Abstract
In the absence of telomerase, telomeres progressively shorten with every round of DNA replication, leading to replicative senescence. In telomerase-deficient Saccharomyces cerevisiae, the shortest telomere triggers the onset of senescence by activating the DNA damage checkpoint and recruiting homologous recombination (HR) factors. Yet, the molecular structures that trigger this checkpoint and the mechanisms of repair have remained elusive. By tracking individual telomeres, we show that telomeres are subjected to different pathways depending on their length. We first demonstrate a progressive accumulation of subtelomeric single-stranded DNA (ssDNA) through 5′-3′ resection as telomeres shorten. Thus, exposure of subtelomeric ssDNA could be the signal for cell cycle arrest in senescence. Strikingly, early after loss of telomerase, HR counteracts subtelomeric ssDNA accumulation rather than elongates telomeres. We then asked whether replication repair pathways contribute to this mechanism. We uncovered that Rad5, a DNA helicase/Ubiquitin ligase of the error-free branch of the DNA damage tolerance (DDT) pathway, associates with native telomeres and cooperates with HR in senescent cells. We propose that DDT acts in a length-independent manner, whereas an HR-based repair using the sister chromatid as a template buffers precocious 5′-3′ resection at the shortest telomeres.
INTRODUCTION
Telomeres are nucleoprotein structures at the ends of linear chromosomes. They ensure genome stability by protecting DNA extremities from fusions and degradations (1). In many eukaryotes, telomeres consist of TG-rich sequences and end with a single-stranded DNA (ssDNA) tail on the strand containing the 3′ extremity. Telomere-binding protein complexes cap both the double- and single-stranded telomeric repeats (2). This unusual structure is subject to the DNA end replication problem, which results in telomere shortening with each replication cycle and is counteracted by a specific cellular reverse transcriptase, the telomerase (3,4). Although telomerase is expressed in unicellular eukaryotes for their long-term maintenance, its expression is downregulated in most somatic tissues of many metazoa. Accordingly, telomere length was found to decrease with age in human individuals (5), suggesting a link between telomere length and aging. In human fibroblasts in culture, a lack of telomerase expression leads to progressive telomere shortening, and cells cease to divide in a process called replicative senescence (6–8). In Saccharomyces cerevisiae, when telomerase activity is removed experimentally, telomeres also shorten progressively and cells gradually lose viability in ∼60–80 population doublings (9,10). In both settings, eroded telomeres were found to activate a DNA damage checkpoint, indicating that the early response to the absence of telomerase is conserved in eukaryotes (11–14). The current model is that, as telomeres shorten, their protective cap declines and they may become recognized as accidental DNA ends, such as double-strand breaks (DSBs). Although some components of this checkpoint have been identified, the structures adopted by the short telomeres that activate them in the absence of telomerase remain to be defined. The issue is particularly obscured by the fact that even when telomerase is present, telomeres recruit many factors usually involved in the response to accidental DNA damage, specifically in DSB processing.
The activation of a DSB-like processing at wild-type telomeres is best described in S. cerevisiae but is also shown to occur in several other eukaryotes. The replication fork passage through telomeres is accompanied by 5′-3′ resection and fill-in (15–19). In budding yeast, these activities promote telomere elongation by telomerase at the shortest telomeres (20–25). Similar to the initial response to a DSB, short telomeres stabilize the MRX complex composed of Mre11, Rad50 and Xrs2, and the phospho-inositide-3 kinase-related protein kinase (PIKK) Tel1. This induces the serial action of the nucleases and helicases Sae2, Sgs1-Dna2 and Exo1, the activity of which is highly stimulated in S and G2/M phases (16,26,27). Accordingly, a DSB placed next to a short telomeric tract displays higher levels of 5′-end resection, compared with one placed next to a longer telomeric tract (28–30). However, direct evidence for a length-dependent processing at native telomeres has been lacking. Nonetheless, in contrast to an accidental DSB that is repaired by homologous recombination (HR) in S and G2/M phases, telomeres are elongated by telomerase.
In telomerase-negative budding yeast cells, the DSB-like response at short telomeres appears to be more complete. Similar to a DSB in S or G2/M phase (31), telomeres recruit Tel1, the major ssDNA-binding protein replication protein A (RPA), and the other PIKK checkpoint kinase Mec1 and its interacting factor Ddc2 (32,33). Rad9, Chk1 and Rad53 are also activated, leading to an effective G2/M arrest (13,34). Moreover, the deletion of DSB processing factors such as Tel1, Rad50 and Xrs2 delays senescence (33,35,36), and the deletion of checkpoint components such as Mec1 or Mec3 abolishes the G2/M cell cycle arrest found in cells of senescing yeast cultures (12,13). This indicates that eroded telomeres indeed activate a full checkpoint similar to the DSB checkpoint. However, in contrast to a DSB, the senescence checkpoint appears to be permanent, and telomeres are not efficiently repaired.
Another consequence of telomerase loss is the rapid recruitment of repair factors that are usually excluded from wild-type telomeres. Rad52, an essential HR factor in S. cerevisiae, relocalizes into telomere-colocalizing foci in most cells depleted of telomerase (32). Together with other HR factors, Rad52 function is critical for both the short- and long-term survival of telomerase-negative cells (13,33,36–43). However, despite the presence of many homologous telomeric repeats, evidence for massive telomeric rearrangements has not been found in short-term cultures of senescing cells (33). Only a small subset of cells, termed post-senescence survivors, arises in late-senescent cultures (37,44). In these cells, telomeres are extended either by long and heterogeneous telomeric tracts or by Y′ subtelomeric elements through HR-based pathways for which the mechanisms remain difficult to uncover. The exact role of HR components during the first part of senescence is even less understood.
Several lines of evidence suggest the simultaneous activation of the replication checkpoint in telomerase-negative budding yeast cells. It was shown that Mrc1, a component of the replication stress checkpoint, is phosphorylated and required, together with Rad9, for Rad53 activation in senescent cells (34). Similarly, Mms1, a subunit of an E3 ubiquitin ligase complex involved in replication repair and stabilization of the replication fork during replication stress, also contributes to the viability of cells in the absence of telomerase (33). These results indicate that replication stress protection and signalling is important in telomerase-negative cells. However, although telomere replication appears to be constitutively difficult and requires telomeric proteins to proceed (45–48), evidence that an additional replicative stress emanates from telomeres after the deletion of telomerase remains controversial (40,49,50).
At stalled replication forks, a myriad of proteins stabilize the forks, activate the replication checkpoint and restart or repair the forks during or after S phase. For instance, the DNA damage tolerance (DDT) pathway, also termed post-replicative repair, allows the bypass of DNA damages by the replication fork and partially requires the replication checkpoint to be activated (51–53). In this pathway, Proliferating Cell Nuclear Antigen (PCNA) is mono-ubiquitylated in response to the fork encountering the damage, thereby recruiting translesional polymerases and promoting an error-prone mode of repair. Alternatively, subsequent poly-ubiquitylation of PCNA by the E2 ubiquitin-conjugating complex Mms2-Ubc13 and the E3 ligase/helicase Rad5 orients the repair to a template-switch mode. The intact sister chromatid serves as a template for DNA synthesis, resulting in an error-free mode of repair (54–56). This template switch generates X-shaped replication intermediates that are resolved mainly by Sgs1-Top3 (57). The HR factors Rad51 and Rad52 are also potentially involved in the generation of these intermediates, but the relationship between the HR and DDT pathways remains unclear (58–62). Although telomeres are difficult replication substrates and the replication checkpoint appears to be activated in telomerase-negative cells, the involvement of the DDT pathway in telomere biology has not yet been explored.
The idea, suggested by in vivo studies, that the senescence process is initiated at the shortest telomere(s) in mammals has been challenged by cell culture analysis (63–66). We demonstrated using formal genetic analysis that the onset of senescence is determined by a single dominant telomere in budding yeast, likely the shortest one (67). Accordingly, generating a single artificial very short telomere accelerates the onset of senescence (33). Our results showed an enrichment of Tel1 and Mec1 at this telomere, indicating that cells sense this short telomere as DNA damage. Interestingly, MEC1 deletion abolished the acceleration of senescence in the presence of a critically short telomere, suggesting that Mec1 recognition of the shortest telomere initiates the senescence process. However, the exact structure that activates Mec1 in this context was not determined. This telomere also rapidly colocalizes with Rad52 after telomerase loss, and strains depended on RAD52 and MMS1 for short-term survival (32,33). Thus, the DNA damage checkpoint and HR are activated earlier in the presence of a very short telomere in the absence of telomerase, but it is not known whether the permanent arrest of senescence is achieved by the accumulation of several such telomeres.
In this work, we took advantage of the cellular setting in which a single very short telomere is generated to address the question of the structure of the shortest telomere that induces senescence. Genetic data combined with direct analysis of telomeres having different lengths in the same cell population deprived of telomerase activity demonstrate that 5′-3′ resection is stimulated at the shortest telomeres, exposing subtelomeric ssDNA and promoting the onset of senescence. HR factors, which are recruited in a Tel1-dependent manner to these short telomeres, counteract this resection, explaining their role in delaying senescence at these early time points after telomerase loss. We also found that factors of the DDT pathway are required to maintain the viability of pre-senescent cells. To our surprise, Rad5 localizes at wild-type telomeres, supporting the notion that semi-conservative DNA replication through telomeric repeats is challenging. We propose that whereas replication stress factors operate in a length-independent manner, HR acts at short telomeres by specifically compensating for 5′-3′ resection and thus limiting the telomere shortening rate. Hence, distinct mechanisms operate at short and relatively longer telomeres.
MATERIALS AND METHODS
Materials
All enzymes were purchased from New England Biolabs, chemicals came from Sigma-Aldrich and yeast media were from Difco. Oligonucleotides listed in Supplementary Table S3 were synthesized by Eurofins MWG Operon.
Yeast strains
Most yeast strains used in this study were derivatives of yT136 and yT138 diploids that contained the Control or the VST versions of the telomere VIIL on both chromosomes VII and bore one copy of the tlc1::PrαNat allele in the W303 background (33). Additional gene deletions or epitope tagging were introduced as described (68) using oligonucleotides listed in Supplementary Table S3 and appropriate crossings. See Supplementary Table S1 for the full list of strains.
Solid senescence assay
Senescence was analysed with spot assays as described in Supplementary Figure S1 and in (33,67). Briefly, diploid strains homozygous for the construct at telomere VIIL and heterozygous for TLC1 and relevant genes were sporulated on 2% potassium acetate for 3–5 days at 30°C. Spores were germinated for 6–9 h at 30°C in YPGal-Raff (2% galactose; 1% raffinose) to induce the excision of the URA3-containing circle. Cells were subsequently plated on YPD containing Nourseothricin and left to grow for 1.5 days to select for tlc1Δ. A small portion of the colonies was used for genotyping in appropriate selective media. Then 8–16 independent clones with the relevant genotypes were spotted (4000 cells and 10-fold serial dilutions) on Nourseothricin-containing rich media and grown for 2 days at 30°C for each passage. Plates were scanned on an EPSON Perfection V750 Pro scanner, and cells from the most concentrated spots were then respotted until senescence was observed. DNA was prepared for telomere length determination by telomere-polymerase chain reaction (telomere-PCR) using oligonucleotides oT182 (Y′ telomeres), oT156 (VIIL telomeres) and oT531 (VIR telomeres; (69)) [Supplementary Table S3].
Quantification and statistical analysis of senescence
Quantification of cell viability was performed as in (67). For each clone, the signal intensity of each spot was measured using the ‘Microarray Profile’ plugin of ImageJ 1.46a. Signal intensity versus dilution could then be plotted. A threshold value of the intensity was set to correspond to an exponentially growing strain (here one-fifth of the maximum intensity of cells grown to saturation) and used for all experiments. The viability index is the theoretical dilution necessary to obtain a spot intensity equal to the threshold value. Negative values were obtained by extrapolation of the curve, and the minimum value was limited to −10. Viability indexes for each genotype were plotted as boxplots using R2.15.2. P-values were determined using pairwise Wilcoxon rank-sum tests and adjusted with a false discovery rate correction. At least two complete independent experiments were performed for each pair of diploid starting strains (Control and VST), and one is shown.
Chromatin immunoprecipitation
Immunoprecipitation of chromatin was performed as described (33) using individual clones obtained as described in Supplementary Figure S1 and subjected to the senescence assay described above. Briefly, colonies obtained in germination plates were inoculated in 50 ml of Nourseothricin-containing rich media, grown to log phase, fixed in 1% formaldehyde solution and lysed in Hepes 50 mM, NaCl 140 mM, EDTA 1 mM, Triton X-100 1%, Na-deoxycholate 0.1% containing Mg132 (C2211, Sigma) and Complete Mini, EDTA-free (Roche) with glass beads (4 × 30s at 6.5 m/s in a cooled FastPrep, MP Bio). Chromatin was sonicated (Bioruptor UCD-200, Diagenode) and immunoprecipitated with anti-c-Myc (9E10 purchased from Sigma), anti-Rad52 or anti-Rad51 (obtained from R. Rothstein and A. Shinohara, respectively). Purified DNA was analysed by quantitative PCR (qPCR) using FastStart Sybr Green master detection (Roche) and primers oT310-oT311 (subtelomere VIIL), oT312-oT313 (subtelomere VIR), oT570-oT571 (subtelomere Y′) and oT314-oT315 (ARO1). Fold increase values are the ratio of a given telomere enrichment (immunoprecipitated/input) over the enrichment of the ARO1 locus (immunoprecipitated/input) unless otherwise stated. Error bars indicate SEM values obtained from at least three independent cultures.
Telomere structure analysis
To obtain a sufficient amount of DNA to further analyse telomere structure, we produced tlc1Δ cells as described above and in Supplementary Figure S1. Relevant starting diploid strains were germinated as described above, and then plated to obtain 2.5 × 106 colonies on plates that contained galactose and Nourseothricin. To simultaneously select for additional mutations, we combined G418 or selective media lacking histidine if required. Under these conditions, 100 and 90–95% of spores in Control and VST strains, respectively, excised the URA3-containing circle. All cells were resuspended, grown in liquid-rich media containing Nourseothricin and diluted daily to 105 cells/ml until senescence was reached. Each day, at least 5 × 108 cells were collected and stored at −20°C. Genomic DNA was purified by shearing cells with glass beads and phenol-chlorophorm extraction according to (70). To determine telomere length and subtelomeric structure, we performed Southern blot by digesting DNA with Xho I (telomeres Y′) or Stu I (telomere VIIL). Fragments were separated by electrophoresis, transferred to a membrane and hybridized with suitable radioactively labelled probes according to a protocol adapted from (71). Probes were generated by random priming on a purified oT316-oT318 PCR fragment (telomere Y′) or a SalI-NcoI digestion of pVIIL URA3-TEL containing adh4-ura3 (telomere VIIL; (72)). Images were obtained with Typhoon Trio and lengths measured with the MolWt macro in ImageJ 1.46a. Subtelomeric ssDNA was quantified according to (73) using primers listed in Supplementary Table S3. To test the effect of resection factors on ssDNA accumulation, colonies with the relevant genotypes were obtained as in the solid senescence assay and grown for 1 day in liquid-rich media containing Nourseothricin. Genomic DNA was purified with glass beads and phenol-chlorophorm extraction, and subtelomeric ssDNA was quantified according to (73) using primers listed in Supplementary Table S3.
Microscope analysis
Yeast cells were grown and processed for fluorescence microscopy as described previously (74). Fluorophores were visualized on a Deltavision Elite microscope (Applied Precision) equipped with a 100× objective lens (Olympus U-PLAN S-APO, NA 1.4), a cooled Evolve 512 EMCCD camera (Photometrics, Japan), and an Insight solid state illumination source (Applied Precision). Images were acquired using softWoRx (Applied Precision) software and processed with Volocity software (PerkinElmer).
RESULTS
5′-3′ resection factors promote the onset of senescence when one single critically short telomere is present
We have previously shown that Tel1 and Mec1 accumulate at the shortest telomere in telomerase-deficient cells and promote the onset of senescence (33). Because Mec1 can be recruited at ssDNA on the 5′-3′ resection of DSB ends, we wondered whether subtelomeric ssDNA accumulates at the shortest telomere in the absence of telomerase.
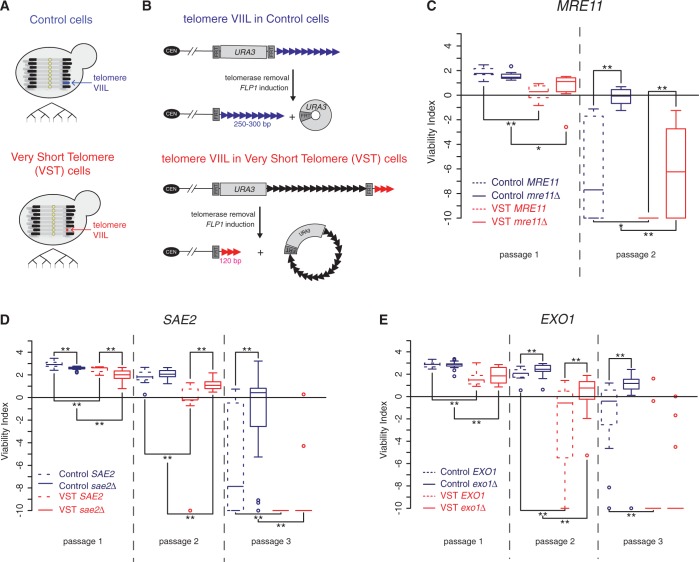
To determine this, we used our previously described system to generate a very short telomere that we could track in several independent clones deprived of telomerase activity [Figure 1A; see Supplementary Figure S1 and ‘Materials and Methods’ section (33)]. This was achieved by the mass sporulation of a diploid heterozygous for TLC1 encoding the template RNA of telomerase and the selection of tlc1Δ clones using a haploid-specific selective marker. The very short telomere was a modified version of the telomere VIIL in which the subtelomeric region was replaced by an artificial construct that allowed the reduction of terminal telomeric repeats in cells termed VST cells (Figure 1B). Control cells contained a control construct in which the VIIL telomere was of nearly wild-type length (Figure 1A and B). To cope with the heterogeneity of the senescence phenotype, we used a sensitive spot assay in which the growth of many independent clones could be assessed in parallel in serial passages performed every 2 days of growth on plates at 30°C in a single full experiment [see Supplementary Figure S1 and ‘Materials and Methods’ section (67)]. With this system, VST cells, which contained a very short VIIL telomere, senesced earlier than Control cells, which had a near–wild-type–length VIIL telomere, as described previously (33).
Figure 1.
5′-3′ resection factors promote senescence. (A and B) Experimental setting used in this study. (A) Two sets of yeast strains deleted for telomerase activity (tlc1Δ) and containing a modified version of the telomere VIIL were used [adapted from (33,75)]. In Control cells the VIIL telomere is of wild-type length, and in VST cells the VIIL telomere is very short. The proliferation capacity of several independent clones of such strains, obtained through the procedure described in Supplementary Figure S1, was compared. (B) Telomeric constructs. The VST strain contains an artificial VIIL telomere, such that a URA3 selection marker and 600 nt of TG1-3 repeats were inserted between two flipase recognition target (FRT) sequences close to the chromosome end. These internal repeats negatively regulated telomerase activity via the protein-counting mechanism (76), resulting in a short array of terminal telomeric repeats after the second FRT. The Control strain did not display internal telomeric sequences, resulting in normal homeostasis of the number of distal telomeric repeats. The induction of Flp1, a site-directed recombinase, resulted in the excision of the fragment between the two FRTs, leaving only the terminal telomeric repeats. (C) Quantitative analysis of senescence by serial spot assays in the presence (VST) or absence (Control) of a very short VIIL telomere was performed on 8 MRE11 tlc1Δ (dotted lines) and 8 mre11Δ tlc1Δ (full lines) independent spores derived from yT235 (Control cells) and yT236 (VST cells; see Supplementary Figure S1 and ‘Materials and Methods’ section). Adjusted P-values were obtained by Wilcoxon rank-sum test with a false discovery rate correction. *P < 0.1; **P < 0.05. See Supplementary Table S2 for detailed P-values. (D) Same as in (C) for 16 SAE2 tlc1Δ and sae2Δ tlc1Δ independent spores derived from yT231 and yT232. (E) Same as in (C) for 16 EXO1 tlc1Δ and 16 exo1Δ tlc1Δ independent spores derived from yT403 and yT404.
To test for the role of DSB processing factors, we deleted their corresponding genes in this cellular setting and derived tlc1Δ spores containing or not the additional mutations from the same diploids. Comparing all genotypes within a single full experiment was important because telomere length and/or structure can be altered by the deletions in the parental strains, affecting senescence rates of all derivatives. For instance, as observed for TEL1, XRS2 and RAD50 (33,35,36), mre11Δ/MRE11 diploids displayed shorter telomeres providing a plausible explanation for the faster senescence of tlc1Δ MRE11 derivatives compared with tlc1Δ clones obtained from MRE11/MRE11 diploids (Supplementary Figure S2 and Figure 1). Nonetheless, in Control cells, the loss of Mre11 resulted in a delay of senescence in the second passage (Figure 1C, blue lines), similar to observations in tlc1Δ xrs2Δ and tlc1Δ rad50Δ mutants described recently (36). In the presence of a single very short VIIL telomere, MRE11 deletion delayed senescence from the first passage (VST cells; Figure 1C, red lines). This is in accordance with the idea that displaying a very short telomere from the moment telomerase is lost anticipates the phenotype observed in Control cells. Thus, Mre11 promotes the establishment of senescence, probably via the action of the MRX complex at the shortest telomere.
At a DSB and at telomeres maintained by telomerase, the MRX complex, along with Sae2 nuclease, allows extended 5′-3′ resection by Exo1 or Sgs1-Dna2 (16,77,78). Deletion of SAE2 in our cellular system initially accelerated senescence in both Control and VST cells (Figure 1D, passage 1) but later delayed it (Figure 1D, passages 2–3). As for mre11Δ, the delay in senescence was more pronounced in cells displaying a very short telomere, which argues for a more prominent action of Sae2 in the presence of a short telomere. EXO1 deletion delayed senescence from the second passage in both Control and VST cells (Figure 1E). In this case, no loss of viability was observed in the first passage as compared with sae2Δ senescent cells. Thus, Mre11, Sae2 and Exo1 display similar phenotypes, suggesting that they act in the same pathway. The accelerated senescence phenotype observed in the presence of a single very short telomere suggests in addition that these factors may act preferentially on short telomeres to promote senescence.
Subtelomeric ssDNA accumulates as telomere shortens in the absence of telomerase
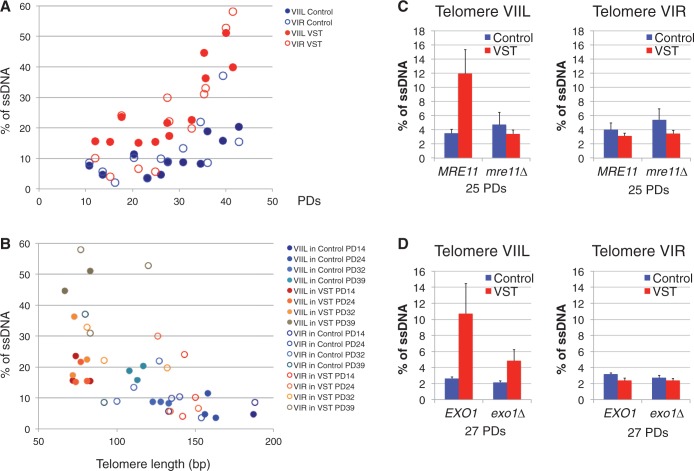
Because all of these factors are involved in the 5′-3′ resection of DSBs, we hypothesized that their effects in promoting senescence could be due to a more prominent action at the shortest telomeres. We thus compared the level of ssDNA at telomere-proximal regions of the artificial VIIL telomere and of the independent native VIR telomere in the same populations of Control and VST cells. To obtain sufficient cells for the analysis, we mixed all independent colonies resulting from the mass sporulation and tlc1Δ selection and grew them in liquid cultures for 4 days until they reached senescence and before the appearance of post-senescence survivors. DNA was prepared each day and ssDNA at ∼100 bp upstream of the telomeric tract was quantified by quantitative amplification of ssDNA [QAOS, (73), Figure 2].
Figure 2.
The accumulation of subtelomeric ssDNA by end resection contributes to senescence. (A) A mixture of tlc1Δ colonies derived from yT136 or yT138 was grown in liquid-rich medium with daily dilutions. DNA was prepared every day, and for each culture the number of PDs grown in liquid culture was estimated. Genomic DNA was prepared and ssDNA quantity was monitored by QAOS using a probe located 56 nt away from the telomeric repeats of telomere VIR (open circles) and 139 nt away from the telomeric repeats of telomere VIIL (full circles) in Control (blue circles) and VST (red circles) tlc1Δ cells and plotted according to the PDs since the loss of telomerase. (B) DNA samples from (A) were used to determine the telomere length of VIR and VIIL by telomere-PCR. ssDNA determined as above was plotted according to telomere length. Circle colors indicate whether DNA was from Control cells (blue circles) or VST cells (red-brown circles). Full or open circles indicate whether data correspond to VIR (open circles) or VIIL (full circles) telomeres. PD indicates the number of estimated PDs in liquid cultures after mass sporulation and colony formation. (C) MRE11 tlc1Δ and mre11Δ tlc1Δ colonies derived from yT235 or yT236 were grown in liquid-rich medium for ∼25 population doublings. Genomic DNA was prepared from Control (blue) and VST (red) cells and ssDNA quantity was monitored by QAOS as in (A) (n = 3–4 clones per genotype). (D) Same as in (C) with EXO1 tlc1Δ and exo1Δ tlc1Δ colonies derived from yT403 or yT404 grown for ∼27 population doublings (n = 3–4 clones per genotype).
We found that ssDNA at the subtelomere of chromosome VIR increased from ∼6–12% at early time points in both Control and VST cells to 20–40% in late cultures (Figure 2A and Supplementary Figure S3A). This indicates that ssDNA progressively accumulates at subtelomeres as cells divide in the absence of telomerase and that one-third of cells presents subtelomeric ssDNA at the VIR telomere at the moment of senescence. A similar profile was observed for the telomere-proximal region of the modified VIIL telomere in Control cells (Figure 2A and Supplementary Figure S3B), showing that the lack of natural subtelomeric sequences did not grossly influence the processing of this nearly wild-type-length telomere in the absence of telomerase. In contrast, in VST cells, the short VIIL telomere showed higher levels of ssDNA (∼18%) in the telomere-proximal region before 30 population doublings (PDs) compared with the same region in Control cells and compared with the VIR subtelomere in the same cell population (Figure 2A and Supplementary Figure S3B). During the course of the culture, these ssDNA levels increased to 40%. To test whether the progressive accumulation of subtelomeric ssDNA as cells divide reflects its preferential accumulation at shorter telomeres, we determined telomere length of each individual sample and plotted ssDNA as a function of telomere length. Strikingly, the amount of subtelomeric ssDNA tended to increase as telomeres shortened (Figure 2B). Thus, native telomeres likely become better substrates for 5′-3′ resection as they become short. Moreover, at the VIIL telomere in VST cells, at a constant short length, the amount of subtelomeric ssDNA increased with the number of PDs (Figure 2B, full red-brown circles). We can thus suggest that the number of telomeres presenting subtelomeric ssDNA increases as telomeres shorten and as cell cultures reach senescence. Thus, cells in crisis probably have several short telomeres with long ssDNA extremities.
To verify that this accumulation of ssDNA at short telomeres is indeed due to 5′-3′ resection, we measured the amount of ssDNA at telomeres in the absence of resection factors. To be able to compare cells derived from the same diploid, we selected MRE11 tlc1Δ and mre11Δ tlc1Δ clones and measured ssDNA ∼25 PDs after the loss of telomerase. We found a similar low level of 3–4% of ssDNA at VIR subtelomeres in Control and VST cells both in MRE11 and mre11Δ cells (Figure 2C and Supplementary Figure S3C). In contrast, the accumulation of ssDNA at the subtelomere of a very short VIIL telomere was lost in mre11Δ tlc1Δ cells (Figure 2C and Supplementary Figure S3C), indicating that the increase in ssDNA specifically at short telomeres depends on Mre11. Likewise, the accumulation of ssDNA at the VIIL subtelomere in VST cells was decreased on EXO1 deletion (Figure 2D and Supplementary Figure S3D), in accordance with the notion that parallel pathways act at telomeres for 5′-3′ resection (16). Importantly, these findings strongly support that the DSB 5′-3′ resection pathway acts to erode telomeres thereby generating long ssDNA tails.
We conclude that an excessive amount of ssDNA at telomeres could be the signal stemming from short telomeres that triggers Mec1-dependent checkpoint activation and cell cycle arrest. Based on this, and the fact that 5′-3′ resection factors promote the onset of senescence, we propose that as telomeres shorten, they become more prone to the action of these resection factors and thus accumulate ssDNA at their subtelomeres, exposing non-telomeric ssDNA that will activate the Mec1 checkpoint.
HR counteracts subtelomeric ssDNA exposure at short telomeres in telomerase-negative cells
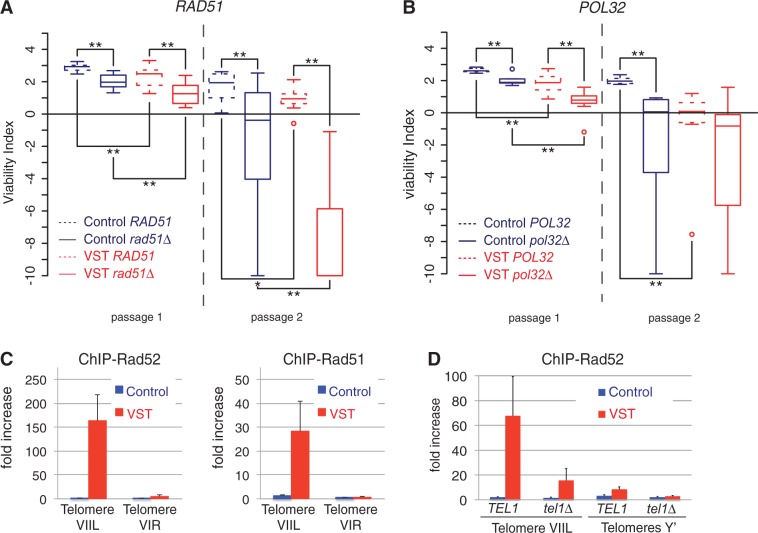
The 3′ extended ssDNA tails created by 5′-3′ resection can be the substrate for HR. Because HR factors such as Rad51 and Rad52 were shown to contribute to the viability of cells in the absence of telomerase, we investigated their roles at the telomeres of senescent cells. We have previously shown that RAD52 deletion accelerates senescence in both Control and VST cells and that Rad52 forms foci at the shortened VIIL telomere (32,33). Here we examined whether RAD51 deletion had the same effect. In a similar manner, RAD51 deletion accelerated the onset of senescence in both Control and VST strains (Figure 3A).
Figure 3.
HR counteracts senescence. (A) The deletion of RAD51 accelerates senescence. Quantitative analysis of senescence as in Figure 1C on 8 RAD51 tlc1Δ and rad51Δ tlc1Δ colonies derived from yT347 and yT348. Adjusted P-values were obtained by Wilcoxon rank-sum test with a false discovery rate correction. *P < 0.1; **P < 0.05. See Supplementary Table S2 for detailed P-values. (B) The deletion of POL32 accelerates senescence. Quantitative analysis of senescence as in Figure 1C on 8 POL32 tlc1Δ and 8 pol32Δ tlc1Δ colonies derived from yT225 and yT226. (C) Rad52 and Rad51 associate preferentially with short telomeres in the absence of telomerase. Cultures from independent tlc1Δ spores derived from yT136 or yT138 were grown in liquid YPD for ∼30 generations after sporulation in the absence (Control cells) or presence (VST cells) of a short VIIL telomere. Chromatin was immunoprecipitated using primary antibodies directed against Rad52 (left panel) or Rad51 (right panel). The association of each protein with the VIR or VIIL telomeres or with an internal locus (ARO1) was quantified by qPCR, and the fold increase in telomere enrichment over ARO1 is indicated. Error bars indicate the SEM from three independent spores. (D) Rad52 association with the short telomere depends on Tel1. ChIP of Rad52 was performed as in (B) in tlc1Δ independent spores derived yT174 or yT176 with the indicated genotypes. Association of each protein with the Y′ telomeres or VIIL telomeres or to an internal locus (ARO1) was quantified by qPCR, and fold increase of telomere enrichment over ARO1 is indicated. Error bars indicate SEM from at least three independent spores.
The DNA polymerase δ is the major polymerase of lagging strand synthesis within the replisome but is also involved in many DNA repair processes requiring DNA synthesis (60,79–81). Pol32 is a non-essential subunit of DNA polymerase δ, the function of which is revealed at low temperatures, where it is required for efficient replication, and in processes such as break-induced replication and telomere maintenance in post-senescence survivors (82–84). To test whether HR-coupled DNA synthesis was required to maintain senescent cells before the establishment of post-senescence survivors, we tested how senescence was affected in the absence of POL32. We found that POL32 deletion resulted in a significant acceleration of the onset of senescence in both the presence and absence of a very short telomere (Figure 3B). This supports the notion that efficient replication and/or DNA repair–coupled DNA synthesis is required to maintain viability before senescence.
The genetic effects were probably due to a direct action of HR at the short telomere, as both Rad52 and Rad51 proteins were found to be enriched at the short VIIL telomere in VST cells compared with other telomeres in the same cell population or to the VIIL telomere in Control cells (Figure 3C). Therefore, the mild increase in recombination factors binding to chromosome end sequences in the absence of telomerase described previously (41) is probably due to its highly preferential binding to the shortest telomere(s) in the cell. In accordance with this idea, Rad51 and Rad52 association with telomeres is dependent on telomere length (Supplementary Figure S4A). In addition, we found that Rad52 preferential recruitment to the short telomere is partially lost in cells lacking Tel1 (Figure 3D). This is in accordance with Tel1 promoting senescence in the same pathway as MRX (33,36), probably through its ability to assist in the formation of long ssDNA tails at short telomeres (85), allowing the loading of Rad52. We therefore speculate that the long ssDNA tails, which accumulate at short subtelomere(s) in senescent cells as a consequence of 5′-3′ resection, are probably coated with RPA, then with Rad51 with the assistance of Rad52 and at the same time activate Mec1.
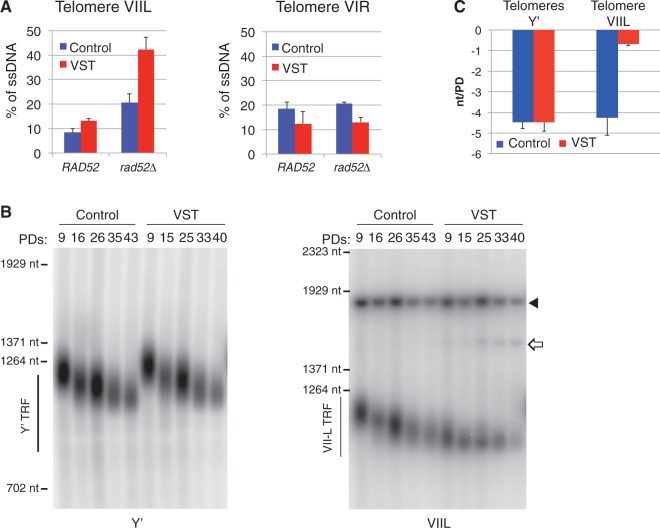
Because short telomeres accumulated both ssDNA and HR factors, we asked whether HR was affecting the amount of ssDNA at telomeres. We observed that the ssDNA at native VIR subtelomeres in VST cells was not affected by the loss of RAD52. In contrast, as early as 9 PDs, VIIL subtelomeres showed a slight increase in ssDNA in rad52Δ Control cells compared with RAD52 (Figure 4A and B). Remarkably, the amount of ssDNA at very short VIIL subtelomeres in VST cells increased to 42%, a level previously observed only in cells late in senescence (see Figure 2). Hence, Rad52 restricts the accumulation of subtelomeric ssDNA at a short telomere at early time points after the loss of telomerase. One hypothesis is that Rad52 counteracts resection by promoting strand invasion and DNA synthesis from a homologous template.
Figure 4.
Rad52 counteracts the accumulation of subtelomeric ssDNA. (A) Rad52 counteracts ssDNA accumulation at subtelomeres of the very short telomere. ssDNA at VIIL (left panel) and VIR (right panel) subtelomeres from Control or VST cells was measured by QAOS as described for Figure 2A ∼9 PDs after the loss of TLC1 in RAD52 (derived from yT136 and yT138) or rad52Δ (derived from yT143 and yT145) cells. Mean ± SEM for three independent experiments. (B) Lengths of Y′ (left panel) and VIIL (right panel) telomeres in Control and VST cells were measured by Southern blot at the indicated PD after the loss of TLC1. Full arrow: internal ura3 locus; open arrow: fragment of the VIIL when excision did not occur. (C) Mean shortening rate per PD was determined from Southern blot as in (B) between 9 and 33–35 PDs for Y′ and VIIL telomeres from three independent cultures. Mean ± SEM.
Because an increased resection at the shortest telomere is expected to increase the telomere shortening rate because of the end replication problem (3), we next asked whether the telomere shortening rate was affected by initial telomere length. We thus measured the length of the short VIIL telomere in VST cells and compared it with that of the VIIL telomere in Control cells and with that of Y′ telomeres in VST and Control cells at several time points in senescing cultures. We found that the latter control telomeres shortened at the expected rate of 3–5 nt per PD [Figure 4B and C and Supplementary Figure S4B (75)]. In contrast, in VST cultures, the length of the short VIIL telomere became stable at an average of 70–80 nt. An increased shortening of the shortest telomere could not be detected. Instead, either an active maintenance of this telomere is operating, or cells with an extremely short telomere—or even no telomeric repeats left—do exist but are rapidly counterselected in liquid cultures [see the spread and loss of the smear of VIIL terminal fragment in VST cells of Figure 4B and (33)], or both. Interestingly, we often could observe VIIL telomeres shorter than this 70–80 nt limit in cells deleted for MRE11 (Supplementary Figure S3C), suggesting that it is the increased resection at short telomeres rather that their length that triggers cell cycle arrest.
Given the involvement of HR factors specifically at the short VIIL telomere, we investigated how RAD52 deletion affected this telomere length-dependent maintenance in VST cultures. Whereas the short VIIL telomere was detected in VST cells at eight PDs, only longer VIIL telomeres were detected in subsequent PDs (Supplementary Figure S4C). This was associated with the growth of a small subset of cells from the initial population that regained higher viability (Supplementary Figure S4D and E) in accordance with strong selection being at work in liquid cultures of rad52Δ tlc1Δ VST cells. We concluded that the very short telomere could not be maintained as short in the absence of Rad52.
In accordance with the sequence of events occurring at the DSB, we propose that HR protects short telomeres by promoting DNA synthesis from a homologous region to counteract extended resection, effectively delaying senescence. This pathway, however, does not result in net telomere elongation, as the length of the VIIL telomere in VST cultures does not increase [Figure 4B (33)]. One possibility is that most of the recombination events occur using the replicated sister chromatid as a template, perhaps coupled with the semi-conservative DNA replication through telomeric repeats. To test for this possibility, we went on testing the effect of replication repair pathways on senescence.
The error-free DDT pathway modulates the rate of senescence
Along with the existence of substantial evidence for a short telomere being recognized as a DSB, some data argue that a short or dysfunctional telomere can be sensed as a replication stress. For example, Mec1 is activated both in the case of a DSB and during replication stress, and some replication-specific factors, such as Mrc1, Mms1 and Pol32, interfere with the rate of senescence (33,34,86) (Figure 3B). We thus investigated how known factors that allow replication through damaged templates control senescence.
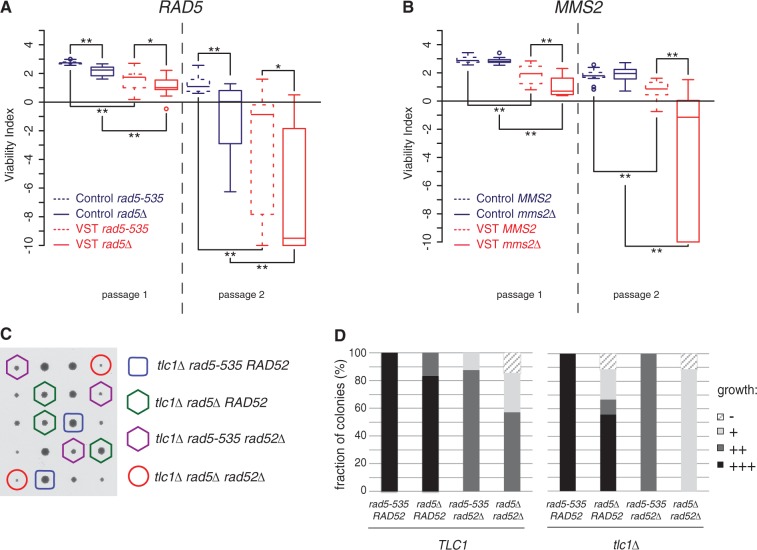
Rad5, a key component of the error-free DDT pathway, is both a RING finger-containing ubiquitin ligase required for the poly-ubiquitylation of PCNA and a DNA helicase with an in vitro activity of fork regression (59,87). All of our work has been performed in the original W303 background, which contains the rad5-535 allele of RAD5. Although this allele displays only mild defects, it corresponds to a point mutation positioned in one of the seven Rad5 helicase domains (88). We verified that senescence in a RAD5 background was also accelerated by the presence of a very short telomere, suggesting that the Rad5-535 protein retains most Rad5 functions in senescence (Supplementary Figure S5A). In contrast, when Rad5 was totally absent, a significant acceleration in senescence was observed in both Control and VST cells (Figure 5A), showing that this ubiquitin ligase/DNA helicase promotes the viability of cells in the absence of telomerase.
Figure 5.
DDT and HR act independently to sustain the viability of senescent cells. (A and B) DDT factors sustain the viability of senescent cells. Quantitative analysis of senescence as in Figure 1C on 16 rad5-535 tlc1Δ and rad5Δ tlc1Δ derivatives of yT299 and yT300 (A) and 16 MMS2 tlc1Δ and mms2Δ tlc1Δ derivatives of yT303 and yT304 (B). Adjusted P-values were obtained by Wilcoxon rank-sum test with a false discovery rate correction. *P < 0.1; **P < 0.05. See Supplementary Table S2 for detailed P-values. (C and D) rad5Δ and rad52Δ display synthetic slow growth phenotypes in TLC1 or tlc1Δ background. TLC1/tlc1Δ RAD52/rad52Δ rad5-535/rad5Δ diploids (yT567) were sporulated and meiosis products dissected and analysed after 2 days of growth on rich medium, photographed and genotyped. (C) Representative tetrads are shown, and genotypes of the tlc1Δ colonies are indicated. (D) Spore colonies of each genotype were given an index according to their size. −: non-growing spore or microcolony, +: small colony, ++: medium colony, +++: normal size colony. n = 15 tetrads/60 spore colonies were analysed.
This result indicates that the error-free branch of the DDT is potentially involved in the maintenance of senescent cells. To check this, we tested the role of MMS2, which encodes a ubiquitin-conjugating component that cooperates with Rad5 for the poly-ubiquitylation of PCNA in the DDT. MMS2 deletion accelerated senescence, but only in the presence of a very short telomere (Figure 5B). This suggests a specific role of Mms2 in the maintenance of a very short telomere, while Rad5 would have a role in both strains, perhaps through a helicase-dependent and ubiquitin-independent function. Alternatively, mms2Δ might confer a milder phenotype than rad5Δ, revealed by our assay that exacerbates phenotypic differences in senescence, as observed for other DDT assays (58).
As a telomerase-positive rad5Δ strain displays a slightly increased telomere length compared with RAD5 [(89) and Supplementary Figure S2], we wondered whether the effects of Rad5 in senescence could be related to telomere length homeostasis. However, we saw no significant difference in Y′ and VIIL telomere shortening rates between rad5-535 and rad5Δ in Control and VST cells (Supplementary Figure S5B and C). Hence, the acceleration of senescence of rad5Δ cells is not simply due to faster erosion of telomeres.
Increasing evidence points to the idea that both DDT and HR factors act at damaged forks to promote replication repair and restart (58–61). However, how these pathways cooperate remains unclear, perhaps because different stresses may trigger different mechanisms. To investigate how these two pathways cooperate at telomeres in the context of senescence, we combined deletions of RAD5, RAD52 and TLC1 in heterozygous diploids. Spore analysis of meiosis products showed a strong synthetic growth defect between rad5Δ and rad52Δ in TLC1 that was exacerbated in tlc1Δ cells (Figure 5C and D). We thus concluded that Rad5-535 and Rad52 act in at least partially non-overlapping pathways to maintain the viability of telomerase-negative cells.
Rad5 localizes to a subset of telomeres in S/G2 phase in telomerase-positive cells
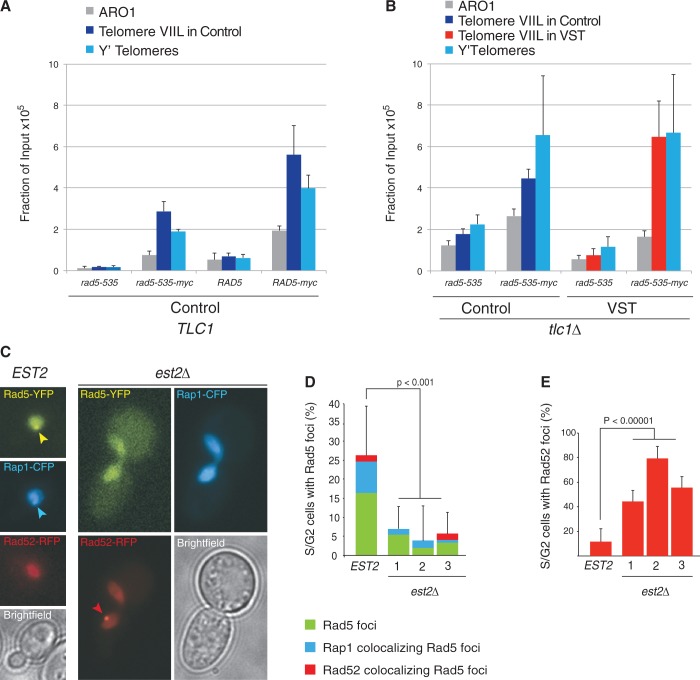
The involvement of Rad5 in the control of senescence prompted us to explore its association with telomeres. We expressed Myc-tagged versions of both Rad5 and Rad5-535 and immunoprecipitated chromatin from telomerase-positive cells. Epitope-tagging these proteins only slightly increased telomere length (Supplementary Figure S2C). We saw a significant enrichment of both Myc-tagged Rad5-535 and Myc-tagged Rad5 over the background for all loci analysed. These included an internal locus (ARO1 gene) as well as two telomere-associated loci, the VIIL subtelomere in the context of a wild-type regulated telomere and the VIR native subtelomere (Figure 6A). Strikingly, the enrichment at telomeric loci was significantly higher compared with the internal locus, suggesting that Rad5 could have a specific role at telomeres in the presence of telomerase.
Figure 6.
Rad5 localizes to a subset of telomeres in wild-type cells. (A) Rad5-535 and Rad5 are enriched at telomeres. Telomerase-positive cells expressing Rad5-535 (yT580), a myc-tagged version of Rad5-535 (yT581), Rad5 (yT583) or a myc-tagged version of Rad5 (yT582) and a wild-type length telomere VIIL (Control construct after excision of the URA3-containing circle) were submitted to ChIP using an anti-myc antibody. The association of Rad5-535-myc and Rad5-myc with the VIIL telomere, the VIR telomere or the internal ARO1 locus was quantified by qPCR. The mean fraction of input ± SEM was calculated from four independent cultures. (B) ChIP of Rad5-535-myc was performed as in (A) in tlc1Δ independent spores derived yT382 or yT383 with the indicated genotypes grown ∼30 PDs after sporulation. Association with the Y′ telomeres or VIIL telomeres or to an internal locus (ARO1) was quantified by qPCR. Error bars indicate SEM from three independent spores. (C-E) Rad5 colocalizes with Rap1 in telomerase-positive cells. Rad5 foci were examined by fluorescence microscopy in wild-type cells expressing Rad5-YFP, Rad52-RFP and Rap1-CFP (ML690-16A) and in est2Δ mutant cells (ML691-12C) 1, 2 or 3 streaks after the loss of a plasmid-borne EST2. (C) Representative images of wild-type (left) and est2Δ (right) cells are shown. Scale bar: 3 µm. (D) Percentage of S/G2 cells displaying Rad5 foci. Blue: percentage of S/G2 cells displaying a Rad5 focus co-localizing with Rap1; red: percentage of S/G2 cells displaying a Rad5 focus co-localizing with Rad52. In EST2 cells, ∼30% of Rad5 foci colocalize with Rap1 and 10% colocalize with a Rad52 focus. Rad5 foci are observed exclusively in budded cells (S/G2). A total of 60–120 cells were counted per genotype. Error bars represent 95% confidence intervals. (E) Quantification of Rad52 foci in S/G2 cells in the experiment shown in (B and C). Error bars represent 95% confidence intervals.
To confirm this, we replaced wild-type Rad5 with a YFP-tagged version in cells expressing a CFP-tagged version of Rap1 and a RFP-tagged version of Rad52 to check for colocalization of Rad5 with both telomeres and HR foci, respectively. We observed that Rad5-YFP was able to form spontaneous foci in one-fourth of unchallenged S/G2 cells (Figure 6C and D, EST2). These foci are only observed in S/G2 cells, when Rad5 activity has been shown to be required for DDT (80). Strikingly, one-third of these foci associated with the telomeric protein Rap1-CFP, indicating that Rad5 foci were indeed found at a subset of wild-type telomeres. Thus, Rad5 and perhaps the error-free DDT pathway might assist in telomere replication, thereby ensuring telomere length homeostasis.
When telomerase activity was removed, the ability of Rad5 to accumulate at telomeres persisted (Figure 6B). Notably, no preference for shorter telomeres was detected because the VIIL telomere in VST cells was not enriched in Rad5-535-myc immunoprecipitates, compared with the VIIL telomere in Control cells or the Y′ telomeres in VST or Control cells. Surprisingly, telomerase removal by the loss of a plasmid-borne copy of EST2 resulted in a significant decrease in Rad5-YFP foci as early as 25 PDs after the loss of telomerase (Figure 6C and D, est2Δ). This decrease included the Rap1-associated Rad5 foci, suggesting that, although present, Rad5 loses its ability to form foci at telomeres and also at other sites in the absence of telomerase. In contrast, Rad52-RFP foci, previously described to associate with short telomeres, increased [Figure 6E (32)]. These results suggested that the interaction between Rad5 molecules at telomeres changed after loss of telomerase. We then asked whether the increase in ssDNA telomeric tails as telomeres shortened explained the inhibition of Rad5-YFP foci. In restrictive conditions, cdc17-1, a conditional mutation of a DNA polymerase α-primase component gene, leads to telomerase-dependent overextension of single-stranded G-tails. However, this genetic background did not affect the ability of Rad5-YFP to form foci in S/G2, indicating that modification of the primary structure of telomeres does not alter the ability of Rad5-YFP to form foci at telomeres (Supplementary Figure S5D). An intriguing possibility could be that telomerase or the cellular checkpoint state controls Rad5 foci formation. An alternative explanation is that the telomeric, and perhaps genomic, chromatin structure or nuclear organization is altered in est2Δ cells in a way that prevents Rad5-YFP from forming foci. In accordance with this hypothesis, an alteration of global chromatin structure after the loss of telomerase was recently proposed (90).
Mus81 and Sgs1 affect senescence through distinct mechanisms
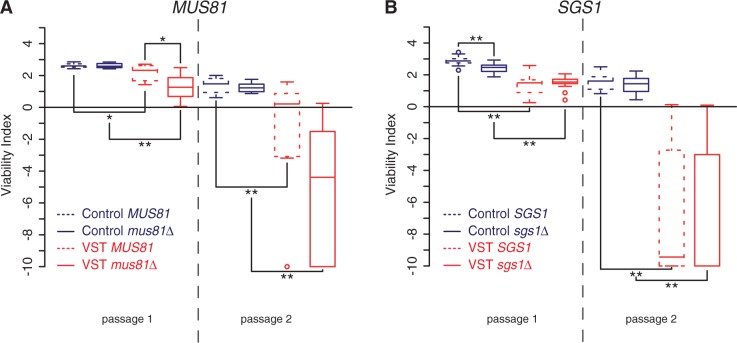
D-loops or Holliday junction intermediates generated by HR and the error-free DDT pathway are expected to be resolved after DNA synthesis by several enzyme complexes. Of these complexes, the Mus81-containing complex has specificity for resolving D-loops and nicked Holliday junctions that are potentially formed by HR and template switching at telomeres (91). Accordingly, Mus81 was recently found at the telomeres of human cells that maintain telomeres through a recombination-based mechanism (92). Confirming previous results, deletion of MUS81 did not affect senescence in Control cells (39); (Figure 7A). In contrast, we found that mus81Δ VST cells displayed faster senescence compared with MUS81 VST cells. This suggests that Mus81 may have a prominent role at critically short telomeres, revealed when at least a single one of these is present when telomerase is lost.
Figure 7.
Distinct effects of Mus81 and Sgs1 in senescence triggered by a very short telomere. Quantitative analysis of senescence as in Figure 1C of 8 MUS81 tlc1Δ and mus81Δ tlc1Δ derivatives of yT355 and yT356 (A) and 16 SGS1 tlc1Δ and sgs1Δ tlc1Δ derivatives of yT257 and yT258 (B). Adjusted P-values were obtained by Wilcoxon rank-sum test with a false discovery rate correction. *P < 0.1; **P < 0.05. See Supplementary Table S2 for detailed P-values.
Sgs1 is a RecQ helicase that, as part of a complex with Rmi1 and Top3, also resolves HR and DDT intermediates (57,93,94) and has long been implicated in telomere biology (40). In addition, Sgs1 is also involved in DSB and telomeric 5′-3′ resection (16,78), shown here to promote senescence. Thus, Sgs1 potentially contributes to opposing phenotypes during senescence. Confirming the literature (95–97), we found that SGS1 deletion accelerated senescence in Control cells. In contrast, in VST cells with a short telomere, an absence of Sgs1 did not significantly affect the onset of senescence (Figure 7B). We conclude that either Sgs1 has no effect in the presence of a single very short telomere or this complex phenotype reflects the multiplicity of Sgs1 activities. We propose that, in sgs1Δ cells, 5′-3′ resection at short telomeres may be limited, decreasing both checkpoint activation and the formation of substrates resolved by Sgs1 itself.
DISCUSSION
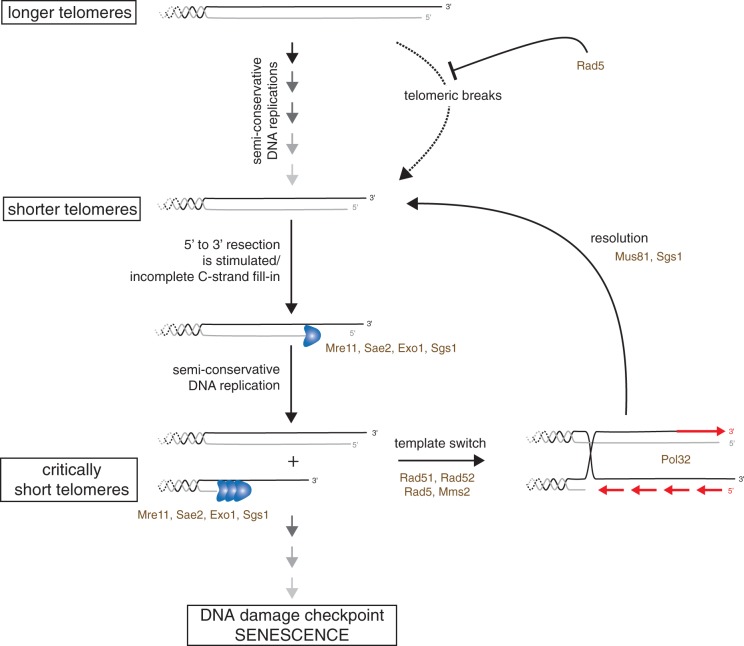
Increasing evidence indicates that in many eukaryotes telomere length dictates the proliferation capacity of a cell lineage in the absence of telomerase. More specifically, the length of the shortest telomere is probably the major determinant of the onset of senescence. Here, using a cellular setting in which we could distinguish a very short telomere from the remaining ones, we have shown that, as they erode, telomeres are subjected to different processing pathways. This is attested by the direct characterization of telomere primary structure, the differential enrichment of several DNA repair factors and the requirement of distinct gene products for cell growth according to telomere length. Namely, as telomeres shorten, we observe a switch between a mode of telomere maintenance that involves Rad5 and a DSB-like processing with increasing accumulation of subtelomeric ssDNA (Figure 8). Thus, it appears that telomeres maintain genome integrity possibly through distinct mechanisms according to their length and that these pathways may simultaneously operate at different telomeres in a given cell.
Figure 8.
Model for telomere maintenance in the absence of telomerase. Factors studied in this work are indicated. In the presence of telomerase or early after the loss of telomerase, a Rad5-dependent mechanism could contribute to the semi-conservative DNA replication of telomeres. As they become short in the absence of telomerase, telomeres are subject to progressive increased resection and/or incomplete replication, increasing telomeric overhang length and subsequently increasing the asymmetry in length of replication products. ssDNA may then trigger the recruitment of HR and error-free DDT factors. A recombination-based mechanism, possibly combined with DDT, would limit the increased resection by allowing re-elongation of the shortened strand using the sister chromatid as the template. This would counteract the generation of long overhangs and diminish the activation of Mec1 in a feedback loop. Note that in all these events, a minimal DNA end replication should remain, causing a minimal shortening rate. Permanent cell cycle arrest is thus inexorably expected when telomeric repeats become exhausted.
Increasing exposure of subtelomeric ssDNA as telomeres shorten
Previous results have indicated that Mec1 is activated in the absence of telomerase by recognizing the shortest telomere in the cell, leading to cell cycle arrest in G2/M (12–14,32,33,98). Although RPA enrichment at telomeres increases as cells senesce (32), indicating the presence of ssDNA, the structure sensed by Mec1 remained uncovered. In this work, we directly observed ssDNA accumulation at subtelomeres in the absence of telomerase. Our results show that this accumulation increases as telomeres shorten and as cell cultures progress into crisis. Two non-exclusive mechanisms could lead to such an outcome. In the first one, the nucleases/helicases in charge of DSB 5′-3′ resection act preferentially at short telomeres, and in the second one, the semi-conservative DNA replication is incomplete at short telomeres leaving ssDNA gaps.
In support of the first mechanism, deletion of factors involved in the generation of ssDNA at DSBs, namely Mre11, Rad50, Xrs2, Sae2 and Exo1, all delayed the onset of senescence [this work and (36)], implying that their activities contribute to the signal for senescence. Moreover, for Mre11, Sae2 and Exo1, we demonstrate here that their role is more prominent when a critically short telomere is present in the cell and that the accumulation of ssDNA at this critically short telomere depends at least on Mre11 and Exo1. The same factors are known to act at wild-type telomeres at each replication cycle to transiently increase the G-overhang (16), and their action is repressed at long telomeric tracts placed next to a DSB (28). Besides, Exo1 is the main exonuclease that acts at dysfunctional telomeres lacking telomeric proteins (99). Thus, the increasing exposure of subtelomeric ssDNA at short telomeres could be caused by a de-repressed action of these nuclease activities at chromosome ends with short telomeric tracts. This would be reminiscent of the situation of telomeres maintained by telomerase, in which telomeric proteins are required to limit the action of the MRX-dependent nucleolytic activities (100).
With respect to the second mechanism, several lines of evidence substantiate the idea that DNA replication through telomeric repeats is challenging. Moreover, DNA replication at telomeres is a complex process, since in addition to the semi-conservative DNA replication, a C-strand fill-in is expected to occur after 5′-3′ C-strand resection and after telomerase elongation. This additional DNA synthesis is believed to depend on the recruitment of the DNA polymerase α/primase by the telomeric complex CST [Cdc13-Stn1-Ten1 in budding yeast or Ctc1-Stn1-Ten1 in metazoa; (15,101,102)]. Therefore, our results could also be explained by a delay in or inhibition of C-strand synthesis. Although Cdc13 is shown to frequently form foci at the very short telomere (32), we cannot rule out that accumulation of subtelomeric ssDNA at short telomeres in the absence of telomerase may be caused by an interference with CST function or C-strand polymerization.
Notwithstanding, our results are compatible with the notion that at least part of the activation of Mec1 stems from the exposure of non-telomeric ssDNA after 5′-3′ resection, RPA association and the subsequent recruitment of Ddc2-Mec1 at short telomeres. In addition, the accumulation of subtelomeric ssDNA in senescent cells could have other consequences, such as Rap1 release and loss of subtelomeric heterochromatin formation (90).
The error-free DDT pathway acts at telomeres
We document here that the DDT helicase/ubiquitin ligase Rad5 forms nuclear foci in unperturbed conditions and that one-third of these foci are associated with telomeres. This indicates that Rad5 may have a role during unperturbed replication, which is surprising because Rad5 foci have only been reported in replication stress conditions (103). Yet, Rad5 was initially described to affect the stability of a poly(GT) tract (88), independently of Rad52 (104). Thus, Rad5 could contribute to maintain the stability of TG1-3 telomeric repeats. Accordingly, deletion of RAD5 leads to slightly longer telomeres (89). In addition to its ubiquitin ligase activity on PCNA, Rad5 possesses an in vitro fork regression activity that might work independently (59,105). An attractive hypothesis is that Rad5 assists in C-strand synthesis after the C-strand resection via the fork reversal activity. Alternatively, the error-free DDT pathway could rescue unscheduled precociously arrested replication forks at telomeres, protecting telomeres from rare shortening events that would be catastrophic in the absence of telomerase to re-elongate them (Figure 8). In support of this, our genetic data point to a role of Rad5 in the maintenance of the viability of telomerase-negative cells. Yet, after the loss of the catalytic subunit of telomerase, the ability of Rad5 to form foci decreases. We thus suggest that the mode of action of Rad5 may change after telomerase loss in such a way that it acts without forming foci.
Furthermore, MMS2 and RAD5 deletions have slightly different phenotypes regarding senescence, with MMS2 effects being restricted to cells bearing a very short telomere. The Ubc13-Mms2 complex is cytoplasmic and relocalizes to the nucleus in response to DNA damage (106). Thus, early activation of the checkpoints by the presence of a very short telomere could result in Mms2-Ubc13 import to the nucleus, explaining the more prominent role of MMS2 in the presence of a very short telomere. This also supports the notion that the role of Rad5 at wild-type telomeres and early after the loss of telomerase is probably independent of the poly-ubiquitylation of PCNA.
DNA repair activities at very short telomeres
Combining our genetic data and ChIP data, we have delineated a critical role for DNA repair activities such as strand invasion, template switching, DNA repair–associated DNA synthesis and resolution at very short telomeres. The activation of these activities is likely due to the presence of long ssDNA stretches because we observe a decrease in Rad52 recruitment in the absence of Tel1, a checkpoint kinase known to contribute to 5′-3′ resection at DSBs and telomeres (107). ssDNA is also the substrate for strand invasion by Rad52 and Rad51 activities and potentially for template switching by the error-free DDT pathway (60). Although the exact role of the known error-free DDT factors in template switching is not well defined, HR and DDT cooperate (58,60). We found that the lack of both pathways leads to an additive effect on the growth of cells, in particular in cells deprived of telomerase activity. An increased synthetic growth defect between these genes has also been observed on ultraviolet light or methyl methanesulfonate treatment (108,109), suggesting that the loss of TLC1 causes similar DNA damage and lethality in rad5Δ rad52Δ cells. Our results suggest an intimate connection of these pathways at short telomeres that remains to be elucidated.
In mitotic cells, recombination intermediates are mainly resolved by the cooperative activities of the helicase Sgs1, the topoisomerase Top3, and Rmi1, which dissolve Holliday junctions (110). Although the interpretation of our genetic results may be confounded by the potential involvement of Sgs1 in the generation of ssDNA at short telomeres, the data are compatible with the Sgs1-dependent resolution of intermediates at telomeres in the absence of telomerase. This interpretation is supported by previous evidence that, in the absence of Sgs1 and telomerase, telomeres accumulate Rad52-dependent secondary structures that resemble hemicatenanes (39,40). Notwithstanding, it is clear that the viability of cells displaying a very short telomere depends on Mus81, suggesting a role for this endonuclease specifically at very short telomeres. An attractive possibility is that the intermediates of very short telomeres preferentially adopt a structure resembling nicked Holliday junctions, as depicted in Figure 8, which is the preferred substrate for the Mus81–Mms4 complex (91). Moreover, a recent report indicated that the action of Mus81 is restricted to G2/M (111). This phase of the cell cycle is when telomeres are subjected to the most profound remodelling events—semi-conservative DNA replication, 5′–3′ resection and telomerase elongation. Thus, although we cannot exclude a role for Sgs1 at this stage, short telomeres may be eminent substrates of the Mus81–Mms4 complex in the context of senescence.
In conclusion, in this work we distinguish the events operating at relatively long and short telomeres in the absence of telomerase and before the emergence of post-senescence survivors. We expect that in this period, which is critical for human health, many of the factors we describe have key roles in the stability of human chromosome ends and consequently the whole genome. Specifically, the repair activities promote mainly sister chromatid exchanges, buffering the DNA end replication problem and the telomere shortening rate, by modulating the overhang length. It is important to note that they do not result in a net increase in telomere length. Hence, these repair pathways may be under tight control to avoid an unscheduled change in the proliferation capacity of a cell lineage. In support of this, most of the factors described have human orthologues that are well-known tumour suppressors. BLM and WRN, the human orthologues of Sgs1, are found to be mutated in aging syndromes characterized by a high incidence of cancer. The Rad5 human orthologues HLTF and SHPRH have likewise been identified as potential tumour suppressors (56). Hence, our work provides a new framework for testing their role in cancer progression through their potential effects on telomere replication and integrity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council [ERC-2010-StG 260906—D-END to M.T.T.]; the Mairie de Paris (Programme Emergences to M.T.T.); ITMO Cancer (to M.T.T.); the ‘Initiative d'Excellence’ program from the French State [‘DYNAMO’, ANR-11-LABX-0011-01]; the Danish Agency for Science, Technology and Innovation (DFF to M.L.); the Villum Kann Rasmussen Foundation (to M.L.); the European Research Council (ERC to M.L.); the Ligue Nationale contre le Cancer (to E.G., Equipe labélisée); the Agence Nationale de la Recherche [TELOREP to E.G.]. Funding for open access charge: European Research Council [ERC-2010-StG 260906—D-END].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Rothstein and A. Shinohara for primary antibodies. We would like to thank Teixeira lab members and B. Llorente for fruitful discussions and critical reading of the manuscript. We thank all members of the FRE3354 unit for technical support.
REFERENCES
- 1.Jain D, Cooper JP. Telomeric strategies: means to an end. Annu. Rev. Genet. 2010;44:243–269. doi: 10.1146/annurev-genet-102108-134841. [DOI] [PubMed] [Google Scholar]
- 2.Giraud-Panis MJ, Pisano S, Poulet A, Le Du MH, Gilson E. Structural identity of telomeric complexes. FEBS Lett. 2010;584:3785–3799. doi: 10.1016/j.febslet.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Lingner J, Cooper JP, Cech TR. Telomerase and DNA end replication: no longer a lagging strand problem? Science. 1995;269:1533–1534. doi: 10.1126/science.7545310. [DOI] [PubMed] [Google Scholar]
- 4.Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 5.Lansdorp PM. Telomeres, stem cells, and hematology. Blood. 2008;111:1759–1766. doi: 10.1182/blood-2007-09-084913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 7.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 8.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 9.Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 10.Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 11.d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 12.Enomoto S, Glowczewski L, Berman J. MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2626–2638. doi: 10.1091/mbc.02-02-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ijpma AS, Greider CW. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:987–1001. doi: 10.1091/mbc.02-04-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teixeira MT. Saccharomyces cerevisiae as a model to study replicative senescence triggered by telomere shortening. Front Oncol. 2013;3:101. doi: 10.3389/fonc.2013.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giraud-Panis MJ, Teixeira MT, Geli V, Gilson E. CST meets shelterin to keep telomeres in check. Mol. Cell. 2010;39:665–676. doi: 10.1016/j.molcel.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 16.Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP. Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol. Cell. 2009;35:70–81. doi: 10.1016/j.molcel.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Larrivee M, LeBel C, Wellinger RJ. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 2004;18:1391–1396. doi: 10.1101/gad.1199404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 19.Wu P, Takai H, de Lange T. Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell. 2012;150:39–52. doi: 10.1016/j.cell.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol. Cell. 2007;27:851–858. doi: 10.1016/j.molcel.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Sabourin M, Tuzon CT, Zakian VA. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol. Cell. 2007;27:550–561. doi: 10.1016/j.molcel.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arneric M, Lingner J. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep. 2007;8:1080–1085. doi: 10.1038/sj.embor.7401082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bianchi A, Shore D. Increased association of telomerase with short telomeres in yeast. Genes Dev. 2007;21:1726–1730. doi: 10.1101/gad.438907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGee JS, Phillips JA, Chan A, Sabourin M, Paeschke K, Zakian VA. Reduced Rif2 and lack of Mec1 target short telomeres for elongation rather than double-strand break repair. Nat. Struct. Mol. Biol. 2010;17:1438–1445. doi: 10.1038/nsmb.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goudsouzian LK, Tuzon CT, Zakian VA. S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol. Cell. 2006;24:603–610. doi: 10.1016/j.molcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Frank CJ, Hyde M, Greider CW. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol. Cell. 2006;24:423–432. doi: 10.1016/j.molcel.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 27.Vodenicharov MD, Wellinger RJ. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol. Cell. 2006;24:127–137. doi: 10.1016/j.molcel.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 28.Negrini S, Ribaud V, Bianchi A, Shore D. DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev. 2007;21:292–302. doi: 10.1101/gad.400907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michelson RJ, Rosenstein S, Weinert T. A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Genes Dev. 2005;19:2546–2559. doi: 10.1101/gad.1293805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ribeyre C, Shore D. Anticheckpoint pathways at telomeres in yeast. Nat. Struct. Mol. Biol. 2012;19:307–313. doi: 10.1038/nsmb.2225. [DOI] [PubMed] [Google Scholar]
- 31.Finn K, Lowndes NF, Grenon M. Eukaryotic DNA damage checkpoint activation in response to double-strand breaks. Cell. Mol. Life Sci. 2012;69:1447–1473. doi: 10.1007/s00018-011-0875-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khadaroo B, Teixeira MT, Luciano P, Eckert-Boulet N, Germann SM, Simon MN, Gallina I, Abdallah P, Gilson E, Geli V, et al. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat. Cell Biol. 2009;11:980–987. doi: 10.1038/ncb1910. [DOI] [PubMed] [Google Scholar]
- 33.Abdallah P, Luciano P, Runge KW, Lisby M, Geli V, Gilson E, Teixeira MT. A two-step model for senescence triggered by a single critically short telomere. Nat. Cell Biol. 2009;11:988–993. doi: 10.1038/ncb1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grandin N, Bailly A, Charbonneau M. Activation of Mrc1, a mediator of the replication checkpoint, by telomere erosion. Biol. Cell. 2005;97:799–814. doi: 10.1042/BC20040526. [DOI] [PubMed] [Google Scholar]
- 35.Ritchie KB, Mallory JC, Petes TD. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ballew BJ, Lundblad V. Multiple genetic pathways regulate replicative senescence in telomerase-deficient yeast. Aging Cell. 2013;12:719–727. doi: 10.1111/acel.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le S, Moore JK, Haber JE, Greider CW. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Q, Ijpma A, Greider CW. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell Biol. 2001;21:1819–1827. doi: 10.1128/MCB.21.5.1819-1827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azam M, Lee JY, Abraham V, Chanoux R, Schoenly KA, Johnson FB. Evidence that the S. cerevisiae Sgs1 protein facilitates recombinational repair of telomeres during senescence. Nucleic Acids Res. 2006;34:506–516. doi: 10.1093/nar/gkj452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JY, Kozak M, Martin JD, Pennock E, Johnson FB. Evidence that RecQ helicase slows senescence by resolving recombining telomeres. PLoS Biol. 2007;5:1334–1344. doi: 10.1371/journal.pbio.0050160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer DH, Bailis AM. Telomerase deficiency affects the formation of chromosomal translocations by homologous recombination in Saccharomyces cerevisiae. PLoS One. 2008;3:e3318. doi: 10.1371/journal.pone.0003318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lebel C, Rosonina E, Sealey DC, Pryde F, Lydall D, Maringele L, Harrington LA. Telomere maintenance and survival in Saccharomyces cerevisiae in the absence of telomerase and RAD52. Genetics. 2009;182:671–684. doi: 10.1534/genetics.109.102939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin YH, Chang CC, Wong CW, Teng SC. Recruitment of Rad51 and Rad52 to short telomeres triggers a Mec1-mediated hypersensitivity to double-stranded DNA breaks in senescent budding yeast. PLoS One. 2009;4:e8224. doi: 10.1371/journal.pone.0008224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 45.Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- 46.Ye J, Lenain C, Bauwens S, Rizzo A, Saint-Leger A, Poulet A, Benarroch D, Magdinier F, Morere J, Amiard S, et al. TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell. 2010;142:230–242. doi: 10.1016/j.cell.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 47.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makovets S, Herskowitz I, Blackburn EH. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol. Cell Biol. 2004;24:4019–4031. doi: 10.1128/MCB.24.9.4019-4031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chavez A, George V, Agrawal V, Johnson FB. Sumoylation and the structural maintenance of chromosomes (Smc) 5/6 complex slow senescence through recombination intermediate resolution. J. Biol. Chem. 2010;285:11922–11930. doi: 10.1074/jbc.M109.041277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Branzei D. Ubiquitin family modifications and template switching. FEBS Lett. 2011;585:2810–2817. doi: 10.1016/j.febslet.2011.04.053. [DOI] [PubMed] [Google Scholar]
- 52.Branzei D, Foiani M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amst) 2007;6:994–1003. doi: 10.1016/j.dnarep.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 53.Gangavarapu V, Santa Maria SR, Prakash S, Prakash L. Requirement of replication checkpoint protein kinases Mec1/Rad53 for postreplication repair in yeast. MBio. 2011;2:e00079–e00011. doi: 10.1128/mBio.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 55.Xiao W, Chow BL, Broomfield S, Hanna M. The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplication repair pathways. Genetics. 2000;155:1633–1641. doi: 10.1093/genetics/155.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Unk I, Hajdu I, Blastyak A, Haracska L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst) 2010;9:257–267. doi: 10.1016/j.dnarep.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 57.Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gangavarapu V, Prakash S, Prakash L. Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell Biol. 2007;27:7758–7764. doi: 10.1128/MCB.01331-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanoli F, Fumasoni M, Szakal B, Maloisel L, Branzei D. Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. PLoS Genet. 2010;6:e1001205. doi: 10.1371/journal.pgen.1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gonzalez-Prieto R, Munoz-Cabello AM, Cabello-Lobato MJ, Prado F. Rad51 replication fork recruitment is required for DNA damage tolerance. EMBO J. 2013;32:1307–1321. doi: 10.1038/emboj.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barlow JH, Rothstein R. Rad52 recruitment is DNA replication independent and regulated by Cdc28 and the Mec1 kinase. EMBO J. 2009;28:1121–1130. doi: 10.1038/emboj.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hemann MT, Strong MA, Hao LY, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 64.Jeyapalan JC, Ferreira M, Sedivy JM, Herbig U. Accumulation of senescent cells in mitotic tissue of aging primates. Mech. Ageing Dev. 2007;128:36–44. doi: 10.1016/j.mad.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- 66.Kaul Z, Cesare AJ, Huschtscha LI, Neumann AA, Reddel RR. Five dysfunctional telomeres predict onset of senescence in human cells. EMBO Rep. 2011;13:52–59. doi: 10.1038/embor.2011.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu Z, Dao Duc K, Holcman D, Teixeira MT. The length of the shortest telomere as the major determinant of the onset of replicative senescence. Genetics. 2013;194:847–857. doi: 10.1534/genetics.113.152322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Longtine MS, McKenzie AI, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 69.Forstemann K, Hoss M, Lingner J. Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res. 2000;28:2690–2694. doi: 10.1093/nar/28.14.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- 71.Church GM, Gilbert W. Genomic sequencing. Proc. Natl Acad. Sci. USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 73.Booth C, Griffith E, Brady G, Lydall D. Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res. 2001;29:4414–4422. doi: 10.1093/nar/29.21.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eckert-Boulet N, Rothstein R, Lisby M. Cell biology of homologous recombination in yeast. Methods Mol Biol. 2011;745:523–536. doi: 10.1007/978-1-61779-129-1_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marcand S, Brevet V, Gilson E. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 1999;18:3509–3519. doi: 10.1093/emboj/18.12.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 77.Iglesias N, Lingner J. Related mechanisms for end processing at telomeres and DNA double-strand breaks. Mol. Cell. 2009;35:137–138. doi: 10.1016/j.molcel.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 78.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johansson E, Garg P, Burgers PM. The Pol32 subunit of DNA polymerase delta contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 2004;279:1907–1915. doi: 10.1074/jbc.M310362200. [DOI] [PubMed] [Google Scholar]
- 80.Maloisel L, Fabre F, Gangloff S. DNA polymerase delta is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Mol. Cell Biol. 2008;28:1373–1382. doi: 10.1128/MCB.01651-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li J, Holzschu DL, Sugiyama T. PCNA is efficiently loaded on the DNA recombination intermediate to modulate polymerase delta, eta, and zeta activities. Proc. Natl Acad. Sci. USA. 2013;110:7672–7677. doi: 10.1073/pnas.1222241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 83.Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 84.Lydeard JR, Lipkin-Moore Z, Sheu YJ, Stillman B, Burgers PM, Haber JE. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 2010;24:1133–1144. doi: 10.1101/gad.1922610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martina M, Clerici M, Baldo V, Bonetti D, Lucchini G, Longhese MP. A balance between Tel1 and Rif2 activities regulates nucleolytic processing and elongation at telomeres. Mol. Cell Biol. 2012;32:1604–1617. doi: 10.1128/MCB.06547-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grandin N, Charbonneau M. Mrc1, a non-essential DNA replication protein, is required for telomere end protection following loss of capping by Cdc13, Yku or telomerase. Mol. Genet. Genomics. 2007;277:685–699. doi: 10.1007/s00438-007-0218-0. [DOI] [PubMed] [Google Scholar]
- 87.Minca EC, Kowalski D. Multiple Rad5 activities mediate sister chromatid recombination to bypass DNA damage at stalled replication forks. Mol. Cell. 2010;38:649–661. doi: 10.1016/j.molcel.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Johnson RE, Henderson ST, Petes TD, Prakash S, Bankmann M, Prakash L. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol. Cell Biol. 1992;12:3807–3818. doi: 10.1128/mcb.12.9.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gatbonton T, Imbesi M, Nelson M, Akey JM, Ruderfer DM, Kruglyak L, Simon JA, Bedalov A. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2006;2:e35. doi: 10.1371/journal.pgen.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Platt JM, Ryvkin P, Wanat JJ, Donahue G, Ricketts MD, Barrett SP, Waters HJ, Song S, Chavez A, Abdallah KO, et al. Rap1 relocalization contributes to the chromatin-mediated gene expression profile and pace of cell senescence. Genes Dev. 2013;27:1406–1420. doi: 10.1101/gad.218776.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schwartz EK, Heyer WD. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma. 2011;120:109–127. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zeng S, Xiang T, Pandita TK, Gonzalez-Suarez I, Gonzalo S, Harris CC, Yang Q. Telomere recombination requires the MUS81 endonuclease. Nat. Cell Biol. 2009;11:616–623. doi: 10.1038/ncb1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 94.Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat. Struct. Mol. Biol. 2010;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johnson FB, Marciniak RA, McVey M, Stewart SA, Hahn WC, Guarente L. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 2001;20:905–913. doi: 10.1093/emboj/20.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cohen H, Sinclair DA. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl Acad. Sci. USA. 2001;98:3174–3179. doi: 10.1073/pnas.061579598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang P, Pryde FE, Lester D, Maddison RL, Borts RH, Hickson ID, Louis EJ. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 2001;11:125–129. doi: 10.1016/s0960-9822(01)00021-5. [DOI] [PubMed] [Google Scholar]
- 98.Eckert-Boulet N, Lisby M. Regulation of homologous recombination at telomeres in budding yeast. FEBS Lett. 2010;584:3696–3702. doi: 10.1016/j.febslet.2010.05.037. [DOI] [PubMed] [Google Scholar]
- 99.Dewar JM, Lydall D. Similarities and differences between “uncapped” telomeres and DNA double-strand breaks. Chromosoma. 2011;121:117–130. doi: 10.1007/s00412-011-0357-2. [DOI] [PubMed] [Google Scholar]
- 100.Bonetti D, Clerici M, Anbalagan S, Martina M, Lucchini G, Longhese MP. Shelterin-like proteins and Yku inhibit nucleolytic processing of Saccharomyces cerevisiae telomeres. PLoS Genet. 2010;6:e1000966. doi: 10.1371/journal.pgen.1000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang F, Stewart JA, Kasbek C, Zhao Y, Wright WE, Price CM. Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Rep. 2012;2:1096–1103. doi: 10.1016/j.celrep.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Diede SJ, Gottschling DE. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- 103.Tkach JM, Yimit A, Lee AY, Riffle M, Costanzo M, Jaschob D, Hendry JA, Ou J, Moffat J, Boone C, et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 2012;14:966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Henderson ST, Petes TD. Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol. Cell Biol. 1992;12:2749–2757. doi: 10.1128/mcb.12.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gangavarapu V, Haracska L, Unk I, Johnson RE, Prakash S, Prakash L. Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol. Cell Biol. 2006;26:7783–7790. doi: 10.1128/MCB.01260-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Longhese MP, Bonetti D, Manfrini N, Clerici M. Mechanisms and regulation of DNA end resection. EMBO J. 2010;29:2864–2874. doi: 10.1038/emboj.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Halas A, Podlaska A, Derkacz J, McIntyre J, Skoneczna A, Sledziewska-Gojska E. The roles of PCNA SUMOylation, Mms2-Ubc13 and Rad5 in translesion DNA synthesis in Saccharomyces cerevisiae. Mol. Microbiol. 2011;80:786–797. doi: 10.1111/j.1365-2958.2011.07610.x. [DOI] [PubMed] [Google Scholar]
- 109.Motegi A, Kuntz K, Majeed A, Smith S, Myung K. Regulation of gross chromosomal rearrangements by ubiquitin and SUMO ligases in Saccharomyces cerevisiae. Mol. Cell Biol. 2006;26:1424–1433. doi: 10.1128/MCB.26.4.1424-1433.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hickson ID, Mankouri HW. Processing of homologous recombination repair intermediates by the Sgs1-Top3-Rmi1 and Mus81-Mms4 complexes. Cell Cycle. 2011;10:3078–3085. doi: 10.4161/cc.10.18.16919. [DOI] [PubMed] [Google Scholar]
- 111.Szakal B, Branzei D. Premature Cdk1/Cdc5/Mus81 pathway activation induces aberrant replication and deleterious crossover. EMBO J. 2013;32:1155–1167. doi: 10.1038/emboj.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.