Abstract
The Puf family of RNA-binding proteins regulates gene expression primarily by interacting with the 3′ untranslated region (3′ UTR) of targeted mRNAs and inhibiting translation and/or stimulating decay. Physical association and computational analyses of yeast Puf3p identified >150 potential mRNA targets involved in mitochondrial function. However, only COX17 has been established as a target of Puf3p-mediated deadenylation and decapping. We have identified 10 new targets that are rapidly degraded in a Puf3p-dependent manner. We also observed changes in Puf3p activity in response to environmental conditions. Puf3p promotes rapid degradation of mRNA targets in the fermentable carbon source dextrose. However, Puf3p-mediated decay activity is inhibited in carbon sources that require mitochondrial function for efficient cell growth. In addition, the activity of Puf3p is rapidly altered by changing the carbon source. PUF3 expression is not decreased at the RNA or protein level by different carbon sources and localization is not significantly altered, suggesting that Puf3p activity is regulated posttranslationally. Finally, under conditions when Puf3p is unable to stimulate decay, Puf3p can still bind its target mRNAs. Together, these experiments provide insight into the carbon source-specific control of Puf3p activity and how such alterations allow Puf3p to dynamically regulate mitochondrial function.
INTRODUCTION
In eukaryotes, regulation of mRNA decay is critical for controlling protein production not only in somatic cells, but in several stages of development as early as the transition from maternal to zygotic mRNA expression (1). The Puf family of proteins regulates diverse cellular processes such as gametogenesis, embryonic development and memory formation by promoting translational repression and/or degradation of targeted mRNAs. At the molecular level, Puf proteins bind conserved UGU sequences within the 3′ untranslated region (3′ UTR) of transcripts, typically resulting in the disruption of translation initiation complex interactions or stimulation of mRNA degradation by recruitment of decay complexes (2). In a few instances, Puf proteins can stabilize mRNA targets by promoting translation (3).
Several factors contribute to mRNA-specific rates of deadenylation, decapping and subsequent decay, such as the presence of cis-acting regulatory sequences found within the 3′ UTR (4,5). Saccharomyces cerevisiae Puf3p specifically binds two CNUGUANAUA elements within the COX17 mRNA 3′ UTR and accelerates deadenylation and subsequent decay of the transcript in the presence of dextrose (6,7). One mechanism of action involves recruitment of the Ccr4p-Pop2p-Notp deadenylation complex to COX17 mRNA by direct interaction of Puf3p with Ccr4p (8). Yeast Puf4p and Puf5p demonstrate direct interactions with Pop2p, which together with Ccr4p results in deadenylation of the mRNA target (9–11). Puf3p most likely recruits decapping factors to COX17 as well, as Puf5p-Pop2p complexes also interact with the decapping factor Dcp1p and the RNA helicase Dhh1p (9–11). Alternatively, Puf3p can promote rapid deadenylation in a Ccr4p-independent manner by altering the conformation of the poly(A) binding protein-mRNP structure (8).
Structurally, Puf3p-COX17 binding interactions are mediated by the repeat domain (RD) region, which is composed of eight imperfect repeats (R1–R8), plus short flanking sequences (R1’ and R8’) (12). Each Puf repeat contains three α helices, with repeats stacking to adopt an overall curved structure (13–15). Individual RNA bases within the Puf binding site are recognized by successive repeats through stacking interactions and base-specific contacts (12,15–23). Similar to other Puf proteins, Puf3RDp binds the 8 nt UGUANAUA sequence in a one base to one repeat manner, in which the conserved UGU bases bind repeats R8, R7 and R6, respectively (12). Moreover, the specificity of S. cerevisiae Puf3p binding its mRNA target uses a cytosine located two bases upstream of the conserved UGUA due to a unique binding pocket in the Puf3p RD (12).
Global analysis of Puf3p-mRNA interactions reveals that Puf3p physically associates with 220 transcripts, 162 of which are nuclear-transcribed mRNAs that encode mitochondrial proteins (24). The consensus Puf3p binding motif derived from the global analysis, (C/U)(A/C/U)UGUA(A/U)AUA, is present in 174 of the transcripts physically associated with Puf3p (24) and is enriched in the 3′ UTRs of many S. cerevisiae mRNAs (25). Only one of these transcripts, COX17 that encodes a mitochondrial copper shuttle (26,27), has been experimentally validated as a target of Puf3p-mediated decay and exhibits conditional regulation of its stability (6,28). Examination of mitochondrial protein steady-state levels reveals that Puf3p is required for decreasing mitochondrial DNA-encoded Cox2p (29), as well as nuclear-encoded Pet100p, Cox4p (29) and Pet123p (30) levels in dextrose conditions. Additionally, Pet123p levels were increased in a nonfermentable carbon source (30). However, it was unclear if Puf3p modulates the mRNA stabilities of these putative targets in a carbon source-dependent manner.
The expression levels of mitochondrial mRNAs containing the Puf3p binding element are tightly regulated both spatially and temporally within the yeast cell. For example, they are coordinately controlled in response to ∼750 environmental stresses, including heat shock, starvation and carbon source (25,31). These mitochondrial transcripts are expressed early in the reductive/building phase of the metabolic transcriptional cycle (32). Furthermore, computational analysis using microarray data sets confirms that expression of putative Puf3p targets is coordinately downregulated in repressing carbon source conditions such as dextrose, and upregulated in nonrepressing conditions such as galactose, raffinose and ethanol (28). These carbon source-dependent changes in gene expression are consistent with the observation that glycolytic/fermentative metabolism of dextrose results in translational repression of genes required for metabolism of alternative carbon sources such as galactose, ethanol and glycerol (33). Specifically, dextrose triggers the repression of genes involved in several aspects of mitochondrial adenosine triphosphate (ATP) production, such as mitochondrial electron export and oxidative phosphorylation (34–36). For example, the cytochrome complex subunit COX6 is dextrose repressible (37). Alternatively, when yeast cells require additional processing of nonrepressing carbon sources and therefore increased production of mitochondria, the translation of nuclear-encoded mitochondrial mRNAs is upregulated.
It is proposed that Puf3p conditionally regulates mitochondrial biogenesis, inheritance (30) and oxidative phosphorylation (29) by stimulating decay of mitochondrial mRNAs in fermentative growth conditions. COX17 mRNA is rapidly degraded in dextrose in a Puf3p-dependent manner (6,7). Alternatively, when yeast cells require additional processing of nonrepressing carbon sources for metabolic functions and therefore increased production of mitochondria, the translation of nuclear-encoded mitochondrial mRNAs is upregulated. Consistent with this observation, COX17 mRNA is stabilized in ethanol conditions to the same extent as a puf3Δ strain (28). In addition to a role in mRNA decay, Puf3p may regulate the localization of mitochondrial mRNAs. Puf3p is required for the association of nuclear-transcribed mitochondrial transcripts containing a Puf3p binding site to the mitochondria in galactose (38) and glucose (39). Puf3p colocalizes with the mitochondrial outer membrane and displays physical interactions with the Mdm12p mitochondrial outer membrane complex (30). Moreover, mitochondrial localization of Puf3p target mRNAs requires Tom20p, which is involved in mitochondrial protein import (40). These data support the model that Puf3p can function as an mRNA shuttle to the mitochondria.
To examine the possibility that Puf3p modulates the stabilities of its targets in a carbon source-dependent manner, we sought to identify additional mRNAs that are regulated by Puf3p. Hundreds of potential mRNA targets of Puf3p were determined in two independent microarray and computational studies (24,28). These studies were valuable for identifying mRNA targets that coprecipitated with Puf3p, contained potential Puf3p binding sites and whose steady-state levels were coordinately regulated under different conditions. However, these genome-wide analyses did not directly analyze the effect of Puf3p on the stability of the transcripts. To experimentally validate these mRNAs as targets of Puf3p-mediated decay, we monitored the stability of 15 transcripts that were top candidates from the genome-wide studies. Transcript stabilities were evaluated in wild-type (WT) and puf3Δ strains, resulting in the identification of 10 new mRNA targets of Puf3p-mediated decay. From this work, we show that Puf3p regulates a class of transcripts encoding mitochondrial proteins, and the extent of Puf3p regulation is dependent on the conservation of predicted consensus binding sites. We further determined that a single Puf3p binding site in an mRNA is sufficient to stimulate both deadenylation and decapping steps of decay. The turnover of Puf3p targets is condition-specific, whereby Puf3p activity is inhibited in the presence of ethanol, galactose or raffinose. The status of Puf3p activity can be rapidly altered by changes in the available carbon source. Analysis of PUF3 mRNA and protein levels from yeast grown in different carbon sources revealed that decreases in Puf3p activity do not result from decreased expression levels. In addition, Puf3p localization is not significantly altered in different carbon sources, suggesting that Puf3p activity is likely modulated by posttranslational modifications. Finally, we determined that target mRNAs can physically associate with Puf3p regardless of the carbon source, indicating that inactivation of Puf3p does not disrupt mRNA binding but instead disrupts decay stimulation of bound targets.
MATERIALS AND METHODS
Yeast strains
The genotypes of all S. cerevisiae strains used in the study are as follows: yWO7 MATα, leu2-3,112, ura3-52, rpb1-1 (yRP693) (41); yWO43 MATα, his4-539, leu2-3,112, trp1-1, ura3-52, cup1::LEU2/PGK1pG/MFA2pG, rpb1-1, puf3::NEOr (yRP1360) (6); yWO73 MATa, leu2-3,112, trp1-1, ura3-52, his4, cup1::LEU2/PGK1pG/MFA2pG, PUF3-TAP (TRP1) (yRP1675); yWO185 MATa leu2-3,112, trp1, ura3-52, his4, cup1::LEU2/PGK1pG/MFA2pG, PUF3-GFP (NEO) (yRP1732) (42).
In vivo decay analysis
Steady-state transcriptional shut off experiments were performed essentially as described (41) on yeast strains yWO7 (WT) and yWO43 (puf3Δ), which contain the temperature-sensitive rpb1-1 allele for RNA polymerase II. The strains were grown in standard yeast extract/peptone (YEP) containing 2% dextrose at 24°C to an OD600 of 0.4. Transcription was quickly repressed by shifting the culture to 37°C. Total RNA was isolated from yeast as described (41), separated on 1.25% agarose gels and transferred to nylon membrane for probing with radiolabeled oligos complementary to the ATP11, COX17, CYT2, MAS6, MRP1, MRP21, MRPL6, MRPL39, PET123, RSM10, RSM19 and TUF1 mRNAs (Supplementary Table S1). Northern blots were normalized for loading by probing for scRI RNA (Supplementary Table S1), an RNA polymerase III transcript (43). All quantification of RNA was accomplished using ImageQuant software (Molecular Dynamics). RNA half-life, T1/2, was determined as the time required for 50% of initial pool of RNA to degrade.
Steady-state transcriptional shut off experiments were also performed using strains yWO7 and yWO43, each transformed with plasmids expressing MFA2 RNA (pRP485), MFA2-CYT2 3′ UTR RNA (pWO97) or MFA2-CYT2mut 3′ UTR (pWO98). pRP485 was created as described (44), with the MFA2 RNA expressed under the control of the GAL UAS. The MFA2 3′ UTR was replaced by the CYT2 3′ UTR to produce pWO97 as follows: a 464-bp polymerase chain reaction (PCR) product containing 448 bp of CYT2 sequence 3′ of the stop codon and flanking BglII and HindIII restriction sites was amplified from genomic DNA using primers oWO394 and oWO395, then inserted into pRP485 between BglII and HindIII, creating pWO97. A mutant version of this plasmid, pMFA2-CYT2mut (pWO98) was generated by site-directed mutagenesis of pWO97 using the QuickChange XL Kit (Stratagene) with oWO415 and oWO416, then verified by sequencing. Yeast strains transformed with pWO97 and pWO98 were grown in standard YEP media containing 2% galactose to an OD600 of 0.4. Transcription was rapidly repressed by simultaneously shifting the culture to 37°C and adding dextrose to 4%. Total RNA was isolated, separated on 1.25% agarose gels and transferred to nylon membrane for probing with radiolabeled oligos complementary to MFA2 (oWO315) or the MFA2-CYT2 3′ UTR hybrid RNA (oWO433) at the MFA2-CYT2 junction (Supplementary Table S1). Northern blots were normalized for loading using the scRI RNA (43).
Modified steady-state transcriptional shut off experiments were performed in which yeast strains yWO7 (WT, rbp1-1) and yWO43 (puf3Δ, rpb1-1) were grown in standard YEP media containing 2% ethanol, galactose, raffinose or dextrose to an OD600 of 0.4. Total RNA was isolated and northern analysis performed using radiolabeled probes for COX17, CYT2 and TUF1 mRNAs as described above. To detect STE3 mRNA, total RNA was prepared by hot phenol extraction as described (45), and northern blots probed using radiolabeled oWO523 (Supplementary Table S1). Northern blots were normalized for loading using the scRI RNA (43).
To assess carbon source changes on mRNA decay, steady-state transcriptional shut off experiments were performed in which yeast strains yWO7 and yWO43 were grown in YEP media supplemented with 2% dextrose, galactose or raffinose to an OD600 of 0.4. The cultures were then centrifuged at 4000 rpm to pellet cells, and the cells were resuspended in YEP media containing 2% of a different carbon source. Cultures were incubated at 24°C for 10 or 2 min and promptly centrifuged at 4000 rpm to pellet cells. The cells were immediately shifted to YEP media heated to 37°C to rapidly repress transcription. Northern analysis was performed using radiolabeled probes for CYT2 and TUF1 as described above. Northern blots were normalized for loading using the scRI RNA (43).
Transcriptional pulse chase experiments were performed essentially as described (44) on strains yWO7 (WT, rbp1-1) and yWO43 (puf3Δ, rpb1-1). Regulated expression of WT MFA2-CYT2 3′ UTR hybrid RNA was accomplished by transformation of yWO7 or yWO43 with pWO97, in which the expression of the chimeric RNA is under the control of the GAL UAS. Cultures were grown in raffinose, which does not induce transcription of the fusion RNA, to an OD600 of 0.4. Cultures were then incubated with galactose to induce transcription of this RNA for 8 min to create a pulse of newly transcribed RNAs. Finally, dextrose was added to the media to repress transcription of this RNA. Poly(A) tail lengths were monitored using RNaseH reactions as previously described (4) with an oligo complementary to a sequence just upstream of the MFA2 stop codon (oWO432, Supplementary Table S1). Total RNA was separated on 6% denaturing polyacrylamide gels at 300V for 6 h, then transferred to nylon membrane for probing with a radiolabeled oligo to the MFA2-CYT2 junction (oWO433).
Steady-State PUF3 mRNA analysis
Yeast strains yWO7 (WT) and yWO43 (puf3Δ) were grown in standard YEP containing 2% dextrose, galactose, raffinose or ethanol at 24°C to an OD600 of 0.4. Total RNA was isolated, separated on 1.25% agarose gels and transferred to nylon membrane for probing with a radiolabeled oligo complementary to PUF3 mRNA (oWO124, Supplementary Table S1). Northern blots were normalized for loading by ethidium bromide staining the 28S and 18S rRNAs.
Puf3p western analysis
Protein extracts were prepared from 25 ml of yeast cultures of yWO7 grown in standard YEP containing 2% dextrose, galactose, raffinose or ethanol at 24°C to an OD600 of 0.4. Harvested cells were resuspended in 0.25 ml of sample buffer [125 mM Tris–HCl, pH 6.8, 1% sodium dodecyl sulphate (SDS), 2% glycerol, 10% beta-mercaptoethanol (BME)], lysed with glass beads and extract collected by poking a hole in the bottom of the microfuge tube and spinning into a 15-ml centrifuge tube. Equal OD600 units of total protein were loaded onto a 10% Tris–glycine polyacrylamide gel (Lonza). Resulting gels were blotted to nitrocellulose and probed with a 1:1000 dilution of anti-Puf3p antibodies. Cross-reacting proteins were visualized by a secondary reaction with a 1:5000 dilution of anti-rabbit IgG antibodies. Blots were stripped and reprobed with a 1:10 000 dilution of anti-Tfp1p antibodies. Cross-reacting proteins were visualized by a secondary reaction with a 1:10 000 dilution of anti-mouse IgG antibodies. Loading of total protein was normalized by staining the blot with 0.5% Ponceau S.
Confocal fluorescent microscopy
Endogenously green fluorescent protein (GFP)-tagged Puf3p cells (yWO185) were grown at 30°C to an OD600 of 0.4 in YEP supplemented with 2% dextrose, galactose or ethanol. Cells were then incubated with 50 nM MitoTracker Deep Red FM (Invitrogen Cat#M22426) for 30 min, then fixed with 3.7% formaldehyde for 1 h. The cells were washed twice with 1× phosphate buffered saline (PBS) and resuspended in 8 ml of 1× PBS. One milliliter of cells was loaded onto a circular glass slide coated with 1% polyethyleneimine (SigmaAldrich Cat# P3143-100ML) and cells were allowed to settle for 5 min. The cell solution was aspirated from the edge of the slide, and the slide was then dipped in 1× PBS twice to remove nonadherent cells. A coverslip was applied and the cells were immediately visualized with a Zeiss LSM-700 confocal microscope on the 100× oil immersion objective for eGFP and mRFP fluorescence. Ten slices comprising 6.42 µm were taken through the Z plane of the cells. The slices were flattened using Fiji Is Just ImageJ (FIJI) Z project at maximum intensity, and the intensity of GFP signal and mRFP signal was adjusted such that the maximum signal for cells grown in YEP + 2% ethanol was set as the maximum for cells grown in dextrose and galactose.
Quantitative real-time PCR of RNA associated with Puf3p
RNA immunoprecipitation (IP) was performed essentially as described (24), with minor alterations. Endogenously TAP-tagged Puf3 cells (yWO73) were grown at 30°C in 2L YEP supplemented with 2% dextrose or galactose to an OD600 of 0.4. The cells were pelleted, washed twice with 25 ml of ice-cold Buffer A (20 mM Tris–HCl [pH 8.0], 140 mM KCl, 1.8 mM MgCl2, 0.1% Nonidet P-40, 0.02 mg/ml heparin) and frozen at −80°C. The following day, cells were thawed on ice and resuspended in 5 ml of Buffer B (Buffer A with 0.5 mM dithiothreitol (DTT), 1× Complete Mini Protease Inhibitors [Roche Diagnostics Ref#11-836-153-001], 40 U/ml Recombinant RNasin® Ribonuclease Inhibitor [Promega Cat#N2511], 0.2 mg/ml heparin). Cells were vortexed in the presence of glass beads for 1 min and placed on ice for 1 min for a total of five times. Lysates were clarified by centrifuging at 7000g for 5 min. The supernatant was collected and protein concentration was measured using a standard Bradford assay. Lysates were normalized to contain 1.625 mg in a volume of 5 ml, and bovine serum albumin (BSA) to 1% and 50 µg yeast tRNA were added. The lysates were incubated with 400 µl of 50% IgG Sepharose 6 Fast Flow (GE Healthcare Cat# 17-0969-010) that had been blocked for 1 h in 1 ml of Buffer B plus 1% BSA and 50 µg of yeast tRNA. Beads were incubated at 4°C for 2 h, washed once with 5 ml of Buffer B for 15 min and three times with 5 ml of Buffer C (20 mM Tris–HCl [pH 8.0], 140 mM KCl, 1.8 mM MgCl2, 0.01% Nonidet P-40, 0.5 mM DTT, 12 U/ml Recombinant RNasin® Ribonuclease Inhibitor) for 15 min each. Beads were resuspended in 400 µl of Buffer C and 80 U ProTEV Plus (Promega Cat#V6101) was added. Beads were incubated for 2 h at 16°C. The resultant supernatant was drawn off and total RNA was isolated via hot phenol extraction (46). Glycogen (20 µg) was added to the final aqueous phase to assist precipitation of RNA before the addition of one-tenth volume 3M NaOAc and 2.5 volumes ethanol.
The entirety of immunoprecipitated RNA was subjected to DNase treatment via manufacturer’s instructions (Ambion Turbo DNase). Reverse transcription of RNA was performed according to manufacturer’s specifications (Biorad iScript). Quantitative PCR (qPCR) was optimized and performed on a Biorad CFX96 Real-Time system using SYBR Green detection chemistry (Biorad SSO advanced). Gene-specific qPCR primers are provided in Supplementary Table S1. All RT-qPCR experiments were conducted in biological and technical triplicates.
RESULTS
Puf3p stimulates decay of a class of transcripts encoding mitochondrial proteins
To examine the prevalence of condition-specific regulation of mRNAs by Puf3p, we first sought to identify additional mRNAs whose stabilities are regulated by Puf3p. Hundreds of potential mRNA targets of Puf3p were evaluated by cross-referencing data from two independent microarray and computational studies (24,28). In an effort to refine our search for bona fide mRNA targets of Puf3p-mediated decay, we selected 15 mRNAs with the following characteristics: (i) mRNAs coprecipitated most frequently with Puf3p and were expressed at greater than one copy per yeast cell; (ii) mRNAs were transcribed in the nucleus and encoded mitochondrial proteins; and (iii) mRNAs contained one or more conserved (C/U/A)(A/C/U)UGUA(A/U)AUA elements within a region 300-nt downstream of the ORF.
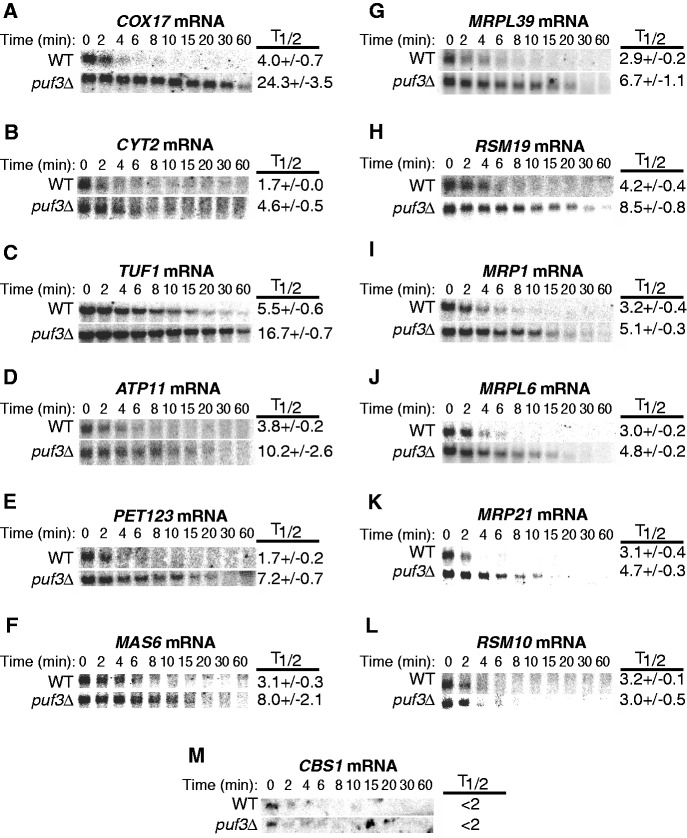
To determine if the stabilities of these 15 mRNAs are regulated by Puf3p, we examined their half-lives in WT and puf3Δ strains, along with COX17 mRNA as a positive target of Puf3p regulation (Figure 1A). These strains harbor a temperature-sensitive lesion in RNA polymerase II (rbp1-1), which allows rapid transcriptional repression on shifting the yeast cells to a nonpermissive temperature (47). If the decay of an mRNA is stimulated by Puf3p, we would expect the half-life of the mRNA to be extended in the puf3Δ strain. Of the 15 mRNAs selected for decay analysis, four transcripts could not be detected by northern blotting (MSF1, CTR1, MRF1 and MRPL7). Analysis of the remaining 11 transcripts support Puf3p’s role as a stimulator of mRNA decay, as 10 transcripts exhibited longer half-lives in the absence of Puf3p (Figure 1B–K). In particular, Pet123 protein levels have previously been observed as inversely affected by Puf3p levels, providing supporting evidence of Puf3p as a downregulator of PET123 (30). As shown in Table 1, the fold stimulation of decay by Puf3p ranged from 1.5 to 4.2. In comparison, the decay of the originally identified Puf3p target, COX17 mRNA, was stimulated 6-fold, similar to results previously described (6). Analysis of the potential Puf3p binding sites revealed that 8 of the 11 newly tested mRNAs contain two or more potential Puf3p binding sites, yet none of the mRNAs contain more than one fully conserved site (Table 1). Only COX17 contains two fully conserved binding sites, which may account for the decay of this mRNA being more highly stimulated by Puf3p than any other target. However, there does not seem to be any correlation between fold stimulation of decay by Puf3p and the number of less-conserved sites beyond a single fully conserved site. For example, ATP11 and CYT2 each have only one potential Puf3p binding site, yet the 2.7-fold stimulation of their decay by Puf3p is greater than five other mRNAs that contain multiple potential Puf3p binding sites but only one fully conserved site. In contrast, TUF1 contains three potential Puf3p binding sites, none of which are fully conserved, yet Puf3p stimulates TUF1 mRNA decay more than most of the other targets (Table 1). The stability of only one transcript, RSM10, was not altered by loss of Puf3p, even though it contains one fully conserved binding site and a second less-conserved site (Table 1). Regulation of mitochondrial-encoding transcripts by Puf3p is nonetheless dependent on the presence of at least one Puf3p binding site, as the decay of CBS1 mRNA lacking Puf3p binding sites is not altered in the puf3Δ strain (Figure 1M and Table 1). The fact that decay of CBS1 is rapid even in the absence of Puf3p suggests that there are other mechanisms to stimulate decay of such mitochondrial transcripts.
Figure 1.
Puf3p promotes rapid decay of multiple mRNA targets. (A–M) Representative northern blot analyses of the decay of each mRNA from a WT strain or a puf3Δ strain are shown. Minutes following transcriptional repression are indicated above each set of blots, with the half-lives (T1/2) and standard error of the mean (SEM) determined from multiple experiments.
Table 1.
RNAs tested for Puf3p-mediated regulation of mRNA decay
| mRNAa | Mitochondrial protein functionb | (C/U)(A/C/U)UGUA(A/U)AUA binding elements located within 300-nt downstream of stop codonc | Ratioe of T1/2 puf3Δ T1/2 WT |
|---|---|---|---|
| Nucleotide positiond | |||
| -2 -1 1 2 3 4 5 6 7 8 | |||
| COX17 | Copper metallochaperone involved in cytochrome c oxidase function | CUUGUAUAUA | 6.0 |
| CCUGUAAAUA | |||
| PET123 | Small ribosomal subunit protein | CAUGUAUUCG | 4.2 |
| CAUGUAUAUA | |||
| TUF1 | Translation elongation factor Tu | CGUGUAAAUA | 3.0 |
| UAUGUAUAUGUAAAGA | |||
| ATP11 | Chaperone involved in F1F0 ATP synthase assembly | CCUGUAAAUA | 2.7 |
| CYT2 | Cytochrome c1 heme lyase involved in ubiquinol-cytochrome-c reductase function | CCUGUAAAUA | 2.7 |
| MAS6 | Component of TIM23 complex that is involved in protein import into matrix and inner membrane | CUUGUAUAUA | 2.6 |
| CAUGUAUGUGUAGAUAUGUACAUA | |||
| CUUGUAUGUU | |||
| UUUGUACUGU | |||
| MRPL39 | Large ribosomal subunit protein | CCUGUAAAUA | 2.3 |
| UUUGUAUACG | |||
| RSM19 | Small ribosomal subunit protein | AGUGUAUAUA | 2.0 |
| CAUGUAAAUA | |||
| GCUGUAUAUA | |||
| MRP1 | Small ribosomal subunit protein | UCUGUAAAUA | 1.6 |
| AUUGUAGCGC | |||
| MRPL6 | Large ribosomal subunit protein | CAUGUAUCCU | 1.6 |
| CUUGUAAAUA | |||
| MRP21 | Small ribosomal subunit protein | UUUGUAUAUU | 1.5 |
| RSM10 | Essential small ribosomal subunit protein | CUUGUAAAUA | 0.9 |
| UUUGUACUAA | |||
| CBS1 | Positive regulator of mitochondrial translation | 1 | |
amRNAs listed were analyzed by transcriptional shut off experiments.
bGene ontology and functional descriptions of the mitochondrial proteins encoded by each mRNA as obtained from the Saccharomyces Genome Database.
cPredicted 10 nt Puf3p binding elements were identified within 300 bp downstream of the translational stop codon, with the minimal Puf element UGUA underlined.
dThe nucleotide positions of the Puf elements are numbered, starting with the UGUA element. Positions upstream of the core UGUA are denoted by negative numbers. Puf3p sequences found within mRNAs are compared with the predicted Puf3p regulatory element (C/U)(A/C/U)UGUA(A/U)AUA (24). Positions conserved within the predicted sequence are in black, while positions that are not conserved are in gray.
eFor each transcript, the fold difference between the average half-life in the puf3Δ versus WT strain is shown.
These results establish 10 new mRNA targets of Puf3p-mediated decay and experimentally demonstrate that Puf3p is a transcript-specific regulator of a class of nuclear-transcribed mRNAs that encode mitochondrial proteins. The data also highlight the importance of experimentally testing candidate transcripts identified through large-scale genomic microarray analyses, as at least one mRNA showed no differential decay in the presence or absence of Puf3p despite the presence of a highly conserved Puf3p binding site in its 3′ UTR. Moreover, the extent of Puf3p regulation on each transcript is not simply defined by the number of potential Puf3p binding sites in the 3′ UTR beyond one required site, but likely depends on sequence context extending outside the 10-nt binding site, as well as other cis or trans factors that control the stability of the mRNA. Physiologically, the variability in Puf3p regulation on different mRNA targets ultimately allows fine-tuned control of protein production that is distinct for each protein and its function.
The CYT2 3′ UTR is sufficient for in vivo Puf3p regulation and requires a single UGUA element
Most of the Puf3p target mRNAs evaluated contain at least two potential Puf3p binding sites, and we have previously shown that full Puf3p regulation of COX17 mRNA requires binding to its two UGUA sites (7). In contrast, ATP11 and CYT2 mRNAs contain only one UGUA site in their 3′ UTRs, yet show higher-fold decay stimulation by Puf3p than many other targets containing multiple sites. We therefore examined the CYT2 mRNA to determine whether its single UGUA site is responsible for complete Puf3p regulation, or whether there are other sequence elements within or outside the 3′ UTR that contribute to Puf3p regulation.
To first determine whether the CYT2 3′ UTR is sufficient to mediate Puf3p regulation, we fused the 3′ UTR of CYT2 behind the coding region of MFA2, which is not regulated by Puf3p (Figure 2A). This fusion RNA was expressed under the control of the GAL UAS in rpb1-1 strains, allowing both temperature shift and galactose to dextrose sugar shift to repress transcription (44). The MFA2/CYT2 3′ UTR transcript was rapidly degraded in the presence of Puf3p with a half-life of 3.0 ± 0.4 min, whereas in the puf3Δ strain, the half-life was increased 2-fold to 6.1 ± 1.0 min (Figure 2B). As a negative control, the WT MFA2 mRNA was not stabilized in the absence of Puf3p. These results demonstrate that the CYT2 3′ UTR alone is sufficient to promote Puf3p-mediated decay.
Figure 2.
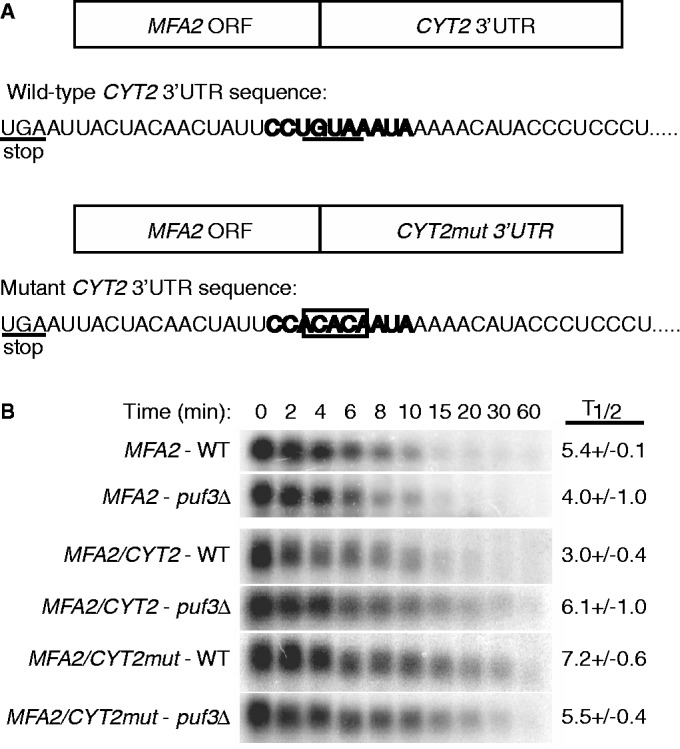
The CYT2 3′ UTR is sufficient to promote Puf3p-mediated mRNA decay, and the conserved UGUA element is required for Puf3p regulation. (A) CYT2 3′ UTR sequences immediately downstream of the translational stop codon for the WT MFA2/CYT2 and mutant MFA2/CYT2mut mRNAs. The conserved 10 nt Puf3p element is highlighted in bold text, with the UGUA element underlined in the WT transcript. In the mutant transcript, the UGUA element is mutated to ACAC, which is boxed. (B) Representative northern blots of the decay of MFA2, MFA2/CYT2 and MFA2/CYT2mut mRNAs expressed from a WT strain or a puf3Δ strain are shown. Minutes following transcriptional repression are indicated above each set of blots, with the half-lives (T1/2) and standard error of the mean (SEM) determined from multiple experiments.
We next examined whether the single Puf3p binding site in the CYT2 3′ UTR is solely responsible for the recruitment of Puf3p and subsequent degradation of CYT2. The core UGUA tetranucleotide of the Puf3p binding site was mutated to ACAC (Figure 2A), which has previously been shown to prevent Puf3p binding and regulation of COX17 mRNA (7). When expressed in a WT strain, the MFA2/CYT2mut 3′ UTR transcript (Figure 2A) was stabilized 2-fold to a half-life of 7.2 ± 0.7 min, mirroring the decay profile of the MFA2/CYT2 3′ UTR expressed in a puf3Δ strain (Figure 2B). To verify the lack of Puf3p activity on the mutant transcript, the MFA2/CYT2mut 3′ UTR was also expressed in a puf3Δ strain. As expected, there was no further extension of the mutant transcript half-life (Figure 2B). Together, these results suggest that the single UGUA-containing binding site located within the 3′ UTR of CYT2 is solely responsible for Puf3p binding and regulation.
Puf3p stimulates deadenylation and subsequent degradation of CYT2 mRNA
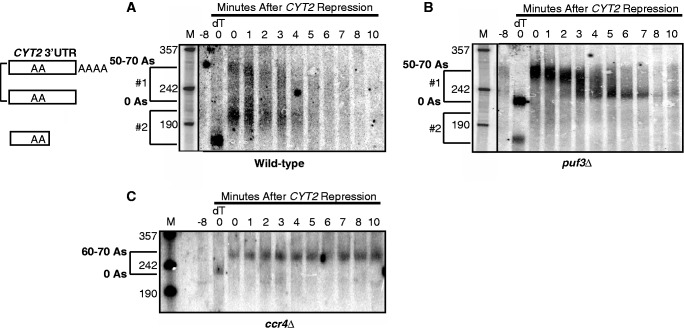
Puf3p promotes the rapid decay of COX17 by stimulating both the deadenylation phase of decay as well as subsequent decapping steps (6). The COX17 3′ UTR contains two Puf3p binding sites that are required for the rapid deadenylation and subsequent decay of the transcript. Since CYT2 contains only one site responsible for Puf3p regulation, we wished to determine if Puf3p could stimulate both deadenylation and subsequent decay steps of this mRNA as well. Transcriptional pulse-chase experiments were performed in WT, puf3Δ and ccr4Δ strains containing the MFA2/CYT2 3′ UTR fusion gene under the control of the inducible/repressible GAL10 UAS (Figure 3A–C). An initial pulse of MFA2/CYT2 3′ UTR transcription is induced by adding galactose to cell cultures and incubating for 8 min, then repressed by the addition of dextrose. This pulse-chase creates a pool of newly transcribed RNAs that can be monitored for both deadenylation and subsequent decay over time. Before northern analysis, the CYT2 3′ UTR is cleaved from the MFA2 coding region using an oligo-directed RNase H reaction, which assures visualization of decay events from only the 3′ end of the mRNA.
Figure 3.
Puf3p promotes rapid deadenylation and decay of CYT2 mRNA. Shown are representative northern blot analyses of transcriptional pulse-chase experiments examining decay of CYT2 mRNA from a WT (A) puf3Δ (B) or ccr4Δ strain (C). Minutes following transcriptional repression are indicated above each blot. All mRNAs were cleaved just downstream of the stop codon via oligo-directed RNaseH reactions before gel loading. The 0dT lane in each blot corresponds to mRNA from the 0 min time point in which the poly(A) tail was removed by RNaseH cleavage with oligo(dT). The −8 lane in each blot corresponds to background levels of mRNA expression before galactose induction of CYT2. Size markers (lanes M) are given in nucleotides. The top brackets in each panel (#1 brackets in A and B, sole bracket in C) correspond to mRNAs with full-length 3′ UTRs and poly(A) tail lengths ranging from 0 to 70 adenosines. The #2 brackets in A and B encompass either alternatively cleaved and polyadenylated 3′ UTR species, or transcripts that are both deadenylated and trimmed into the 3′ UTR (top of bracket #2) and species resulting from an additional cleavage product of RNaseH in the 0dT lanes resulting from a poly(A) tract located within the CYT2 3′ UTR (bottom of bracket #2).
Puf protein stimulation of deadenylation has previously been shown to act at least partially through the deadenylase Ccr4p (8,10). Our analysis of CYT2 in a ccr4Δ strain shows that the transcript is initially synthesized with a poly(A) tail length of 60–70 adenosines as measured by the difference in size between RNA species in the 0 min time point and the 0 min time point treated with oligo dT (Figure 3C, bracket). In the presence of RNase H and oligo dT, the poly(A) tail is cleaved off the 3′ UTR species, thus establishing the size of the 3′ UTR. Following transcriptional repression, there is no shortening of the poly(A) tail through 10 min, indicating that Ccr4p is the primary deadenylase acting on CYT2 (Figure 3C).
In the WT strain, CYT2 mRNA initially displayed three populations of transcript sizes following the induction of transcription. The top population contains the full poly(A) tail length of 50–70 As (Figure 3A, 0 min lane, top of bracket #1). After transcriptional repression, the poly(A) tails on this population are shortened within 3–4 min, followed by rapid degradation of CYT2 within 5–6 min. A minor population of transcripts is fully deadenylated at the 0 min time point as indicated at the bottom of bracket #1. However, the majority of CYT2 following transcriptional induction is found in the bottom population of transcripts that are shorter than the fully deadenylated species (Figure 3A, 0 min lane, top of bracket #2). This species could represent RNAs trimmed into the 3′ UTR following deadenylation, or it could represent polyadenylated transcripts from an alternative internal cleavage site in the 3′ UTR. In the 0 min time point treated with oligo dT, we observe that most of the transcripts are cut into an even shorter population (Figure 3A, 0dT lane, bottom of bracket #2). Analysis of the CYT2 3′ UTR revealed that the size of this short population corresponds to the distance from the beginning of the 3′ UTR to a large internal poly(A) tract. It is possible that oligo dT is simply binding this internal poly(A) tract to induce RNase H cleavage at this site. Alternatively, the short population may represent an alternative 3′ end. We performed RNase protection assays to analyze CYT2 3′ ends and detected both the full-length 3′ end and the alternative shorter 3′ end at similar levels (data not shown). Thus, the species at the top of bracket #2 may represent the polyadenylated version of the alternative 3′ end, which degrades rapidly within 5–6 min. Together, these results demonstrate that CYT2 transcripts, regardless of the cleavage/polyadenylation site, undergo rapid deadenylation and subsequent degradation in the presence of Puf3p.
In contrast, following the induction of transcription in the puf3Δ strain, CYT2 is found in one major population of transcripts with poly(A) tail lengths of 50–70 As (Figure 3B, 0 lane, top of bracket #1). The deficiency of deadenylated transcripts at this time point as compared with the WT strain indicates slower deadenylation in the absence of Puf3p. The majority of transcripts are deadenylated within 4–5 min, but then persist as a deadenylated species through 10 min following transcriptional repression. The persistence of deadenylated transcripts as compared with their rapid decay in the WT strain suggests that a step of decay following deadenylation is also slowed in the absence of Puf3p. The most likely explanation is that the decapping step following deadenylation is slowed, as seen previously with COX17 (6). Cleavage of the poly(A) tails off the mRNAs shows that the majority of transcripts contain the full-length 3′ UTR fragment, while only a small fraction are of the smaller sized 3′ end (Figure 3B, 0dT lane, bottom of bracket #2). Our RNase protection assays detected both 3′ ends in the puf3Δ strain at equal levels to the WT strain (data not shown), indicating that the absence of Puf3p does not alter the distribution of cleavage/polyadenylation sites. A small, but detectable, population of transcripts that may represent polyadenylated forms of the smaller 3′ end exist in the 0 min lane, but disappear rapidly within 4 min, similar to the WT strain (Figure 3B, top of bracket #2). These observations suggest that regardless of the presence of Puf3p, CYT2 transcripts with the smaller 3′ end are rapidly degraded. However, the full-length transcript is dependent on Puf3p to promote rapid deadenylation and decapping, supporting a model whereby a single Puf3 protein stimulates multiple steps of decay through a single 3′ UTR binding site. If the bottom species represent an alternate cleavage/polyadenylation site, it is unclear why it escapes Puf3p decay stimulation, as it still contains the Puf3p binding site. In addition, we do not detect the alternative shorter 3′ UTR species in the ccr4Δ strain.
Regulation of Puf3p targets is condition-specific
Having experimentally confirmed the new mRNA targets of Puf3p regulation, we next analyzed whether these mRNAs are regulated in a condition-specific manner. Computational analyses found that the levels of mRNAs containing Puf3p binding elements are coordinately regulated under different carbon source conditions (28), which suggests that the activity of Puf3p to promote rapid decay of its targets is regulated by the available carbon source. In support of this prediction, previous experiments demonstrated that COX17 mRNA is stabilized when WT yeast is grown in ethanol, exhibiting a decay phenotype identical to that observed in a puf3Δ strain (28). Here, we wanted to test whether the mRNA decay activity of Puf3p is also inactivated by galactose and raffinose conditions, and if so, if these sugars induce partial or complete inactivation of Puf3p. We also wanted to test whether different mRNA targets are affected equally by condition-induced changes in Puf3p activity. For these experiments, we used the Puf3p target mRNAs CYT2, TUF1 and COX17 as reporters to monitor Puf3p activity in ethanol, galactose and raffinose conditions.
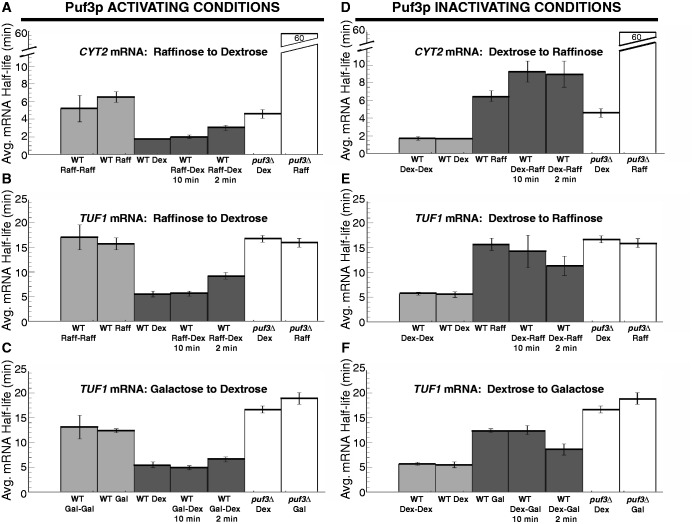
Steady-state transcriptional shut-off experiments were performed with WT and puf3Δ yeast strains grown continuously in ethanol, galactose or raffinose to monitor the decay of the three Puf3p target mRNAs. Growth in ethanol caused all three mRNAs to decay with extended half-lives that were similar between WT and puf3Δ strains, demonstrating complete inactivation of Puf3p activity in this carbon source (Figures 4A, 5A and 6A). For example, the half-life of CYT2 is extended from 1.7 ± 0.1 min in WT-dextrose conditions to 11.8 ± 1.2 min in WT-ethanol conditions (Figure 4A). Given that the half-life of CYT2 in puf3Δ-dextrose conditions is 4.6 ± 0.5 min, it appears that ethanol has a Puf3p-independent stabilizing effect on CYT2 decay. However, a half-life of 13.6 ± 1.9 min in puf3Δ-ethanol conditions shows that Puf3p makes no contribution to CYT2 decay in ethanol, supporting complete inactivation of Puf3p (Figure 4A). For TUF1 mRNA, growth in ethanol extended the half-life from 5.5 ± 0.6 min in WT-dextrose to 17.9 ± 0.9 min in WT-ethanol, which is similar to the half-lives determined in puf3Δ-dextrose and puf3Δ-ethanol conditions (Figure 5A). These data again support complete inhibition of Puf3p-mediated decay activity in ethanol. Finally, as described previously, COX17 mRNA is stabilized from a half-life of 4.0 ± 0.7 min in WT-dextrose to 15.0 ± 1.0 min in WT-ethanol conditions, similar to that found in puf3Δ-ethanol conditions (Figure 6A). Together, these results demonstrate that ethanol fully inhibits the activity of Puf3p to stimulate decay of these target mRNAs.
Figure 4.
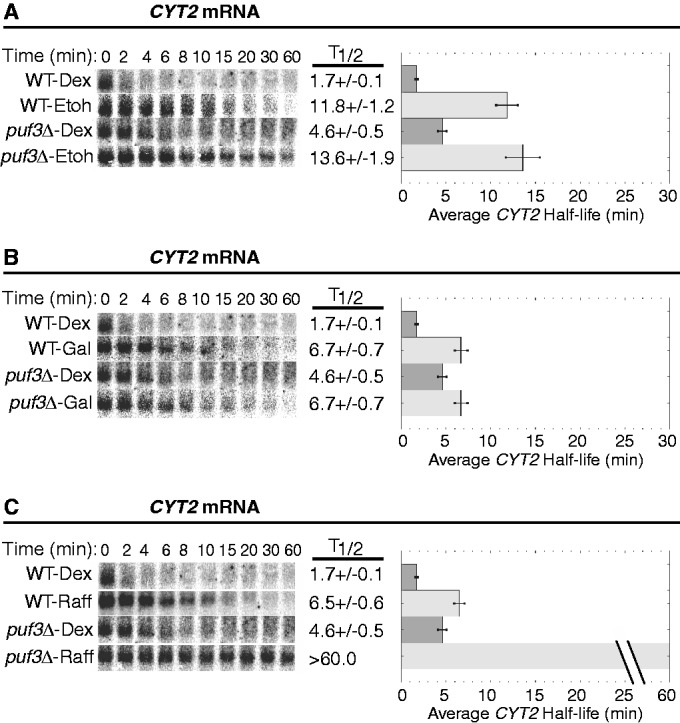
Puf3p-mediated decay of CYT2 mRNA is conditionally regulated. Shown are representative northern blot analyses of the decay of the CYT2 transcript from a WT and a puf3Δ strain grown in media supplemented with dextrose (Dex), ethanol (EtOH), galactose (Gal) or raffinose (Raff). Minutes following transcriptional repression are indicated above each set of blots, with the half-lives (T1/2) and standard error of the mean (SEM) determined from multiple experiments shown graphically to the right of each set of blots.
Figure 5.
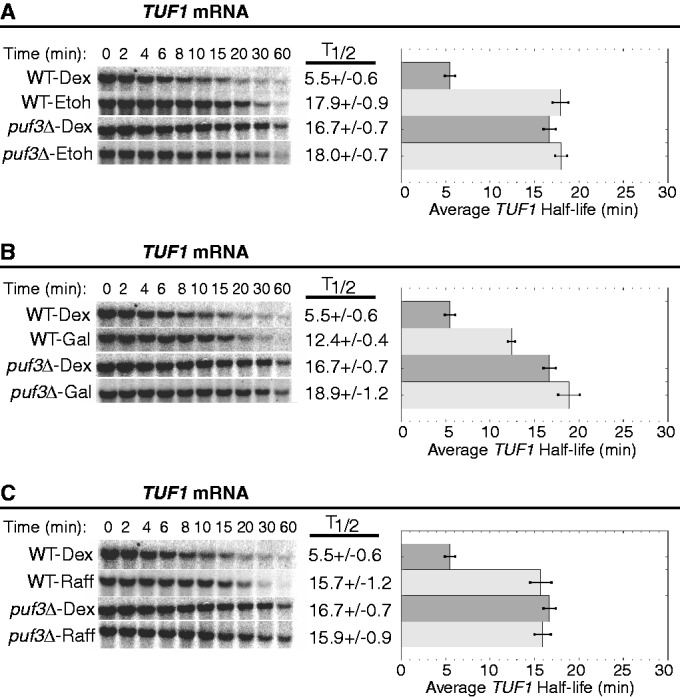
Puf3p-mediated decay of TUF1 mRNA is conditionally regulated. Shown are representative northern blot analyses of the decay of the TUF1 transcript from a WT strain and a puf3Δ strain grown in media supplemented with dextrose (Dex), ethanol (EtOH), galactose (Gal) or raffinose (Raff). Minutes following transcriptional repression are indicated above each set of blots, with the half-lives (T1/2) and standard error of the mean (SEM) determined from multiple experiments shown graphically to the right of each set of blots.
Figure 6.
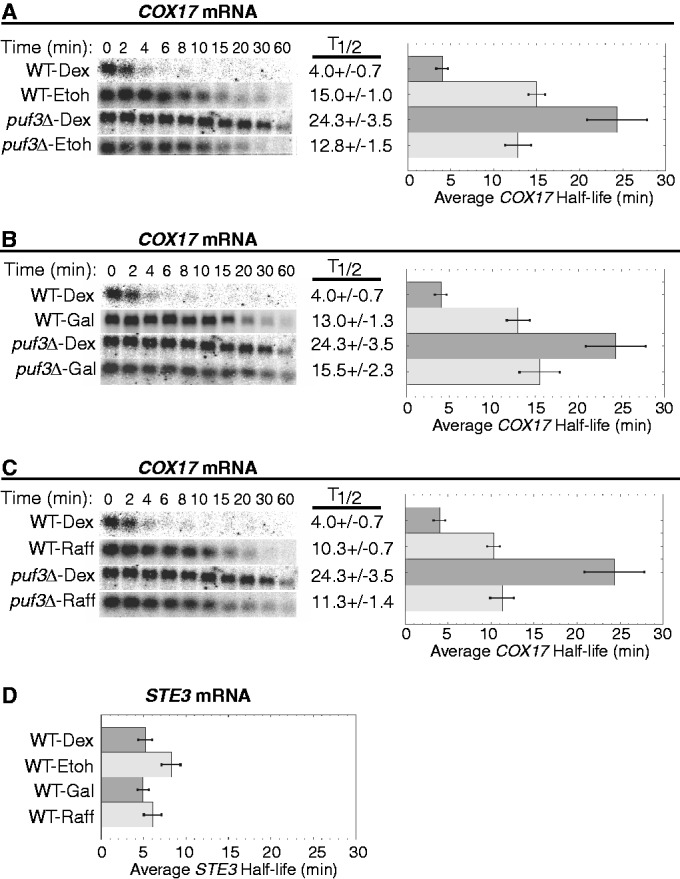
Puf3p-mediated decay of COX17 mRNA is conditionally regulated. Shown are representative northern blot analyses of the decay of COX17 (A–C) and STE3 (D) transcripts from a WT strain and a puf3Δ strain grown in media supplemented with dextrose (Dex), ethanol (EtOH), galactose (Gal) or raffinose (Raff). Minutes following transcriptional repression are indicated above each set of blots, with the half-lives (T1/2) and standard error of the mean (SEM) determined from multiple experiments shown graphically to the right of each set of blots. The average half-life of STE3 mRNA is represented graphically with SEM determined from multiple experiments.
Next, we examined the effect of galactose on Puf3p activity. This carbon source was the only one of the three tested that did not induce complete inhibition of Puf3p activity on all three mRNA reporters (Figures 4B, 5B and 6B). For both CYT2 mRNA and COX17 mRNA, Puf3p was unable to stimulate decay in galactose conditions. CYT2 mRNA was stabilized 3.9-fold by galactose compared with dextrose and decayed with the identical half-life of 6.7 ± 0.7 min in WT-galactose and puf3Δ-galactose conditions (Figure 4B). COX17 mRNA was stabilized 3.2-fold by galactose versus dextrose and decayed with similar half-lives of 13.0 ± 1.3 min in WT-galactose and 15.5 ± 2.3 min in puf3Δ-galactose (Figure 6B). In contrast, Puf3p was able to partially stimulate TUF1 mRNA decay in galactose. Compared with WT-dextrose, TUF1 mRNA was stabilized in WT-galactose 2.2-fold to a half-life of 12.4 ± 0.4 min, while in puf3Δ-galactose, TUF1 mRNA was stabilized 3.4-fold to a half-life of 18.9 ± 1.2 min (Figure 5B). Thus, galactose severely inhibits Puf3p activity, but target mRNAs are not affected equally by the residual Puf3p activity in this carbon source.
Finally, the effect of raffinose on Puf3p activity was analyzed. Like ethanol, raffinose appears to completely inactivate Puf3p for all three mRNAs tested (Figures 4C, 5C and 6C). In WT-raffinose versus dextrose, TUF1 mRNA was stabilized 2.8-fold to a half-life of 15.7 ± 1.2 min, which was indistinguishable from half-lives in puf3Δ-dextrose and puf3Δ-raffinose (Figure 5C). Likewise, COX17 mRNA was stabilized 2.6-fold in WT-raffinose (10.3 ± 0.7 min), which was similar to puf3Δ-raffinose (Figure 6C). For CYT2, raffinose also stabilized this mRNA 3.8-fold to a half-life of 6.5 ± 0.6 min, similar to the stabilization by galactose and the puf3Δ-dextrose conditions (Figure 4C), thus supporting complete inactivation of Puf3p activity in raffinose. However, unexplainably CYT2 mRNA was extremely stable in puf3Δ-raffinose conditions. As determined from multiple experiments, this transcript displayed a half-life >60 min, suggesting a transcript-specific effect of this combination on CYT2 stability.
To further ensure that the stabilization of CYT2, TUF1 and COX17 mRNAs in ethanol, galactose and raffinose conditions is attributed to altered Puf3p activity as opposed to nonspecific indirect effects of the carbon sources, we also monitored the decay of STE3 mRNA, an unstable transcript that is not regulated by Puf3p. As shown in Figure 6D, STE3 rapidly degraded in dextrose, galactose and raffinose, with average half-lives between 5 and 6 min in these conditions. The slight 1.6-fold increase of the STE3 mRNA half-life in ethanol compared with dextrose was expected, as previous microarray data sets have shown a 1.8-fold increase in STE3 levels in ethanol versus dextrose (31). We have previously shown that the half-life of another transcript that is not a target of Puf3p regulation, MFA2 mRNA, is not altered in ethanol conditions (28). Together, the data support the hypothesis that mRNAs regulated by Puf3p are specifically stabilized in ethanol, galactose and raffinose conditions owing to decreased Puf3p activity. All Puf3p target mRNAs encode proteins involved in the mitochondria. Therefore, the stabilization of Puf3p targets promotes increased mitochondrial function under those conditions that require mitochondria for efficient cell growth.
Puf3p activity is rapidly altered by carbon source changes
We next wanted to determine how quickly Puf3p could be activated or inactivated by changing the available carbon source. To assess the dynamics of Puf3p activity, we performed modified steady-state transcriptional shut off experiments with WT and puf3Δ strains, each containing the rpb1-1 allele. To examine how quickly Puf3p activity could be activated, cultures were grown in raffinose or galactose (inhibitors of Puf3p activity) and harvested at mid-log growth phase. Cells were then quickly shifted to media containing dextrose (activator of Puf3p activity) and incubated for 2 or 10 min before transcriptional repression by temperature shift. In contrast, to assess how quickly Puf3p could be inactivated, cultures were grown in dextrose to mid-log phase, then shifted to media with galactose or raffinose for an incubation period of 2 or 10 min before transcriptional repression. CYT2 and TUF1 mRNA half-lives were monitored to evaluate Puf3p activity.
For both mRNAs and with all carbon source changes tested, we found that Puf3p can be rapidly activated or inactivated within 2 min. Following a shift from raffinose to dextrose to examine Puf3p activation, the half-life of CYT2 mRNA decreased from 6.5 ± 0.5 min in raffinose alone to 3.0 ± 0.3 min within 2 min of the shift (Figure 7A). Within 10 min of the shift, the CYT2 half-life of 2.0 ± 0.2 min was identical to that in cells grown continuously in dextrose, indicating full Puf3p activation (Figure 7A). Likewise, the half-life of TUF1 mRNA decreased from 15.7 ± 1.2 min in raffinose alone to 9.2 ± 0.7 min within 2 min and 5.6 ± 0.5 min within 10 min of shifting to dextrose, fully mirroring the rapid half-life in continuous dextrose (Figure 7B). A shift from galactose to dextrose promoted a similar activation of Puf3p. The half-life of TUF1 decreased from 12.4 ± 0.4 min in galactose alone to 6.6 ± 0.5 min within 2 min of the shift to dextrose and 4.9 ± 0.4 min within 10 min of the shift, which is identical to the half-life in continuous dextrose (Figure 7C). Therefore, regardless of the initial carbon source in the culture, the addition of dextrose rapidly activates the function of Puf3p to stimulate decay of its target mRNAs, with significant activation occurring in only 2 min.
Figure 7.
Puf3p-mediated decay is rapidly activated or inactivated by altering the available carbon source. Shown are graphical representations of the average CYT2 and TUF1 mRNA half-lives as determined by multiple steady-state transcriptional repression experiments. For Puf3p activating conditions (A–C), WT cells were grown in raffinose (Raff) or galactose (Gal) conditions, then briefly incubated in raffinose, galactose or dextrose (Dex) for 2 or 10 min. For Puf3p inactivating conditions (D–F), WT cells were grown in dextrose conditions, then briefly incubated in dextrose, raffinose or galactose for 2 or 10 min. To control for possible alterations to mRNA stabilities due to the resuspension procedure, Raff-Raff, Gal-Gal and Dex-Dex experiments were performed in cells grown continuously in raffinose, galactose or dextrose conditions, harvested, and then incubated with the respective carbon source for 2 min.
To ensure that the observed changes in mRNA half-life reflect changes in Puf3p activity and not an indirect effect of the carbon source resuspension process, WT cells grown in media supplemented with raffinose or galactose were harvested then resuspended in fresh media containing the respective raffinose or galactose for 2 min before transcriptional repression. For both CYT2 and TUF1 mRNAs, half-lives remained constant between cells grown continuously in raffinose and cells that underwent the raffinose to raffinose resuspension treatment (Figure 7A and B). Similarly, the half-life of TUF1 was unchanged between cells grown continuously in galactose and cells that underwent the galactose to galactose resuspension treatment (Figure 7C). These results eliminate the possibility that shifting the cells to fresh media causes physiological changes that indirectly alter mRNA stability.
To examine Puf3p inactivation, we first monitored the decay of CYT2 mRNA following a shift from dextrose to raffinose. Within 2 min of the shift, the half-life of CYT2 was already extended from 1.7 ± 0.1 min in dextrose alone to 9.0 ± 1.5 min (Figure 7D). A longer incubation to 10 min following the shift showed no further extension of the half-life, suggesting that Puf3p is inactivated within 2 min of raffinose addition. A similar phenotype was seen for TUF1 mRNA, as its half-life was extended from 5.5 ± 0.6 min in dextrose alone to 11.3 ± 2.0 min within 2 min following the shift to raffinose and 14.2 ± 3.3 min within 10 min of the shift, mirroring the extended half-life measured in continuous raffinose (Figure 7E). Analysis of TUF1 mRNA following a shift from dextrose to galactose also supported rapid inactivation of Puf3p. The TUF1 half-life was extended to 8.6 ± 1.1 min within 2 min of the shift and 12.5 ± 0.9 min within 10 min of the shift, which matches the half-life measured in continuous galactose (Figure 7F). As a control, equivalent half-lives of both CYT2 and TUF1 mRNAs were determined between cells grown continuously in dextrose and those that had been resuspended in fresh dextrose media for 2 min before transcriptional repression (Figure 7D–F). These data again suggests that the resuspension process does not indirectly cause the changes in mRNA stabilities. Changes in the available carbon source cause rapid activation or inactivation of Puf3p’s capacity to promote mRNA decay.
Condition-specific inactivation of Puf3p is not due to decreased expression or altered localization
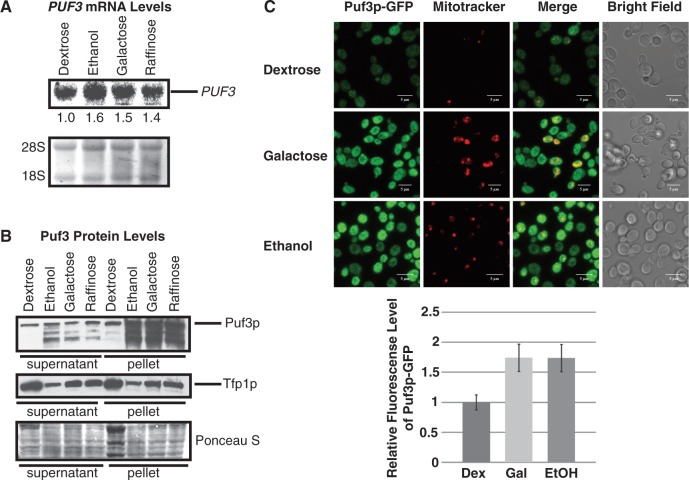
One mechanism by which Puf3p activity could be altered by environmental conditions is by changes in PUF3 expression, either at the transcriptional or translational level. Based on our observations that Puf3p activity is inhibited by galactose, raffinose and ethanol, this hypothesis would suggest that PUF3 expression is downregulated in these conditions. To test this hypothesis, steady-state levels of PUF3 mRNA and Puf3 protein were monitored in WT yeast strains grown in different carbon sources. As shown in Figure 8A, steady-state PUF3 mRNA levels in ethanol, galactose and raffinose conditions were slightly elevated as compared with PUF3 levels in dextrose conditions, suggesting that inhibition of Puf3p activity by ethanol, galactose and raffinose is not a consequence of reduced transcription. Similarly, Puf3 protein levels in ethanol, galactose and raffinose conditions were not decreased as predicted, but were increased, especially in the insoluble pellet fractions, when compared with Puf3p levels in dextrose (Figure 8B, top panel). To control for loading, we analyzed levels of Tfp1p, a subunit of the vaculolar ATPase V1 domain, which is minimally affected in dextrose, ethanol, galactose and raffinose conditions (<1.3-fold variation) according to microarray data sets provided at http://genome-www.stanford.edu/yeast_stress/explorer.shtml (31). Furthermore, the 3′ UTR of TFP1 mRNA does not contain any putative Puf3p binding sites. Tpf1p levels were reduced in ethanol, galactose and raffinose conditions when compared with dextrose conditions, which is opposite to the pattern of Puf3p levels (Figure 8B, middle panel). Ponceau S total protein staining further controlled for loading, showing a similar pattern to the Tfp1p levels (Figure 8B, bottom panel). It is not apparent why Puf3p levels increase under inactivating conditions. However, these results clearly demonstrate that inactivation of Puf3p by ethanol, galactose and raffinose is not accomplished by reduction of transcription or translation in these conditions, but may result from other regulatory mechanisms such as posttranslational modification or altered localization.
Figure 8.
Ethanol, galactose and raffinose do not decrease PUF3 expression levels or alter localization. (A) Northern blot analysis of PUF3 mRNA levels from WT cells grown in YEP media supplemented with 2% dextrose, ethanol, galactose or raffinose is shown with normalized fold changes in expression levels relative to dextrose indicated, along with ethidium bromide staining of 28S and 18S rRNA. (B) Puf3p from WT cells grown in YEP media supplemented with 2% dextrose, ethanol, galactose and raffinose was visualized in the top panel by western blot analysis using antibodies against Puf3p. Western blots were stripped and Tfp1p was visualized in the second panel using antibodies against Tfp1p. Equal OD600 units of cells before preparation of protein extracts were calculated for loading onto SDS polyacrylamide gels. Total protein loading was visualized in the bottom panel by Ponceau S staining. (C) Yeast expressing endogenously GFP-tagged Puf3p were grown in YEP media supplemented with 2% dextrose, galactose or ethanol. Each image represents 10 flattened Z plane slices through fixed cells using a confocal fluorescence microsope. Puf3p-GFP is shown in green, while mitochondria stained with Mitotracker Deep Red are shown in red. Merge indicates merger of the Puf3p-GFP and Mitotracker windows. Bright field images of each cell field are shown. The bar equals 5 µm. Fluorescence levels of Puf3p-GFP, quantified across cell fields and normalized to levels in dextrose, are graphically represented.
To analyze Puf3p localization under different conditions, endogenously GFP-tagged Puf3p was visualized in cells grown in dextrose, galactose or ethanol conditions. Similar to the results seen by western analysis, Puf3p-GFP levels detected microscopically were increased >1.5-fold in galactose and ethanol as compared with dextrose (Figure 8C). Despite differences in expression levels, Puf3p-GFP was diffusely localized across the cytoplasm with multiple small foci in all conditions, demonstrating that localization is not markedly altered by inactivating conditions. Previous work has shown a similar localization pattern in dextrose (24). Specifically, Puf3p does not exclusively localize to mitochondria in galactose or ethanol, though some Puf3p does appear to localize to the perimeter of mitochondria as seen by the yellow rings surrounding the red mitochondrial foci in the merged images between Puf3p-GFP and Mitotracker red (Figure 8C). This result supports previous studies indicating that Puf3p localizes to the cytosolic face of mitochondria (30).
Condition-specific inactivation of Puf3p does not disrupt Puf3p-mRNA interactions
The condition-specific regulation of Puf3p could be altering its ability to associate with its mRNA targets or its ability to functionally stimulate decay of the targets. To address whether Puf3p-mRNA interactions are disrupted under inactivating conditions, qPCR was used to quantitate the amount of mRNA that copurified with endogenously TAP-tagged Puf3p from cells grown in dextrose or galactose. We tested COX17 mRNA as a positive target of Puf3p regulation, as well as CBS1 and STE3 mRNAs as negative controls to ensure we specifically enriched for Puf3p binding targets after the IP process. As shown in Figure 9A and B, COX17 mRNA was enriched 200- to 800-fold following Puf3p IP from both galactose and dextrose conditions as compared with input mRNA levels, while there was no enrichment of either CBS1 or STE3 mRNA. Having demonstrated specificity of Puf3p binding, we next compared levels of COX17 that copurified with Puf3p from dextrose and galactose conditions. COX17 mRNA levels from the IP were >2-fold higher in galactose versus dextrose, indicating that Puf3p-mRNA interactions are not disrupted under conditions that inactivate Puf3p-medicated decay stimulation (Figure 9C). The higher level of COX17 mRNA that copurified with Puf3p in galactose likely reflects the increased amount of Puf3p expressed in galactose conditions (Figure 9D). These results support the hypothesis from previous studies that Puf3p binds and traffics its mRNA targets to the mitochondria (38,39), and our results demonstrate that such binding occurs regardless of the conditions. In contrast, the inactivation of Puf3p is likely disrupting its ability to stimulate decay of the bound targets.
Figure 9.
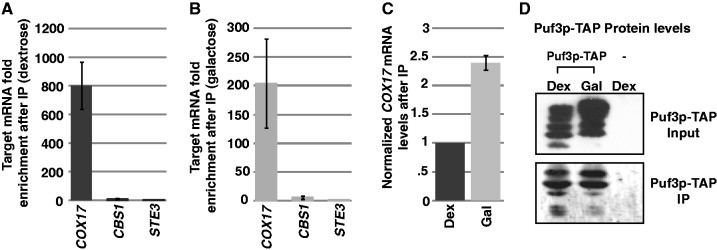
Puf3p binds its target mRNAs in both activating and inactivating conditions. Endogenously TAP-tagged Puf3p was immunoprecipitated (IP) from yeast grown in YEP media supplemented with 2% dextrose or galactose using IgG-Sepharose, and RNAs associated with Puf3p were analyzed by qPCR. Fold mRNA enrichment after IP from dextrose (A) or galactose (B) conditions was calculated by comparing average Cq values of mRNAs isolated after IP versus average Cq values of the respective mRNAs isolated from total cell lysate before IP. (C) Average Cq values of mRNAs isolated after IP were compared between dextrose and galactose conditions, with levels normalized to that found in dextrose. (D) Puf3p-TAP protein levels from dextrose and galactose conditions in total cell lysates (Input) and after IP were assessed by western blot using anti-TAP antibodies. A strain expressing nontagged Puf3p (-lane) is also shown.
DISCUSSION
Yeast Puf3p is a member of the Puf class of eukaryotic proteins that bind conserved 3′ UTR sequences to promote rapid decay and/or repress translation of the bound transcript. This study has identified 10 new mRNA targets of Puf3p-mediated decay, all of which are nuclear-transcribed mRNAs encoding proteins involved in mitochondrial function (Figures 1–3). In addition, the activity of Puf3p to promote decay of these transcripts is rapidly altered by changes in the available carbon source (Figures 4–7). Puf3p is active to degrade its target mRNAs in dextrose conditions when mitochondria are not needed; however, Puf3p is inhibited by ethanol, galactose and raffinose conditions when mitochondrial function is required for efficient growth, resulting in stabilization of the target mRNAs. These rapid changes in Puf3p activity are not due to decreased transcription or translation, or altered localization under inactivating conditions (Figure 8). Puf3p has previously been shown to regulate COX17 mRNA decay, interact with the cytoplasmic face of mitochondrial membranes, regulate mitochondrial biogenesis and inheritance, as well as promote the asymmetric localization of nuclear-encoded mRNAs that are translated near the mitochondria (30,38,39). We demonstrate that Puf3p can associate with its target mRNA regardless of the available carbon source (Figure 9), permitting Puf3p to traffic mRNAs to the mitochondria even when it cannot stimulate their decay. Together, these observations support Puf3p’s comprehensive role as a global and dynamic regulator of mitochondrial function.
Our results indicate that only those Puf3p binding sites that perfectly conform to the consensus (C/U)(A/C/U)UGUA(A/U)AUA motif contribute to Puf3p-mediated decay (Table 1). Many of the mRNA targets contain one perfect site plus one or more imperfect sites, yet their fold stimulation of decay is equal to or less than the decay stimulation of targets with only one perfect Puf3p site. These data imply that the imperfect sites, even if only lacking conservation at a single nucleotide outside the core UGUA, contribute little to Puf3p recruitment and decay stimulation. In contrast, binding by Puf4p and Puf5p appears to be more promiscuous because these proteins can flip out nonconserved nucleotides away from the binding surface (45). Previous binding experiments have demonstrated that mutations altering the consensus Puf3p sites in COX17 reduce binding affinity to Puf3p (7). Our decay analysis of multiple mRNA targets suggests that only the presence of a sequence that perfectly conforms to the Puf3p consensus motif can predict the extent of Puf3p decay stimulation of an mRNA. mRNAs lacking a Puf3p binding site, including other nuclear-encoded transcripts encoding mitochondrial proteins, are not regulated by Puf3p. Even so, experimental validation is still necessary to validate targets containing perfect Puf3p motifs, as RSM10 with one perfect and one imperfect site shows no decay stimulation by Puf3p. Sequences flanking the Puf3p motif or other trans factors may influence the accessibility of the site to Puf3p.
The consensus Puf3p binding motif derived from global association studies places either a cytosine or uracil at the −2 position upstream of the core UGUA sequence. However, the crystal structure of yeast Puf3p bound to the conserved motifs from the COX17 3′ UTR demonstrated that a cytosine at the −2 position is important for high-affinity binding as well as decay regulation (12). Our analysis of Puf3p binding motifs in multiple target mRNAs demonstrates a similar finding, in that most of the fully conserved sites contain a cytosine at −2 (Table 1). The MRP1 and MRP21 mRNAs that contain uracils at the −2 positions of their conserved sites display a <2-fold stimulation of decay by Puf3p, suggesting reduced binding to these motifs.
A common mechanism by which Puf proteins promote mRNA decay is through recruitment of deadenylase enzymes (2). In yeast, Puf3p promotes both deadenylation and decapping of COX17 mRNA (6), Puf4p and Puf5p promote deadenylation of HO mRNA, and Puf5p interacts with decapping factors (9–11). In both of the above situations, two Puf proteins act together to stimulate decay of a target mRNA, as COX17 binds two Puf3 proteins. Other mRNAs identified as targets of yeast Puf proteins are also regulated by combinations of two or three Pufs (48). These observations might suggest that multiple Puf proteins are required for full decay regulation of an mRNA. However, this study shows that Puf3p regulation of the CYT2 mRNA occurs through a single Puf3p binding site in the 3′ UTR of CYT2 (Figure 2). Our analysis of CYT2 decay also demonstrates that Puf3p promotes both rapid deadenylation and decapping of this transcript (Figure 3). Thus, a single Puf3p can promote multiple steps of decay, presumably through interaction with multiple decay factor complexes.
The kinetics of CYT2 deadenylation and decay are extremely rapid. Previously we found that in the presence of Puf3p, a pool of adenylated COX17 is deadenylated within 4 min following transcriptional repression, then decapped and degraded within 10 min (6). In the absence of Puf3p, COX17 deadenylates slowly for 10 min and persists for 30 min. In contrast, in the presence of Puf3p the majority of CYT2 transcripts are already fully deadenylated at the time of transcriptional repression following the 8-min pulse of transcription. Subsequent decapping and degradation occurs within 5 min. In the absence of Puf3p, the pool of CYT2 is still fully adenylated at the end of the transcriptional pulse period. The pool deadenylates for 4 min following transcriptional repression, then persists as a deadenylated species for 10 min. In the presence of Puf3p, a species smaller than the deadenylated form predominates at the time of transcriptional repression. This species could represent mRNA from an alternative internal cleavage and polyadenylation site, as dT treatment to remove poly(A) tails resulted in an even smaller species. These species were also seen in the absence of Puf3p, but at a much lower fraction of the total population. If these species are indeed from an alternative polyadenylation site, the poly(A) tail would be ∼25 As, which is abnormally short. In addition, these species are not stabilized in the puf3Δ strain, even though the Puf3p binding site is still present. The size of the smallest species in the dT lanes also corresponds to cleavage at a large internal poly(A) tract within the 3′ UTR, which could simply be a result of the dT binding this region for cleavage by RNase H. RNase protection assays to evaluate 3′ ends revealed both the full-length 3′ UTR as well as a smaller end corresponding to the internal poly(A) tract, but these species were found equally represented between the WT and puf3Δ strains. Thus, Puf3p does not influence cleavage at the internal site. In fact, it is possible that we saw the smaller species in the RNase protection assays simply because large poly(A) tracts do not hybridize well, so RNase could attack this region. An alternative explanation for the smaller species detected primarily in the WT strain at the time of transcriptional repression is that Puf3p stimulates 3′ trimming into the 3′ UTR. Such trimming is typically not seen for other mRNAs owing to slower kinetics of such 3′ to 5′ decay compared with the kinetics of decapping and 5′ to 3′ decay (49). However, 3′ to 5′ trimming can be seen with COX17 in the presence of Puf3p when decapping is blocked (W.M.O., unpublished observation). The rapid 3′ to 5′ trimming of CYT2 could result from stimulation of the exosome or hyperstimulation of the Ccr4p deadenylase by Puf3p. We believe the latter possibility may be the case because an exosome mutant does not block the 3′ to 5′ trimming of COX17 in the absence of decapping (J.R., unpublished observation).
While Puf3p stimulates deadenylation and subsequent decay of both CYT2 and COX17, the default rate of CYT2 decay in the absence of Puf3p is faster than the default rate of COX17 decay (half-lives of 4.6 versus 24.3 without Puf3p, respectively). We believe this may be due to additional elements in the 3′ UTR of CYT2 that modulate decay. Specifically, the CYT2 3′ UTR contains three AU-rich elements, which have been shown to mediate either stabilizing or destabilizing effects on yeast mRNAs depending on the type of AU-rich sequence (4,50). At least one of these elements is likely acting independently of Puf3p to promote rapid decay of CYT2 mRNA. The presence of these elements may also explain the >13-fold stabilization of CYT2 in the absence of Puf3p in raffinose versus dextrose (Figure 4). Some AU-rich elements stabilize mRNAs only in certain carbon sources due to differential protein binding (50). We hypothesize that in the absence of Puf3p, at least one of the AU-rich elements becomes accessible for binding to a stabilizing protein that is active in raffinose conditions. Analysis of the mitochondrial-encoding transcripts in general demonstrates that these mRNAs are degraded rapidly even in the absence of Puf3p, suggesting that many of the mRNAs use multiple mechanisms, including Puf3p, to regulate their stability.
The competence of Puf3p to mediate rapid mRNA decay is dependent on the environmental growth conditions. For the CYT2, TUF1 and COX17 mRNA targets tested, Puf3p is only fully active to stimulate rapid decay in dextrose (Figures 4–6). In the presence of ethanol or raffinose, the mRNA targets are stabilized, with their extended half-lives indistinguishable between WT and puf3Δ strains (except for CYT2 in raffinose as described above), suggesting complete inactivation of Puf3p by these carbon sources. In galactose conditions, Puf3p’s decay activity is also inhibited, but some transcripts are still affected by a residual amount of Puf3p activity. The half-lives of CYT2 and COX17 are similarly extended between WT and puf3Δ strains in galactose, demonstrating no decay stimulation by Puf3p. For TUF1, galactose extends the half-life over 2-fold, but deletion of PUF3 results in a further 1.5-fold extension in the half-life. Because steady-state levels of TUF1 mRNA are higher than either CYT2 or COX17 mRNA, we hypothesize that the residual amount of active Puf3p in galactose is more likely to bind mRNAs that are more abundant.
Analysis of the dynamics of condition-specific Puf3p activity revealed that decay stimulation is quickly altered by changing the available carbon source. For all carbon sources tested, Puf3p is fully activated or inactivated within 2–10 min of changing the carbon source to or from dextrose (Figure 7). We also observed no reduction in PUF3 mRNA or protein levels in conditions that inactivate Puf3p (Figure 8). In fact, Puf3p levels appear to be increased under all inactivating conditions, especially in the insoluble fraction of the protein extract. Similar results were seen previously, with PUF3 cDNA levels significantly higher in ethanol conditions than in dextrose (51). One previous study did report a decrease of Puf3p levels in lactate (30), but that result is likely unique to that carbon source where many proteins are downregulated, and not representative of other Puf3p inactivating conditions. Together, these data suggest that changes in Puf3p decay activity are regulated posttranslationally. One possibility is that Puf3p is sequestered into aggregates or its localization is otherwise altered to render it inactive. However, our results indicate that localization is not significantly altered between conditions, being diffusely localized with multiple small foci throughout the cytoplasm (Figure 8). A second possibility is that phosphorylation modulates Puf3p activity, as Puf6p-mediated translational repression is regulated by CK2 phosphorylation (52). Activity may also be altered by changes in proteins that interact with Puf3p.
In addition to its role in stimulating mRNA decay, Puf3p stimulates mitochondrial localization of nuclear-transcribed mRNAs containing Puf3p binding sites. In a puf3Δ strain grown in galactose (38) or glucose (39), these transcripts have decreased association with the mitochondria. It is hypothesized that Puf3p shuttles mRNA targets to the mitochondrial outer membrane surface, where they are translated and imported into the mitochondria. This hypothesis is supported by physical interactions between Puf3p and Mdm12p, a mitochondrial outer membrane protein (30). Tom20p, a component of the translocase of the mitochondrial outer membrane complex, is also required for mitochondrial localization of Puf3p target mRNAs (40). Our results demonstrating that Puf3p physically associates with its target mRNA in both dextrose and galactose conditions further supports the shuttling hypothesis (Figure 9). The following model accounts for the dual condition-specific functions of Puf3p. In dextrose conditions, yeast cells can readily perform fermentation, so mitochondria are not required and are in low abundance. To limit mitochondria, expression of mitochondrial proteins is downregulated. One mechanism of such repression involves Puf3p binding to mRNAs such as COX17, TUF1 and CYT2 and acting to mediate rapid degradation of the transcripts, presumably by recruiting deadenylase and decapping factors. In ethanol, galactose and raffinose conditions when yeast require additional metabolism via the mitochondria, Puf3p’s ability to stimulate decay is turned off, but its ability to bind its target mRNAs remains. With the bound mRNA now stabilized, Puf3p can increasingly shuttle the mRNA to the mitochondria for translation and import, though mRNAs could also be shuttled in dextrose conditions before degradation. This role of Puf3p in mRNA localization explains why Puf3p is not downregulated in nonfermenting conditions. Posttranslational modifications such as phosphorylation of either Puf3p or Puf3p binding partners may serve as the molecular switch that inhibits the decay activity of Puf3p. Future work will elucidate the mechanism of these activity changes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation [MCB-0848078 to W.M.O.]. Funding for open access charge: National Science Foundation [MCB-0848078].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Colin MacDiarmid for providing anti-Tfp1p antibodies, Carolyn Decker and Roy Parker for providing the Puf3p TAP-tagged strain and Puf3p GFP-tagged strain and Marc Spingola, Mindy Steiniger and members of the Olivas lab for helpful discussions and review of the manuscript.
REFERENCES
- 1.Schier AF. The maternal-zygotic transition: death and birth of RNAs. Science. 2007;316:406–407. doi: 10.1126/science.1140693. [DOI] [PubMed] [Google Scholar]
- 2.Miller MA, Olivas WM. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip. Rev. RNA. 2011;2:471–492. doi: 10.1002/wrna.69. [DOI] [PubMed] [Google Scholar]
- 3.Quenault T, Lithgow T, Traven A. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 2011;21:104–112. doi: 10.1016/j.tcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Muhlrad D, Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 5.Ross J. mRNA stability in mammalian cells. Microbiol. Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olivas W, Parker R. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 2000;19:6602–6611. doi: 10.1093/emboj/19.23.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson JS, Jr, Houshmandi SS, Lopez Leban F, Olivas WM. Recruitment of the Puf3 protein to its mRNA target for regulation of mRNA decay in yeast. RNA. 2004;10:1625–1636. doi: 10.1261/rna.7270204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee D, Ohn T, Chiang Y, Quigley G, Yao G, Liu Y, Denis C. PUF3 acceleration of deadenylation in vivo can operate independently of CCR4 activity, possibly involving effects on the PAB1-mRNP structure. J. Mol. Biol. 2010;399:562–575. doi: 10.1016/j.jmb.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 10.Goldstrohm AC, Seay DJ, Hook BA, Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- 11.Hook BA, Goldstrohm AC, Seay DJ, Wickens M. Two yeast PUF proteins negatively regulate a single mRNA. J. Biol. Chem. 2007;282:15430–15438. doi: 10.1074/jbc.M611253200. [DOI] [PubMed] [Google Scholar]
- 12.Zhu D, Stumpf CR, Krahn JM, Wickens M, Hall TM. A 5' cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc. Natl Acad. Sci. USA. 2009;106:20192–20197. doi: 10.1073/pnas.0812079106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–289. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol. Cell. 2001;7:855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 16.Opperman L, Hook B, DeFino M, Bernstein DS, Wickens M. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat. Struct. Mol. Biol. 2005;12:945–951. doi: 10.1038/nsmb1010. [DOI] [PubMed] [Google Scholar]
- 17.Cheong CG, Hall TM. Engineering RNA sequence specificity of Pumilio repeats. Proc. Natl Acad. Sci. USA. 2006;103:13635–13639. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta YK, Nair DT, Wharton RP, Aggarwal AK. Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity. Structure. 2008;16:549–557. doi: 10.1016/j.str.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 19.Miller MT, Higgin JJ, Hall TM. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat. Struct. Mol. Biol. 2008;15:397–402. doi: 10.1038/nsmb.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stumpf CR, Kimble J, Wickens M. A Caenorhabditis elegans PUF protein family with distinct RNA binding specificity. RNA. 2008;14:1550–1557. doi: 10.1261/rna.1095908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta YK, Lee TH, Edwards TA, Escalante CR, Kadyrova LY, Wharton RP, Aggarwal AK. Co-occupancy of two Pumilio molecules on a single hunchback NRE. RNA. 2009;15:1029–1035. doi: 10.1261/rna.1327609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koh YY, Opperman L, Stumpf C, Mandan A, Keles S, Wickens M. A single C. elegans PUF protein binds RNA in multiple modes. RNA. 2009;15:1090–1099. doi: 10.1261/rna.1545309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Opperman L, Wickens M, Hall TM. Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc. Natl Acad. Sci. USA. 2009;106:20186–20191. doi: 10.1073/pnas.0812076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riordan DP, Herschlag D, Brown PO. Identification of RNA recognition elements in the Saccharomyces cerevisiae transcriptome. Nucleic Acids Res. 2011;39:1501–1509. doi: 10.1093/nar/gkq920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glerum DM, Shtanko A, Tzagoloff A. Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 1996;271:14504–14509. doi: 10.1074/jbc.271.24.14504. [DOI] [PubMed] [Google Scholar]
- 27.Beers J, Glerum DM, Tzagoloff A. Purification, characterization, and localization of yeast Cox17p, a mitochondrial copper shuttle. J. Biol. Chem. 1997;272:33191–33196. doi: 10.1074/jbc.272.52.33191. [DOI] [PubMed] [Google Scholar]
- 28.Foat BC, Houshmandi SS, Olivas WM, Bussemaker HJ. Profiling condition-specific, genome-wide regulation of mRNA stability in yeast. Proc. Natl Acad. Sci. USA. 2005;102:17675–17680. doi: 10.1073/pnas.0503803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatenay-Lapointe M, Shadel GS. Repression of mitochondrial translation, respiration and a metabolic cycle-regulated gene, SLF1, by the yeast Pumilio-family protein Puf3p. PLoS One. 2011;6:e20441. doi: 10.1371/journal.pone.0020441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Rodriguez LJ, Gay AC, Pon LA. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J. Cell Biol. 2007;176:197–207. doi: 10.1083/jcb.200606054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lelandais G, Saint-Georges Y, Geneix C, Al-Shikhley L, Dujardin G, Jacq C. Spatio-temporal dynamics of yeast mitochondrial biogenesis: transcriptional and post-transcriptonal mRNA oscillatory modules. PLoS Comput. Biol. 2009;5:e1000409. doi: 10.1371/journal.pcbi.1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol. Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guarente L, Lalonde B, Gifford P, Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984;36:503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- 35.Entian KD. Glucose repression: a complex regulatory system in yeast. Microbiol. Sci. 1986;3:366–671. [PubMed] [Google Scholar]
- 36.Gancedo JM, Gancedo C. Catabolite repression mutants of yeast. FEMS Miocrobiol. Lett. 1986;32:179–187. [Google Scholar]
- 37.Trawick JD, Rogness C, Poyton RO. Identification of an upstream activation sequence and other cis-acting elements required for transcription of COX6 from Saccharomyces cerevisiae. Mol. Cell. Biol. 1989;9:5350–5358. doi: 10.1128/mcb.9.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saint-Georges Y, Garcia M, Delaveau T, Jourdren L, Le Crom S, Lemoine S, Tanty V, Devaux F, Jacq C. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS One. 2008;3:e2293. doi: 10.1371/journal.pone.0002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gadir N, Haim-Vilmovsky L, Kraut-Cohen J, Gerst JE. Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. RNA. 2011;17:1551–1565. doi: 10.1261/rna.2621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eliyahu E, Pnueli L, Melamed D, Scherrer T, Gerber AP, Pines O, Rapaport D, Arava Y. Tom20 mediates localization of mRNAs to mitochondria in a translation-dependent manner. Mol. Cell. Biol. 2010;30:284–294. doi: 10.1128/MCB.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caponigro G, Muhlrad D, Parker R. A small segment of the MATa1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol. 1993;13:5141–5148. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Felici F, Cesareni G, Hughes JMX. The most abundant small cytoplasmic RNA of Saccharomyces cerevisiae has an important function required for normal cell growth. Mol. Cell. Biol. 1989;9:3260–3268. doi: 10.1128/mcb.9.8.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 45.Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 46.Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ulbricht RJ, Olivas WM. Puf1p acts in combination with other yeast Puf proteins to control mRNA stability. RNA. 2008;14:246–262. doi: 10.1261/rna.847408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muhlrad D, Decker CJ, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vasudevan S, Peltz SW. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol. Cell. 2001;7:1191–1200. doi: 10.1016/s1097-2765(01)00279-9. [DOI] [PubMed] [Google Scholar]
- 51.Jiang H, Guan W, Gu Z. Tinkering Evolution of Post-Transcriptional RNA Regulons: Puf3p in Fungi as an example. PLoS Genet. 2010;6:e1001030. doi: 10.1371/journal.pgen.1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng Y, Singer RH, Gu W. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev. 2008;22:1037–1050. doi: 10.1101/gad.1611308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.