Abstract
Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) in association with CRISPR-associated (Cas) proteins constitutes a formidable defense system against mobile genetic elements in prokaryotes. In type I-C, the ribonucleoprotein surveillance complex comprises only three Cas proteins, namely, Cas5d, Csd1 and Csd2. Unlike type I-E that uses Cse3/CasE for metal-independent CRISPR RNA maturation, type I-C that lacks this deputes Cas5d to process the pre-crRNA. Here, we report the promiscuous DNase activity of Cas5d in presence of divalent metals. Remarkably, the active site that renders RNA hydrolysis may be tuned by metal to act on DNA substrates too. Further, the realization that Csd1 is a fusion of its functional homolog Cse1/CasA and Cse2/CasB forecasts that the stoichiometry of the constituents of the surveillance complex in type I-C may differ from type I-E. Although Csd2 seems to be inert, Csd1 too exhibits RNase and metal-dependent DNase activity. Thus, in addition to their proposed functions, the DNase activity of Cas5d and Csd1 may also enable them to be co-opted in adaptation and interference stages of CRISPR immunity wherein interaction with DNA substrates is involved.
INTRODUCTION
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) and CRISPR-associated (Cas) genes bestow an adaptive and heritable immune system to bacteria and archaea (1–4). This immune system targets the invading mobile genetic elements by acquiring a fragment of invading genome—referred as protospacer element—and inserting it into the CRISPR array, which is populated by tandem arrangement of redundant repeats and unique spacers (4). The spacer transcript is subsequently processed by endoribonucleases to produce the mature CRIPSR RNA (crRNA). In association with a set of Cas proteins, the crRNA directs the recognition and cleavage of the target genome via base complementarity (5–9).
The functions of CRISPR-Cas system can be broadly categorized under three stages: (i) acquisition of spacers from genome invaders, (ii) expression and maturation of small crRNA and (iii) interference of invading genetic elements. There are mechanistic differences in the way the aforementioned aspects of CRISPR-Cas system operate across microbial species and based on this, it has been categorized as type I, type II and type III (10). The maturation of pre-crRNA in type I is mediated by an endonuclease, namely, Cse3 in Escherichia coli/Thermus thermophilus HB8 (type I-E), Csy4 in Pseudomonas aeruginosa (type I-F) and Cas5d in Bacillus halodurans/T. thermophilus HB27 (type I-C). These are shown to recognize the structured form of repeat RNA and cleave at the base of the stem-loop (11–16), whereas the type III-specific Cas6 from Pyrococcus furiosus recognizes the unstructured form by wrapping around the repeat RNA (17–19). Notwithstanding the mode of RNA recognition that seems to vary between type I and III, the endoRNases seem to follow a metal-independent acid–base hydrolysis mechanism producing a 2′–3′ cyclic intermediate and the final product having 5′-OH and 3′-P ends (12,13,19). Further in type I, as exemplified in E. coli, Cse3/CasE seems to function in association with Cse1/CasA, Cse2/CasB, Cse4/CasC, Cas5e/CasD and crRNA to form a ribonucleoprotein assembly termed as Cascade (CRISPR-associated complex for antiviral defense) that eventually recruits Cas3, which performs the target cleavage (5,8,10,20–22). The maturation in type II occurs via an unusual mechanism facilitated by the combined action of Csn1/Cas9, RNaseIII and a tracr-RNA (9,23).
The crRNA maturation in type I-C seems to be unusual from the other type I systems. This subtype lacks the Cse3/CasE counterpart and instead its role is adopted by Cas5d. Cse3/CasE possesses duplicated RNA recognition motif (RRM) domain where the C-terminal RRM domain is involved in RNA recognition and N-terminal domain that harbors the catalytic residues participates in the cleavage (12,15). In contravention to this, the structure of Cas5d shows a single RRM domain, and despite this variation it appears to be adept at performing both the RNA recognition and cleavage (11,14,24). Like other subtype-specific endoRNases, Cas5d too processes the pre-crRNA in a metal-independent manner to produce the mature crRNA. However, the crRNA recognition was shown to be stronger toward the 3′-region of the stem (14). In E. coli, Cse3/CasE seems to process the pre-crRNA as part of the Cascade complex; however, the recent study hints at the possibility of Cas5d mediating this process as a standalone protein (11,14). In association with crRNA and the type I-C-specific Csd1 and Csd2, Cas5d is also shown to assemble into a Cascade-like complex (14). Intuitively, owing to the absence of Cse3/CasE, the copy number of the assembly components in type I-C is likely to differ from type I-E.
To gain more insight into type I-C-specific crRNA maturation, we probed for additional functionalities of the Cas proteins. Here, we report that Cas5d, in addition to being an endoRNase, also exhibits metal-dependent endoDNase activity. The active site center for DNase and RNase activity seems to be overlapping. Unexpectedly, Csd1 is identified to be a fusion protein of two subunits, namely, Cse1/CasA and Cse2/CasB. Further, Csd1 too exhibits endoRNase activity that is independent of metal and endoDNase activity that is promoted in presence of metal. Although there seems to be sequence- and structure-specific recognition of repeat RNA substrates, Cas5d and Csd1 apparently lack such specificity toward the DNA substrates. We have discussed the possible consequences in the light of these additional functionalities in the context of CRISPR immunity in type I-C system.
MATERIALS AND METHODS
Cloning, expression and purification
Genes encoding cas5d, csd1 and csd2 were amplified from B. halodurans genomic DNA using gene-specific primers (see Supplementary Table S1) with Pfu DNA polymerase (Fermentas). Amplicons of cas5d and csd1 were cloned into pQE2 using the restriction sites for NdeI and PstI and that of csd2 in modified pET23a using the restriction sites for NdeI and EcoRI (New England Biolabs). Constructs that were cloned in pQE2 harbors an N-terminal (His)6 tag, and the one in modified pET23a harbors a C-terminal strep tag. Point mutants (Y35F, K39A, H169A, W47F, W187F, Y46F, K116A, H117A, E4A, D56A, H98A and E100A) of Cas5d were generated by mega primer-based PCR method. All mutants were cloned in pQE2 except H98A and H117A, which were cloned in Ligation Independent Cloning (LIC) vector (Addgene ID: 29717) having a pET backbone and N-terminal StrepII tag. The cloned constructs were verified by sequencing. Two random mutations (G158R and Y162H) have been identified in Cas5d wild-type construct; however, these point mutations are located distantly from the catalytic triad and have no apparent effect on the nuclease activity. All the reported point mutants were generated on this genetic background.
Expression was performed in E. coli BL21 (DE3) by growing the cells in LB medium supplemented with ampicillin (100 μg/ml) at 37°C until OD at 600 nm reached 0.7. The temperature was then reduced to 20°C for 20 min, and protein expression was induced by the addition of 0.2 mM isopropyl β-D-1-thiogalactopyranoside followed by incubation at 20°C overnight. The cells were harvested by centrifugation and resuspended in buffer A containing 20 mM Tris–HCl (pH 8.0), 500 mM NaCl, 6 mM β-mercaptoethanol and 0.1 mM phenylmethanesulfonyl fluoride. After sonication, the lysate was clarified by centrifugation at 36 500g for 30 min. The supernatant was treated with RNase to remove any bound RNA and then loaded onto a 5 ml HiTrap IMAC HP column (GE Healthcare) pre-equilibrated with buffer B containing 20 mM Tris–HCl (pH 8.0), 500 mM NaCl and 6 mM β-mercaptoethanol. After washing the column with buffer C containing 20 mM Tris–HCl (pH 8.0), 500 mM NaCl, 6 mM β-mercaptoethanol and 40 mM imidazole, the bound protein was eluted using a linear gradient of imidazole (upto 500 mM) in buffer C. For strep-tagged proteins, the elution was carried out with buffer C that contained 2.5 mM d-desthiobiotin in place of imidazole. For some of the preparations of Cas5d and Csd1, anion-exchange chromatography was used wherein the protein was eluted in a linear gradient using 1 M NaCl. The eluted protein (see Supplementary Figure S16) was incubated with 10 mM EDTA for 1 h to remove the bound metal ions if any and then dialyzed against buffer D containing 20 mM Tris–HCl (pH 8.0), 200 mM NaCl and 6 mM β-mercaptoethanol. Subsequently, the proteins were aliquoted, snap frozen in liquid nitrogen and stored at −80°C until required.
Preparation of CRISPR repeat RNA and DNA
Pre-crRNA containing only the repeat sequence was chemically synthesized and labeled with a 3′-FAM (Integrated DNA Technologies (IDT)). The DNA sequences corresponding to the CRISPR repeat with a T7 promoter were synthesized from IDT and 3′-end labeled with fluorescein using deoxy terminal transferase (New England Biolabs). The pQE2 or pEGFP plasmid was used as circular DNA, and pQE2 linearized with KpnI served as linear substrate. Single-stranded circular M13mp18 phage DNA was obtained from New England Biolabs.
Nuclease activity assays
All pre-crRNA processing reactions were performed at 37°C for 1 h. Time-dependent studies were done at room temperature. The 3′-FAM-labeled pre-crRNA repeat at 0.2 µM concentration was incubated with Cas5d (2 µM) in 20 mM Tris–HCl (pH 8), 100 mM KCl and 6 mM β-mercaptoethanol. RNase activity was also tested in the presence of 10 mM Mg2+. Cleavage products were analyzed on 15% (w/v) denaturing urea PAGE.
DNase activity assays were performed with double stranded, linear or circular and single stranded DNA at 37°C for 1 h in the buffer containing 20 mM Tris–HCl (pH 8.0), 100 mM KCl, 10 mM MgCl2 and 2 μM of the respective protein. Time-dependent nuclease activity was performed at 37°C, and samples were taken out at the indicated time intervals. The reaction was stopped using 50 mM EDTA (pH 8.0) and the products were analyzed on 0.8% agarose gel and visualized by ethidium bromide staining. Metal-dependent DNase activity was measured in the presence of 10 mM divalent cation (Mg2+, Mn2+, Zn2+, Ca2+ and Ni2+) and 50 mM EDTA (if indicated).
Analysis of metal binding sites
The nature of metal binding is probed using Hill plot analysis in the absence of DNA using the following form of the equation:
| (1) |
where nH is the Hill coefficient and Kd is the apparent dissociation constant. ΔF = F0−F, where F0 denotes the fluorescence intensity in the absence of Mg2+, F is the fluorescence intensity at a particular concentration of Mg2+ and ΔFmax represents the difference between F0 and F at the infinite concentration of Mg2+.
The dependence of the DNA cleavage activity on the concentration of Mg2+ was analyzed using the following form of the Hill equation:
| (2) |
where nH is the Hill coefficient and Kd is the apparent dissociation constant, b is the activity of enzyme in the absence of Mg2+ and x is the percentage of activity in the presence of varying concentrations of Mg2+. The maximal percentage activity of DNA cleavage was taken as 100%. The data were fit using SigmaPlot version 12.5.
Intrinsic fluorescence emission spectrum of the protein was measured at 26°C by using a Fluoromax spectrofluorometer (HORIBA Jobin Yvon). To probe the tryptophan environment, the excitation wavelength used was 295 nm and emission scan was done from wavelength 310–500 nm. The concentration of protein used was 10 µM in 20 mM Tris-Cl (pH8.0). The spectrum generated is an average of three scans after baseline correction. The slit width used for excitation was 1 nm and 9 nm for emission.
RESULTS
Cas5d shows metal-dependent DNase activity
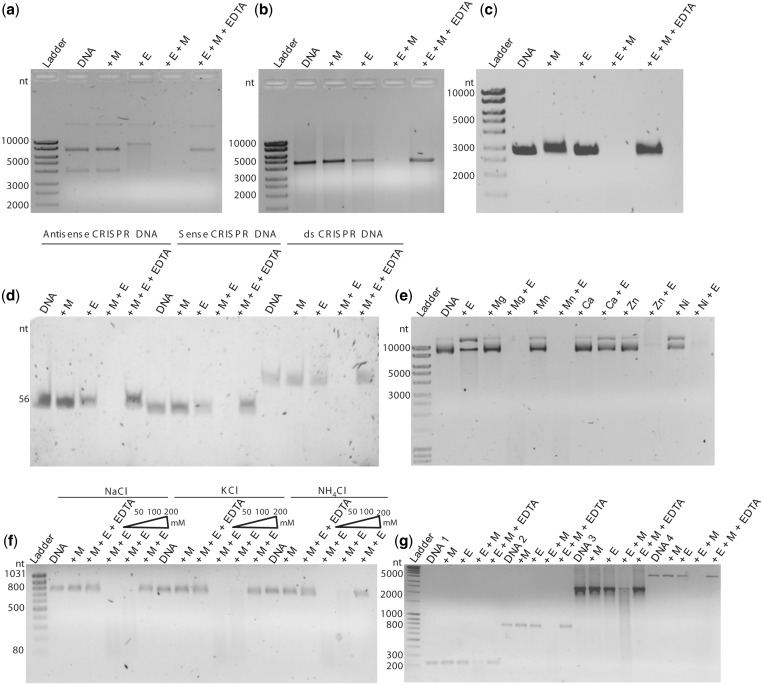
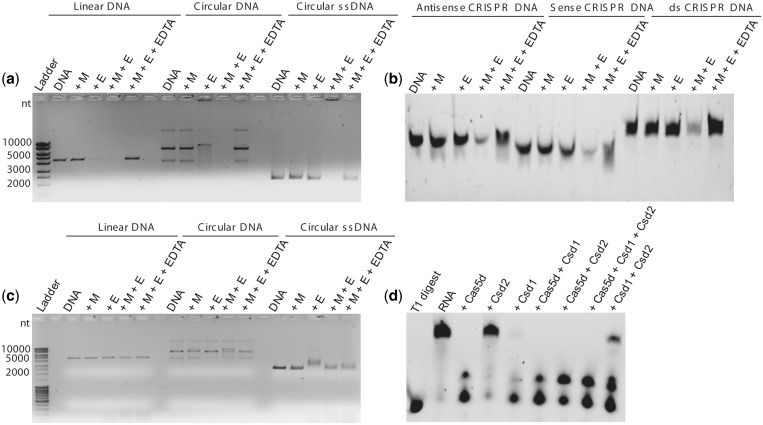
Cas5d was shown to cleave CRISPR repeat RNA recognizing the structured region of the repeat. Recently, it was also reported that Cas5d orthologs from Streptococcus pyogenes and Xanthomonas oryzae bind DNA but do not cleave it (24). This prompted us to investigate the factors that typically influence the DNase activity. RNA has an inbuilt nucleophile in the form of 2′-OH group, and therefore the catalysis may be initiated in the absence of an external nucleophile. DNA lacks this group and consequently most of the DNases use metal ions as cofactors for the cleavage activity. Therefore, we intended to test whether metal ions could stimulate the DNA cleavage. We used pQE2 plasmid as substrate to test the nuclease activity. It was observed that while Cas5d cleaved the circular DNA, the cleavage was more dramatic in the presence of a divalent metal ion (Figure 1a). The plasmid preparation showed polymorphism in its mobility that is typical for a circular DNA. Cas5d showed no preference for these, and all of them were digested with no traces of DNA left behind (Figure 1a). This suggested that Cas5d exhibits endodeoxyribonuclease activity that is stimulated in the presence of a divalent metal. Encouraged by this, we asked whether it is adept at acting on linear substrate too. Therefore, we linearized the pQE2 and repeated the assay. Here too, the activity was seen prominently in the presence of a divalent metal ion (Figure 1b). The aforementioned experiments were performed using double-stranded DNA substrates. This raised the question whether Cas5d could act on single-stranded DNA as well. Hence, we used the single-stranded circular DNA from M13mp18 phage, and here too we noticed the activity that was discernable when a divalent metal ion was present (Figure 1c).
Figure 1.
Cas5d is a metal-dependent DNase. In all panels, E represents Cas5d and M denotes Mg2+. The presence of DNA and EDTA in the respective lanes is indicated. The lane where the ladder is loaded is shown. The DNase activity of Cas5d was assayed in the presence of (a) double-stranded circular DNA, (b) double-stranded linear DNA, (c) M13mp18 phage single-stranded circular DNA and (d) the antisense, the sense and the sense and antisense duplex CRISPR repeat DNA. (e) Nuclease activity in presence of 10 mM Mg2+, Mn2+, Ca2+, Zn2+ and Ni2+ is shown. (f) Assay in the presence of NaCl, KCl and NH4Cl is shown, and the corresponding salt in the lanes is indicated above. The increasing concentration (50, 100 and 200 mM) of the salt in the corresponding lanes is depicted as a triangle. (g) Activity in presence of the substrate with varied length is shown. DNA 1 (260 bp), DNA 2 (800 bp), DNA 3 (2500 bp) and DNA 4 (4800 bp) are indicated.
The hallmark of the RNase activity of Cas5d is its ability to recognize the structured form of RNA. Nam et al. (14) systematically varied the sequences of the repeat RNA and identified that the recognition is stronger at the base of the stem and 3′-end overhang. Prompted by this, we asked whether the DNA recognition too is structure specific. We used the sense, the antisense and the duplex forms of the CRISPR repeat DNA to clarify whether the sequence is recognized and cleaved in a manner similar to RNA. Both the sense and antisense showed the presence of stem and loop when subjected to the fold prediction using MFOLD (see Supplementary Figure S1). We observed that Cas5d cleaved sense, antisense and the duplex forms preferentially in the presence of the divalent metal (Figure 1d). Unlike the CRISPR repeat RNA where a single cleavage is shown to occur between the positions G21 and U22, there seemed to be no such preferential cleavage of the DNA substrates (Figure 1d). This corroborates that Cas5d displays sequence-independent endonuclease activity against both single- and double-stranded DNA substrates that is stimulated in the presence of the divalent metal ion.
Factors modulating the DNase activity of Cas5d
Motivated by the nuclease activity against circular and linear DNA substrates, we investigated whether Cas5d shows preference toward a particular type of metals. To examine this, the reaction was conducted in the presence of Mg2+, Mn2+, Zn2+, Ca2+ and Ni2+, which are often found to be associated with nucleic acid binding proteins.. Activity was observed in the presence of all metals except Ca2+ (Figure 1e). This suggests that perhaps there is a metal selectivity filter within the structure, which determines the specificity for a particular kind of a metal. We also tested the influence of the nature and concentration of salts on the nuclease activity. Hence, the assay was conducted in the presence of 50, 100 and 200 mM of NaCl, KCl and NH4Cl. It was observed that the activity remained unaffected at 50 and 100 mM, respectively, for all type of salts, which suggests that the nature of the salt does not have any apparent effect on the activity. However, when the concentration was increased to 200 mM, in all three salts, the activity got reduced to a significant extent (Figure 1f). This also suggests that the DNA recognition could involve significant electrostatic interaction. Next, we set out to ask whether there is any length dependence toward the DNA substrates. To address this, we used linear DNA substrates of varied sizes (260, 795, 2500 and 4800 bp) and sequences. Incision of the DNA substrates was observed prominently when Mg2+ was included (Figure 1g). This suggests that the nuclease activity against DNA seems to be promiscuous and independent of the length and sequences of the substrates.
Probing the nature of the metal binding in Cas5d
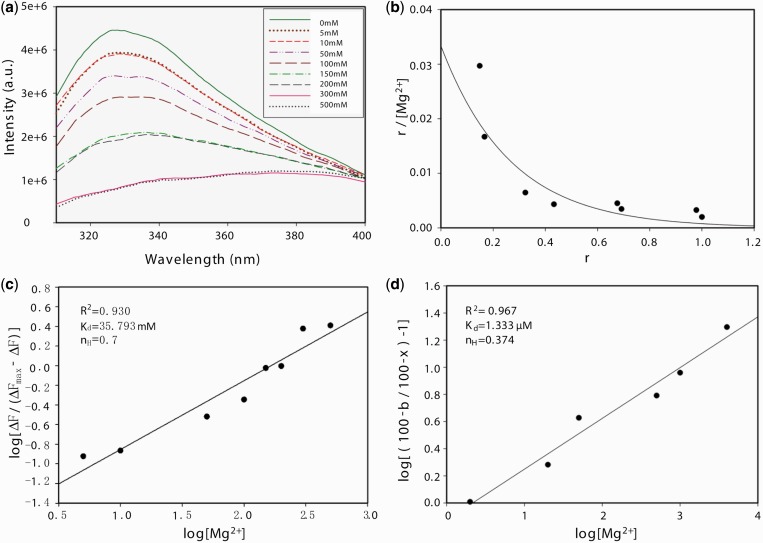
The metal-dependent DNase activity instigated us to probe the metal binding sites using the intrinsic tryptophan fluorescence. Cas5d encompasses two tryptophan residues: W47 is exposed to solvent (relative surface accessibility = 39.9%) and W187 is largely buried (relative surface accessibility = 10.2%). Addition of increasing amounts of Mg2+ ensues quenching of tryptophan fluorescence suggesting that one of them may get exposed to the solvent, or the metal binding sites are situated closer to it (Figure 2a). The buried tryptophan (perhaps W187) getting exposed to the solvent is possible if it undergoes conformational changes on metal binding. In other words, it may be inferred that metal binding may induce a localized conformational transitions in Cas5d. To probe the metal binding sites further, we resorted to the Scatchard plot analysis that showed a concave-up curve, suggesting that either there are multiple classes of metal binding sites or the presence of cooperativity (Figure 2c). To clarify this, we used Hill plot analysis that showed a linear trend with the Hill coefficient (nH) of 0.7, and the apparent dissociation constant (Kd) in the absence of DNA turns out to be 35.79 mM (Figure2c). This, together with Scatchard and Klotz plot analysis, allows us to negate the presence of multiple classes of metal binding sites and to propose the possibility of negative cooperativity in binding the metal (Figure 2b and c; see Supplementary Figure S2). Interestingly, the Hill plot analysis in the presence of DNA suggests that the affinity for Mg2+ (Kd) is found to be 1.33 µM with the Hill coefficient (nH) of 0.37 (Figure 2d; see Supplementary Figure S15). This, in turn, implies that the DNA binding increases the affinity for metal or vice versa.
Figure 2.
Fluorescence studies to probe the link between metal binding and DNase activity. (a) Cas5d tryptophan fluorescence undergoes quenching with increasing Mg2+ concentration (0–500 mM). The concentration of Mg2+ for the corresponding curve is indicated in the inset. The fluorescence intensity is shown in arbitrary units (a. u.). (b) Scatchard plot shows a concave-up curve. Here, r is represented as the ratio of ΔF and ΔFmax, where ΔFmax is the differential intensity at the infinite concentration of Mg2+. (c) Hill plot in the absence of DNA is shown. The linear trend suggests the existence of cooperativity. (d) The dependence of DNA cleavage on Mg2+ was analyzed by Hills plot. The maximal activity is taken as 100%. The percentage activity in the absence of Mg2+ is b, and x is the percentage activity in the presence of varying concentrations of Mg2+. For (c) and (d), R2 represents the square of the goodness of fit, (nH) denotes the Hill coefficient and Kd indicates the apparent dissociation constant.
Tunable DNase activity of Cas5d
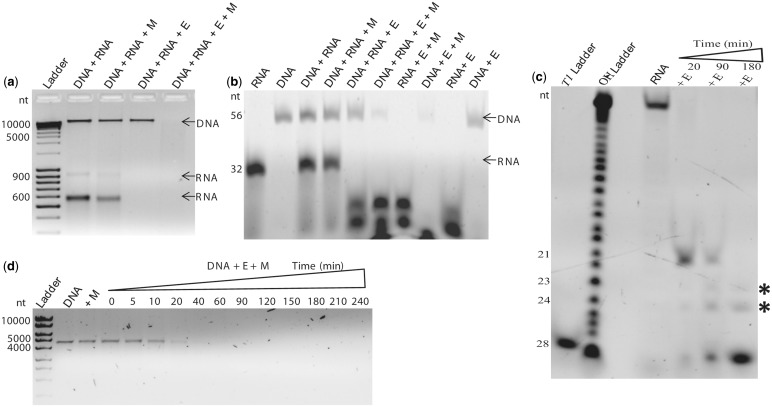
Cas5d was shown to selectively process the pre-crRNA leading to the crRNA maturation in a metal-independent manner. Here, we have shown that it also exhibits a metal-dependent DNase activity. Because it seems to act against both RNA and DNA substrates, it presents an interesting question whether there is any preference between the two. This was tested against a mixture of RNA and DNA substrates under two different conditions: (i) non-specific substrates and (ii) cognate CRISPR repeat RNA and DNA. When metal was not present, Cas5d acted on RNA alone, and when metal was added, DNA cleavage was observed, while the RNase activity remained unaffected. Thus, DNA and RNA were simultaneously acted on in presence of metal (Figure 3a and b). This suggests that the metal bestows a new functionality to Cas5d (i.e. DNase activity) even in the presence of RNA. Because Cas5d exhibits metal-dependent DNase activity, even in the presence of RNA, we did time-dependent studies on both the substrates to probe its RNase and DNase activity further. When a 3′-end-labeled repeat RNA was incubated with Cas5d for different time intervals, we observed differences in the product size over time. Initially, a single band was observed that could have been resulted from the cleavage between G21and U22 as shown earlier (Figure 3c; see Supplementary Figure S3). A longer incubation ensued the appearance of a smaller sized band, whose cleavage site was deciphered by comparing the cleavage pattern obtained from RNase T1 digestion and alkaline hydrolysis ladder (Figure 3c; see Supplementary Figure S3). In the 3′-end of the repeat RNA that appears to be single stranded, three potential cleavage sites, namely, G23, G24 and G28 were noted for T1. Complete cleavage occurring at both G23/24 and G28 is expected to produce two fragments of ∼4 nt each, of which G28 fragment is likely to be visible on the gel, as it is fluorescently tagged at the 3′-end (see Supplementary Figure S4). Comparing the Cas5d cleavage products with OH and T1 ladders, it seems that Cas5d processes the 3′-end of the CRISPR repeat further via intermediates (presumably by cleavage at G21, G23 and G24) to produce a final product of ∼4 nt in size (via cleavage at G28) with time (Figure 3c; see Supplementary Figures S3 and S4). Under this circumstance, the crRNA will have a 4 nt psi-tag followed by the spacer element as against the 11 nt psi-tag. This sequential processing of repeat RNA provoked us to assess the DNA processing over long time period. To test this, we performed time-dependent assay with DNA in presence of metal and found no distinct intermediate accumulation (Figure 3d).
Figure 3.
Metal tunable DNase activity. In all panels, E represents Cas5d and M denotes Mg2+. The presence of DNA and RNA in the respective lanes is indicated. (a) Activity was tested with a mixture of non-cognate RNA and DNA and in the presence and absence of the metal. Two populations of RNA of varied sizes were used, and their locations are indicated by an arrow. (b) Nuclease activity on the cognate CRISPR repeat RNA and DNA, respectively, is shown. The DNA substrate has antisense T7 promoter region, and hence its size is larger. This enables to distinguish the DNA and RNA substrates. (c) Shorter incubation of ∼20 min with repeat RNA at room temperature produces a fragment of ∼21 nt, but longer incubation of ∼180 min produces a fragment of ∼4 nt that is similar in size as that of the T1 digest. Asterisk denotes the intermediate products. (d) Time-dependent assay with double-stranded linear DNA substrate is shown. The time points (0–240 min) are indicated above by a triangle.
Active site plasticity in Cas5d
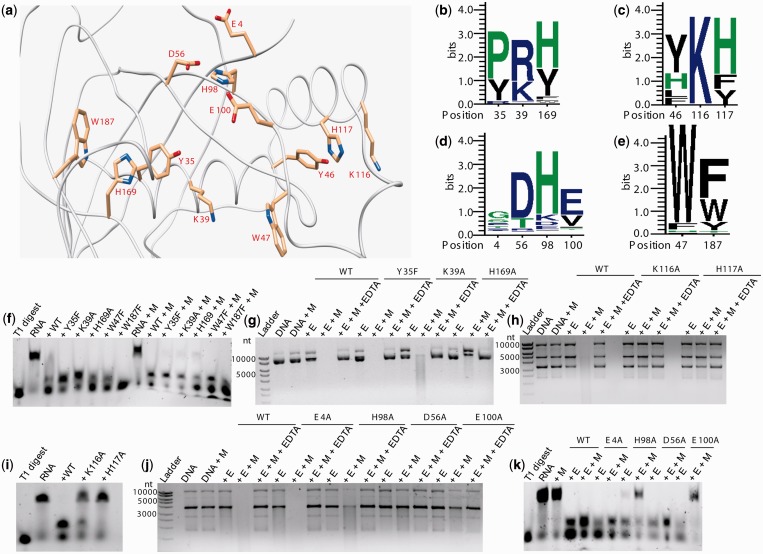
Cas5d displays a single RRM domain with a distinct C-terminal β-sheet extension. Y46, K116 and H117 were shown to play critical roles in endoribonuclease activity. Inspection of the structure enabled us to identify Y35, K39 and H169 to be located geometrically in a conformation analogous to Y46, K116 and H117 (Figure 4a). Remarkably, like W47, which is located adjacent to the triad—Y46, K116 and H117—we spotted W187 in proximity to Y35, K39 and H169 (Figure 4a). Looking at the sequence conservation profiles of these residues across the type I-C organisms, it appears that the chemical nature of the residues seems to be preserved over their identity (Figure 4b, c and e). This has driven us to hypothesize that the triad comprising Y35, K39 and H169 and possibly W187 may be involved in catalysis. To unravel this, we generated point mutants Y35F, K39A, H169A, W47F and W187F and investigated their role in the cleavage of the repeat RNA. W47F, H169A and W187F mutants produced the smaller fragment as that of the wild-type (Figure 4f; see Supplementary Figure S12). Intriguingly, Y35F and K39A showed accumulation of the larger fragment, suggesting that they may possibly involve in further processing of the RNA (Figure 4f). The addition of metal had not significantly altered the cleavage pattern of the aforementioned mutants. Led by the differential activities of Y35F, K39A and H169A, we set out to test their involvement in the DNase activity too. Surprisingly, we found that H169A mutant that was active against the RNA substrate showed loss of DNase activity even in the presence of metal (Figure 4g; see Supplementary Figure S5). K39A showed reduced activity, and Y35F was as active as the wild-type (Figure 4g; see Supplementary Figure S5). This suggests that H169A participates in the DNase activity. We also assessed the DNase activity of K116A and H117A, which were known to be involved in RNA processing. Intriguingly, in addition to their participation in the RNA hydrolysis, these mutations abrogated the DNase activity too (Figure 4h and i; see Supplementary Figure S6).
Figure 4.
Effect of mutations on Cas5d nuclease activity. In all panels, M denotes Mg2+ and E denotes the presence of enzyme. WT represents the wild-type Cas5d, and the respective mutants are shown at the top. The lane that has substrate as control is denoted as DNA or RNA. The lane that contained EDTA is indicated. The lane having the ladder or T1 digest of RNA is shown. The RNA is 3′-end labeled with FAM. (a) Structure of Cas5d (PDB ID: 4F3M) displaying the position of residues that are proposed to be involved in substrate binding and/or nuclease activity. The residues are represented as sticks with the carbon atom in light orange, nitrogen in blue and oxygen in red. The sequence number of the corresponding residue is indicated. This figure was rendered using Chimera (25). (b–e) The sequence logo depicting the conservation of these residues across type I-C orthologs is shown. The residue number is indicated below the logo. The height of a residue (in bits) represents the extent of conservation. Assays to monitor the effect on (f) RNase and (g) DNase activity of the point mutants Y35F, K39A, H169A, W47F and W187F are shown. The involvement of the alanine scanning mutants K116 and H117 in the (h) DNase and (i) RNase activity is presented. The nuclease assays against (j) DNA and (k) RNA to probe the role of residues in binding the metal are shown.
To examine the residues involved in metal coordination, we analyzed the Cas5d structure and identified E4, D56, H98 and E100 as prospective candidates owing to the propensity of these residues to coordinate the metal ligands and their clustered location (Figure 4a). Among the identified residues, D56, H98 and E100 seem to be highly conserved across the Cas5d orthologs (Figure 4d). Alanine scanning mutagenesis of these residues showed that DNase activity of E4A remained unaffected and H98A showed marginal reduction in the activity (Figure 4j). However, D56A and E100A mutations drastically affected the activity (Figure 4j). This suggests that these residues may be involved in coordinating the metal or binding the DNA. Inquisitively, we also tested these residues for their effect on RNase activity. Although E4A and D56A had no effect on RNase activity, H98A showed reduced activity that was further inhibited in the presence of metal. E100A exhibited RNase activity that was severely impaired in the presence of metal (Figure 4k). The experiments with the aforementioned mutants allowed us to segregate their involvement in hydrolyzing the DNA and the RNA, respectively. Remarkably, the mutational analysis of K116A and H117A that affects both DNase and RNase activity led us to suggest the possibility that the catalytic site for processing the DNA and the RNA is interrelated.
Features of the constituents of the Cascade-like complex in type I-C
Cas5d together with other subtype I-C-specific proteins, namely, Csd1 and Csd2 assembles into a Cascade-like complex. In this complex, Cas5d is proposed to function equivalently to Cse3/CasE and Cas5e/CasD and that Csd1 and Csd2 are proposed to play a role analogous to Cse1/CasA and Cse2/CasB, respectively, in type I-E system. Although looking at the sequence length of Csd1, we noted that it is longer than the Cse1/CasA (627 versus 502 amino acids). This prompted us to probe whether the additional region at the C-terminus can exist as a separate domain in Csd1. Therefore, we subjected this region to fold recognition using FFAS sever (26), which predicted that this region could be a separate domain and showed similarity to Cse2/CasB in T. thermophilus (see Supplementary Figure S7). Secondary structure prediction using JPRED (27) suggested that the C-terminal region (554–627 aa) is largely α-helical as seen in Cse2/CasB. This points at the likelihood that Csd1 could be a fusion protein of its functional counterparts Cse1/CasA and Cse2/CasB. Encouraged by this, we further investigated whether Csd1 and Csd2 possess nuclease activity. Csd1 incised non-specific linear and circular DNA in the presence of metal (Figure 5a). However, in the absence of metal, reduced cleavage of DNA was observed. Although no cleavage of single-stranded circular M13mp18 phage DNA was seen, it exhibited binding to DNA in the presence of metal (Figure 5a). Assessing the activity against the cognate CRISPR repeat sense, antisense and duplex DNA showed incision only in the presence of metal (Figure 5b). Intriguingly, though there was no cleavage against the circular single-stranded DNA, the complementarity within the cognate CRISPR repeat DNA presumably could have activated the DNase activity or it is selectively inactive against the single stranded circular DNA. On the other hand, Csd2 seemed to be inert against the DNA (Figure 5c). Encouraged by the DNase activity of Csd1, we tested its activity against the 3′-end-labeled repeat RNA in the absence of metal. The pattern of cleavage was similar to that obtained for Cas5d suggesting that the point of cleavage may also be similar (Figure 5d; see Supplementary Figure S13). In line with its inactivity against DNA, Csd2 seems to be inert against the RNA substrate too (Figure 5d). In conjunction with its inertness against RNA, whenever Csd2 associates with Cas5d and Csd1, it appears to slow down their RNase activity (Figure 5d; see Supplementary Figure S14). This led us to posit the possibility that Csd2 may downregulate the RNase activity of Cas5d and Csd1 when they assemble into the Cascade-like complex.
Figure 5.
Nuclease activity assays with Csd1 and Csd2. In all panels, M denotes Mg2+ and E represents the corresponding protein. The presence of DNA and RNA in the respective lanes is indicated. The lane that contained EDTA or the ladder is indicated. The RNA and CRISPR repeat DNA is 3′-end labeled with fluorescein. (a) The DNase activity of Csd1 was assayed in the presence of double-stranded linear, double-stranded circular and single-stranded circular DNA substrates as indicated above the gel. An upward shift of single-stranded circular DNA in the presence of Csd1 and metal is noted, which suggests binding as against the cleavage. (b) Csd1 activity against the antisense, the sense and duplex CRISPR repeat DNA is presented. (c) Assay to probe the nuclease activity of Csd2 against double-stranded linear, double-stranded circular and single-stranded circular DNA is shown. (d) Combined effect of Cas5d, Csd1 and Csd2 activity on repeat RNA is presented.
DISCUSSION
Factors contributing to metal-dependent substrate switching
The role of Cas5d was shown to be an RNA processing enzyme akin to Cse3/CasE and Cas6 in type I and III, respectively. Our work reveals that it also plays a role of DNA targeting enzyme in the presence of a divalent metal ion. Hence, it appears that the nuclease activity can be tweaked toward the DNA substrates by the use of a metal cofactor. However, it is also evident that there is preference for certain kinds of metals, namely, Mg2+, Mn2+, Ni2+ and Zn2+ over Ca2+ (Figure 1e). Because the size of these metal ions differ, it is possible to attribute the differences in the activity to differences in the size (ionic radius) of the metal ions—Ni2+ (83 pm); Mn2+ (81 pm); Zn2+ (88 pm); Mg2+ (86 pm); and Ca2+ (114 pm). Therefore, it is likely that the active site that harbors the metal may accommodate those metals with ionic radius ranging from 80 to 90 pm. This also explains why Ca2+ with an ionic radius well beyond the aforementioned range does not elicit a response. Owing to this opposing nature, it seems probable that Mg2+ and Ca2+ can compete against each other to regulate the DNase activity (see Supplementary Figure S8).
The Hill analysis hints at the possibility of negative cooperativity in binding the metal in Cas5d. Though Cas5d binds metal, in the absence of DNA, the affinity toward metal seems to be weak (kd = 35.79 mM). However, when DNA is present, the affinity seems to be strong (kd = 1.33 µM), suggesting that the DNA too contributes to metal binding. It is possible that one of the ligands that coordinate the metal may be contributed by the DNA itself as seen in several DNA binding metalloenzymes (28). The cellular concentration of magnesium is around 30 mM (29), and hence when the DNA is not in the vicinity of Cas5d, the low affinity for metal would render it to be an RNase, thus enabling it to facilitate the crRNA maturation. However, proximity to DNA, which might be brought about during the stages of CRISPR immunity, is likely to enhance the metal affinity (kd = 1.33 µM), thus transforming it to be a DNase too.
The mutational analysis on Cas5d indicates that D56, H98, E100 and H169 are involved in the DNase activity either by contributing to DNA binding or by coordinating the metal ion. Though the mutations D56A, E100A and H169A render Cas5d incompetent against DNA, the activity against RNA is retained (Figure 4f, g, j and k). Thus these represent the separation-of-function mutants. Intriguingly, when metal is provided, the RNase activity of H98A and E100A is drastically reduced (Figure 4k). Although the functional roles of these residues in processing the RNA and DNA targets remain to be examined, this mutational analysis raises the possibility that there is a considerable functional overlap of residues involved in processing the DNA and RNA substrates. Further, the intrinsic tryptophan fluorescent studies hint at the possibility of conformational changes on metal/RNA binding (Figure 2a; see Supplementary Figure S11). This suggests that residues that are located away from each other may be brought closer for binding and/or catalysis by inducing conformational changes on metal and/or nucleic acid binding.
The simultaneous occurrence of RNA and DNA hydrolysis in presence of metal ion seems to suggest the possibility of harboring a single active site with tunable target selectivity (Figure 3a and b). Our work together with earlier demonstration by Nam et al. (14) suggests that K116 and H117 participate in hydrolyzing the RNA (Figure 4i). The alanine scanning mutagenesis of K116 and H117 suggests that the mutation of these residues abrogates the DNase activity too (Figure 4h). Y46, K116 and H117 seem to be attractive candidates to assume the analogous role as proposed for the equivalent residues in Cas6 [Y31: K52: H46] and tRNA intron splicing endonucleases [Y246: K287: H257] (19,30). The archetypal enzyme RNaseA too exhibits similar triad [H12:K41:H119] in catalysis wherein the Tyr is replaced by His12 (31). This reinforces the notion that the structurally unrelated enzyme may exhibit similar catalytic mechanism by means of convergent evolution (32). Subscribing to this view, it is possible to propose that Y46 in Cas5d may play a role of a base deprotonating the 2′-OH of G21 for inline nucleophilic attack on the scissile phosphate. This proposition is also supported by the observation that substituting this 2′-OH with deoxy derivative and/or mutating this Tyr to Ala abolishes the cleavage (14,19). K116 is a likely candidate to stabilize the negatively charged transition state, and H117 may protonate the leaving group akin to K41 and H119, respectively, in RNaseA (31). In line with this, the roles of K116 and H117 seem to be apt in satisfying the requirements for hydrolyzing the DNA too. However, for the DNA hydrolysis, the role of a nucleophile activator, Y46, may be taken over by a metal ion, as the nucleophile is most likely a water molecule here and not an intrinsic 2′-OH group (28). Therefore, this drives us to hypothesize that the active site that promotes RNA hydrolysis may also have the potential to participate in hydrolyzing the DNA too.
Additional roles of Cas5d and Csd1 in CRISPR Interference
The CRISPR-Cas system of type I and III appears to use multi-protein assemblies to facilitate the crRNA maturation and/or target interference (5–8,33). Though there seems to be no appreciable sequence similarity between the type I and III Cascade-like complex, the genome architecture and synteny of the cas operon suggests that both these systems are recognized with contextual semblance and are found to have minimal components comprising a large subunit (Cas8/CasA in type I, and Cas10/Csm1 in type III), a small unit (CasB in type I, and Csm2 in type III), a backbone subunit (Cas7/CasC in type I, and Csm3 in type III) and Cas5 and Cas6 (34). The large subunit in type III harbors motifs that are reminiscent of a palm domain that are found in polymerases, and the equivalent subunit in type I shows inactivated polymerase domain (34). However, the large subunit Csd1 in B. halodurans that also belong to type I, though seems to have inactivated polymerase domain, exhibits nuclease activity against DNA and RNA substrates. The ortholog of Csd1 in Methanothermobacter thermoautotrophicus (see Supplementary Figure S9), i.e. referred as Nar71 (MTH1090) is also reported to display nuclease activity (35). Though the region harboring the nuclease activity in Csd1 and its effect on the functionality of Cascade-like complex needs to be investigated further, it appears that the Csd1 has undergone adaptation compared with its counterpart in other type I-E and type III systems, perhaps, in conformity with the requirement of type I-C immune response.
The Cascade complex in E. coli participates in the crRNA maturation and target interference (5,8,33). Based on the similarity between the protein components, it was envisaged that the B. halodurans Cascade-like complex may also play a similar role (14). However, in the case of E. coli where both Cas5 (referred as Cas5e/CasD) and Cse3/CasE are present, Cas5e/CasD appears to be catalytically inactive and plays only a structural role (8). The catalytic residues identified in Cas5d were absent at the equivalent position in Cas5e (see Supplementary Figure S10). Though Cas5e and Cas5d share an RRM domain, there seems to be considerable adaptation in Cas5d to offset the absence of Cse3/CasE.
The constituents of the Cascade complex in E. coli are shown to exist in a defined stoichiometry: Cas[A1:B2:E1:C6:D2] (5,33). Similar studies on B. halodurans showed that the complex comprises [Csd11:Csd26:Cas5d2] (14) wherein the subscript denotes the copy number of the respective protein. We identified Csd1 to be a fusion protein of its functional homolog Cse1/CasA and Cse2/CasB. Owing to this covalent linkage, the copy number of the Cse2/CasB that is fused to Cse1/CasA in Csd1 is expected to differ, and therefore the functionalities of Cascade-like complex in type I-C may deviate from its counterpart in type I-E. In line with this, except Csd2, none of the other constituents are able to functionally complement the respective subunits in E. coli Cascade complex (14). Our conjecture is that the copy number of Csd1 may not be attuned to the copy number of Cse1/CasA and Cse2/CasB in E. coli, thereby accommodating Csd1 in E. coli Cascade complex may not be possible. Although Cse4/CasC and Csd2, owing to their existence as a separate entity, could complement each other, this scenario presents a tempting proposition that perhaps the absence of Cse3/CasE in B. halodurans and Cas5d, adopting its role may be to offset this variation in the structural composition of the Cascade-like complex.
Although the processing of CRISPR repeat RNA seems to proceed with specificity, the DNase activity is apparently non-specific. This raises questions on the possible role(s) of this promiscuous DNase activity in type I-C. One can consider the promiscuous DNase activity exhibited by the constituents of Cascade-like complex, namely, Cas5d and Csd1 in B. halodurans as an evolutionary adaptation. Recent studies have shown that promiscuous restriction is conferring selective advantage by increasing the survival fitness of bacteria to cope up against invading phages (36). This characteristic feature has the potential to enhance the outreach of the defense strategy by countering the phages that escapes the bacterial defense with reduced restriction sites and/or modification of phage genome. Therefore, the promiscuous DNase activity of Cascade-like complex, in association with Cas3, may elicit a rapid action response for target degradation during CRISPR interference. Yet another aspect of non-specific DNA targeting could be its involvement during the adaptation stage as well. Acquisition of new spacer from the invading genome requires fragmentation of the nucleic acids and subsequent incorporation of this short fragment into the CRISPR locus. Previously, this process was shown to proceed with the involvement of Cas1 and Cas2 alone (37); however, recent studies have highlighted the participation of Cascade complex and Cas3 along with Cas1 and Cas2 (38). This association primes the acquisition of more spacers from multiple regions of the escape phage genome, which has accumulated mutations in protospacer/PAM sequence for mounting defense response during subsequent infection of the escape phage (38). This is undoubtedly beneficial to the host by allowing it to adapt and become resistant to these escape phages. Therefore, the promiscuous DNase activity of Cas5d and Csd1 in the Cascade-like complex could come in handy either during the adaptation and/or the interference stage of the CRISPR immunity pathway during which direct interaction with DNA is envisaged. Although the precise role of this nuclease activity during these processes needs further investigation, this observation allows one to forecast that lineage-specific functional variations operate in CRISPR-Cas systems across diverse microbial species that may confer selective advantage for niche-specific adaptation in protecting against genome predators.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Biotechnology [BT/PR14402/BRB/10/830/2010, BT/341/NE/TBP/2012 and BT/PR5511/MED/29/631/2012] and Board of Research in Nuclear Sciences [2011/20/37B/15/BRNS]. Funding for open access charge: Department of Biotechnology, Government of India.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Drs B. Prakash, N. Srinivasan, M.R.N. Murthy, U. Varshney and S. Ramaswamy for their constant encouragement and enthusiastic support. They acknowledge Dr Scott Gradia for the LIC vector (2R-T). They thank all their group members for their critical comments and suggestions on this work.
REFERENCES
- 1.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 2.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 4.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 5.Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 6.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garside EL, Schellenberg MJ, Gesner EM, Bonanno JB, Sauder JM, Burley SK, Almo SC, Mehta G, MacMillan AM. Cas5d processes pre-crRNA and is a member of a larger family of CRISPR RNA endonucleases. RNA. 2012;18:2020–2028. doi: 10.1261/rna.033100.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat. Struct. Mol. Biol. 2011;18:688–692. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- 13.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nam KH, Haitjema C, Liu X, Ding F, Wang H, DeLisa MP, Ke A. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR-Cas system. Structure. 2012;20:1574–1584. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat. Struct. Mol. Biol. 2011;18:680–687. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- 16.Sternberg SH, Haurwitz RE, Doudna JA. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA. 2012;18:661–672. doi: 10.1261/rna.030882.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–264. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–2188. doi: 10.1261/rna.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lintner NG, Frankel KA, Tsutakawa SE, Alsbury DL, Copie V, Young MJ, Tainer JA, Lawrence CM. The structure of the CRISPR-associated protein Csa3 provides insight into the regulation of the CRISPR/Cas system. J. Mol. Biol. 2011;405:939–955. doi: 10.1016/j.jmb.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P, Siksnys V. In vitro reconstitution of cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koo Y, Ka D, Kim EJ, Suh N, Bae E. Conservation and variability in the structure and function of the Cas5d endoribonuclease in the CRISPR-mediated microbial immune system. J. Mol. Biol. 2013;425:3799–3810. doi: 10.1016/j.jmb.2013.02.032. [DOI] [PubMed] [Google Scholar]
- 25.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 26.Jaroszewski L, Li Z, Cai XH, Weber C, Godzik A. FFAS server: novel features and applications. Nucleic Acids Res. 2011;39:W38–W44. doi: 10.1093/nar/gkr441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Maguire ME, Cowan JA. Magnesium chemistry and biochemistry. Biometals. 2002;15:203–210. doi: 10.1023/a:1016058229972. [DOI] [PubMed] [Google Scholar]
- 30.Calvin K, Li H. RNA-splicing endonuclease structure and function. Cell Mol. Life Sci. 2008;65:1176–1185. doi: 10.1007/s00018-008-7393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raines RT. Ribonuclease A. Chem. Rev. 1998;98:1045–1066. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 32.Galperin MY, Koonin EV. Divergence and convergence in enzyme evolution. J. Biol. Chem. 2012;287:21–28. doi: 10.1074/jbc.R111.241976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guy CP, Majernik AI, Chong JP, Bolt EL. A novel nuclease-ATPase (Nar71) from archaea is part of a proposed thermophilic DNA repair system. Nucleic Acids Res. 2004;32:6176–6186. doi: 10.1093/nar/gkh960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vasu K, Nagamalleswari E, Nagaraja V. Promiscuous restriction is a cellular defense strategy that confers fitness advantage to bacteria. Proc. Natl Acad. Sci. USA. 2012;109:E1287–E1293. doi: 10.1073/pnas.1119226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.