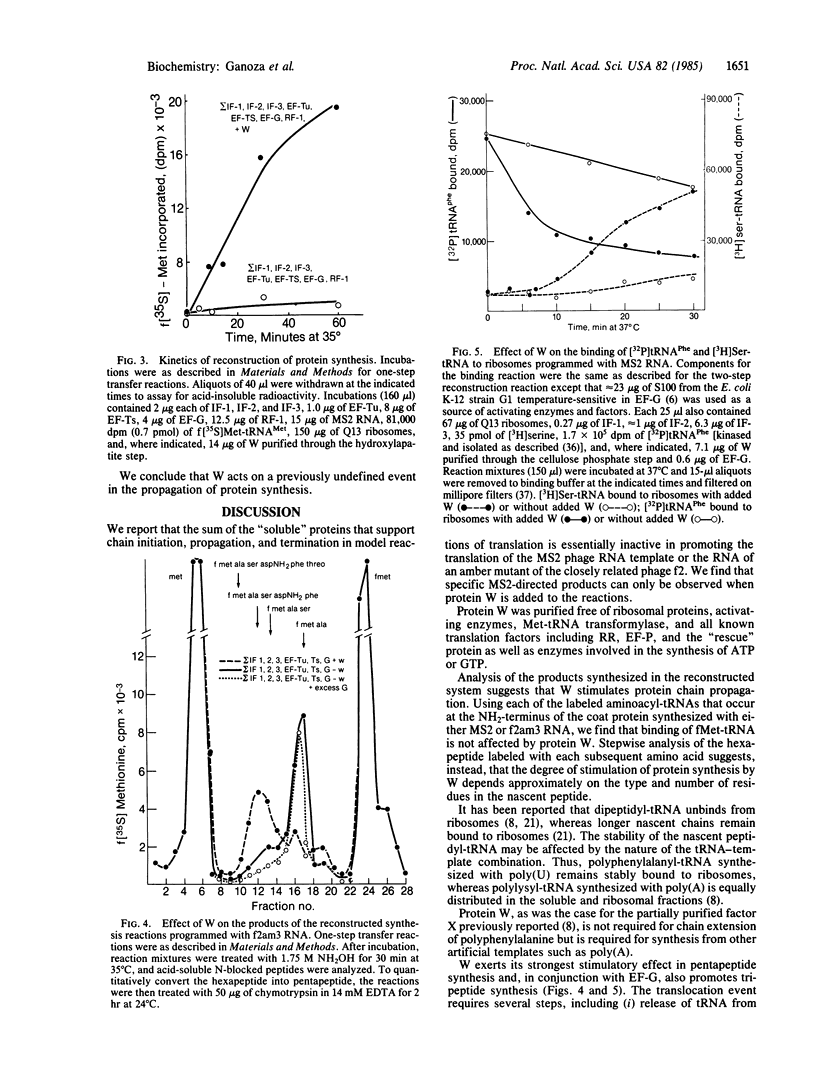
To study the mechanism of translation we have attempted to reconstruct the process from purified components. Protein synthesis was programmed by the RNAs of wild-type or amber mutants of bacteriophages f2 or MS2. Translation programmed by MS2 or f2am3 RNA does not occur using ribosomes, precharged aminoacyl-tRNAs, and the sum of the purified proteins involved in initiation (initiation factors; IF-1, IF-2, and IF-3), propagation (elongation factors; EF-Tu, EF-Ts, and EF-G) and termination (release factors; RF-1 or RF-2) of protein synthesis. The requirement for a protein called W was demonstrated. Protein W was purified free of all translation factors, activating enzymes, and other proteins such as the RR, "rescue," and EF-P implicated in translation. The stimulation of propagation by W depended on the position of the amino acid residue to be added in the synthesis of the NH2-terminal hexapeptide of the coat protein. In the reconstructed system, with the sum of all translation factors but in the absence of W, only dipeptides and smaller quantities of tripeptides were synthesized under the direction of f2am3 RNA. W stimulated the synthesis of the hexapeptide, fMet-Ala-Ser-AspNH2-Phe-Thr directed by this RNA. In addition, W stimulated ejection of non-cognate tRNAs that bind to ribosomal particles.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arai K. I., Kawakita M., Kaziro Y. Studies on polypeptide elongation factors from Escherichia coli. II. Purification of factors Tu-guanosine diphosphate, Ts, and Tu-Ts, and crystallization of Tu-guanosine diphosphate and Tu-Ts. J Biol Chem. 1972 Nov 10;247(21):7029–7037. [PubMed] [Google Scholar]
- Capecchi M. R. A rapid assay for polypeptide chain termination. Biochem Biophys Res Commun. 1967 Sep 7;28(5):773–778. doi: 10.1016/0006-291x(67)90384-1. [DOI] [PubMed] [Google Scholar]
- Cenatiempo Y., Robakis N., Reid B. R., Weissbach H., Brot N. In vitro expression of Escherichia coli ribosomal protein L 10 gene: tripeptide synthesis as a measure of functional mRNA. Arch Biochem Biophys. 1982 Oct 15;218(2):572–578. doi: 10.1016/0003-9861(82)90381-2. [DOI] [PubMed] [Google Scholar]
- Cuzin F., Kretchmer N., Greenberg R. E., Hurwitz R., Chapeville F. Enzymatic hydrolysis of N-substituted aminoacyl-tRNA. Proc Natl Acad Sci U S A. 1967 Nov;58(5):2079–2086. doi: 10.1073/pnas.58.5.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganoza M. C., Barraclough N., Wong J. T. Purification and properties of an N-formylmethionyl-tRNA hydrolase. Eur J Biochem. 1976 Jun 1;65(2):613–622. doi: 10.1111/j.1432-1033.1976.tb10379.x. [DOI] [PubMed] [Google Scholar]
- Ganoza M. C., Buckingham K., Hader P., Neilson T. Effect of base sequence on in vitro protein-chain termination. J Biol Chem. 1984 Nov 25;259(22):14101–14104. [PubMed] [Google Scholar]
- Ganoza M. C., Fox J. L. Isolation of a soluble factor needed for protein synthesis with various messenger ribonucleic acids other than poly(U). J Biol Chem. 1974 Feb 25;249(4):1037–1043. [PubMed] [Google Scholar]
- Ganoza M. C. The Ayerst Award Lecture 1976. Novel factors in protein biosynthesis. Can J Biochem. 1977 Apr;55(4):267–281. doi: 10.1139/o77-038. [DOI] [PubMed] [Google Scholar]
- Ganoza M. C., Vandermeer J., Debreceni N., Phillips S. L. Mechanism of protein chain termination: further characterization of a mutant defective in a new protein synthesis factor. Proc Natl Acad Sci U S A. 1973 Jan;70(1):31–35. doi: 10.1073/pnas.70.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B. R., Chládek S., Ganoza M. C. Peptide bond formation stimulated by protein synthesis factor EF-P depends on the aminoacyl moiety of the acceptor. Eur J Biochem. 1979 Jun;97(1):23–28. doi: 10.1111/j.1432-1033.1979.tb13081.x. [DOI] [PubMed] [Google Scholar]
- Glick B. R., Ganoza M. C. Identification of a soluble protein that stimulates peptide bond synthesis. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4257–4260. doi: 10.1073/pnas.72.11.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B. R., Green R. M., Ganoza M. C. Purification and Best Department of Medical Research, University of Toronto, Ont., Canada. Can J Biochem. 1979 Jun;57(6):749–757. [PubMed] [Google Scholar]
- Herrington M. B., Doherty M. J., Ganoza M. C. Translation of natural mRNAs in vitro by extracts of a mutant in protein synthesis. Nature. 1975 Aug 21;256(5519):678–679. doi: 10.1038/256678a0. [DOI] [PubMed] [Google Scholar]
- Hershey J. W., Yanov J., Johnston K., Fakunding J. L. Purification and characterization of protein synthesis initiation factors IF1, IF2, and IF3 from Escherichia coli. Arch Biochem Biophys. 1977 Aug;182(2):626–638. doi: 10.1016/0003-9861(77)90543-4. [DOI] [PubMed] [Google Scholar]
- Hirashima A., Kaji A. Purification and properties of ribosome-releasing factor. Biochemistry. 1972 Oct 24;11(22):4037–4044. doi: 10.1021/bi00772a005. [DOI] [PubMed] [Google Scholar]
- Igarashi S. R17 RNA replicase. VI. A large scale preparation of R17 template RNA. J Biochem. 1975 Jun;77(6):1271–1275. [PubMed] [Google Scholar]
- Iwasaki K., Sabol S., Wahba A. J., Ochoa S. Translation of the genetic message. VII. Role of initiation factors in formation of the chain initiation complex with Escherichia coli ribosomes. Arch Biochem Biophys. 1968 May;125(2):542–547. doi: 10.1016/0003-9861(68)90612-7. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt E., Wittmann H. G. Ribosomal proteins. VII. Two-dimensional polyacrylamide gel electrophoresis for fingerprinting of ribosomal proteins. Anal Biochem. 1970 Aug;36(2):401–412. doi: 10.1016/0003-2697(70)90376-3. [DOI] [PubMed] [Google Scholar]
- Kung H. F., Chu F., Caldwell P., Spears C., Treadwell B. V., Eskin B., Brot N., Weissbach H. The mRNA-directed synthesis of the alpha0peptide of beta-galactosidase, ribosomal proteins L12 and L10, and elongation factor Tu, using purified translational factors. Arch Biochem Biophys. 1978 Apr 30;187(2):457–463. doi: 10.1016/0003-9861(78)90057-7. [DOI] [PubMed] [Google Scholar]
- Kung H. F., Treadwell B. V., Spears C., Tai P. C., Weissbach H. DNA-directed synthesis in vitro of beta-galactosidase: requirement for a ribosome release factor. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3217–3221. doi: 10.1073/pnas.74.8.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Menninger J. R. The accumulation as peptidyl-transfer RNA of isoaccepting transfer RNA families in Escherichia coli with temperature-sensitive peptidyl-transfer RNA hydrolase. J Biol Chem. 1978 Oct 10;253(19):6808–6813. [PubMed] [Google Scholar]
- NIRENBERG M., LEDER P. RNA CODEWORDS AND PROTEIN SYNTHESIS. THE EFFECT OF TRINUCLEOTIDES UPON THE BINDING OF SRNA TO RIBOSOMES. Science. 1964 Sep 25;145(3639):1399–1407. doi: 10.1126/science.145.3639.1399. [DOI] [PubMed] [Google Scholar]
- Nierhaus K. H. Structure, assembly, and function of ribosomes. Curr Top Microbiol Immunol. 1982;97:81–155. doi: 10.1007/978-3-642-68318-3_3. [DOI] [PubMed] [Google Scholar]
- Phillips S. L. Termination of messenger RNA translation in a temperature-sensitive mutant of Escherichia coli. J Mol Biol. 1971 Aug 14;59(3):461–472. doi: 10.1016/0022-2836(71)90310-x. [DOI] [PubMed] [Google Scholar]
- Rheinberger H. J., Nierhaus K. H. Testing an alternative model for the ribosomal peptide elongation cycle. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4213–4217. doi: 10.1073/pnas.80.14.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudland P. S., Klemperer H. G. A factor promoting the ejection of deacylated initiator transfer RNA from ribosomes. J Mol Biol. 1971 Oct 28;61(2):377–385. doi: 10.1016/0022-2836(71)90387-1. [DOI] [PubMed] [Google Scholar]
- Ryoji M., Berland R., Kaji A. Reinitiation of translation from the triplet next to the amber termination codon in the absence of ribosome-releasing factor. Proc Natl Acad Sci U S A. 1981 Oct;78(10):5973–5977. doi: 10.1073/pnas.78.10.5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle D. P., Haralson M. A., Ravel J. M. Initiation factor 3 requirement for the formation of initiation complexes with synthetic oligonucleotides. Biochem Biophys Res Commun. 1973 Mar 17;51(2):376–382. doi: 10.1016/0006-291x(73)91268-0. [DOI] [PubMed] [Google Scholar]
- Traub P., Nomura M. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc Natl Acad Sci U S A. 1968 Mar;59(3):777–784. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WALEY S. G., WATSON J. Trypsin-catalysed transpeptidations. Biochem J. 1954 Aug;57(4):529–538. doi: 10.1042/bj0570529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer J. P., Ganoza M. C. Purification and characterization of a new factor which restores protein synthesis in a conditionally lethal mutant of Escherichia coli. Eur J Biochem. 1975 May;54(1):229–237. doi: 10.1111/j.1432-1033.1975.tb04132.x. [DOI] [PubMed] [Google Scholar]