Abstract
A conditional gene expression system that is fast-acting, is tunable and achieves single-gene specificity was recently developed for yeast. A gene placed directly downstream of a modified GAL1 promoter containing six Zif268 binding sequences (with single nucleotide spacing) was shown to be selectively inducible in the presence of β-estradiol, so long as cells express the artificial transcription factor, Z3EV (a fusion of the Zif268 DNA binding domain, the ligand binding domain of the human estrogen receptor and viral protein 16). We show the strength of Z3EV-responsive promoters can be modified using straightforward design principles. By moving Zif268 binding sites toward the transcription start site, expression output can be nearly doubled. Despite the reported requirement of estrogen receptor dimerization for hormone-dependent activation, a single binding site suffices for target gene activation. Target gene expression levels correlate with promoter binding site copy number and we engineer a set of inducible promoter chassis with different input–output characteristics. Finally, the coupling between inducer identity and gene activation is flexible: the ligand specificity of Z3EV can be re-programmed to respond to a non-hormone small molecule with only five amino acid substitutions in the human estrogen receptor domain, which may prove useful for industrial applications.
INTRODUCTION
Numerous strategies have emerged for controlling levels of gene expression in yeast. These include introducing sequences that disfavor nucleosome localization near the transcription start site (1), varying both the affinity and number of binding sites for transcriptional activators in the promoter (2–4) and randomization of core promoter elements (5). Protein levels can also be greatly influenced by the 5′-UTR and 3′-UTR sequences. The sequence ‘AAA’ at positions −1 → −3 of the gene’s initial ATG favor high protein expression, while out-of-frame AUGs in the 5′-UTR near the start codon reduce protein levels (6). In the 3′-UTR, higher A/T content upstream of the polyadenylation site correlates with higher protein expression (7). While promoter strength and protein expression can be tuned in a variety of ways, it is also desirable to selectively introduce or remove selected gene products so that the effects of gain or loss of function can be assayed (3,8–10).
Conditional induction of gene expression has relied heavily on placing genes under the control of nutrient-responsive DNA sequences (11,12). Promoters responsive to the tetracycline analog doxycycline and to hormones such as β-estradiol, both of which are inert with respect physiology, have also been developed (13–15). In the case of β-estradiol, artificial transcription factors (ATFs) containing a DNA binding domain, a hormone binding domain and an activation domain are expressed in the cell. The ATF Gal4dbd.hER.VP16 (GEV) is a fusion of the Saccharomyces cerevisiae Gal4p DNA binding domain, the ligand binding domain of the human estrogen receptor (hER) and the activation domain of viral protein 16 (VP16, from herpes simplex virus) (14,15). In the absence of inducer, GEV is sequestered to the cytoplasm where, through the hER, it interacts with the Hsp90 chaperone complex (15,16). On addition of inducer, GEV dissociates from the Hsp90 chaperone complex, localizes to the nucleus and activates expression of genes placed downstream of promoters containing Gal4p-responsive DNA elements (14,15).
The GEV system has many desirable properties. It is fast-acting, has a graded input–output relationship with respect to inducer concentration, activates expression in all cells and works in diverse media formulations (including those using 2% glucose as a carbon source) (15,17). However, the presence of the Gal4p DNA binding domain results in unwanted activation of many off-target genes (as well as indirect repression of others), resulting in defective growth at moderate-to-high levels of inducer (15). By replacing the DNA binding domain of GEV with others that have few-to-no binding sites in the genome [as detailed in (3,18)], the desirable properties of GEV can be retained while eliminating any unwanted off-target effects. One such protein, Z3EV, is identical to GEV, except for the DNA binding domain, which is replaced with Zif268 (Z3 refers to the fact that Zif268 has three zinc fingers, which results in 9 bp of DNA specificity).
In this manuscript, we tested promoter design principles and expanded the range of Z3EV functionality. Specifically, the effects of 5′-UTR sequence, binding site copy number and binding site spacing on gene expression were tested in Z3EV-responsive promoters. We find that a single binding site is enough to make a promoter Z3EV-inducible. In promoters with multiple binding sites, inter-binding site spacing can affect the level of induction, especially at sub-saturating concentrations of inducer. Whereas previously we exclusively used GAL1 promoters modified to include Z3EV binding sites, here we test the ability of other promoter chassis to be made inducible by Z3EV with a high dynamic range. Finally, we show that changing five amino acids in the hER domain is sufficient to make Z3EV completely non-responsive to β-estradiol, but strongly responsive to a non-steroid small molecule.
MATERIALS AND METHODS
Strains, media and cloning
The parent strain used in the promoter- and ATF-characterization experiments is a derivative of CEN.PK113-7 d (19) in which URA3 was replaced by NatMX. NatMX is flanked by targeting sequences for the endonuclease F-CphI to facilitate its removal if desired. Chemostat and growth experiments were performed using DBY12000, an s288c-derivative with a functional HAP1 allele (8). For batch experiments, yeast cells were cultured in either complete synthetic medium lacking uracil (CSM-U; 2% dextrose) or yeast extract-peptone-dextrose medium (1% yeast extract, 2% bactopeptone and 2% dextrose).
Transformations were performed using a standard lithium acetate method (20). All cloning was performed using DNA gap repair in the yeast strain CEN.PK2-1c, a tryptophan auxotroph (MATa; ura3-52; trp1-289; leu2-3,112; his3Δ 1; MAL2-8C; SUC2). Complete synthetic medium lacking tryptophan (CSM-W; 2% dextrose) was used as selective medium for yeast outgrowth following transformation. Yeast transformations were miniprepped directly from liquid cultures and transformed into XL1-Blue cells (Agilent). Single colonies were selected on LB-CARB (0.5% yeast extract, 1% tryptone, 0.5% NaCl and 0.1 g/l carbenicillin) and proper assemblies were confirmed via restriction digestion and Sanger sequencing as described previously (21). The vector used for assembly is a derivative of the TRP1-marked yeast shuttle vector pRS414 with the LacZ open reading frame disrupted by a pair of linkers, referred to as 0 and 9, between which DNA assemblies were constructed: Linker 0 = GACGGCACGGCCACGCGTTTAAACCGCC; Linker 9 = CGGTGTTTAAACCCCAGCGCCTGGCGGG. Genomic insertions were generated via transformation of two linear DNA fragments with overlap in the URA3 marker and subsequent selection for growth on CSM-U agar medium. Colony polymerase chain reaction was used to confirm proper insertion at the BUD9 locus.
β-estradiol (Sigma-Aldrich) and 4′-4′-dihydroxybenzyl (DHB; Austin Chemical Company) were dissolved in 100% ethanol at 25 mM and stored at 4°C. Synthetic promoters and yeast codon-optimized ATFs were synthesized commercially (SGI-DNA, La Jolla, CA, USA). Codon-optimized sequences were determined using Integrated DNA Technologies’ web interface (Coralville, IA). The fluorescent reporter, GFP-Dasher, was obtained from DNA2.0 (Menlo Park, CA, USA). Restriction enzymes were obtained from New England Biolabs (Ipswich, MA, USA).
Flow cytometry
For dose–response experiments, saturated overnight cultures in CSM-U medium were diluted 1:200 in 5 ml of fresh CSM-U medium. Cells were incubated with different levels of β-estradiol or DHB for ∼12 h. Approximately 107 cells were harvested and re-suspended in phosphate buffered saline supplemented with 0.1% Tween-20. Green fluorescent protein (GFP) fluorescence measurements were made with a BD FACSAria II (BD Biosciences, Sparks, MD, USA). Mean fluorescence values were obtained from a minimum of 10 000 cells in each sample, and the fluorescence detector calibration settings maintained constant between experiments. Positive and negative GFP controls were used to ensure that the instrument maintained proper calibration and to ensure proper normalization. For kinetic experiments, cells were allowed to grow for 4 h before introduction of inducer. For all flow cytometry experiments, data analysis was performed in MATLAB, as described previously (18).
Chemostat experiment
Yeast cells were cultured in a 500-ml chemostat (Sixfors, Infors AG, Bottmingen, Switzerland) under phosphate limitation (20 mg/l). The culture was grown at 30 °C, stirred at 400 rpm, aerated with filtered humidified air, and maintained at a volume of 300 ml. The growth rate was maintained at 0.17 h−1 (culture doubling time = 4.08 h). Batch growth was initiated from a 1:60 dilution of an overnight culture (also grown in phosphate-limited chemostat medium). Cells were grown to steady state (as determined by optical density and cell size) before addition of DHB.
Microarrays
RNA extraction, labeling and hybridization were performed as described previously (22) with slight modifications. Briefly, chemostat samples (∼5 ml) were vacuum-filtered onto 0.45 -µm nylon membranes (Millipore, HNWP02500), placed in 2-ml locking lid tubes (FisherBrand, 02-681-291) and flash-frozen in liquid nitrogen. Crude RNA was extracted with a standard acid–phenol procedure and cleaned with RNeasy mini columns (QIAGEN, Valencia, CA, USA) before mRNA amplification and labeling with the Agilent Quick-Amp Labeling Kit (Part No. 5190-0424). Amplification and labeling were performed according to the manufacturer’s instructions with half the volume of each reagent and 0.6 µl of Cy3 or Cy5 dye. Reference RNA was extracted from cells harvested just before addition of DHB. Agilent Yeast Oligo V2 microarrays (8 × 15 k) were hybridized for 17 h at 65°C on a rotisserie at 20 rpm. Following a series of wash steps, hybridized microarrays were scanned and raw data were extracted with Agilent Feature Extractor Software version 9.5.
RESULTS
For all promoter characterization experiments, a contiguous string of DNA ‘parts’ replaces the entire BUD9 open reading frame. Z3EV is constitutively expressed from the ACT1 promoter, followed sequentially by URA3 (used as a selection marker for genomic integration), the synthetic promoter to be tested and GFP (Figure 1). The level of gene expression from each promoter was measured from three independent cultures (from three independent colonies) via flow cytometry in the absence or presence (medium level = 10 nM or high level = 1 µM) of β-estradiol. A complete list of strains can be found in Table 1, and the sequences of DNA parts can be found in the Supplement. In addition to the figures that illustrate binding site location and spacing, each promoter is assigned a unique number (P1–P15).
Figure 1.
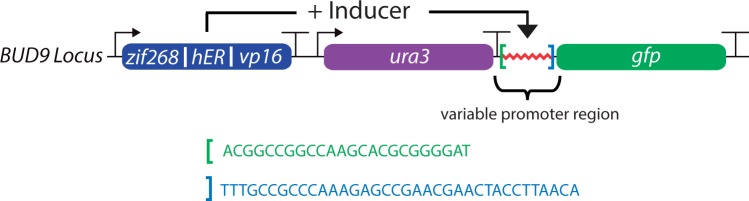
Schematic of BUD9 locus in engineered yeast strains. Universal primer sites (shown in green and blue) flank the variable promoter region in all strains. Gene terminators are represented by ‘T’ in the figure. The ENO2 terminator follows Z3EV and the GAL80 terminator follows GFP. The promoter/terminator pair surrounding URA3 is the same as the native gene.
Table 1.
List of yeast strains used in experiments
| Strain | Genotype | Source |
|---|---|---|
| DBY12000 | GAL2+ s288c with repaired HAP1 allele | (8) |
| DBY19000 | ura3Δ::NatMX | This study |
| DBY19004 | ura3Δ::NatMX, bud9Δ::ACT1pr-Z3EV-ENO2term-URA3-Z3EVpr(P1)-GFP-Gal80term | This study |
| DBY19017 | ura3Δ::NatMX, bud9Δ::ACT1pr-Z3EV-ENO2term-URA3-CM2pr(P15)-GFP-Gal80term | This study |
| DBY19018 | ura3Δ::NatMX, bud9Δ::ACT1pr-Z3EV-ENO2term-URA3-HOP2pr(P12)-GFP-Gal80term | This study |
| DBY19019 | ura3Δ::NatMX, bud9Δ::ACT1pr-Z3EV-ENO2term-URA3-HOP1pr(P11)-GFP-Gal80term | This study |
| DBY19050 | ura3Δ::NatMX, bud9Δ::ACT1pr-Z3EV-ENO2term-URA3-CM1pr(P14)-GFP-Gal80term | This study |
| DBY19052 | ura3Δ::NatMX, bud9Δ::ACT1pr-Z3EV-ENO2term-URA3-Z3EVpr(P2)-GFP-Gal80term | This study |
| DBY19053 | ura3Δ::NatMX, bud9Δ::ACT1pr-Z3EV-ENO2term-URA3-Z3EVpr(P3)-GFP-Gal80term | This study |
| DBY19054 | ura3Δ::NatMX, bud9Δ::ACT1pr-Z3EV-ENO2term-URA3-Z3EVpr(P4)-GFP-Gal80term | This study |
| DBY19055 | ura3Δ::NatMX, bud9Δ::ACT1pr-Z3EV-ENO2term-URA3-Z3EVpr(P6)-GFP-Gal80term | This study |
| DBY19056 | ura3Δ::NatMX, bud9Δ::ACT1pr-Z3EV-ENO2term-URA3-Z3EVpr(P5)-GFP-Gal80term | This study |
| DBY19058 | ura3Δ::NatMX, bud9Δ::ACT1pr-Z3EV-ENO2term-URA3-CrippledCYC1pr(P7)-GFP-Gal80term | This study |
| DBY19059 | ura3Δ::NatMX, bud9Δ::ACT1pr-Z3EV-ENO2term-URA3-CrippledCYC1pr(P8)-GFP-Gal80term | This study |
| DBY19061 | ura3Δ::NatMX, bud9Δ::ACT1pr-Z3EV-ENO2term-URA3-CrippledCYC1pr(P9)-GFP-Gal80term | This study |
| DBY19063 | ura3Δ::NatMX, bud9Δ::ACT1pr-Z3EV-ENO2term-URA3-CrippledCYC1pr(P10)-GFP-Gal80term | This study |
| DBY19065 | ura3Δ::NatMX, bud9Δ::ACT1pr-Z3EV-ENO2term-URA3-DAN1pr(P13)-GFP-Gal80term | This study |
| DBY19068 | ura3Δ::NatMX, bud9Δ::CYC1pr-Z3E(4S)V-YPR052Cterm -URA3-Z3EVpr(P1)-GFP-Gal80term | This study |
| DBY19070 | ura3Δ::NatMX, bud9Δ::ACT1pr-Z3E(4S)V-YPR052Cterm-URA3-Z3EVpr(P1)-GFP-Gal80term | This study |
All 19 000 series yeast strains are in the CEN-PK113-7d background. In the genotypes, ‘pr’ indicates a promoter and ‘term’ indicates a terminator. Z3EVpr indicates a modified version of the GAL1 promoter that contains Zif268 binding sites. All Z3EV-responsive promoters have a unique number (P1, P2, … , P15).
Modified GAL1 promoters
Previously, we removed the three canonical Gal4p activation sequences in the GAL1 promoter and replaced them with six Zif268 binding sites with single nucleotide spacing (Figure 2A, P1) (3). A modified GAL1 promoter in which five Zif628 binding sites have 9-bp spacing remains inducible at both 10 nM and 1 µM β-estradiol (Figure 2A, P2), demonstrating that there is some flexibility over the exact choice of binding site spacing.
Figure 2.
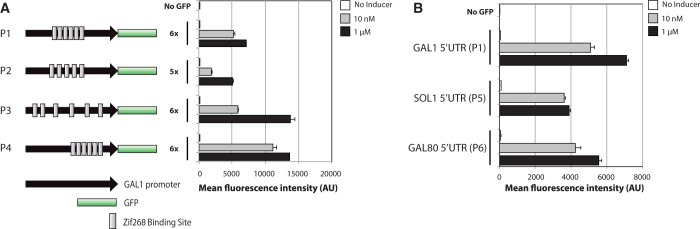
Engineered GAL1 promoter variants. (A) Measuring the effects of Zif268 binding site location and spacing on Z3EV-mediated gene induction. Schematic cartoons (not drawn to scale) illustrate the tested promoter architectures and bar plots are measurements of GFP intensity in the presence or absence of inducer following ∼12 h of induction. (B) Measuring the effects of 5′-UTR sequence on Z3EV-mediated gene induction, by replacing the GAL1 5′-UTR with that of SOL1 or GAL80. Error bars represent the standard deviation of fluorescence from three independent colonies/cultures.
In promoters containing six randomly dispersed binding sites (Figure 2A, P3), or six binding sites with single nucleotide spacing (Figure 2A, P4), expression in the presence of 1 µM β-estradiol nearly doubled with respect to our original promoter design, P1. In both P3 and P4, the binding site closest to the start codon is 235 bp and 242 bp, respectively (∼100 bp upstream of the transcription start state [TSS]). In P1 and P2, the binding site closest to the start codon is 408 and 385 bp (∼250 bp upstream of the TSS), respectively. These results demonstrate that moving binding sites closer to the TSS of modified GAL1 promoters can increase induced expression without increasing basal expression (i.e. in the absence of inducer, the distribution of fluorescence intensities is indistinguishable from cells lacking GFP), leading to an overall increase in the system’s dynamic range.
To test the effect of 5′-UTR sequence on induciblity, the GAL1 5′-UTR was removed and replaced with that of GAL80 or SOL1 (23), both of which encode more stable transcripts than GAL1 in glucose-grown cells (24). In glucose, the average half-life of mRNAs in yeast is 20.1 ± 8.6 min (24). The GAL80 mRNA half-life is 27 min and that of SOL1 is 79 min, whereas the half-life of GAL1 mRNA is 19 min (24). Despite GAL80 and SOL1 being more stable transcripts, we find their 5′-UTR sequence to be slightly deleterious to overall protein production in the context of the modified GAL1 promoter (Figure 2B, P5 and P6).
Altering promoter context and binding site copy number
Different numbers of Zif268 binding sites were placed upstream of a 249-bp crippled CYC1 promoter sequence from (25). From these experiments, we find that that at both 10 nM and 1 µM β-estradiol, induction correlates with binding site copy number (Figure 3A). One binding site is sufficient to make the promoter inducible (Figure 3A, P7). Unlike in the case of the modified GAL1 promoters, increased induction comes at the cost of increased basal expression in the absence of inducer (the basal expression from the promoter with eight binding sites is ∼6-fold greater than that with a single binding site). While going from 4 to 8 binding sites results in a 17% increase in GFP expression at 1 µM β-estradiol, the basal expression doubles. This results in the fold-change of induction being non-monotonic with respect to binding site copy number (Figure 3B).
Figure 3.
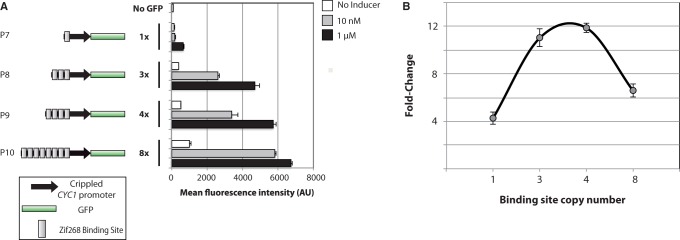
The effect of Zif268 binding site copy number on expression in crippled CYC1 promoters. (A) Measuring the effect of Zif268 binding site copy number on basal and induced expression when placed directly upstream of the crippled CYC1 promoter from (25). (B) The fold-change of expression at 1 µM β-estradiol over uninduced cells as a function of binding site copy number. Error bars represent the standard deviation of fluorescence from three independent colonies/cultures.
Increasing the number of promoter chassis
Based on the aforementioned results, it seemed reasonable that other promoters could be made inducible as well. DAN1, HOP1 and HOP2 are strongly repressed in asexually grown yeast. DAN1 encodes a cell wall mannoprotein that is expressed under anaerobic conditions (26,27). HOP1 and HOP2 gene products are both involved in the regulation of meiosis (28,29). Whereas a DAN1 promoter containing Zif268 binding sequences was made inducible by β-estradiol (Figure 4, P13), the modified HOP1 and HOP2 promoters were not (Figure 4, P11 and P12). ‘CYC1 Minimal’ promoters (CM1 and CM2), whose expression was attenuated based on results from Guarente and colleagues (30), were also made strongly inducible by the addition of Zif268 binding sites (Figure 4, P14 and P15).
Figure 4.
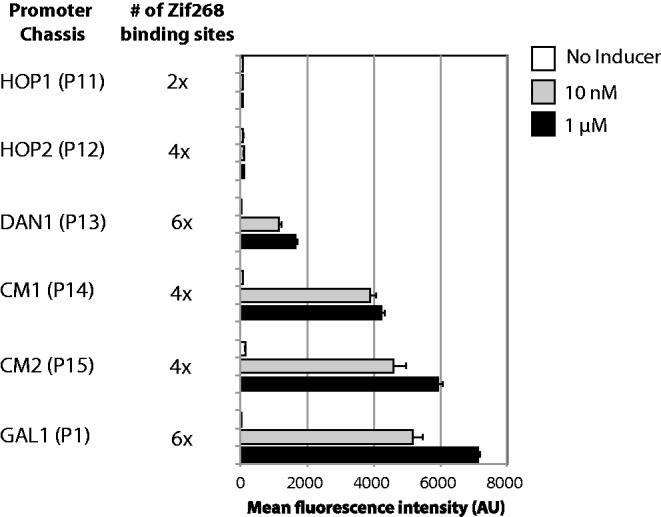
Testing the ability of alternative promoter chassis to be made inducible. Data are shown for six different promoters, including our initial modified GAL1 promoter design (P1). CM1 and CM2 are two different variants of ‘CYC1 Minimal’ promoters we designed to remove basal activity from the wild-type CYC1 promoter. Error bars represent the standard deviation of fluorescence from three independent colonies/cultures.
Changing the ligand specificity of Z3EV
After finding that Z3EV-responsive promoters could be engineered in a straightforward fashion, we wanted to see if they could be made to respond to a non-hormone small molecule. It has previously been demonstrated that ligand specificity of hER can be altered through directed evolution using two-hybrid screening in yeast (31–34). Indeed, mutant estrogen receptors have been engineered with enhanced specificity for resveratrol-like compounds (33), androgens (34), and synthetic small molecules, including DHB and 2,4-di(4-hydroxyphenyl)-5-ethylthiazole (L9) (31,32). Based on results from (31), we synthesized a codon-optimized variant of Z3EV, called Z3E(4S)V, that contains five hER domain amino acid substitutions. Z3EV and Z3E(4S)V contain amino acids 282–576 of the alpha subunit of the hER. According to this numbering, Z3E(4S)V contains the hER substitutions L346I, A350M, M388Q, G521S and Y526D.
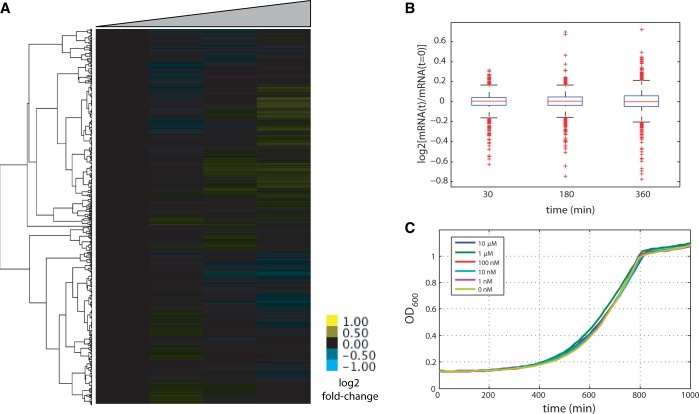
We find that the wild-type Z3EV construct is non-responsive with respect to the small molecule DHB at or below concentrations of 1 µM (Figure 5). Z3E(4S)V is induced by 100 nM DHB, but is non-responsive to any of the tested concentrations of β-estradiol. For both Z3EV and Z3E(4S)V, the response to inducer is graded and the coefficient of variation in GFP intensities remains nearly unchanged in induced cells (i.e. the shape of the distribution of expression intensities remains unchanged) (Supplementary Figure S1). Altering the ligand specificity did, however, increase the leakiness of Z3E(4S)V over the original Z3EV construct in the absence of inducer during log-phase growth (Supplementary Figure S2). The leakiness was reduced by decreasing the promoter strength driving Z3E(4S)V, but it is not clear from our results if this problem is completely avoidable. Changing ligand specificity does not affect the time it takes to see induction of GFP (Supplementary Figure S2), and similar to β-estradiol, DHB has no measured effect on gene expression (Figure 6A and B) or growth (Figure 6C).
Figure 5.
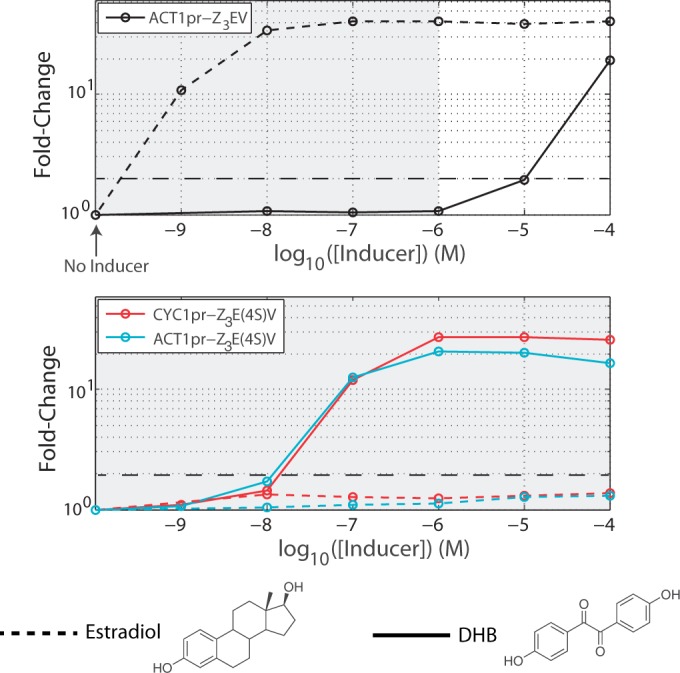
Dose–response curves for wild-type and mutant Z3EV variants. Responses to β-estradiol and DHB (short-dashes and solid lines, respectively) are shown for Z3EV (top) and Z3E(4S)V (bottom). A horizontal line (long dashes) indicates a 2-fold change in expression of GFP in each plot. Gray shading indicates concentrations of DHB to which Z3EV is not activated (top) or concentrations of β-estradiol to which Z3E(4S)V is not activated (bottom). Fluorescence measurements were made using flow cytometry.
Figure 6.
DHB has no effect on gene expression or growth. 1 µM DHB was introduced to phosphate-limited chemostat culture of strain DBY12000 at steady state. Samples were harvested at t = 0, 30, 180 and 360 min following DHB addition. (A) Hierarchical clustering of global gene expression response to DHB. Data are normalized to the t = 0 sample. (B) Data from (A) represented as a box plot. (C) DBY12000 grown in the presence of different amounts of DHB.
DISCUSSION
The presented experiments sought to identify basic design principles for engineering Z3EV-responsive promoters, develop and characterize a toolkit for making their implementation straightforward and illustrate that the ligand specificity of Z3EV can be altered to respond to a non-hormone small molecule. As S. cerevisiae is important in both basic and applied research, these tools have many potential applications, such as engineering orthogonal pathways into yeast for making target molecules: the fact the Z3EV system is titratible should enable facile testing of the relationship between target gene expression levels and product yield. We envision that the different strength promoters developed here will enable simultaneous expression of multiple gene products to different levels in the presence of a single concentration of inducer, making them effective tools for metabolic engineering and pathway optimization.
Finding cheaper non-hormone ligands
While inert with respect to yeast physiology, the use of hormone-based genetic switches is limited to academic or small- to moderate-scale experiments due to their potential for negatively impacting the environment. Only a handful of mutations are required for making a Z3EV variant that is strongly activated in the presence of a non-hormone small molecule, DHB; this molecule, however, is quite expensive, costing ∼$2000/g to synthesize commercially. A less soluble, cheaper DHB-like molecule in which the hydroxyl groups are replaced with methyl groups failed to induce Z3E(4S)V (data not shown). To make Z3EV a system that can be used at industrial scales, variants need to be engineered that respond not just to any non-hormone small molecules, but to ones that are (i) cheap to synthesize in large quantities and (ii) retain high specificity for the mutant receptor. Such mutant receptors may be identifiable using the previously published yeast two-hybrid approach (34), or perhaps a new selection scheme using the Z3EV system.
Increasing sequence diversity
DNA assembly methods facilitate stitching large numbers of individual DNA ‘parts’ to form larger DNA assemblies. Generating these assemblies through gap-repair cloning in yeast (35) can facilitate engineering biochemical pathways (36) as well as entire genomes (37) on a single plasmid. However, all DNA assembly methods require sufficient DNA sequence diversity for parts to be properly assembled (large stretches of degenerate sequences increases the number of statistically likely assemblies).
In this manuscript, we identified several different promoter chassis that can be made inducible. Therefore, if one wants to assemble a pathway in which each gene is inducible to different levels (and is expressed at different basal levels), the promoters we identified can be readily used. In the case where more than a handful of genes require unique basal/induced expression levels, it may be necessary to find additional promoter chassis, whose basal expression can be predicted via publicly available gene expression microarray data sets. Promoters that are repressed during asexual growth (such as HOP1 and HOP2) may not become inducible simply by adding Zif268 binding sites, which we anticipate is due to the presence of strong repressor elements; however, we have not completely ruled out the possibility that such promoters could become inducible with (i) additional Zif268 binding sites and/or (ii) Zif268 binding sites in different locations.
Conclusion
We have presented synthetic biology tools that enable dynamic perturbation of gene expression levels in S. cerevisiae using the ATF Z3EV. We demonstrate that the ligand specificity of Z3EV can be readily re-programmed using just a handful of amino acid substitutions in the hER domain. In contrast to other published methods, these tools can be immediately applied to cells growing under diverse physiological conditions without otherwise perturbing the cellular physiology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We would like to acknowledge support provided by the Automated Strain Engineering group and Lab Services team at Amyris, Inc. We thank Chris Paddon, Tim Geistlinger, and Xin Wang for many lively discussions, David Pincus for providing the crippled CYC1 promoter sequence, and finally, Zhongxia Wang and Yuzhong Wang for their hospitality while this paper was being written.
FUNDING
National Institutes of Health grant [GM046406 to D.B.]; NIGMS Center for Quantitative Biology grant [GM071508 to D.B.]. Funding for open access charge: National Institutes of Health grant [GM046406 to D.B.].
Conflict of interest statement. P.A. Gibney, S.S. Chandran, and K.R. Benjamin own shares of Amyris, Inc. D. Botstein is a member of the Scientific Advisory Board for Amyris, Inc.
REFERENCES
- 1.Raveh-Sadka T, Levo M, Shabi U, Shany B, Keren L, Lotan-Pompan M, Zeevi D, Sharon E, Weinberger A, Segal E. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat. Genet. 2012;44:743–750. doi: 10.1038/ng.2305. [DOI] [PubMed] [Google Scholar]
- 2.Khalil AS, Lu TK, Bashor CJ, Ramirez CL, Pyenson NC, Joung JK, Collins JJ. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McIsaac RS, Oakes BL, Wang X, Dummit KA, Botstein D, Noyes MB. Synthetic gene expression perturbation systems with rapid, tunable, single-gene specificity in yeast. Nucleic Acids Res. 2013;41:e57. doi: 10.1093/nar/gks1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharon E, Kalma Y, Sharp A, Raveh-Sadka T, Levo M, Zeevi D, Keren L, Yakhini Z, Weinberger A, Segal E. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat. Biotechnol. 2012;30:521–530. doi: 10.1038/nbt.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blount BA, Weenink T, Vasylechko S, Ellis T. Rational diversification of a promoter providing fine-tuned expression and orthogonal regulation for synthetic biology. PLoS One. 2012;7:e33279. doi: 10.1371/journal.pone.0033279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dvir S, Velten L, Sharon E, Zeevi D, Carey LB, Weinberger A, Segal E. Deciphering the rules by which 5′-UTR sequences affect protein expression in yeast. Proc. Natl Acad. Sci. USA. 2013;110:E2792–E2801. doi: 10.1073/pnas.1222534110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shalem O, Carey L, Zeevi D, Sharon E, Keren L, Weinberger A, Dahan O, Pilpel Y, Segal E. Measurements of the impact of 3′ end sequences on gene expression reveal wide range and sequence dependent effects. PLoS Comput. Biol. 2013;9:e1002934. doi: 10.1371/journal.pcbi.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hickman MJ, Petti AA, Ho-Shing O, Silverman SJ, McIsaac RS, Lee TA, Botstein D. Coordinated regulation of sulfur and phospholipid metabolism reflects the importance of methylation in the growth of yeast. Mol. Biol. Cell. 2011;22:4192–4204. doi: 10.1091/mbc.E11-05-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McIsaac RS, Petti AA, Bussemaker HJ, Botstein D. Perturbation-based analysis and modeling of combinatorial regulation in the yeast sulfur assimilation pathway. Mol. Biol. Cell. 2012;23:2993–3007. doi: 10.1091/mbc.E12-03-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglas AC, Smith AM, Sharifpoor S, Yan Z, Durbic T, Heisler LE, Lee AY, Ryan O, Gottert H, Surendra A, et al. Functional analysis with a barcoder yeast gene overexpression system. G3. 2012;2:1279–1289. doi: 10.1534/g3.112.003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labbe S, Thiele DJ. Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 1999;306:145–153. doi: 10.1016/s0076-6879(99)06010-3. [DOI] [PubMed] [Google Scholar]
- 12.Hovland P, Flick J, Johnston M, Sclafani RA. Galactose as a gratuitous inducer of GAL gene expression in yeasts growing on glucose. Gene. 1989;83:57–64. doi: 10.1016/0378-1119(89)90403-4. [DOI] [PubMed] [Google Scholar]
- 13.Belli G, Gari E, Piedrafita L, Aldea M, Herrero E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998;26:942–947. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louvion JF, Havaux-Copf B, Picard D. Fusion of GAL4-VP16 to a steroid-binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene. 1993;131:129–134. doi: 10.1016/0378-1119(93)90681-r. [DOI] [PubMed] [Google Scholar]
- 15.McIsaac RS, Silverman SJ, McClean MN, Gibney PA, Macinskas J, Hickman MJ, Petti AA, Botstein D. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol. Biol. Cell. 2011;22:4447–4459. doi: 10.1091/mbc.E11-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fliss AE, Benzeno S, Rao J, Caplan AJ. Control of estrogen receptor ligand binding by Hsp90. J. Steroid Biochem. Mol. Biol. 2000;72:223–230. doi: 10.1016/s0960-0760(00)00037-6. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi S, Pryciak PM. Membrane localization of scaffold proteins promotes graded signaling in the yeast MAP kinase cascade. Curr. Biol. 2008;18:1184–1191. doi: 10.1016/j.cub.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McIsaac RS, Oakes BL, Botstein D, Noyes MB. Rapid synthesis and screening of chemically activated transcription factors with GFP-based reporters. J. Vis. Exp. 2013;81:e51153. doi: 10.3791/51153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nijkamp JF, van den Broek M, Datema E, de Kok S, Bosman L, Luttik MA, Daran-Lapujade P, Vongsangnak W, Nielsen J, Heijne WH, et al. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb. Cell Fact. 2012; 11:36. doi: 10.1186/1475-2859-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 21.Dharmadi Y, Patel K, Shapland E, Hollis D, Slaby T, Klinkner N, Dean J, Chandran SS. High-throughput, cost-effective verification of structural DNA assembly. Nucleic Acids Res. 2013;42:e22. doi: 10.1093/nar/gkt1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol. Biol. Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munchel SE, Shultzaberger RK, Takizawa N, Weis K. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol. Biol. Cell. 2011;22:2787–2795. doi: 10.1091/mbc.E11-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mrsa V, Ecker M, Strahl-Bolsinger S, Nimtz M, Lehle L, Tanner W. Deletion of new covalently linked cell wall glycoproteins alters the electrophoretic mobility of phosphorylated wall components of Saccharomyces cerevisiae. J. Bacteriol. 1999;181:3076–3086. doi: 10.1128/jb.181.10.3076-3086.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sertil O, Cohen BD, Davies KJ, Lowry CV. The DAN1 gene of S. cerevisiae is regulated in parallel with the hypoxic genes, but by a different mechanism. Gene. 1997;192:199–205. doi: 10.1016/s0378-1119(97)00028-0. [DOI] [PubMed] [Google Scholar]
- 28.Hollingsworth NM, Byers B. HOP1: a yeast meiotic pairing gene. Genetics. 1989;121:445–462. doi: 10.1093/genetics/121.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leu JY, Chua PR, Roeder GS. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell. 1998;94:375–386. doi: 10.1016/s0092-8674(00)81480-4. [DOI] [PubMed] [Google Scholar]
- 30.Guarente L, Lalonde B, Gifford P, Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984;36:503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- 31.Chockalingam K, Chen Z, Katzenellenbogen JA, Zhao H. Directed evolution of specific receptor-ligand pairs for use in the creation of gene switches. Proc. Natl Acad. Sci. USA. 2005;102:5691–5696. doi: 10.1073/pnas.0409206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLachlan MJ, Chockalingam K, Lai KC, Zhao H. Directed evolution of orthogonal ligand specificity in a single scaffold. Angew. Chem. Int. Ed. Engl. 2009;48:7783–7786. doi: 10.1002/anie.200903413. [DOI] [PubMed] [Google Scholar]
- 33.Islam KM, Dilcher M, Thurow C, Vock C, Krimmelbein IK, Tietze LF, Gonzalez V, Zhao H, Gatz C. Directed evolution of estrogen receptor proteins with altered ligand-binding specificities. Protein Eng. Des. Sel. 2009;22:45–52. doi: 10.1093/protein/gzn067. [DOI] [PubMed] [Google Scholar]
- 34.Chen Z, Katzenellenbogen BS, Katzenellenbogen JA, Zhao H. Directed evolution of human estrogen receptor variants with significantly enhanced androgen specificity and affinity. J. Biol. Chem. 2004;279:33855–33864. doi: 10.1074/jbc.M402118200. [DOI] [PubMed] [Google Scholar]
- 35.Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 36.Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, Hutchison CA., III One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc. Natl Acad. Sci. USA. 2008;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.