Abstract
Understanding the relationships between regulatory factor binding, chromatin structure, cis-regulatory elements and RNA-regulation mechanisms relies on accurate information about transcription start sites (TSS) and polyadenylation sites (PAS). Although several approaches have identified transcript ends in yeast, limitations of resolution and coverage have remained, and definitive identification of TSS and PAS with single-nucleotide resolution has not yet been achieved. We developed SMORE-seq (simultaneous mapping of RNA ends by sequencing) and used it to simultaneously identify the strongest TSS for 5207 (90%) genes and PAS for 5277 (91%) genes. The new transcript annotations identified by SMORE-seq showed improved distance relationships with TATA-like regulatory elements, nucleosome positions and active RNA polymerase. We found 150 genes whose TSS were downstream of the annotated start codon, and additional analysis of evolutionary conservation and ribosome footprinting suggests that these protein-coding sequences are likely to be mis-annotated. SMORE-seq detected short non-coding RNAs transcribed divergently from more than a thousand promoters in wild-type cells under normal conditions. These divergent non-coding RNAs were less evident at promoters containing canonical TATA boxes, suggesting a model where transcription initiation at promoters by RNAPII is bidirectional, with TATA elements serving to constrain the directionality of initiation.
INTRODUCTION
Transcription initiation depends on interactions between general transcription factors (TFs) and RNA polymerase with promoter sequences and nucleosomes near the transcription start site (TSS) (1), while posttranscriptional regulation typically depends on sequences in 5′- and 3′-untranslated regions (UTRs) of mRNAs (2). Understanding the overall relationships between these aspects of gene regulation requires knowledge of TSS and transcript ends, or polyadenylation sites (PAS), of mRNAs genome-wide at single-nucleotide resolution. Although genes often have multiple TSS and PAS, identifying the most prominent transcript ends is useful for revealing their relationships to cis elements like TATA boxes, polyadenylation control sequences and other features like positioned nucleosomes. Definitive annotation of transcript ends is also critical for accurate mapping of reads generated by next-generation sequencing (NGS) to reference transcriptomes. High-resolution tiling microarrays and NGS methods are increasingly used for transcript analysis, but even for a well-studied model organism like Saccharomyces cerevisiae, currently available and commonly used transcript annotations remain inaccurate and potentially obscure relationships between the aforementioned aspects of gene regulation.
Saccharomyces cerevisiae has many qualities that make it an ideal model organism for studying gene expression and chromatin architecture, including a relatively small number of genes and a compact genome. The first high-throughput TSS identification in yeast was based on Sanger sequencing of 5′-end tags from cDNAs, and mapped 2231 TSS with single-nucleotide resolution (3). A subsequent study used a ‘vector-capping’ approach with Sanger sequencing to identify TSS, but coverage was limited to only ∼60% of all genes (4). More importantly, although the Sanger sequencing provided single-nucleotide resolution, the number of sequence tags counting towards a given TSS was low. This inherently low sampling of ends with Sanger sequencing makes it difficult to assign one prominent TSS for a gene with high confidence, especially for genes with low transcript levels.
Subsequent approaches used tiling oligonucleotide microarrays to study the yeast transcriptome at high resolution and defined TSS of mRNAs and non-coding RNAs (ncRNAs) (5,6). However, in these studies, which are the highest resolution microarray analyses of transcripts carried out to date in any organism, the resolution was limited to 8 nucleotides (nt), the distance between adjacent probes interrogating transcripts from each strand of genomic DNA. This 8-nt resolution is apparent for both TSS and PAS, in a comparison of independently published datasets using the same microarray platform (Supplementary Figure S1). Although these TSS and PAS have been used in many recent landmark analyses of TF and nucleosome localization datasets (7,8), the 8-nt resolution remains a limitation. RNA-seq can potentially identify TSS and PAS at single-nucleotide resolution (9). However, RNA-seq signals are complex and do not necessarily show an easily identifiable boundary corresponding to transcript ends. In addition, this strategy will tend to identify the most distal 5′- or 3′-ends, which may not be the site most frequently used in vivo.
In order to overcome these limitations, refinements of NGS-based methods have been developed to map TSS and PAS. One approach involves the use of tobacco acid pyrophosphatase (TAP) to remove the 5′ cap and allow ligation of a sequencing adapter specifically to the 5′-end of the RNA (10–12). To map PAS, methods based on oligo(dT) priming or poly(A) capture have been used (13–19). Existing methods work well to map TSS or PAS but only identify one end of transcripts. The recently introduced TIF-seq method can be utilized to simultaneously map TSS and PAS (20), but this study focused on the diversity of transcript isoforms in yeast and did not define canonical TSS and PAS. Thus, none of these methods has been employed to identify a definitive set of TSS and PAS in yeast, which has the most extensive complementary data on the location of the general transcription machinery (7,21) and nucleosome positions (8).
Here, we describe SMORE-seq (simultaneous mapping of RNA ends with sequencing), a method for identifying both mRNA TSS and PAS from a common set of experimental data with single-nucleotide resolution. We demonstrate that SMORE-seq maps TSS and PAS more accurately and efficiently than existing methods. The improved annotations of transcript ends revealed a significant fraction of likely mis-annotated protein-coding sequences in the genome, and showed sharper relationships between cis-regulatory elements, chromatin features and transcript ends. SMORE-seq also revealed pervasive bidirectional transcription from most promoters, and our analysis suggests that the TATA element serves to constrain the direction of transcription initiation by RNA polymerase.
MATERIALS AND METHODS
Yeast growth and RNA preparation
The S. cerevisiae strain used in this study was BY4741, and cells were grown in yeast extract-peptone-dextrose (YPD, Difco) at 30°C to an A600 OD of 0.8. We harvested the cells by centrifugation at 3000 rcf for 5 min, and the cell pellets were frozen in liquid nitrogen after discarding supernatant. Total RNA was extracted with a standard hot phenol method (22).
Construction of SMORE-seq libraries
Poly(A)+ RNA was purified from yeast total RNA using the MicroPoly(A) Purist kit from Life Technologies. 500 ng poly(A) RNA was mixed with 5 units (1 μl) TAP (Epicentre) and 2.5 µl 10 x TAP buffer in a 25 -μl total volume. A parallel reaction without TAP enzyme was also performed. TAP reactions were carried out at 37°C for 1 h, followed by heat inactivation at 65°C for 5 min. RNA was purified with the RNEasy MinElute kit (Qiagen) and eluted in 26 μl of water. 23.5 μl of this RNA was combined with 1 μl of a 1/2 dilution of 5′ SR Adaptor, 3 μl 10 x Ligation Reaction Buffer and 2.5 ul 5′ Ligase Enzyme Mix (for descriptions of these components see NEBNext Small RNA Library Prep Set for Illumina). This reaction was incubated one hour at 25°C, followed by purification with Agencourt AmPure XP beads (Beckman Coulter) following manufacturer’s instructions at a 1.5× concentration and elution in 18 μl water. This RNA was then fragmented for 4 min at 94°C using NEB fragmentation reagent, followed by cleanup with AmPure XP (1.8×) and elution in 10 μl of water. This RNA was then ligated to a 3′-sequencing adapter as described in the manufacturer’s protocol (NEBNext Small RNA Library Prep Set for Illumina), followed by reverse transcription and 10 cycles of PCR according to manufacturer’s instructions. PCR products of ∼250 bp were selected by E-gel (Invitrogen) and subjected to another eight cycles of PCR. The resulting libraries were verified on an Agilent Bioanalyzer and sequenced on an Illumina HiSeq 2000 with single-end or paired-end 100 base reads.
Analysis of sequencing reads
Alignment of sequencing reads was performed with bwa (version 0.6.2) using default options for paired end or single end libraries, as appropriate (23). The reference genome was sacCer3 (April 2011) from UCSC, derived from the Saccharomyces Genome Database. The 100-bp read sequences were trimmed to 50 bp before alignment. Aligned R1 (5′ reads) were extracted from the resulting BAM files using samtools (version 0.1.18) (24) and merged for the three TAP+ and TAP– replicates, respectively. Reads that mapped to snRNA and rRNA were removed. Plus (Watson) and minus (Crick) strand aligned reads were then extracted and processed separately for TSS calling.
TSS calling algorithm
According to previous studies that mapped TSS in yeast, the estimated median 5′-UTR length is 50–60 bp, and ∼90% of 5′-UTRs are <300 bp (3,5,9,25). For each verified and uncharacterized gene, we searched for TSS within a window ranging from 300-bp upstream of the annotated ORF to the midpoint of the ORF downstream from its annotated start codon. In order to correct TAP+ by TAP–, Gaussian-kernel-density estimation was utilized for peak calling, and a bandwidth of 5 and a read threshold of 2 were applied. When TAP+ peaks were present within ±50 bp of TAP– peaks, the peaks were corrected. Then, the base position with the highest read stack within the highest corrected peak was called as the TSS. Manual curation was mainly aimed at calling TSS for the genes with a 5′-UTR >300 bp. In addition to recovering TSS with long 5′-UTRs, potential TSS that showed the following examples were dropped during manual curation: evenly distributed peaks with a low number of reads, TSS adjacent to tRNA, snRNA or rRNA, TSS overlapping with a neighboring gene, and TSS in close proximity to a neighboring gene.
TATA element data processing
The ChIP-exo technique previously identified the TATA box as well as TATA-like elements at ‘TATA-less’ promoters (7). In this study, a canonical TATA represents a TATA-box with no mismatches, and TATA-elements include canonical TATA with 0,1 or 2 mismatches. TATA element data for sacCer3 were downloaded from the SGD Genome Browser (http://browse.yeastgenome.org/fgb2/gbrowse/scgenome/).
High resolution tiling array data processing
Although these data are available in SGD, the data were downloaded from the journals where the papers were originally published because the authors assigned gene names but SGD provided only segment information (5,6,26). The data were lifted over into sacCer3 from the genome version the authors originally used.
RNAPII Ser 5-P and nucleosome localization
Of wild-type (WT) cells, 150 ml were grown in YPD, and harvested at 0.8 A600 OD for each sample. Cells were cross-linked with formaldehyde to a final concentration of 1% for 15 min, then quenched with glycine to a final concentration of 125 mM. For ChIP, cells were resuspended in 2 ml of lysis buffer, and were lysed by glass bead beating for 9 min. Chromatin was sheared with a probe-sonicator to 150–200-bp fragments. After pre-clearing with protein A-agarose beads (Roche), the fragmented chromatin was incubated with 8 μg of RNAPII Ser 5-P specific antibody (Abcam, cat.# ab5131) overnight, then further incubated with 100 μl protein A beads. Serial washing was performed, and finally DNA was reverse-crosslinked at 65°C overnight, then collected by ethanol precipitation. For mononucleosome isolation, we followed the protocol described in (27). Briefly, cells were resuspended 20 ml of zymolyase buffer, and spheroplasts were made with 250 μg of zymolyase. The spheroplasts were spun down and resuspended in 2 ml NP buffer. Then, micrococcal nuclease (MNase) was added at a concentration from 40 U-100 U for 10 min at 37°C. Digested chromatin was reverse-crosslinked with Proteinase K in 1% SDS and 10 mM EDTA solution at 65°C overnight. After RNase A treatment, DNA was purified by phenol chloroform extraction followed by ethanol precipitation. Finally, DNA fragments of ∼147 bp were size-selected with an E-gel system (Invitrogen). Sequencing libraries for both ChIP and mono-nucleosomes were prepared using NEB Library Prep Kit and Bioo multiplex adapter for Illumina, and then sequenced by paired-end sequencing. In order to profile occupancy, coordinates of mapped reads were shifted toward the center of the insert DNA by a distance equal to half of the insert size, then reads were counted in bins of 5 bp.
Conservation and ribosome footprinting analysis
WIG files of conservation scores were downloaded from the UCSC Genome browser (http://hgdownload.cse.ucsc.edu/goldenPath/sacCer3/phastCons7way/). Using a customized python script, we extracted base-by-base conservation scores near annotated start codons of all genes and internal TSS genes.
Raw sequencing data of ribosome footprinting in rich media were downloaded from Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) (accession number GSE13750) (28). Only the first 21 nt were mapped onto sacCer3 with bwa using default options, and for any given gene, only reads that mapped to the sense strand were considered.
PAS analysis
The sequenced read fastq files from both TAP+ and TAP– (three replicates each) were first processed to remove 3′ adapter sequences with cutadapt version 1.2.1 (29). Each resulting sequence set was filtered, retaining only the R1 sequences with at least 35 bases followed by a stretch of at least 8 A bases within 5 bp of the adapter-trimmed 3′-end. For each resulting poly(A) selected sequence set, a corresponding trimmed version was created such that only bases 5′ of the poly(A) stretch were retained. The poly(A) selected full length and poly(A) selected trimmed fastq files were then single-end aligned to sacCer3 with bwa as described above (Supplementary Table S1).
PAS data in sacCer3 were downloaded from the SGD Genome Browser (14). Since Ozsolak et al. (14) provided the genomic coordinates of the read clusters and the scores of the clusters as the read counts that support the highest peak, we processed the data to call one poly(A) site per gene. In order to process the data in the same way as the SMORE-seq, we assigned the clusters into ranges from annotated stop codons to 300-bp downstream. Among the clusters per gene, the position that had the highest read count was defined as the poly(A) site for the gene.
RESULTS
SMORE-seq identifies 5′ cap sites of mRNAs with single-nucleotide resolution
We constructed SMORE-seq libraries according to the flowchart shown in Figure 1A. Two technical replicate libraries and one biological replicate library were prepared, with a control library that was not treated with the TAP enzyme prepared in parallel for each sample. In total, we produced 12 652 059 and 11 161 171 single-end, 100-base reads for the TAP+ and TAP– samples, respectively, after filtering reads mapping to snRNA and rRNA regions. In the TAP+ libraries, 7 622 443 (60.2%) reads were mapped within 300-bp region upstream of ORF start codons, whereas only 890 128 (8.0%) reads were mapped to those regions in the TAP– libraries. Most reads mapped to or near genes in both TAP+ and TAP–, and the strongest read signals were observed just upstream of annotated start codons in TAP+, whereas relatively few reads in TAP– mapped to these locations (Figure 1B). This difference in the read pattern in the TAP+ and TAP– libraries suggests that our TAP+ library was selective for the TSS.
Figure 1.
(A) Overview of the SMORE-seq method. TAP enzyme is used to convert mRNA 5′ caps into phosphates, followed by 5′ adapter ligation, fragmentation, 3′ adapter ligation, RT-PCR, size selection and additional PCR. (B) Heat map representation of SMORE-seq read data. Genes are sorted by ORF length. The arrow represents the positions of start codons in SGD annotation, and genes are aligned by the start codon. Color scale is read count per 10 bp. (C) Comparison of SMORE-seq to standard RNA-seq and ChIP-seq data, visualized in the UCSC Genome Browser mirror. TAP+ has similar background signal as TAP–, including appreciable signal at the 3′-end, indicating that correction by TAP– is necessary for TSS identification. The peak shape of TF-binding sites in ChIP-seq is different from that of TSS peaks in TAP+.
To identify candidate TSS, we employed a modified version of our peak-finding algorithm based on Gaussian-kernel-density estimation, followed by correction of the TAP+ data by the TAP– control. Although TAP+ reads were highly enriched in 5′-UTRs, there was appreciable background signal within ORFs and 3′-UTRs, necessitating its correction by TAP– in order to reduce false positives. We adapted the parameters of our peak finding algorithm to exploit the characteristics of the SMORE-seq data, which was distinct from standard RNA-seq and ChIP-seq data in terms of its strand-specificity, localization relative to ORFs and sharpness (Figure 1C). This procedure resulted in the identification of 138 352 candidate TSS that were defined by two or more reads. To identify the most prominent TSS for a gene, we assigned the corrected peaks at 5′-ends to genes, then determined the position of the most abundant read stack within the most significant peak for each gene. By doing so, we obtained the most frequently used TSS for a gene rather than the most upstream TSS identified in previous studies (5,6). We applied this procedure independently to the three replicates, and ascertained that the identification of TSS with single-base resolution was highly reproducible across replicates (Supplementary Figure S2A).
The major cause of non-identical TSS calls between replicates was low read coverage in genes with low expression (Supplementary Figure S2B); therefore to increase coverage, we combined all replicates, then identified TSS again as described above using the combined datasets. These computationally defined TSS were further manually curated by visual inspection of the raw-read data in our UCSC genome browser mirror. For a small fraction of cases (289/5207 or 5.6%), our computational procedure had missed the TSS that was evident by visual inspection of the data; these were therefore manually corrected (Supplementary Figure S3). This rate of manual correction is significantly lower than in previous studies (6), and could potentially be reduced further by incorporating steps in our algorithms tailored to address the main reasons for erroneous assignment that we observed during curation. Based on our TSS annotations, we determined that the median and mean 5′-UTR lengths in yeast are 52 and 84 nt, respectively (n = 5203, Supplementary Figure S4). Thus, SMORE-seq provides a systematic framework to reproducibly identify TSS with single-nucleotide resolution in a largely automated manner. These and all subsequent analyses in this study are based on the primary TSS that we identified for each gene. However, we also used our data to determine the extent of utilization of additional TSS for a given gene. As expected, secondary TSS tend to be used less than the primary TSS, but their distribution of relative utilization varied over a broad range, indicating that for some genes, the secondary TSS are used at rates comparable to the primary one (Supplementary Figure S5).
SMORE-seq TSS show sharper relationships with other transcriptional features
Currently, the most complete and widely utilized yeast TSS annotations are based on the study of Xu et al. (6,30–32), because the data are strand specific, manually curated, replicated several times, generated under various perturbation conditions and cover almost all genes. We therefore assessed the accuracy of the TSS from SMORE-seq (S-TSS) by comparison to the TSS from Xu et al. (X-TSS). The S-TSS and X-TSS were generally in agreement, with 80% of the S-TSS located within 40 bp of the X-TSS (Figure 2A, B and C). Globally, 5′-UTRs from S-TSS were shorter than X-TSS by a median of 11 nt. Our algorithm was designed to pick the nucleotide position with the strongest read signal as the TSS whereas Xu et al. picked the upstream coordinate of the 8-nt tile containing the most upstream signal for a given gene as its TSS. Because of this systematic difference, the finding that our S-TSS were closer to the start codon with a median difference of 11 nt is likely due to the improved accuracy of our TSS calls. However, to independently verify the accuracy of S-TSS, we evaluated both sets of TSS calls with regard to TATA element positions, consensus sequences at TSS, nucleosome positions near the TSS and localization of active RNAPII phosphorylated at Serine 5.
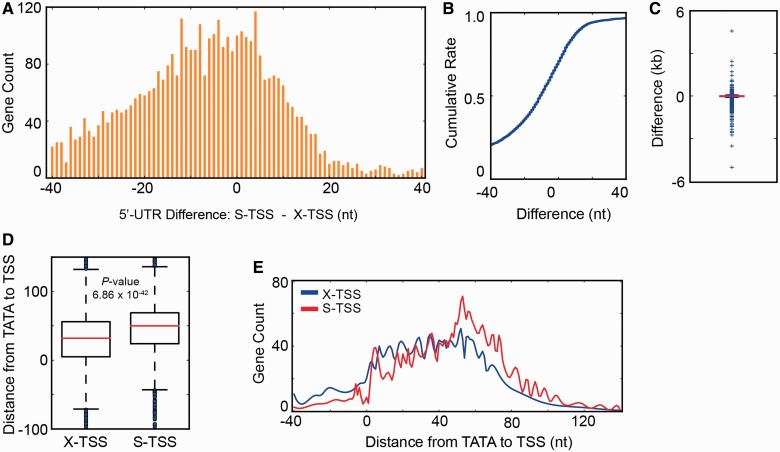
Figure 2.
(A–C) Comparison of SMORE-seq TSS coordinates (S-TSS) with the commonly referenced TSS coordinates reported by Xu et al. (X-TSS) (6) by histogram (A), cumulative distribution (B) and box plot (C), demonstrating that S-TSS and X-TSS are in high agreement. Overall, 5′-UTRs of S-TSS are shorter (S-TSS are more downstream). (D and E) Distance between TATA-like elements in TATA-less genes (n = 4065) (7) and S-TSS or X-TSS. S-TSS shows a narrower distribution with a larger average distance.
Interaction between TATA-boxes or TATA-like elements in promoters and general TFs serves to recruit RNAPII and initiate transcription some distance away (33). Distances between the TSS and canonical TATA boxes are believed to be distributed in a narrow range of 45–125 bp for most yeast genes (3,34). Compared to X-TSS, S-TSS showed a narrower distance distribution from canonical TATA boxes (n = 716) (Supplementary Figure S6). A similar pattern was also observed for the distance between TATA-like elements and the TSS in TATA-less genes (n = 4065) (Figures 2D and E). Together, these results suggest that initiation of transcription within a narrow distance window from TATA elements generates the sharper distribution of distances between TATA elements and S-TSS, supporting the higher accuracy of SMORE-seq. A consensus sequence of PyA has previously been identified at TSS in yeast (3,34). This sequence was readily identifiable in TSS identified by SMORE-seq, but could not be identified using X-TSS coordinates or a typical RNA-seq data set (Figure 3A).
Figure 3.
(A) Consensus sequence around TSS identified by SMORE-seq, Xu et al., and RNA-seq data (6,9), visualized by WebLogo (35). The consensus motif identified in S-TSS matches what has been previously described (3). (B) Nucleosome occupancy profiles relative to the TSS in each of four groups of 1000 yeast genes, arranged by the distance between S-TSS and X-TSS in descending order. Nucleosome positions relative to X-TSS (blue line) differ between the groups whereas their positions relative to S-TSS (red line) are constant. Nucleosomes also show the expected periodicity in group 1 (top) relative to S-TSS but not X-TSS. (C) Localization of RNAPII Ser 5-P in the same groups as in (B).
A core promoter in yeast is situated within a nucleosome-depleted region (NDR) and is followed by a well-aligned array of nucleosomes starting from the TSS (36). This property of nucleosome organization allowed us to test the accuracy of TSS calls by examining their relationship to nucleosome profiles. We generated nucleosome occupancy maps using MNase-seq and plotted their profiles for each of four groups of 1000 genes formed in descending order of absolute difference between S-TSS and X-TSS coordinates. Interestingly, the centers of the nucleosomes relative to the TSS did not change between these gene groups when using S-TSS coordinates. In contrast, when using X-TSS coordinates, nucleosomes appeared to be shifted downstream with decreasing rank of the gene groups. Because the rank of the groups had no prior relationship to nucleosome positions, the S-TSS coordinates, which yielded a similar nucleosome occupancy pattern across all four groups are likely to be more accurate. Moreover, the nucleosome profile in group 1 was flatter and poorly defined relative to X-TSS, whereas the S-TSS coordinates showed a more characteristic NDR and nucleosome periodicity. Thus, the inaccuracy of X-TSS leads to lower definition and offset of nucleosome occupancy profiles for a subset of genes.
Phosphorylation of RNAPII at Serine 5 (Ser 5-P) is a marker of transcription initiation and early elongation (37). RNAPII Ser 5-P occupancy is therefore expected to start at the TSS and increase toward mid-ORF. We used ChIP-seq to measure the localization of RNAPII Ser 5-P relative to S-TSS and X-TSS, in the same four groups of genes as for the nucleosome analysis above. RNAPII Ser 5-P occupancy increased from the TSS to 200-bp downstream in all four groups, but as with the nucleosome profiles, its pattern of occupancy relative to S-TSS was more invariant than the occupancy relative to X-TSS (Figure 3C). Thus, S-TSS shows a more consistent relationship with a genome-wide mark of transcription initiation. Taken together, these analyses show that the genome-wide TSS identified by SMORE-seq are not merely more downstream than TSS identified by other global methods, but show more clear-cut relationships to biological features of transcription initiation and are therefore likely to be more accurate.
SMORE-seq identifies mis-annotated start codons
We identified 222 genes with TSS downstream of their annotated ATG start codons: we refer to these as internal TSS. We defined the putative start codon of these genes as the first ATG downstream of the TSS. Of the 222 genes, 127 had a putative start codon in frame with the annotated ORF, 91 had a putative start codon out of frame with the annotated ORF and four had no start codon between the TSS and the annotated stop codon. We reexamined these 95 genes with an out of frame or no start codon and flagged 72 genes because they either had a secondary, well-represented upstream TSS that agreed with the SGD start codon, an apparently incorrect TSS call, or low, ambiguous signal that prevented a confident TSS call. The 23 genes that were not flagged were grouped with the 127 that contained an in frame start codon, and the 123 verified genes out of this combined group of 150 were used for further analysis.
Although previous studies have reported internal TSS and confirmed several by quantitative PCR (qPCR) and primer extension assays (3,5,9), the veracity of such internal TSS, which would be indicative of potentially mis-annotated protein-coding regions, has not been systematically evaluated. We evaluated whether these internal TSS were indeed the bonafide TSS by examining the propensity of such genes to have an alternative start codon downstream of the annotated start codon, evolutionary conservation, ribosome footprinting profiles, and presence of a preferred Kozak consensus sequence for translation initiation.
We observed that internal TSS genes tended to have an in-frame methionine codon just downstream of the internal TSS (Figure 4A). For most genes, the likelihood of having an internal methionine downstream of the annotated start codon is expected to increase monotonically with increasing distance from the start codon. Indeed, all verified genes showed this expected pattern (Figure 4B). However, internal TSS genes showed a markedly steeper increase, indicative of a higher likelihood of having another methionine shortly downstream of the annotated start codon. This distinctive behavior suggests that the internal TSS of this subset of genes could be the true TSS, with translation initiating from an internal methionine to generate a polypeptide that is truncated at the N-terminus relative to the currently annotated protein-coding sequence.
Figure 4.
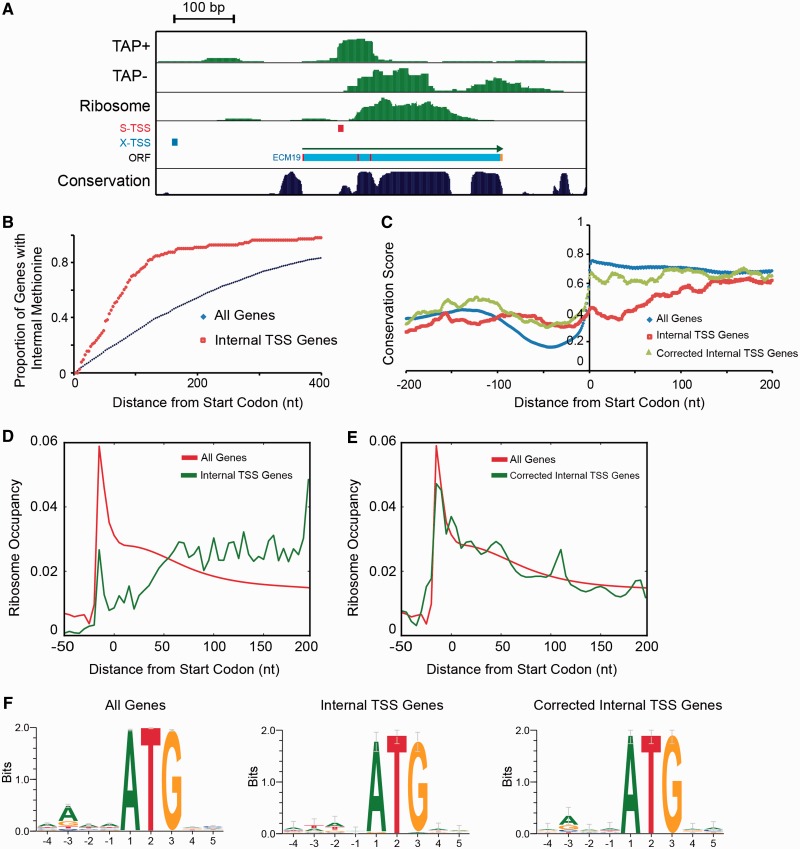
(A) Example of an internal TSS downstream of the annotated start codon. Ribosome footprinting and conservation score are visualized in the UCSC Genome Browser mirror (28,38). In-frame methionine codons are indicated in red within the ORF track. (B) Cumulative proportion of genes that have an in-frame methionine at the indicated distance from the SGD annotated start codon, for each of the indicated groups. (C) Seven-species yeast conservation near start codons of all verified genes according to SGD annotations (all genes), internal TSS genes according to SGD (internal TSS genes) and internal TSS genes with start codon predicted based on SMORE-seq (corrected internal TSS genes). (D and E) Ribosome profiles near start codons as predicted in (C). Ribosome-profiling data was taken from (28) and plotted as the average proportion of reads. (F) Consensus sequence upstream of the start codon of the indicated gene sets, where the start codon used was as described in (C).
Protein-coding regions of yeast genes show significantly higher evolutionary conservation than non-coding regions (38–40). To determine if this conservation could shed light on the potential usage of internal TSS, we analyzed conservation around the start codon between seven yeast species. The set of all genes showed a sharp increase in conservation downstream of the start codon. This increase in conservation was not seen in the internal TSS genes when using the SGD start codon (Figure 4C). However, if we used the first methionine downstream of our internal S-TSS as the start codon, conservation just downstream of the start was restored for this set of genes. This data strongly suggests that the internal methionine downstream of the internal S-TSS is the true start of the protein-coding region for these genes, rather than the currently annotated start codon.
Next, we analyzed published genome-wide ribosome footprinting data to obtain experimental evidence regarding translation at either annotated or internal start codons (28). Ribosome footprinting measures occupancy of ribosomes along mRNAs, and has shown that there is high ribosome occupancy 12–13-nt upstream of start codons (28). We analyzed ribosome footprints from the previously published study in the three groups of genes described above. The set of all genes showed a strong ribosome occupancy peak 12–13-nt upstream of the start codon. This peak was largely absent near the SGD-annotated start codons of internal TSS genes (Figure 4D), but was clearly restored when we used start codons downstream of the internal TSS predicted by SMORE-seq (Figure 4E). This analysis provides strong evidence of the accuracy of SMORE-seq TSS coordinates and start codon predictions for internal TSS genes.
Consensus sequence analysis of the regions near annotated start codons for all genes showed strong enrichment of A residues at the –3 position relative to the ATG start codon, which is a characteristic of the Kozak consensus sequence in yeast (41,42) (Figure 4F). In contrast, enrichment of A at the –3 position was not observed for internal TSS genes, indicating that the annotated start codons are unlikely to be used for translation initiation. Strikingly, the Kozak consensus sequence was restored at the corrected, internal start codon for the internal TSS genes. Thus, start codons predicted by SMORE-seq for internal TSS genes have a more appropriate sequence context for initiation of translation than the current SGD annotations.
SMORE-seq identifies PASs
Visual inspection of SMORE-seq data indicated a large number of reads mapped to 3′-regions of mRNAs, near ORF stop codons (Figure 1B and 5B). Because of the 3′ bias of these reads and their abundance in both TAP+ and TAP– samples, we hypothesized that these reads originated from mRNA degradation products. One of the main mRNA-degradation pathways in eukaryotes starts with shortening of poly(A) tails to ∼10–15 A bases, followed by decapping and 5′–3′ exonuclease-mediated degradation (43). These degradation products have a 5′ phosphate that is amenable to ligation during the SMORE-seq protocol, and thus would be represented in both TAP+ and TAP– samples. Because some of these reads would also be expected to contain the PAS, the site where the mRNA is cleaved and an untemplated stretch of A residues is added, we reasoned that these reads could be used to map PAS.
Figure 5.
(A) Strategy used to extract PAS-containing reads, those with a 3′ stretch of untemplated As, from SMORE-seq data. Reads that ended in a stretch of A residues were selected, and those that mapped to the genome only after removal of the 3′ poly(A) stretch were retained as PAS reads. (B and C) SMORE-seq reads near ORF stop codons (vertical line) before and after applying filtering described in (A). PAS reads mostly mapped just downstream of stop codons. (D) PAS reads in all genes sorted by ORF length and aligned by start codon (arrow), demonstrating that few PAS reads mapped within ORFs. (E) Occurrence of the polyadenylation efficiency element (EE) and positioning element (PE), elements utilized for PAS selection, relative to PAS identified by SMORE-seq. (F) Difference between SMORE-seq PAS and those identified by Ozsolak et al. (14) using Helicos NGS-based method. The inset shows the cumulative difference profile.
We used a simple but effective workflow to obtain reads representing potential PAS in our data (Figure 5A). We first selected all reads ending in a string of As (see Materials and methods section). We then mapped these reads to the yeast genome and sorted the results into unmapped or mapped groups, with the expectation that reads with an untemplated stretch of As, representing a potential PAS, would be unmapped, whereas those reads that mapped represented a genomic poly(A) stretch and should be discarded. We then trimmed the poly(A) stretch off the unmapped reads and mapped these trimmed reads again, with the expectation that the reads that mapped after trimming represented PAS. This set of reads, which we called PAS reads, mapped almost exclusively to likely 3′-UTR regions of mRNAs (Figure 5B, C and D), indicating that our strategy was effective in identifying PAS. This procedure yielded a total of 55 419 candidate PAS where each PAS was defined by at least two reads. In order to identify a dominant PAS for each gene, we determined the base position with the highest read stack in the PAS reads in the range from the gene’s stop codon to 300-bases downstream. We were able to identify a PAS for 5277 (91%) yeast genes using this strategy. Based on SMORE-seq PAS annotations, the median and mean 3′-UTR lengths in yeast are 120 and 137 nt, respectively (n = 5277, Supplementary Figure S4).
Sequence elements that contribute to PAS selection have been discovered in yeast, and although these elements are less conserved and less well-defined than in higher eukaryotes, a PE with sequence AAWAAA and an EE with sequence TAYRTA have been identified ∼10–30-nt and 25–75-nt upstream of PAS, respectively (44). A search for these elements in the sequences surrounding PAS as determined by SMORE-seq revealed enrichment of these sequences with expected positioning relative to PAS, indicating that SMORE-seq was successful in determining correct PAS (Figure 5E).
PASs have been previously measured in yeast with a specialized deep-sequencing based strategy (14). To further verify the accuracy of SMORE-seq PAS, we compared our results to this study. In order to define PAS with single-nucleotide resolution from the published data, which reported PAS regions rather than a single base position, we downloaded their data and found the position with the highest read stack as described above (see Materials and methods section). We could identify a PAS for 5314 genes in the published data, and of these genes, 5119 also had a PAS identified by SMORE-seq. There was striking agreement between PAS identified by the two methods, with almost 80% of PAS within 30 bases and almost 800 genes showing an identical PAS between samples (Figure 5F). Thus, SMORE-seq can accurately map both TSS and PAS from the same sequencing dataset with single-nucleotide resolution. Similar to TSS, many genes also showed alternative PAS, which were used at rates lower than the primary PAS (Supplementary Figure S5).
SMORE-seq reveals widespread bidirectional transcription initiation from yeast promoters
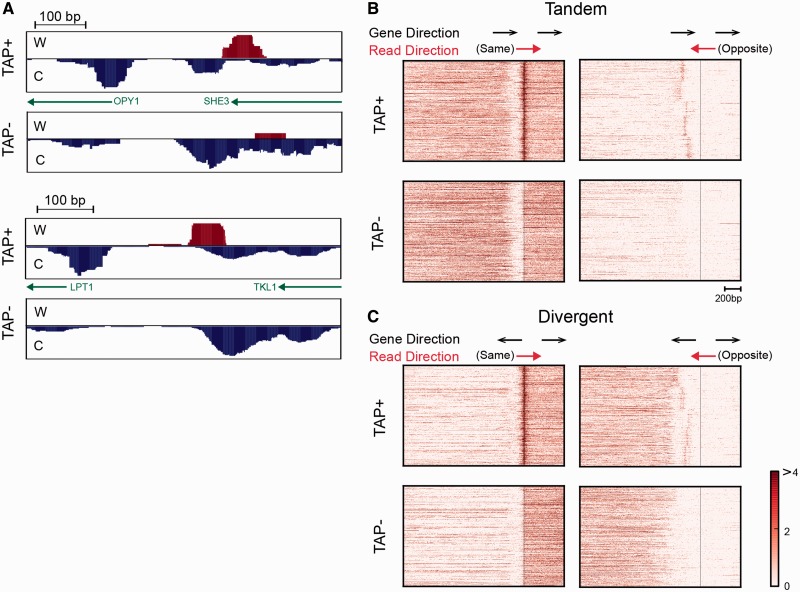
We observed more than a thousand regions where reads aligning in the opposite direction of the coding strand were concentrated in a region 50–300-bp upstream of the S-TSS, indicating ncRNA transcripts resulting from bidirectional promoters. Previous studies have reported ncRNAs at bidirectional promoters only in strains deleted for genes associated with gene looping or the nuclear exosome (6,45), as the directionality of transcription was thought to be tightly regulated and antisense ncRNAs rapidly degraded in WT strains. For example, the promoter-associated ncRNA at the bidirectional promoter between OPY1 and SHE3 was previously identified only in an ssu72-2 mutant and therefore interpreted as arising due to disruption of a gene loop (45). However, this RNA was readily identifiable by SMORE-seq in a WT strain, likely due to the higher sensitivity of our method (Figure 6A). SMORE-seq identified more than a thousand new bidirectional promoter-associated ncRNAs (Figure 6A). Here, we refer to the antisense ncRNAs detected by SMORE-seq at promoters as bncRNAs (bidirectional ncRNAs).
Figure 6.
(A) A previously known ssu72-restricted transcript (SRT) in the promoter of OPY1 is detected by SMORE-seq in WT cells under normal growth conditions (45) (top two panels). A novel antisense ncRNA that may share a bidirectional promoter with LPT1 is shown below. (B and C) Widespread occurrence of bncRNAs (antisense ncRNAs at bidirectional promoters). Genes were clustered by K-means clustering (K = 5, repeat = 1000) of bncRNA signal in a range 300 to 50 bp upstream of TSS. Genes in the indicated tandem arrangement are shown in (B), and (C), in the divergent arrangement. Divergent genes whose TSS are closer than 300 bp are excluded in (C). The vertical line represents the TSS of downstream genes. The number of tandem genes and divergent genes in this heat map are 2401 and 1635, respectively.
In order to visualize the prevalence of bncRNAs in WT cells under normal growth conditions, we separately plotted the SMORE-seq reads aligning to each strand near promoters, split according to the orientation of two adjacent genes. The terms ‘same’ and ‘opposite’ for read directionality are defined with respect to the downstream gene, and ‘tandem’ and ‘divergent’ define the orientation of the upstream gene (Figure 6B and C). Interestingly, opposite reads showed a strong signal 50–300-bp upstream from the S-TSS of the downstream genes (Figure 6B). In particular, the widespread signal from opposite reads in tandem genes, where this signal is unequivocally independent from the TSS of the upstream gene, shows that products of bidirectional transcription are much more pervasive than previously appreciated in WT yeast cells.
A canonical TATA-box element suppresses bidirectional transcription
Previous studies reporting the expression of promoter-associated ncRNAs in mutants defective in RNA processing have noted that highly expressed genes show higher levels of the promoter-associated ncRNAs (45). In order to assess the correlation between the bncRNAs identified by SMORE-seq and downstream gene expression, we generated heat maps showing bncRNAs with their downstream genes sorted by mRNA abundance (46) (Supplementary Figure S7). The intensity of the bncRNA signal did not appear to correlate with expression of the downstream gene. The correlation coefficient between levels of bncRNAs and downstream gene expression was close to zero (Spearman rank r = –0.02), indicating that expression of the downstream gene is unrelated to bncRNA levels.
Mutation of the TATA-box in the TPI1 promoter has been reported to increase antisense transcription from its bidirectional promoter (26). We hypothesized that the presence of a TATA-box in promoters correlates genome-wide with levels of bncRNAs. To test this hypothesis, we separated tandem genes based on whether the downstream gene contained a canonical TATA-box, then plotted reads arising from the opposite strand in a heat map (Figure 7A). The signal from bncRNAs in TATA-box containing genes was significantly lower compared to TATA-less genes (Figure 7B, P = 3.67 × 10−7 by Welch’s t-test). Moreover, the proportion of TATA-containing genes was lower for genes with higher levels of bncRNA transcription (Supplementry Figure S8). Thus, promoters lacking a canonical TATA box, or TATA-less promoters, have a higher chance of giving rise to a bidirectional ncRNA in the opposite direction. Additional evidence in favor of the TATA-box model for bncRNA transcription comes from nucleosome localization data. A well-positioned + 1 nucleosome is believed to help form the pre-initiation complex and recruit RNAPII at TATA-less promoters (7,47). If bncRNA transcription used the same mechanism as normal initiation, the –1 nucleosome with respect to sense genes could act as the +1 nucleosome with respect to bncRNA, and similarly facilitate bncRNA transcription. Supporting this hypothesis, TATA-less genes, which have high bncRNA expression, have well-positioned –1 nucleosomes (Supplementary Figure S9A), and the highly expressed bncRNAs have a more well-defined +1 nucleosome (Supplementary Figure S9B).
Figure 7.
(A) Opposite reads in tandem genes grouped by presence or absence of a canonical TATA-box in the gene’s promoter. TATA-less tandem genes (n = 2031) show stronger bncRNA signal than TATA-box-containing tandem genes (n = 370). (B) Average proportion of reads in this window, demonstrating that TATA-less genes have higher bncRNA expression. The P-value for the difference in bncRNA signals between TATA (blue) and TATA-less (red) genes at –200 was 3.67 × 10–7 by Welch's t-test.
DISCUSSION
We have shown that the high accuracy and sensitivity of mapping transcript 5′- and 3′-ends using SMORE-seq reveals more well-defined relationships of transcript ends with cis-elements and chromatin structure, identifies widespread bidirectional transcriptional initiation and suggests a novel role for a canonical TATA-elements in orienting transcription initiation. The singular advantage of SMORE-seq is that it can identify TSS and PAS using the same deep sequencing data derived from a single RNA-seq library, allowing the investigation of transcription initiation as well as termination/polyadenylation in the same RNA sample. Despite this gain in efficiency, SMORE-seq is also a relatively simple method. Other comparable approaches to map TSS using NGS, are generally more tedious. For example, CAGE (cap analysis of gene expression), which has been adapted for deep sequencing, is a relatively cumbersome procedure that involves biotinylated oligos and contains 18–25 major steps spread over 8–14 days to generate a sequencing library (48). Various NGS-based methods to map PAS have been recently utilized to map PAS (49). Some PAS mapping methods involve the use of specialized primers, and others require deep sequencing technologies that are not commonly available (14,19,50). While these methods map PAS with single-nucleotide resolution, they provide no data that can be used to map TSS. In contrast, SMORE-seq avoids any specialized primers and can be completed by one researcher in one day using standard reagents and deep sequencing kits. The improved efficiency of SMORE-seq will be valuable in situations where there is a limited amount of material available, such as human patient samples or microbial species that are difficult to propagate.
During preparation of this manuscript, a study using another method to simultaneously map TSS and PAS, TIF-seq, was published (20). SMORE-seq and TIF-seq generate complementary data, but there are a few noteworthy differences. TIF-seq simultaneously sequences the TSS and PAS of the same mRNA molecule, whereas SMORE-seq identifies TSS and PAS separately for the same population of mRNAs. The TIF-seq study provided a comprehensive catalog of all transcript ends and isoforms in yeast, but it did not provide a definitive annotation of the most prominent TSS and PAS for each gene, and therefore did not uncover the same biological insights about transcriptional regulation that we were able to with SMORE-seq. Although the two methods use a similar strategy to ligate a 5′ adapter at mRNA cap sites, TIF-seq follows this step with reverse transcription using a modified oligo(dT) primer. This may result in several potential complications: (i) efficiency of reverse transcription will be biased toward shorter RNA molecules, resulting in overrepresentation of shorter mRNAs and under-representation of longer mRNAs in final libraries, (ii) mRNAs with a high degree of secondary structure may not be efficiently reverse transcribed and therefore under-represented, (iii) mis-priming with the modified oligo(dT) primer may result in improper PAS calls and (iv) intact full-length mRNAs are likely to be rare in partially degraded RNA samples, such as those from human patient material. Points 1, 2 and 3 are addressed in SMORE-seq by direct ligation of sequencing adapters to both 5′- and 3′- ends of RNA molecules, whereas point 4 is a weakness of both methods. This weakness can be easily addressed in SMORE-seq by using ribosomal RNA depletion rather than poly(A) selection in the first step, and although the data would be noisier and contain more ncRNA signal, this could largely be addressed through deeper sequencing. Another minor weakness of the TIF-seq method is that 30 total cycles of PCR were necessary compared to just 18 cycles in SMORE-seq, likely due to the additional steps in the TIF-seq protocol. However, TIF-seq provides single-molecule data that SMORE-seq cannot. We compared transcript annotations generated by SMORE-seq with the major TSS and PAS sites identified in the TIF-seq study and found strong concordance between both methods (Supplementary Figure S10). This study also identified a set of genes with TSS downstream of the annotated start codon, similar to what we reported (Figure 4). There is strong and significant overlap of the two sets of genes with internal TSS genes (Supplementary Figure S11). We believe that the existence of these complementary methods will assist researchers by allowing them to choose the one best suited to their research goals and conditions.
It is noteworthy that the dominant TSS of at least 150 genes is downstream of the annotated start codon, resulting in protein sequences that differ from SGD annotations. In 127 of these genes the start codon predicted by SMORE-seq is in frame with the annotated start codon, resulting in truncation of the encoded proteins at the N-terminus, with implications for protein function and construction of N-terminal fusion derivatives in experimental studies. For 22 genes, our predicted start codon is not in frame with the annotated start codon, resulting in either a protein with a completely different sequence or a short ORF that is unlikely to encode a functional protein. Interestingly, the TSS and predicted start codon are very close in many of these genes, which may prevent the ribosome from binding to this ATG and allowing initiation of translation at a downstream ATG that is in frame with the annotated protein. Another possibility is that these loci encode ncRNAs with regulatory, enzymatic or structural function.
The enrichment in SMORE-seq data of reads at the 3′-ends of mRNAs likely results from the sequencing of degradation products created by deadenylation and de-capping dependent 5′ to 3′ degradation. mRNA poly(A) tails are shortened to ∼10–20 A residues by the Ccr4-Caf1 deadenylase complex, followed by decapping by Dcp1-Dcp2 and 5′ to 3′ exonucleolytic degradation by Xrn1 (43). Although reads resulting from such degradation products might be expected to map along the entire length of the mRNAs, we propose two explanations for the observed 3′ enrichment of reads: (i) short poly(A) tails of degradation products do not support hybridization of long mRNA-degradation products to oligo(dT) beads during poly(A) selection, and/or (ii) kinetics of degradation result in accumulation of smaller degradation products. Either of these scenarios would result in the observed abundance of reads representing 3′ regions and PASs of mRNAs. Notably, the presence of these reads in almost all genes indicates that degradation of the vast majority of yeast mRNAs depends at least partially on decapping and 5′ to 3′ decay, although further experimentation will be needed to confirm this hypothesis. It is also noteworthy that other TSS-mapping methods treat RNA with a phosphatase enzyme before TAP (11,12), but we were able to recover degradation intermediates used to map PAS only because we did not use phosphatase pre-treatment.
Previous studies have reported antisense ncRNAs (6,26), but their transcriptional regulatory mechanisms are largely unknown. The observation that bncRNAs were detected in TAP+ samples but not in TAP– (Figure 6B and C) strongly indicates that they are 5′-capped. The presence of these RNAs following poly(A) selection also indicates either that these RNAs had poly(A) tails or that they were recovered via hybridization to sense transcripts during poly(A) selection. A recent study indicates that bidirectionally transcribed, promoter-associated RNAs are indeed polyadenylated in human cells (51), supporting the former possibility. However it is not known whether this is also true in yeast. One study suggested that highly expressed genes also show higher levels of promoter ncRNA transcription, although the evidence for this relationship was modest (45). Another model suggested that a TATA-box in a sense promoter could suppress antisense transcription (26). Since highly transcribed genes in yeast generally contain a canonical TATA-box within their promoter (1), these two models are contradictory. We observed no correlation between bncRNA and sense RNA abundance, but we did observe high expression of bncRNAs in TATA-less promoters of sense genes (Figure 7), supporting the latter model. The low correlation between bncRNA and sense RNA abundance is consistent with previous studies showing that distinct pre-initiation complexes are responsible for sense and antisense transcription, and that antisense transcripts are independently regulated (7,52,53). The relationships that we observed between TATA elements, nucleosomes and bncRNAs support a model where the presence of a TATA-box strongly influences the directionality of transcription. We anticipate that the use of SMORE-seq in conjunction with other genomic assays of chromatin structure in different species and cellular states will shed new light on the genome-wide mechanisms of transcriptional control.
ACCESSION NUMBERS
The SMORE-seq data from this study have been deposited in NCBI GEO under accession number GSE49026. The MNase-seq data for nucleosome mapping is also available from GEO under accession number GSE52355.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health [CA095548 to V.R.I., (in part)]; Cancer Prevention and Research Institute of Texas [RP120194 to V.R.I., (in part)]. Funding for open access charge: NIH [CA095548] and CPRIT [RP120194] grant funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dia Bagchi, Yunyun Ni and Damon Polioudakis for discussions and helpful comments, and Yaelim Lee for providing total RNA for an independent replicate experiment. We also acknowledge the University of Texas Genomic Sequencing and Analysis Facility for sequencing.
REFERENCES
- 1.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 2.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-reviews0004. REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z, Dietrich FS. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res. 2005;33:2838–2851. doi: 10.1093/nar/gki583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miura F, Kawaguchi N, Sese J, Toyoda A, Hattori M, Morishita S, Ito T. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc. Natl Acad. Sci. USA. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc. Natl Acad. Sci. USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olivarius S, Plessy C, Carninci P. High-throughput verification of transcriptional starting sites by Deep-RACE. Biotechniques. 2009;46:130–132. doi: 10.2144/000113066. [DOI] [PubMed] [Google Scholar]
- 11.Yamashita R, Sathira NP, Kanai A, Tanimoto K, Arauchi T, Tanaka Y, Hashimoto S, Sugano S, Nakai K, Suzuki Y. Genome-wide characterization of transcriptional start sites in humans by integrative transcriptome analysis. Genome Res. 2011;21:775–789. doi: 10.1101/gr.110254.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu W, Lee HC, Chaves D, Youngman EM, Pazour GJ, Conte D, Jr, Mello CC. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell. 2012;151:1488–1500. doi: 10.1016/j.cell.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, Xu A. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011;21:741–747. doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kuhn U, Menzies FM, Oude Vrielink JA, Bos AJ, Drost J, Rooijers K, et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149:538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 19.Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat. Methods. 2013;10:133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelechano V, Wei W, Steinmetz LM. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497:127–131. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li,H., Handsaker,B., Wysoker,A., Fennell,T., Ruan,J., Homer,N., Marth,G., Abecasis,G., Durbin,R. and Genome Project Data Processing. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed]
- 25.Lipson D, Raz T, Kieu A, Jones DR, Giladi E, Thayer E, Thompson JF, Letovsky S, Milos P, Causey M. Quantification of the yeast transcriptome by single-molecule sequencing. Nat. Biotech. 2009;27:652–658. doi: 10.1038/nbt.1551. [DOI] [PubMed] [Google Scholar]
- 26.Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 27.Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 2011;17:10–12. [Google Scholar]
- 30.Jiang C, Pugh BF. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009;10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan X, Moqtaderi Z, Jin Y, Zhang Y, Liu XS, Struhl K. Nucleosome depletion at yeast terminators is not intrinsic and can occur by a transcriptional mechanism linked to 3′-end formation. Proc. Natl Acad. Sci. USA. 2010;107:17945–17950. doi: 10.1073/pnas.1012674107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Ma H, Pugh BF. Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res. 2011;21:875–884. doi: 10.1101/gr.117465.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemon B, Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- 34.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iyer VR. Nucleosome positioning: bringing order to the eukaryotic genome. Trends Cell Biol. 2012;22:250–256. doi: 10.1016/j.tcb.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat. Struct. Mol. Biol. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 40.Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, Waterston R, Cohen BA, Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 41.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 42.Gingold H, Pilpel Y. Determinants of translation efficiency and accuracy. Mol. Syst. Biol. 2011;7:481. doi: 10.1038/msb.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen CY, Shyu AB. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA. 2011;2:167–183. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan-Wong SM, Zaugg JB, Camblong J, Xu Z, Zhang DW, Mischo HE, Ansari AZ, Luscombe NM, Steinmetz LM, Proudfoot NJ. Gene loops enhance transcriptional directionality. Science. 2012;338:671–675. doi: 10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Dijk EL, Chen CL, d’Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Ne P, Loeillet S, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 47.Nock A, Ascano JM, Barrero MJ, Malik S. Mediator-regulated transcription through the +1 nucleosome. Mol. Cell. 2012;48:837–848. doi: 10.1016/j.molcel.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Itoh M, Kojima M, Nagao-Sato S, Saijo E, Lassmann T, Kanamori-Katayama M, Kaiho A, Lizio M, Kawaji H, Carninci P, et al. Automated workflow for preparation of cDNA for cap analysis of gene expression on a single molecule sequencer. PLoS ONE. 2012;7:e30809. doi: 10.1371/journal.pone.0030809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian B, Manley JL. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem. Sci. 2013;38:312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moqtaderi Z, Geisberg JV, Jin Y, Fan X, Struhl K. Species-specific factors mediate extensive heterogeneity of mRNA 3′ ends in yeasts. Proc. Natl Acad. Sci. USA. 2013;110:11073–11078. doi: 10.1073/pnas.1309384110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–363. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yassour M, Pfiffner J, Levin JZ, Adiconis X, Gnirke A, Nusbaum C, Thompson DA, Friedman N, Regev A. Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol. 2010;11:R87. doi: 10.1186/gb-2010-11-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murray SC, Serra Barros A, Brown DA, Dudek P, Ayling J, Mellor J. A pre-initiation complex at the 3′-end of genes drives antisense transcription independent of divergent sense transcription. Nucleic Acids Res. 2012;40:2432–2444. doi: 10.1093/nar/gkr1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.