Abstract
Mammalian mitochondrial transcription is executed by a single subunit mitochondrial RNA polymerase (Polrmt) and its two accessory factors, mitochondrial transcription factors A and B2 (Tfam and Tfb2m). Polrmt is structurally related to single-subunit phage RNA polymerases, but it also contains a unique N-terminal extension (NTE) of unknown function. We here demonstrate that the NTE functions together with Tfam to ensure promoter-specific transcription. When the NTE is deleted, Polrmt can initiate transcription in the absence of Tfam, both from promoters and non-specific DNA sequences. Additionally, when in presence of Tfam and a mitochondrial promoter, the NTE-deleted mutant has an even higher transcription activity than wild-type polymerase, indicating that the NTE functions as an inhibitory domain. Our studies lead to a model according to which Tfam specifically recruits wild-type Polrmt to promoter sequences, relieving the inhibitory effect of the NTE, as a first step in transcription initiation. In the second step, Tfb2m is recruited into the complex and transcription is initiated.
INTRODUCTION
The mitochondrial genome (mtDNA) encodes 22 tRNAs, 2 rRNAs and 13 proteins required for oxidative phosphorylation. Transcription of the double-stranded circular genome is initiated from two promoter regions, the heavy- and light-strand promoters (HSP1 and LSP), and generates polycistronic transcripts that become processed to produce individual RNA molecules (1). A second site for transcription initiation (HSP2) is located about 100-bp downstream from HSP1, but the sequence requirements and the function of this site in vivo are still debated (2–6).
The mitochondrial transcription machinery is simple. In yeast, the only two factors required are the mitochondrial RNA polymerase (Rpo41) and its accessory factor sc-mtTFB. Mammalian cells contain homologues to these factors, denoted as POLRMT and TFB2M using human nomenclature and Polrmt and Tfb2m for the mouse (1). Mammalian transcription also involves a third factor, transcription factor A (human TFAM and mouse Tfam), which is a high-mobility group-box protein (7,8). TFAM binds sequence specifically to regions immediately upstream from mitochondrial transcription start sites (9,10). TFAM bound at this position introduces a sharp 180° bend in DNA, and this structural change may be an essential step in the transcription initiation process (11–13). TFAM is also a DNA packaging factor that binds throughout the entire mtDNA molecule in a sequence-independent manner and wraps the genome into compact nucleoid structures (14–17).
POLRMT is structurally related to the single-subunit RNA polymerase encoded by bacteriophage T7 (T7 RNAP) (18,19). POLRMT contributes actively to promoter recognition (10), but in contrast to the phage polymerase, it cannot initiate transcription on its own (2,20). TFB2M interacts with POLRMT and functions as a transient component of the catalytic site during transcription initiation (21). The TFB2M protein interacts directly with the priming substrate but is probably lost from POLRMT when transcription enters the elongation phase (21,22). The X-ray structure of human POLRMT (19) revealed a T7-like catalytic domain in the C-terminus of the protein (amino acids 647–1230, Figure 1). In the preceding region, the N-terminal domain (NTD, amino acids 368–647, Figure 1), there is a region that resembles the AT-rich recognition loop of T7 RNAP, which in the phage polymerase is used for promoter binding. There is also a putative intercalating hairpin structure in the region, which, in T7 RNAP, helps to separate double-stranded DNA and to stabilize the single-stranded DNA for RNA synthesis.
Figure 1.
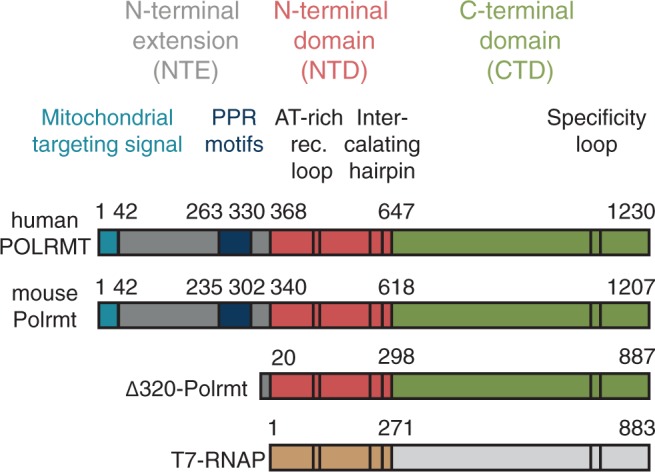
Schematic comparison of human POLRMT, mouse Polrmt, mouse Δ320-Polrmt and T7-RNAP. Major domains (NTE, NTD and CTD), the mitochondrial targeting signal and the PPR motifs are indicated and colour-coded. The AT-rich recognition loop, the intercalating hairpin and the specificity loop that all contribute to promoter recognition and melting in T7-RNAP are indicated in black. The transition between the NTE and the NTD is found at amino acid 368 in human POLRMT and 340 in mouse Polrmt. The N-terminal truncation mutant Δ320-Polrmt is a protein of 887 amino acids retaining only 20 amino acids of the NTE and thereby lacking the PPR motifs found in the wt polymerase.
POLRMT also contains an N-terminal extension (NTE, amino acids 42–368, Figure 1) not present in T7 RNAP. The NTE harbours a pentatricopeptide repeat (PPR) domain, which sequesters the AT-rich recognition loop (19). There is also a large portion of the N-terminal region of the NTE for which there is no structural information available. The yeast homologue of POLRMT, Rpo41, also contains an NTE, but the sequence similarities to mammalian POLRMT are limited. A limited truncation of the NTE in yeast generates a functional polymerase with an increased full length to abortive transcription ratio, whereas a larger truncation generates a polymerase incapable of initiation of transcription from double-stranded DNA (dsDNA) (23).
TFAM helps to recruit the POLRMT/TFB2M complex to mitochondrial promoters, and mutations that impair sequence-specific binding of TFAM upstream of promoters also inactivate promoter activity (10,24). The C-terminal tail of TFAM is likely to make direct physical contact with the POLRMT/TFB2M heterodimer (25). TFAM induces a sharp U-turn in promoter DNA, which places the C-terminal tail next to the transcription start site (11–13). In addition, changes in the distance between the TFAM binding site and start site for transcription impair transcription initiation efficiency (24). Therefore, the current model for transcription initiation in mitochondria states that TFAM interacts with a POLRMT/TFB2M heterodimer and recruits it as a complex to the promoter. The importance of TFAM for transcription initiation has been questioned based on the observation that only POLRMT and TFB2M were required to initiate promoter-specific transcription in vitro (26). However, later studies demonstrated that TFAM-independent transcription was inefficient and only observed under conditions that allowed for promoter breathing (i.e. low salt concentrations) or when negatively supercoiled DNA templates were used (27). Therefore, the situation is somewhat similar to nuclear transcription, where the free energy of supercoiling can circumvent the absolute requirement for some basal transcription factors (TFIIE, TFIIF and TFIIH) (28).
In the current work, we address the role of the NTE in POLRMT function. We conclude that this domain represses transcription initiation, both at promoter elements and unspecific DNA sequences. An important function of TFAM is to overcome this repression specifically at mitochondrial promoters. Our findings also suggest a step-wise assembly of the transcription machinery, according to which TFAM first recruits POLRMT to specific promoters, relieves the inhibitory effect of the NTE and enables TFB2M entry and transcription initiation.
MATERIALS AND METHODS
Förster resonance energy transfer
Measurements were performed as previously described (27). Oligos containing the Förster resonance energy transfer (FRET)-pair consisting of fluorescent cytosine analogue tCO (X) as a donor (D4) and non-emissive cytosine analogue tCnitro (Y) as an acceptor (A4). Oligonucleotide sequences are shown later in the text. D4 5′-d(AAA CTG TGG GGG GTG CXT TTG GGG TTT GGT TGG TTC GGG GTA TGG GGT TAG CAG CGG TGT GTG TGT GCT G)-3′ and A4 3′-d(TTT GAC ACC CCC CAC GGA AAC CCY AAA CCA ACC AAG CCC CAT ACC CCA ATC GTC GCC ACA CAC ACA CGA C)-5′.
Protein expression and purification
Mouse and human proteins were cloned and individually expressed as described (10). We constructed a recombinant baculovirus and expressed Δ320-Polrmt in insect cells. The proteins were purified over HIS-Select Nickel Affinity Gel (Sigma-Aldrich) that was washed with 40 mM imidazole in buffer A [25 mM Tris–HCl pH 8.0, 0.4 M NaCl, 10% glycerol, 10 mM 2-mercaptoethanol and 1 × protease inhibitors (29)] followed by elution by 250 mM imidazole in buffer A. The proteins were purified over a 1 ml HiTrap Heparin column (GE-healthcare) and a 1 ml HiTrap SP column (GE-healthcare). The HiTrap columns were equilibrated in 0.2 M NaCl purification buffer (25 mM potassium phosphate pH 7.0, 10% glycerol, 0.5 mM EDTA, 1 mM DTT) and the proteins were eluted by a 15 ml gradient (0.2–1.2 M NaCl in purification buffer). The proteins were dialyzed against 0.2 M NaCl purification buffer.
DNase I footprinting assay
The footprinting template was produced by PCR using the primer pair 5′ CAT GCT TGT TAG ACA TAA ATG C (forward) and 5′ CAT GAT TTT GTA AAA TTT TTA CAA GTA C (reversed). The reversed primer was labeled with T4 polynucleotide kinase enzyme (New England Biolabs) and γ-32P ATP (3000 Ci/mmol). The labelled template was produced using a plasmid (pCR2.1-TOPO) containing the mouse mitochondrial promoter region (mtDNA positions 15942-341) as template. The footprint reaction was performed in 20 μl containing 25 mM Tris–HCl pH 8.0, 10 mM MgCl2, 1 mM ATP, 100 μg/ml BSA, 1 mM DTT, 40 mM NaCl, labelled template (22 500 counts per minute) and proteins as indicated (0.25 pmoles Tfam and 1 pmole Polrmt and Tfb2m). The mixture was incubated for 20 min at room temperature followed by addition of 2 μl of 30 mU/μl DNase I diluted in 2.5× DNase I buffer with MgCl2 (Thermo Scientific). The DNase I reaction was stopped after 2 min by addition of 20 μl stop buffer [200 mM NaCl, 20 mM EDTA, 1% SDS and 100 μg/ml yeast tRNA (Ambion)] directly followed by incubation on ice. The DNA fragments were recovered with phenol extraction and ethanol precipitation and analysed on 8% denaturing polyacrylamide sequencing gels (1× TBE and 7M urea).
Microscale thermophoresis
The interaction affinity between Tfb2m and Polrmt (wt and Δ320) was studied using microscale thermophoresis (MST) as implemented in the Monolith NT.115 instrument (NanoTemper Technologies GmbH, Munich, Germany). We used hydrophilic capillaries with 80% light emitting diode power and 20% MST power for 40 s at 25°C. The Tfb2m was amine-labelled with 2-fold excess of NT-647 dye (Nanotemper) using the manufacturer’s protocol. Unlabelled Polrmt (wt and Δ320) was diluted in sixteen 2-fold dilution series starting from 1.50 mM and 1.07 mM for wt Polrmt and Δ320-Polrmt, respectively. Either 25 nM or 40 nM of the labelled Tfb2m was titrated to each of the dilution of wt Polrmt and Δ320-Polrmt, and the complex was incubated for 10 min at room temperature in the final buffer containing 20 mM HEPES, pH 7.5, 300 mM NaCl, 0.5 mM TCEP. GraphPad Prism® software was used for apparent Kd calculations and curve fitting using the law of mass action as described in (30). Each experiment was done in quadruplicate. Baseline-corrected, normalized and scaled fluorescence were plotted against the log of the range of Polrmt and Δ320-Polrmt concentrations.
In vitro transcription
The mouse LSP template was produced as previously described where EcoRI restriction sites flank the mitochondrial DNA insert (10). The non-promoter template was produced by digestion of the LSP template with EcoRI followed by gel purification (Quiagen) of the vector fragment. All transcription reaction volumes were 25 μl and contained 25 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 64 mM NaCl, 100 μg/ml BSA, 1 mM DTT, 400 μM ATP, 150 μM GTP, 150 μM CTP 10 μM UTP, 0,02 μM α-32P UTP (3000 Ci/mmol), 4 U RNase inhibitor Murine (New England Biolabs), 90 fmole of indicated template and proteins at indicated concentrations. The reactions were stopped after 30 min at 32°C by the addition of stop buffer (10 mM Tris–HCl, pH 8.0, 0.2 M NaCl, 1 mM EDTA and 100 μg/ml proteinase K) followed by incubation at 42°C for 45 min. The transcripts were purified with ethanol precipitation and the pellets were dissolved in 20 μl gel loading buffer (98% formamide, 10 mM EDTA, 0.025% xylene cyanol and 0.025% bromophenol blue) and heated at 95°C for 5 min. The samples were analysed on 4% denaturing polyacrylamide gels (1× TBE and 7 M urea) followed by exposure on photo film.
RESULTS
Production of a functional mouse Polrmt mutant lacking a major part of the NTE
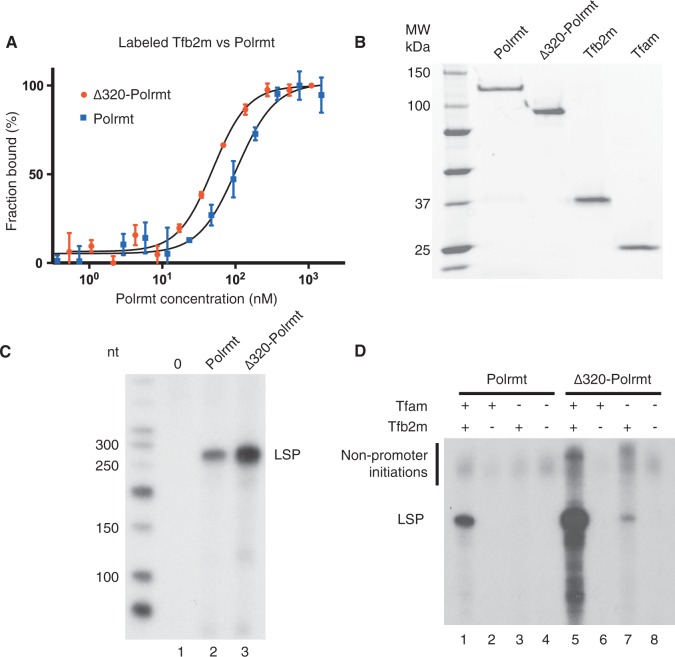
As a part of our structure function analysis of POLRMT, we expressed a number of N-terminally truncated versions of the human and mouse polymerases as well as point mutations disrupting the interactions between the PPR domain and the core polymerase. In agreement with previous studies of human POLRMT (19), both polymerases were sensitive to NTE truncations and mutations and most of our mutant versions could not be expressed as soluble proteins (data not shown). We were, however, able to identify one mutant version of mouse Polrmt, lacking the amino-terminal 320 amino acids, that we could readily express and purify. The Δ320 truncation was interesting, as it removed almost the entire NTE, but still retained all parts responsible for promoter recognition and catalysis of transcription found in the T7 RNA polymerase (Figure 1). Deletion of the NTE in yeast lowers the affinity between Rpo41 and sc-mtTFB (23). To investigate how the NTE truncation of Polrmt affected the Tfb2m interaction, we performed MST. In contrast to the yeast system, our results reveal that the Kd for Tfb2m interactions with Δ320-Polrmt was lower than that with wt Polrmt (Figure 2A). Removal of the NTE of Polrmt thus stabilized interactions with Tfb2m.
Figure 2.
Truncated Polrmt lacking the NTE has higher affinity to Tfb2m and can initiate promoter-specific transcription in the absence of Tfam. (A) MST of fluorescently labelled Tfb2m to Polrmt (wt and Δ320). The concentration of labelled Tfb2m was kept constant, whereas the concentration of Polrmt was varied as described in the ‘Materials and Methods’ section. The estimated fraction bound based on fluorescence is plotted against the concentration of the unlabelled Polrmt. Interaction KD between Tfb2m with wt and Δ320 was determined to be 105 ± 1.2 nM and 49.4 ± 1.1 nM, respectively. (B) Purified proteins used for in vitro transcription. (C) In vitro transcription from a mouse LSP template (90 fmole). Tfam (2.5 pmole) and Tfb2m (1.5 pmole) were added to all reactions. Reactions are no Polrmt in lane 1, wt Polrmt (0.5 pmole) in lane 2 and Δ320-Polrmt (0.5 pmole) in lane 3. Expected run-off product is 270 nt. (D) In vitro transcription from a mouse LSP template (90 fmole) was performed with Tfam (2.5 pmole) and Tfb2m (1.5 pmole) added as indicated. Polrmt (0.5 pmole) was added to reactions in lanes 1–4, and Δ320-Polrmt (0.5 pmole) was added to reactions in lanes 5–8. LSP run-off transcript is indicated.
The NTE together with Tfam ensures promoter-specific transcription
To address the role of NTE in transcription initiation, we compared Δ320-Polrmt with wt Polrmt in our reconstituted in vitro transcription system (Figure 2B) (10,20). We found that Δ320-Polrmt could initiate transcription from LSP and that the truncated polymerase was even more active than wild-type Polrmt (Figure 2C). We could thus conclude that the NTE in mouse Polrmt is dispensable for promoter recognition, promoter melting and transcription initiation.
We next analysed the transcription factor dependency of Δ320-Polrmt. As expected, both wt Polrmt and Δ320-Polrmt were strictly dependent on Tfb2m for transcription initiation (Figure 2D, lanes 1–2 and 5–6, respectively). Interestingly, loss of the NTE affected the strict requirement of TFAM for transcription of initiation. With the Δ320-Polrmt, we observed low levels of transcription initiation in the absence of Tfam (Figure 2D, lane 7). Transcription was initiated from the LSP promoter, but we also observed other unspecific transcription products in the absence of Tfam. We assumed that these products were formed by transcription initiation from non-promoter sequences in the linearized plasmid template. To investigate this possibility, we repeated the experiment using a linearized template without mitochondrial promoter sequences. In contrast to wt Polrmt (Figure 3A, lanes 1 and 2), Δ320-Polrmt was able to initiate transcription of promoter-independent transcription products on this dsDNA template. The activity was further stimulated in the presence of Tfam (Figure 3A, lanes 3 and 4, respectively). Based on our observation, we can thus conclude that the presence of the NTE has three distinct effects on Polrmt activity. First, it renders Polrmt strictly dependent on Tfam for transcription initiation. Second, the NTE decreases Polrmt activity in promoter-specific transcription initiation. And finally, the NTE inhibits transcription initiation at random non-promoter sequences.
Figure 3.
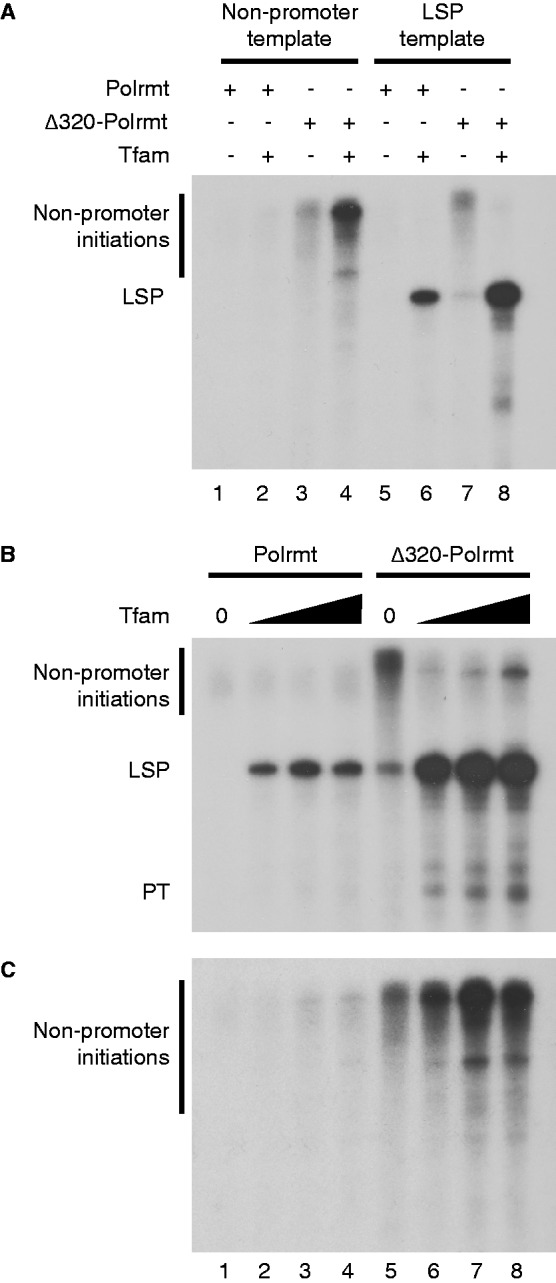
(A) In vitro transcription with either the mouse LSP template or a template lacking the mitochondrial promoter region (named non-promoter template) reveals that Δ320-Polrmt is active on a template lacking a mitochondrial promoter both in the presence and absence of Tfam. Mouse LSP or non-promoter template (90 fmole), Tfam (1.0 pmole), Polrmt (0.5 pmole) or Δ320-Polrmt (0.5 pmole) added as indicated. Tfb2m (1.5 pmole) was added to all reactions. (B) The non-promoter initiations of Δ320-Polrmt are strongly decreased in the presence of Tfam and LSP promoter. In vitro transcription assay from a mouse LSP template (90 fmole) was performed with Polrmt (0.5 pmole), added to reactions in lanes 1–4 and Δ320-Polrmt (0.5 pmole) added to reactions in lanes 5–8. Tfam was omitted in lanes 1 and 5 and added in increasing amounts in lanes 2–4 and 6–8 (0.25, 1 and 4 pmoles). Tfam concentrations correspond to ∼1 Tfam molecule per 1600, 400 and 100 bp of DNA, respectively. LSP run-off transcript and pre-termination (PT) as well as non-promoter initiation products are indicated. Tfb2m (1.5 pmole) was added to all reactions. (C) The non-promoter initiations of Δ320-Polrmt are not inhibited in the presence of Tfam in transcription from a template lacking a mitochondrial promoter region. The same experimental setup as in (B) but with a template lacking the mitochondrial promoter region.
Our results show that Tfam in the presence of a mitochondrial promoter region can reduce non-specific transcription and at the same time stimulate promoter-specific transcription (Figure 3A, compare lanes 7 and 8). To further investigate this effect, we examined how changes in Tfam concentrations could influence Δ320-Polrmt activity. When we analysed Δ320-Polrmt activity using a promoter-containing plasmid, we observed both promoter-specific and -unspecific transcription in the absence of Tfam (Figure 3B, lane 5). Already at low concentrations (1 Tfam/1600 bp of template), Tfam almost completely abolished non-specific transcription products and strongly stimulated promoter-specific transcription (Figure 3B, lane 6). When we analysed transcription from a template lacking promoter sequences, it was clear that wt Polrmt was inactive in all Tfam concentrations analysed (Figure 3C, lanes 1–4). Interestingly, Tfam did not inhibit the transcription of Δ320-Polrmt from this template; instead, the non-promoter DNA initiations seemed to be increased by addition of Tfam in the absence of a mitochondrial promoter (Figure 3C, lanes 5–8). From the results of promoter and non-promoter templates combined, we conclude that the inhibition of transcription initiation from non-promoter DNA by Tfam is not due to protection of the DNA, and that it needs the presence of a mitochondrial promoter region. Our findings instead support a model in which Tfam recruits Δ320-Polrmt to specific promoter sequences and thereby prevents transcription initiation at non-promoter sites. In the absence of a specific promoter sequence, the ability of Tfam to stimulate transcription initiation can also be observed at other DNA sequences, probably as an effect of Tfam increasing DNA flexibility and stimulating DNA breathing (31).
The mitochondrial transcription machinery can be recruited in discrete steps
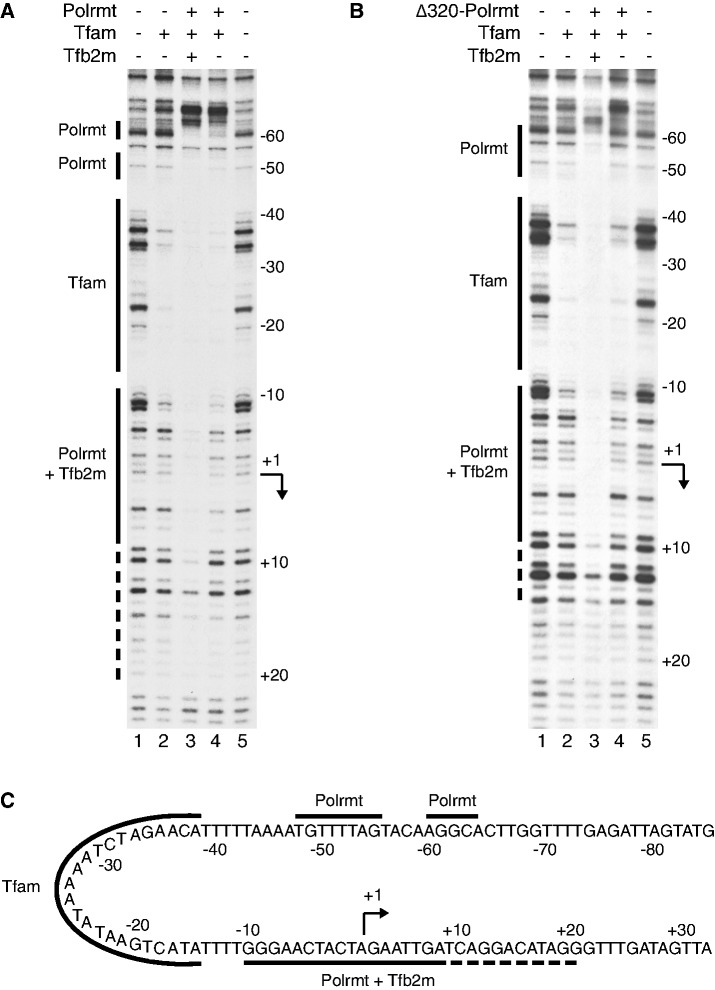
To further address the mechanism by which Tfam recruits other members of the transcription complex and the role of the NTE in this process, we performed DNase I footpriting. In previous experiment, we have showed that human TFAM protects a region upstream from the LSP transcription start site at position −15 to −38. In contrast, POLRMT and TFB2M cannot interact with the promoter when they are added by themselves or, together (10). We found that mouse Tfam leaves a footprint starting at position −11 of mouse LSP (Figure 4A, lane 2). With the complete transcription machinery present (Tfam, Polrmt and Tfb2m), the footprint was dramatically extended, and we observed protection downstream from the Tfam binding site, over transcription start site and a region extending to +9 (Figure 4A, lane 3). So far, our observations were in agreement with the previous analysis of the human mitochondrial transcription machinery (10). We also noted a weaker footprint extending from +10 to +20, which had not been observed with the human transcription machinery, which could indicate that the Polrmt/Tfb2m complex has a larger contact surface with DNA than previously suggested (10). In addition, there were also two distinct, not previously reported, footprints upstream from the Tfam binding site, one around position −50 and the other around position −60. Interesting, POLRMT can be cross-linked to the −50 to −60 region, suggesting that Polrmt interactions with this region may be important during assembly of the mitochondrial transcription machinery (Morozov et al., submitted to NAR). Because Tfam bends the promoter 180°, the Polrmt could interact with this region and still be in close contact with the transcription start site (Figure 4C).
Figure 4.
A complex composed of Tfam and Polrmt forms at the LSP promoter even in the absence of Tfb2m. (A) DNase I footprinting reveals a Tfam footprint (lane 2; solid line marked Tfam) upstream the promoter. Addition of Polrmt and Tfb2m (lane 3) generates additional footprints (solids line marked Polrmt and Polrmt + Tfb2m, respectively), over the promoter and transcription start site as well as upstream the Tfam binding site at position −50 to −60. The dashed line indicates an extended weaker promoter footprint reaching to +20. When only Tfam and Polrmt are added (lane 4), there are still weak footprints over the promoter, and strong footprints over the −50 and −60 region. Also a clear border is visible upstream the −60 footprint (B) Δ320-Polrmt leaves a similar, but not identical, footprint at the promoter and the −50 and −60 region in presence of both transcription factors (lane 3) but almost no footprint in absence of Tfb2m (lane 4). (C) A schematic representation of the protected areas of the mouse LSP promoter region. Tfam protects around −11 to −40, Polrmt protects the promoter and the transcription start site extending to +20 and two smaller regions around −50 and −60. The Polrmt promoter protection is strongly enhanced by Tfb2m. With the 180° bending effect of Tfam, it becomes clear that the different Polrmt-DNA interaction sites are placed close to each other in space.
We next addressed if Tfam could recruit Polrmt in the absence of Tfb2m. When Tfam was added together with Polrmt, but in the absence of Tfb2m, we observed protection not only over the Tfam binding site but also in the −50 region and the −60 region (Figure 4A, lane 4). The observed upstream pattern was similar to that observed with the complete transcription machinery, demonstrating that Tfam can recruit Polrmt to the promoter even in the absence of Tfb2m. In the absence of Tfb2m, no clear footprint was observed over the transcription start site (Figure 4A, compare lanes 3 and 4), demonstrating that interactions with DNA sequences surrounding the transcription start site require Tfb2m. This finding is in agreement with previous studies, which have identified Tfb2m as a transient component of the catalytic site, located near the transcription start site (21). In contrast to Polrmt, Tfb2m could not create an additional footprint in presence of Tfam (Supplementary Figure S1).
Human TFAM interacts with POLRMT, and the interaction surfaces have been mapped to the NTE and other regions of the POLRMT (Morozov et al., 2013, submitted to NAR). We wondered if Tfam could recruit Δ320-Polrmt to specific promoter elements or if interactions with NTE were required for this function. Addition of Δ320-Polrmt together with Tfam did create a footprint in the −50 and −60 regions, albeit weaker than that observed with wt Polrmt (Figure 4B, lane 4). When Tfb2m was added, Tfam together with Δ320-Polrmt produced a footprint over the promoter region that was nearly identical to that observed with wt Polrmt (Figure 4B, lane 3). Therefore, our data demonstrated that NTE is not required for promoter recruitment of the mitochondrial transcription machinery.
Tfb2m is required for promoter melting
Our DNase I footprinting analysis suggested that Tfam could specifically recruit Polrmt to promoter elements, but that Tfb2m was required for tight interactions with the transcription initiation site. To substantiate this idea further and to demonstrate its validity at other mitochondrial promoters, we turned to our human in vitro transcription system. We had previously used FRET to monitor how the mitochondrial transcription machinery influences the DNA structure at the human HSP1 transcription start site. We incorporated the fluorescent cytosine analogue tCO (donor) and the non-emissive cytosine analogue tCnitro (acceptor) close to the human HSP1 transcription initiation site and monitored changes in energy transfer efficiency on addition of TFAM (27). We now used this experimental system but with addition of POLRMT and TFB2M to the reactions (Table 1).
Table 1.
Efficiencies of the tCO-tCnitro FRET-pair in duplex D4-A4 after subsequent addition of POLRMT and TFAM (rows 1–3); POLRMT, TFB2M and TFAM (rows 4–7); or only TFAM (row 8) (EFRET,protein)
 |
FRET efficiency on addition of the corresponding volumes of the buffers in which all three proteins are dissolved is also shown (EFRET,buffer) and the difference in FRET efficiency between addition of buffer only or corresponding protein (ΔEFRET). Error intervals show the standard deviation. D4-A4 duplex is displayed below the table. DNA sequences containing the FRET-pair consisting of fluorescent cytosine analogue tCO (X) as a donor (D) and non-emissive cytosine analogue tCnitro (Y) as an acceptor (A). The high-affinity TFAM binding site is underlined and the transcription start site is highlighted in grey.
Addition of TFAM to this template introduces structural changes in the DNA (Table 1). We have previously demonstrated that the effect of TFAM is not restricted to promoter-containing templates, but due to the protein’s ability to bend and wrap any piece of DNA (27). We now monitored if addition of POLRMT and TFB2M could affect TFAM-dependent changes in energy transfer at the HSP1 transcription start site (Table 1). Interestingly, addition of TFAM together with POLRMT yielded a much smaller change in FRET efficiency compared with TFAM alone, suggesting that the presence of POLRMT could repress structural alterations near the transcription start site (Table 1). The addition of TFB2M reversed this inhibitory effect of POLRMT, and we again observed structural changes near the transcription start site (Table 1). Therefore, the FRET findings further support a model, in which TFAM recruits POLRMT to the promoter in the absence of TFB2M, where the polymerase physically blocks non-specific interactions between TFAM and the transcription start site. TFB2M is required for melting the transcription start site and initiation of transcription, which explains why the energy transfer efficiency increased when TFB2M is added together with POLRMT and TFAM.
DISCUSSION
We here analyse the function of the NTE of Polrmt in mitochondrial transcription and find that this domain is dispensable for transcription initiation in vitro. The Δ320-Polrmt protein displays higher sequence-specific and non–sequence-specific transcription activities, and this truncated polymerase can initiate transcription even in the absence of Tfam. In combination with DNase I footprinting, FRET analyses and in the light of an accompanying report demonstrating cross-linking between TFAM and POLRMT (Morozov et al., 2013, submitted to NAR), our findings allow us to formulate a model for how mitochondrial transcription is initiated in mammalian cells (Figure 5A and B).
Figure 5.
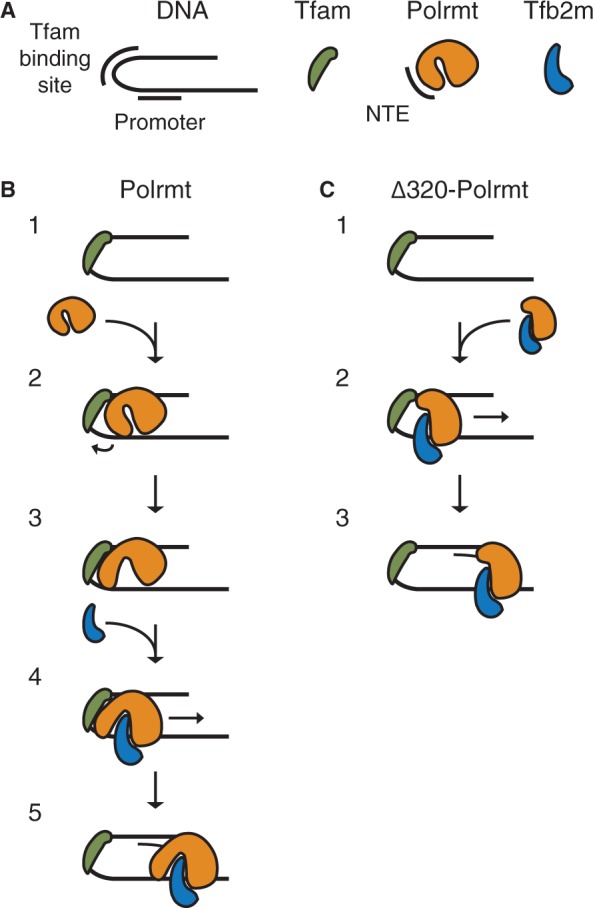
Model of mitochondrial transcription initiation. (A) Symbols. DNA with Tfam binding box and the promoter indicated, Tfam in green, Polrmt in orange with the NTE indicated and Tfb2m in blue. (B) Model of transcription initiation by Polrmt. (1) Tfam binds to a high-affinity site upstream of the mitochondrial promoter and introduce a 180° bend in the DNA. (2) Polrmt is recruited to the promoter region by protein–protein interactions with Tfam and protein–DNA interactions with the promoter and the region upstream of Tfam. (3) Tfam triggers a conformational change that relieves the inhibitory effect of the NTE and creates a conformation more similar to the Δ320-Polrmt protein that enables (4) binding of Tfb2m and formation of the fully assembled initiation complex. (5) Transcription is initiated. (C) Model of transcription initiation by Δ320-Polrmt. (1) Tfam is bound as in (A). (2) Δ320-Polrmt resembles the open Polrmt structure and interacts stronger with Tfb2m in solution. This heterodimer is recruited in a similar way as the wt creating the complete initiation complex. (3) Transcription is initiated.
According to our footprinting findings, the transcription initiation process begins with the binding of Tfam to a high-affinity binding site starting 10–15-bp upstream from the transcription start site. In the next step, Tfam recruits Polrmt to the promoter. Polrmt recruitment depends on direct interactions with Tfam (Morozov et al., 2013, submitted to NAR), but also involves structural changes in the promoter region, which facilitates Polrmt-DNA interactions. In agreement with this notion, conditions that induce promoter breathing can circumvent the strict Tfam requirement for transcription initiation (27). In the absence of Tfb2m, Polrmt-promoter interactions are mainly restricted to sequences around position −50 to −60. This interaction receives support from cross-linking experiments that demonstrated direct contacts between POLRMT and the promoter upstream region (Morozov et al., 2013, submitted to NAR). The contacts formed are explained from structural data demonstrating that Tfam induces a sharp bend in the promoter DNA, which help to juxtapose POLRMT and the −50 to −60 region (11–13). In the next step, Tfb2m enters the transcription complex and the fully assembled initiation complex that encircles the promoter and creates a clear footprint over the transcription start site. This observation is in agreement with our FRET analysis, which demonstrated that Tfb2m is required for structural changes around the transcription start site.
The strict requirement of Tfam for transcription initiation is lost when the NTE is deleted (Figure 5C). The molecular explanation for this effect is still obscure, but an important role of Tfam may be to regulate Polrmt’s ability to interact with promoter sequences, perhaps affecting the NTE structure and relieving its negative effect on transcription. In support of this notion, cross-linking experiments have demonstrated that TFAM interacts directly with regions within the NTE (Morozov et al., 2013, submitted to NAR). Because the Polrmt structure allows only specific deletions without affecting protein stability, we have not been able to study alternative deletions in the NTE. The exact structural motif in the NTE that is responsible for its inhibitory function has therefore not been identified. Rearrangements in the NTE structure triggered by Tfam and/or DNA interactions may also result in a more open Polrmt structure enabling Tfb2m binding.
The NTE of Polrmt helps to prevent initiation of transcription at other sites than the promoters. The observation that Tfam only activates wt Polrmt at specific promoter sites may be explained by previous structural work and single molecule analysis. Structural characterization of human TFAM bound to a mitochondrial promoter fragment (LSP) revealed that TFAM induces a sharp U-turn in the DNA (11–13). The conformation of TFAM bound at its specific binding site together with the stable U-turn may create a structural context that allows TFAM to interact with POLRMT and relieve the inhibitory function of the NTE. In contrast, single-molecule characterization of TFAM interactions with non-specific DNA demonstrated that the protein freely diffuse on DNA and that TFAM can soften the DNA structure, by partially unwinding the double-helix, leading to a compaction of the mtDNA genome (31). These observations suggest that there may be a structural difference between TFAM bound at sequence-specific and non–sequence-specific sequences. This difference can perhaps help to explain why TFAM only activates transcription at specific promoter sites. A stable U-turn at promoter sequences could function as a signal to the transcription machinery and specifically recruit POLRMT and TFB2M for transcription initiation. To clarify the situation, it would be valuable to obtain high-resolution structural data for TFAM bound to non–sequence-specific DNA fragments and compare the results with those already obtained with a LSP promoter fragment.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council grants [2010-2766 to N.G.L., 2011-6510 to B.M.H., 2010-5059 to B.M.H. and 2009-4848 to C.G.]; European Research Council [advanced grant no. 268897]; Swedish Cancer Foundation; and Olle Engkvist Byggmästare Foundation (to M.W.). Funding for open access charge: Vetenskapsrådet (Swedish Research Council).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Ann. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 2.Litonin D, Sologub M, Shi Y, Savkina M, Anikin M, Falkenberg M, Gustafsson CM, Temiakov D. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem. 2010;285:18129–18133. doi: 10.1074/jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lodeiro MF, Uchida A, Bestwick M, Moustafa IM, Arnold JJ, Shadel GS, Cameron CE. Transcription from the second heavy-strand promoter of human mtDNA is repressed by transcription factor A in vitro. Proc. Natl Acad. Sci. USA. 2012;109:6513–6518. doi: 10.1073/pnas.1118710109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc. Natl Acad. Sci. USA. 1982;79:7195–7199. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zollo O, Tiranti V, Sondheimer N. Transcriptional requirements of the distal heavy-strand promoter of mtDNA. Proc. Natl Acad. Sci. USA. 2012;109:6508–6512. doi: 10.1073/pnas.1118594109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terzioglu M, Ruzzenente B, Harmel J, Mourier A, Jemt E, Lopez MD, Kukat C, Stewart JB, Wibom R, Meharg C, et al. MTERF1 binds mtDNA to prevent transcriptional interference at the light-strand promoter but is dispensable for rRNA gene transcription regulation. Cell Metab. 2013;17:618–626. doi: 10.1016/j.cmet.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 9.Fisher RP, Topper JN, Clayton DA. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987;50:247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- 10.Gaspari M, Falkenberg M, Larsson NG, Gustafsson CM. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 2004;23:4606–4614. doi: 10.1038/sj.emboj.7600465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallberg BM, Larsson NG. TFAM forces mtDNA to make a U-turn. Nat. Struct. Mol. Biol. 2011;18:1179–1181. doi: 10.1038/nsmb.2167. [DOI] [PubMed] [Google Scholar]
- 12.Ngo HB, Kaiser JT, Chan DC. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat. Struct. Mol. Biol. 2011;18:1290–1296. doi: 10.1038/nsmb.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubio-Cosials A, Sidow JF, Jimenez-Menendez N, Fernandez-Millan P, Montoya J, Jacobs HT, Coll M, Bernado P, Sola M. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat. Struct. Mol. Biol. 2011;18:1281–1289. doi: 10.1038/nsmb.2160. [DOI] [PubMed] [Google Scholar]
- 14.Diffley JF, Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl Acad. Sci. USA. 1991;88: 7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 16.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parisi MA, Xu B, Clayton DA. A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol. Cell. Biol. 1993;13:1951–1961. doi: 10.1128/mcb.13.3.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masters BS, Stohl LL, Clayton DA. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 19.Ringel R, Sologub M, Morozov YI, Litonin D, Cramer P, Temiakov D. Structure of human mitochondrial RNA polymerase. Nature. 2011;478:269–273. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- 20.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 21.Sologub M, Litonin D, Anikin M, Mustaev A, Temiakov D. TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 2009;139:934–944. doi: 10.1016/j.cell.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mangus DA, Jang SH, Jaehning JA. Release of the yeast mitochondrial RNA polymerase specificity factor from transcription complexes. J. Biol. Chem. 1994;269:26568–26574. [PubMed] [Google Scholar]
- 23.Paratkar S, Deshpande AP, Tang GQ, Patel SS. The N-terminal domain of the yeast mitochondrial RNA polymerase regulates multiple steps of transcription. J. Biol. Chem. 2011;286:16109–16120. doi: 10.1074/jbc.M111.228023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dairaghi DJ, Shadel GS, Clayton DA. Human mitochondrial transcription factor A and promoter spacing integrity are required for transcription initiation. Biochim. Biophys. Acta. 1995;1271:127–134. doi: 10.1016/0925-4439(95)00019-z. [DOI] [PubMed] [Google Scholar]
- 25.Dairaghi DJ, Shadel GS, Clayton DA. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- 26.Shutt TE, Lodeiro MF, Cotney J, Cameron CE, Shadel GS. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc. Natl Acad. Sci. USA. 2010;107:12133–12138. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Y, Dierckx A, Wanrooij PH, Wanrooij S, Larsson NG, Wilhelmsson LM, Falkenberg M, Gustafsson CM. Mammalian transcription factor A is a core component of the mitochondrial transcription machinery. Proc. Natl Acad. Sci. USA. 2012;109:16510–16515. doi: 10.1073/pnas.1119738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parvin JD, Sharp PA. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 29.Myers LC, Leuther K, Bushnell DA, Gustafsson CM, Kornberg RD. Yeast RNA polymerase II transcription reconstituted with purified proteins. Methods. 1997;12:212–216. doi: 10.1006/meth.1997.0473. [DOI] [PubMed] [Google Scholar]
- 30.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 31.Farge G, Laurens N, Broekmans OD, van den Wildenberg SM, Dekker LC, Gaspari M, Gustafsson CM, Peterman EJ, Falkenberg M, Wuite GJ. Protein sliding and DNA denaturation are essential for DNA organization by human mitochondrial transcription factor A. Nat. Commun. 2012;3:1013. doi: 10.1038/ncomms2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.