Abstract
Initiation of transcription in human mitochondria involves two factors, TFAM and TFB2M, in addition to the mitochondrial RNA polymerase, POLRMT. We have investigated the organization of the human mitochondrial transcription initiation complex on the light-strand promoter (LSP) through solution X-ray scattering, electron microscopy (EM) and biochemical studies. Our EM results demonstrate a compact organization of the initiation complex, suggesting that protein–protein interactions might help mediate initiation. We demonstrate that, in the absence of DNA, only POLRMT and TFAM form a stable interaction, albeit one with low affinity. This is consistent with the expected transient nature of the interactions necessary for initiation and implies that the promoter DNA acts as a scaffold that enables formation of the full initiation complex. Docking of known crystal structures into our EM maps results in a model for transcriptional initiation that strongly correlates with new and existing biochemical observations. Our results reveal the organization of TFAM, POLRMT and TFB2M around the LSP and represent the first structural characterization of the entire mitochondrial transcriptional initiation complex.
INTRODUCTION
Mitochondria are responsible for many essential cellular functions, including energy production through oxidative phosphorylation, fatty acid oxidation, heme and phospholipid biosynthesis, apoptosis and signal transduction (1–3). Mitochondria harbor a small genome (16.5 kb in mammals), encoding mitochondrial tRNAs, two rRNAs and 13 components of the respiratory chain (4). Despite its small size, expression of the mitochondrial genome is critical for maintaining normal mitochondrial function. Consistently, defects in mitochondrial gene expression are associated with a myriad of genetic mitochondrial disorders, age-related chronic diseases and the aging process itself (5–7).
The first step in the process of gene expression is transcription of the mitochondrial DNA (mtDNA). The central component of the transcription machinery in eukaryotes is the mitochondrial RNA polymerase (POLRMT), believed to have originated from the T-odd phage family as a result of an ancient phage-integration event (8,9). POLRMTs have a conserved 80 kDa C-terminal domain that shares significant structural homology with their T-odd phage counterparts (10). Despite this considerable homology, the mechanism of mitochondrial transcription differs from that in T-odd phages in some important aspects. For example, although viral RNA polymerases can mediate all steps of transcription, POLRMTs are unable to catalyze promoter-dependent initiation without additional cofactors (11). Accordingly, POLRMTs possess some unique structural features not present in T-odd RNA polymerases (10). In yeast and animals, POLRMTs contain a large divergent N-terminal extension (NTE) of unclear function (12). In human POLRMT, this NTE contains two pentatricopeptide repeat domains that are believed to possess nucleic acid binding ability (13,14) and have been proposed to interact with the DNA substrate (10). In yeast, deletion of the NTE leads to a loss of mitochondrial genome integrity (15) suggesting that the NTE might mediate important interaction(s) between POLRMT, the proteins responsible for RNA processing and translation, (16) and the essential transcription factor Mtf1 (17,18). Experiments with yeast POLRMT mutants that were partially or fully devoid of the NTE demonstrated that it is dispensable for processive RNA elongation, but does influence promoter-specific initiation, presumably by modulating the interaction with Mtf1 (15). Moreover, it was recently shown that deletion of the NTE in human POLRMT similarly precludes promoter-dependent initiation (10).
While in yeast only one cofactor, Mtf1, is required for efficient promoter-dependent initiation of mitochondrial transcription, metazoans require two cofactors: TFB2M (homolog of Mtf1) and TFAM. TFB2M belongs to a family of ribosomal RNA (rRNA) methyltransferases (18,19) and in fact its paralog TFB1M is responsible for an essential modification of the mitochondrial 12S rRNA (20). Crosslinking studies have revealed that, at least in humans, TFB2M interacts with the first incoming ribonucleoside triphosphate, and along with POLRMT, appears to participate in formation of the active site where the initial steps of RNA primer synthesis are catalyzed (21). Moreover, it has been reported that POLRMT and TFB2M can be co-purified (22), further implying that an interaction might exist between both proteins.
TFAM is a high mobility group protein that serves two functions in metazoans, playing a role in both transcription initiation and nucleoid organization (23). The crystal structure of TFAM reveals its ability to bend the promoter DNA, implying that this unique property might be important for transcription initiation (24,25). In addition, a C-terminal tail in TFAM has been shown to be essential for promoter-dependent initiation of transcription, suggesting that this tail might mediate an important protein–protein or even protein–DNA interaction (26–28).
Although TFAM is important for initiation in mammalian mitochondria (29), the yeast ortholog of TFAM, Abf2, does not play a role in activation of transcription (18) and is only responsible for nucleoid packaging (30). Interestingly, it has been reported that human POLRMT, like its yeast homolog, retains some ability to catalyze TFAM-independent initiation under certain conditions (31). Moreover, TFAM-independent transcription activity by POLRMT is critical for generating the RNA primers needed for lagging strand synthesis during mtDNA replication (32)
Despite the substantial understanding of the transcription initiation process we have gained to date, certain key questions remain unanswered. For instance, the specific protein–DNA and protein–protein interactions that govern and regulate the initiation process are still not well understood. Furthermore, current models for the organization of the transcription initiation complex are based on indirect observations and the overall structure of the initiation complex remains unknown. Here, we present a study of the protein–protein interactions that take place during the initiation process, combined with a structural characterization of the initiation complex based on single-particle electron microscopy (EM) reconstructions and solution X-ray scattering analysis. Our results comprise, to our knowledge, the first direct structural characterization of the architecture of the mitochondrial transcription initiation complex.
MATERIALS AND METHODS
Protein expression and purification
Human TFAM (UniProt Q00059; residues 43–246), TFB2M (UniProt Q9H5Q4; residues 20–396) and POLRMT (UniProt O00411; residues 42–1230) were expressed as fusions with his-tagged maltose binding protein and purified as described (33). The POLRMT N-terminal extension (NTE) deletion mutant in which the whole N-terminal domain (residues 1–339) has been eliminated (POLRMTΔNTE; Supplementary Figure S1A) and the TFAM C-terminal extension (CTE) deletion mutant (TFAMΔCTE; residues 43–226; Supplementary Figure S1B) were cloned into pTEV-HMBP3 and over-expressed in Arctic Xpress Escherichia coli (DE3) cells (Stratagene) at 16°C for 20 h. TFAM and TFB2M were purified using ProBond Resin (Invitrogen), followed by overnight tobacco etch virus (TEV) protease cleavage, Heparin, Mono S and size-exclusion chromatography. POLRMT was purified by Heparin chromatography, overnight TEV protease cleavage, followed by a second Heparin, Mono S and size-exclusion chromatography. Proteins were concentrated using a 10 000 MWCO Amicon Ultra-15 device. Concentrated proteins were stored in 20 mM HEPES (pH 8.0), 200 mM KCl, 5% glycerol and 1 mM DTT. All proteins used in this study correspond to the mature form and were cloned without the mitochondrial localization sequence, which was experimentally determined for TFAM (34), predicted with MitoProt (35) for POLRMT and predicted with TargetP (36) for TFB2M. The proteins that were used in the reconstruction of the mitochondrial transcription machinery were 95–99% pure (Supplementary Figure S1C).
Transcription assay
For promoter-dependent transcription, nucleotides 171–470 of the human mitochondrial DNA (containing the light-strand promoter) were cloned between the NcoI and HindIII sites of pET-22 (Novagen). The substrate was linearized using HindIII. Reactions were carried out in 20 μl and contained 30 ng DNA, 20 mM HEPES (pH 8.0), 5% glycerol, 10 mM MgCl2, 150 mM KCl, 1 mM DTT, 100 μg/ml BSA, 0.4 mM ATP, 0.15 mM CTP and GTP, 0.01 mM UTP and 1 μl of [α-32P]UTP. Transcription was initiated by addition of 400 fmol POLRMT or POLRMTΔNTE, 500 fmol TFB2M and 2.5 pmol TFAM. Reactions were incubated for 30 min at 32°C and stopped by adding 100 μl of 1% SDS, 20 mM EDTA, 300 mM NaAc, 20 μg Calf Thymus DNA and 0.12 mg/ml glycogen. Products were phenol extracted, ethanol precipitated, resuspended in 20 μl of loading buffer and analyzed by polyacrylamide gel electrophoresis (PAGE) (Supplementary Figure S1D).
To confirm that our POLRMTΔNTE mutant is stable, we have analyzed its activity using a simple polymerization assay in which the polymerase is faced with a tailed substrate. This assay mimics elongation conditions in which the polymerase is synthesizing the nascent RNA chain while melting and displacing the DNA hybrid. Both POLRMTΔNTE and full-length POLRMT were active in this assay (Supplementary Figure S1E).
Pull-down assays
POLRMT, POLRMTΔNTE, TFAM or TFAMΔCTE were covalently immobilized into NHS-activated Agarose resin (Thermo Scientific) following the manufacturer’s recommendations. The coupling reaction was performed on 50 µl of resin with 50 µl 1 mg/ml protein in an amine-free buffer (20 mM HEPES pH 8.0, 5% glycerol, 150 mM KCl, 1 mM DTT) for 4 h at 4°C. Following immobilization, the remaining sites were blocked with aminoethanol. The resin was subsequently washed and incubated with a 1 mg/ml solution of each protein. After 1-h incubation at 4°C, the resin was washed and eluted with 0.2 M glycine-HCl (pH 2.5–3.0). Fractions were analyzed by SDS gel electrophoresis. To account for non-specific binding, the same amount of deactivated resin without immobilized protein was used as a control.
Isothermal titration calorimetry (ITC) experiments
Experiments were performed with a VP-ITC calorimeter (Microcal) at 25°C with a mixing speed of 302 rpm. POLRMT (70 μM) or POLRMTΔNTE (75 μM) proteins in the sample cell were titrated with 5 -μl injections (first injection 2 μl) of 0.7 mM TFAM, TFAMΔCTE or TFB2M. Samples were prepared by dialyzing all interacting components extensively against a buffer containing 20 mM HEPES (pH 8.0), 200 mM KCl, 5% glycerol and 1 mM EDTA. The heats of dilution of both proteins with buffer were determined and subtracted prior to analysis. Data were analyzed using the ORIGIN software supplied with the instrument according to the one- and two-binding site models.
Size-exclusion chromatography
10 µM POLRMT or POLRMTΔNTE were pre-mixed with 20 µM TFB2M in the absence of DNA or in presence of the LSP27 oligonucleotide (10 µM). LSP27 was made by annealing LSP27-1 (5′-CAA ATT TTA TCT TTT GGC GGT ATG CAC-3′) and LSP27-2 (5′-GTG CAT ACC GCC AAA AGA TAA AAT TTG-3′). A Sephadex 200 10/300 gel-filtration column was used for analytical gel-filtration analysis of the samples. Fractions were analyzed by SDS gel electrophoresis.
SAXS/WAXS measurements
TFB2M samples were prepared in 20 mM HEPES (pH 8.0), 200 mM KCl, 5% glycerol and 1 mM DTT within 24 h of data acquisition and were stored at 4°C. Scattering data were collected at beamline X9 of the National Synchrotron Light Source (NSLS, Upton, New York) using a Pilatus 300 K located 3.4 m from the sample for small angle X-ray data. Wide-angle X-ray scattering data were collected simultaneously with SAXS data using a Photonic Science CCD located 0.47 m from the sample. About 20 μl of sample were continuously flowed through a 1 mm diameter capillary and exposed to a 400 × 200 μm X-ray beam with a wavelength of 0.9184 Å for 30 s of total measurement time. Scattering data for TFB2M were collected at concentrations of 2.5, 5, and 10 mg/ml in triplicate. Normalization for beam intensity, buffer subtraction and merging of data from both detectors were carried out using PRIMUS (37). The radius of gyration (Rg) was calculated using a Guinier approximation, I(q) = I(0) exp(−q2Rg2/3), where a plot of I(q) and q2 is linear for q < 1.3/Rg (38). Six independent scattering trials were averaged. GNOM was used to determine the pair distribution function, P(r) and maximum particle dimension, Dmax (39). The linearity of the Guinier region and the forward scattering intensity were used to validate that the TFB2M samples were monodisperse in solution. The forward scattering intensity, I(0), is the theoretical scattering at a q-value of 0 and is proportional to the molecular weight of the sample (40). I(0)/c, where c is concentration, was identical for all TFB2M measurements. About 10 independent ab initio beads models that describe the experimental data were calculated using DAMMIF (41). The resulting models, which had an average χ2 of 1.65 ± 0.06, were subsequently aligned, averaged and filtered using DAMAVER (42).
EM analysis
POLRMT alone was adsorbed to glow discharged carbon-coated copper grids at 1 µM concentration and stained with 1% (w/v) uranyl acetate. To reconstruct a mitochondrial transcription initiation complex, POLRMT, TFB2M and TFAM were mixed in a 1:1 molar ratio at a concentration of 7 µM in the presence of 45-bp double-stranded substrate containing the mitochondrial light-strand promoter (LSP) and TFAM-binding site (coding strand: 3′-AAT CAA CCC CCC ACT GAC AAT TTT CAC GTA TGG CGG TTT TCT ATT-5′) and ATP and diluted to 0.5 µM with binding buffer (20 mM HEPES pH 8.0, 150 mM KCl, 1 mM EDTA, 10 mM MgCl2 and 5 mM DTT). Immediately after dilution, 3 µl aliquots of protein were applied onto 300-mesh copper grids (Ted Pella) that were coated with carbon film and glow-discharged for 10 s. After 1 min, the protein solution was blotted with filter paper and stained with 1% (w/v) uranyl acetate solution.
The grids with non-promoter DNA (coding strand: 3′-CTA GCC ATG CCA CTG GCT CTT AGA TAA CCC TAT ACA GTG ATT ATG-5′) and the three full-length proteins (type A control grids) and the grids with the LSP promoter DNA, full-length polRMT, TFB2M and C-terminally truncated TFAM (TFAMΔCTE) were prepared according to the same procedure (Supplementary Figure S2).
Grids were inspected on a Tecnai F20 operated at 200 kV with a 4 k × 4 k Tietz CCD camera. Images were acquired at a magnification of ×88 249 corresponding to 3.4 Å/pixel. All image processing and reconstructions steps were carried out using EMAN (43). Briefly, 3764 particles (for POLRMT apoenzyme) and 12 453 (for the full initiation complex) were selected, the contrast transfer function (ctf) of the images was determined and the phases flipped accordingly and an initial model was generated from reference-free class-averages and used for reference-based alignment. For the reconstruction of the mitochondrial initiation complex, a set of 12 453 raw images was subjected to reference-free alignment. Reference-free classes were divided into two subsets based on particle size and two initial models were generated from each subset. One model had a visibly elongated shape, whereas the other was more compact. We utilized both models as starting points for parallel refinement, as implemented in EMAN’s MULTIREFINE routine (43). In addition, a smaller size nearly spherical blob of ∼70 Å diameter was generated as a third model. The latter model was used as a potential scavenger of smaller size fragments. The procedure of parallel refinement yielded three models; two of them showed only minor differences. The size and shape of both models roughly corresponded to POLRMT. A third model was a good candidate for the quaternary initiation complex (POLRMT:TFAM:TFB2:DNA) and was subsequently used for reconstruction of the initiation complex (see below). The raw images were distributed approximately evenly between the first two models and the third one (2875, 3048 and 6530, accordingly). Based on that, we conclude the initiation complex was 50% dissociated before or during grid preparation. Smaller fragments (TFB2M and TFAM) were not detected after reference-free alignment/refinement, probably because the corresponding raw images were poorly visible on the grids, so they were ignored during particle picking.
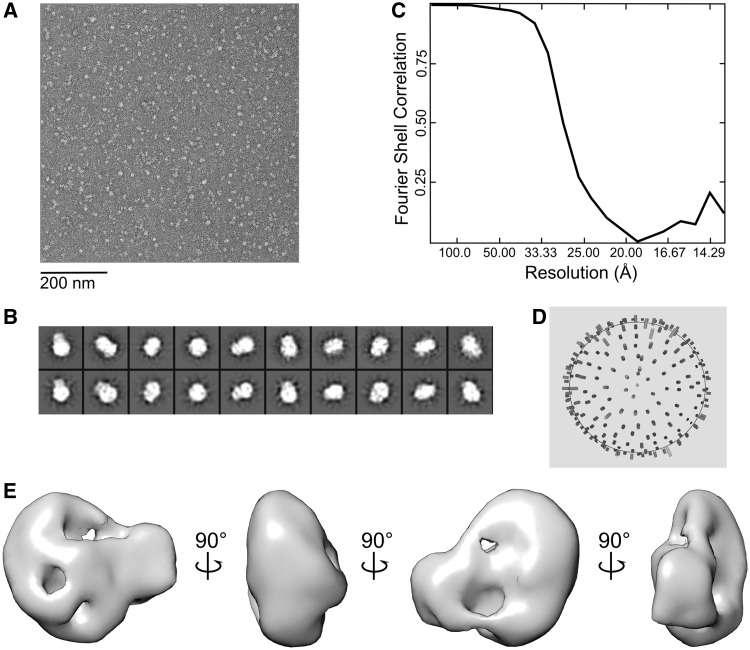
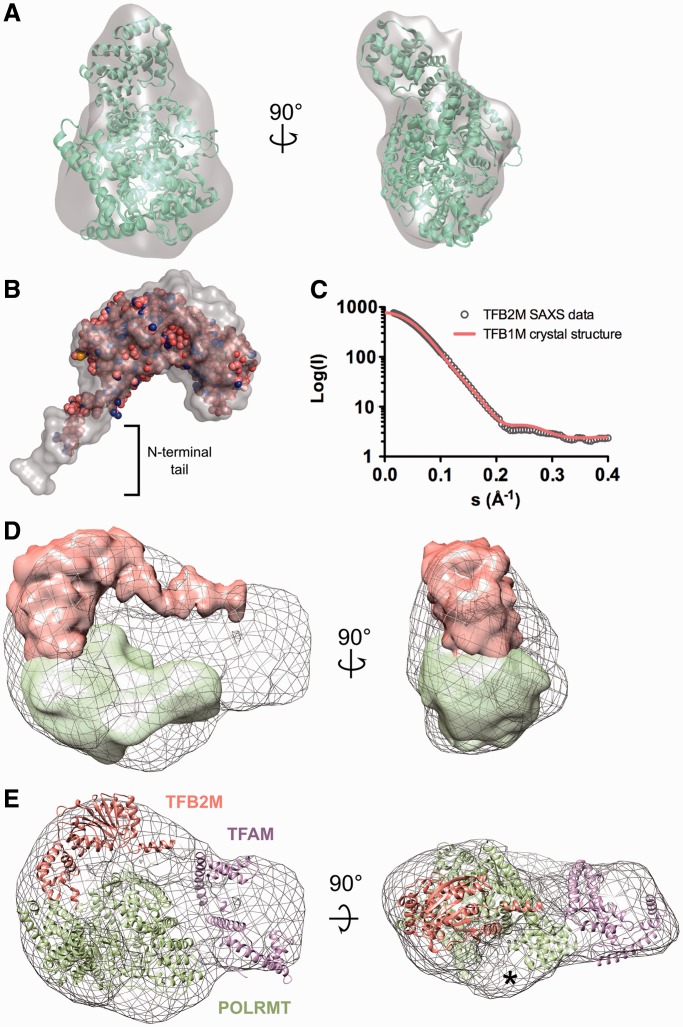
The particle images that grouped with the third model (i.e. the putative initiation complex model) were combined in a separate set and used for further structure refinement, which provided an additional improvement of the EM model (Supplementary Figure S3). To estimate the resolution of the final structure, two structures were calculated on the basis of even or odd halves of the final class set using the EOTEST routine. According to the Fourier shell correlation (FSC) (Figure 1C) curve at a 0.5 threshold value, the resolution of the final structures was ∼25 Å. The volumes for surface rendering were calculated using a known standard protein density of 0.81 Da/Å2 and molecular mass of 206 kDa (for the initiation complex).
Figure 1.
EM reconstruction of the mitochondrial transcription initiation complex. (A) Representative raw micrograph of uranyl-stained grids containing a quaternary POLRMT:TFB2M:TFAM:DNA complex. (B) Representative reference-free class averages (top row) and corresponding re-projections of the final model (bottom row). (C) The final resolution was estimated as 25 Å using the Fourier shell correlation criterion with a cutoff of 0.5. (D) Euler angle distribution plot of POLRMT:TFB2M:TFAM particle projections by the end of the refinement procedure. (E) The final structure of the mitochondrial transcription initiation complex shown in 90° rotations.
Docking
For the POLRMT map, the crystal structure of POLRMT (PDB accession number 3SPA) was positioned as a rigid body automatically using SITUS (44) and our in-house fast Fourier transform (FFT)-based software (45) for systematic search.
For the full initiation complex, we extended our systematic global search protein docking approach (45) for its application to docking of proteins (PDB accession numbers 3SPA, 4GC5 and 3TQ6) into electron density maps. We followed a two-step algorithm: (i) For a given protein, billions of docked conformations were sampled by soft rigid body docking based on a FFT correlation approach (45). The method performs exhaustive evaluation of correlations in the discretized 6D space of mutual orientations of the density map and the protein. The center of the EM density map (or its segment) was fixed at the origin of the coordinate system. The translational space was represented as a grid of 2 Å displacements of the protein center of mass, and the rotational space was quasi-uniformly sampled using 50 000 rotations (45). Correlation was calculated between smoothed EM density grid and the smoothed grid of atomic volumes of the protein (45). (ii) The highest scoring solutions were clustered for redundancy using a simple greedy clustering algorithm. The lowest correlation score solution was reported and all the neighboring solutions within 10 Å root-mean-square deviation (RMSD) were assigned to the same cluster. The members of this cluster were removed and the next lowest correlation structure is selected as the seed of the next cluster. The process is repeated until the desired number of clusters is reported or the current cluster correlation score falls <30% of the top scoring solution.
RESULTS
EM reconstruction of the transcription initiation complex reveals a bi-lobed arrangement
Although the recently solved crystal structures of TFAM and POLRMT have provided significant structural insight into the protein–DNA interactions relevant to transcription initiation (24,25,46) no structural information exists regarding the organization of the initiation complex as a whole. In an attempt to expand our understanding of the overall architecture of the initiation complex and the functional implications of the protein–protein and protein–DNA interactions relevant for transcription, we have carried out a single-particle EM reconstruction of the full initiation complex. Considering the likely importance of the promoter DNA for the stability of the initiation complex, we decided to conduct our reconstruction by combining full-length POLRMT, TFB2M and TFAM with a 45-bp DNA oligonucleotide containing both the mitochondrial LSP and TFAM-binding site, in conditions that mimic those used for initiation assays in vitro (see ‘Materials and Methods’ section). In addition to that, two sets of control grids were prepared. The type A control grids were prepared with the DNA fragment containing no promoter sequence and the three full-length proteins (Supplementary Figures S1A and S2A). The type B grids contained the LSP promoter DNA, full-length polRMT, TFB2M and C-terminally truncated TFAM (TFAMΔCTE; Supplementary Figures S1B and S2B)
The EM images of the grids containing the initiation complex (Figure 1A; Supplementary Figure S2C and E) contained a considerable amount of particles having a bi-lobal shape whose size roughly corresponded to the expected size of the quaternary initiation complex. In addition to that, smaller particles (probably individual components of the initiation complex, including POLRMT) were also observed on the grids, which implied partial dissociation of the initiation complex during or before grid preparation. In contrast, the control A grids contained particles that were highly diverse in size and shape, probably as a result of random aggregation of initiation complex components in the absence of the promoter DNA sequence. The appearance of the control B grids more closely resembled that of the initiation complex grids (Figure 1A and Supplementary Figure S2C). However, reference-free alignment of the particles collected from those grids revealed an almost total absence of well-organized larger size particles (Supplementary Figure S2D). Smaller particles having a compact round shape were observed instead. Taken together, these data confirm the importance of the promoter-containing DNA for assembly of the initiation complex as well as the role the C-terminus of TFAM plays in the assembly of the initiation complex at LSP. We can conclude, therefore, that the particles observed on the initiation complex grids belonged, most likely, to the initiation complex, not to a simple quaternary complex of POLRMT, TFB2M and TFAM non-specifically bound to the DNA molecule.
As a considerable amount of smaller size particles was observed on the initiation complex grids (Figure 1A), the concern arose that those particles could adversely affect our 3D model reconstruction. Therefore, instead of a simple refinement procedure, we decided to utilize a multirefinement protocol for the 3D reconstruction, as implemented in the MULTIREFINE routine of the EMAN package (43). Briefly, three initial models were generated as described in the ‘Materials and Methods’ section, and they were subjected to iterative refinement using the entire image set. During this procedure, each image from the set is used for only one model (out of three). As a rule, when the image set belongs to a structurally homogeneous particle (in other words, different size images correspond to different particle orientations, not to different size particles), all models converge to a similar structure. In our case, however, only the first and the second models converged (Supplementary Figure S3) to a compact round shaped structure, whereas the third model produced a larger, elongated bi-lobal shape. As 6530 (out of 12 453) images corresponded to the latter structure, we concluded that about half of the initiation complex existed in a dissociated from (perhaps resulting from grid preparation). Taking into account that the concentration of the initiation complex in the solution applied to the grids was ∼0.1 μM, such a degree of dissociation is quite reasonable; the complex cannot be highly stable, because such high stability would likely prevent the required transition from initiation to elongation mode. Further refinement using the 6530 image set, which was separated from the initial raw image set, yielded a final model whose re-projections satisfactorily matched representative reference-free class averages (Figure 1B). The final reconstruction was based on 6530 particles and resulted in a 25 Å resolution model of the quaternary POLRMT:TFB2M:TFAM:DNA complex (Figure 1C). The EM map of the initiation complex had a distorted bi-lobed shape with dimensions 140 × 110 × 85 Å (Figure 1E). Brief inspection of the envelope demonstrated that its volume and shape allows accommodation of all three proteins from the transcription initiation complex and its compact shape implies that interactions are likely to take place between the three protein components of the complex. Given the functional role of this complex, these interactions are likely to be transient.
POLRMT physically interacts with TFAM
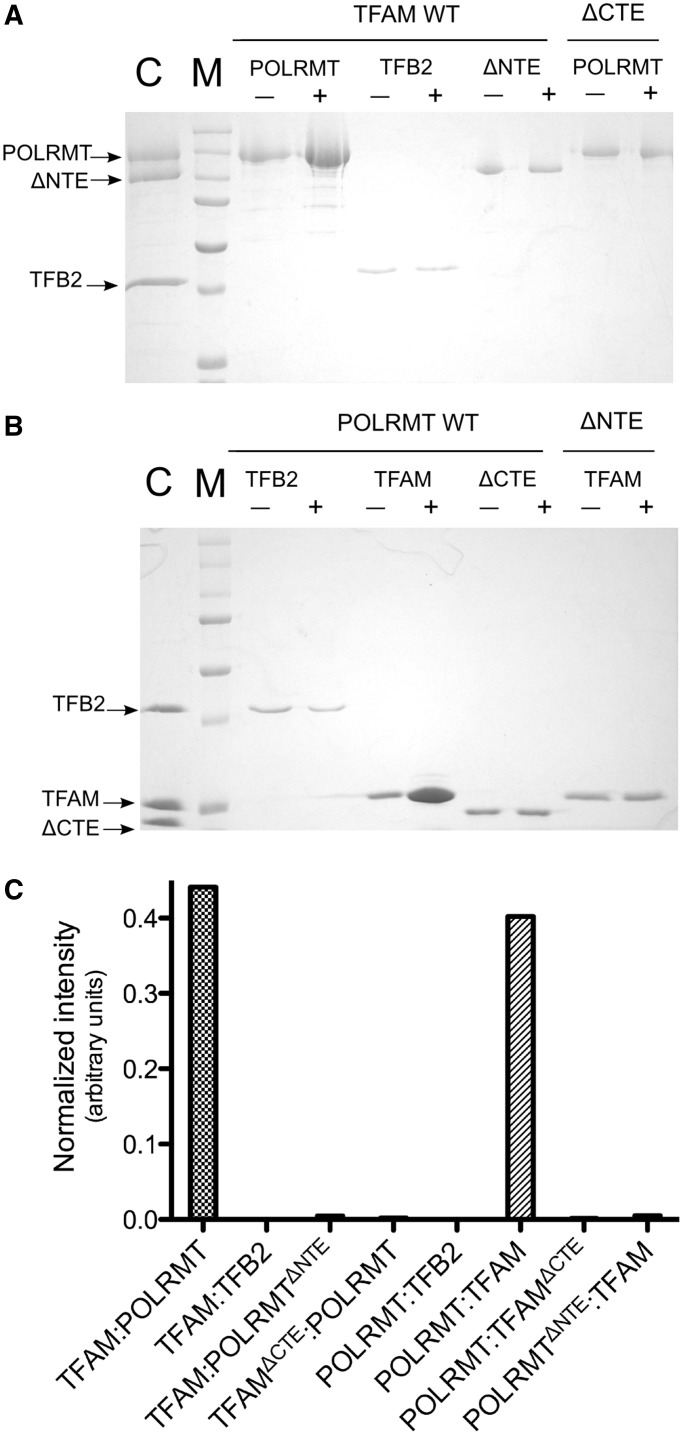
After considering the shape of our EM envelope and the likelihood of protein–protein interactions within the initiation complex at LSP, we decided to probe the nature of the putative interactions within the transcription initiation complex. We performed a pull-down assay using purified un-tagged proteins (see ‘Materials and Methods’ section). First, we covalently immobilized TFAM on N-hydroxy-succinimide (NHS)-activated agarose and used this resin to probe all possible interactions between POLRMT, TBF2M and TFAM. As a control for non-specific binding to the resin, inactivated resin without immobilized protein was used. The only clear evidence of an interaction that was observed corresponded to full-length POLRMT and full-length TFAM (Figure 2A and C).
Figure 2.
POLRMT physically interacts with TFAM. (A) Pull-down assay to analyze interactions with purified untagged TFAM or TFAMΔCTE. The proteins were covalently bound to NHS-activated agarose and incubated with POLRMT, TFB2M or POLRMTΔNTE to test for the presence of a direct physical interaction. As a control for non-specific binding to the resin, inactivated resin without immobilized protein was used. Compared with the control, the only clear interaction observed was between full-length POLRMT and full-length TFAM. (B) A reverse experiment performed by covalently immobilizing full-length POLRMT or POLRMTΔNTE onto the resin. We then probed for interactions with TFB2M, TFAM and TFAMΔCTE. The result further suggests the existence of an interaction between POLRMT and TFAM. However, no evidence was found of a direct interaction between POLRMT and TFB2M or between TFB2M and TFAM. (C) Quantification of the results shown in panels A and B. The Precision Plus Protein Dual Color Standard protein marker (BioRad) shown in panels A and B (indicated with the letter M) contains the following bands: 250 kDa, 150 kDa, 100 kDa, 75 kDa, 50 kDa, 37 kDa and 25 kDa. A control lane in panels A and B is indicated with the letter C and the predicted molecular masses (based on sequence) for the proteins are as follows: 134.5 kDa for POLRMT, 101 kDa for POLRMTΔNTE, 24.4 kDa for TFAM, 21.9 kDa for TFAMΔCTE and 43.3 kDa for TFB2M.
In order to further confirm this observation, we performed the reverse experiment by covalently immobilizing full-length POLRMT onto the resin. We then probed for interactions with TFAM and TFB2M. As shown in Figure 2B, the result clearly suggests the existence of a stable interaction between POLRMT and TFAM. However, no evidence of a direct interaction between POLRMT and TFB2M or between TFB2M and TFAM was observed under our experimental conditions.
The POLRMT–TFAM interaction is of low affinity and depends on the NTE of POLRMT and the C-terminal tail of TFAM
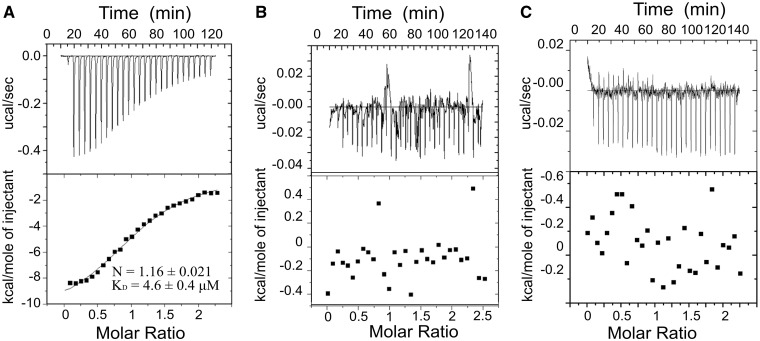
To further investigate the nature of the TFAM–POLRMT interaction we decided to use isothermal titration calorimetry (ITC) to quantitatively characterize the binding equilibrium. ITC measurements demonstrate a clear exothermic (ΔH = 11.1 kcal/mol) interaction between POLRMT and TFAM (KD = 4.6 µM; Figure 3A). This interaction is also characterized by moderately negative binding entropy (ΔS = −11.4 cal/mol·K), suggesting that stabilization of the POLRMT–TFAM complex has predominantly an enthalpic, not entropic nature. We next decided to probe which regions in POLRMT and TFAM might be responsible for this interaction. The non-conserved NTE of POLRMT was an obvious candidate, and consistent with the pull-down results (Figure 2), titration of a ΔNTE mutant of POLRMT exhibited no thermal effect in the ITC experiments (Figure 3B), suggesting that the binary interaction between TFAM and POLRMT is mediated by the NTE. Similarly, we decided to also examine the importance of the C-terminal domain of TFAM (CTE) for this interaction. Like the NTE of POLRMT, the CTE of TFAM is known to be essential for transcription initiation and it does not appear to directly interact with DNA in the TFAM crystal structure. Once again, consistent with pull-down experiments (Figure 2), ITC results using TFAMΔCTE suggest that the C-terminal domain of TFAM plays an essential role in mediating the TFAM–POLRMT interaction (Figure 3C).
Figure 3.
The interaction between POLRMT and TFAM is dependent on the NTE. (A) Isothermal titration calorimetry experiments of TFAM-binding to POLRMT (B) TFAM-binding to POLRMTΔNTE or (C) TFAMΔCTE binding to POLRMT performed at 25°C. In all three cases, the top panel represents the heat signal for injections of TFAM or TFAMΔCTE into the calorimeter cell containing POLRMT or POLRMTΔNTE. The bottom panel displays the integrated heat for each injection after peak integration and subtraction of basal values fitted to a simple single-site binding model (solid line). The stoichiometry (N) and binding constant (KD) for the interaction between TFAM and POLRMT are shown in the bottom panel. No thermal effect was observed for the putative interaction between TFAM and POLRMTΔNTE, or between TFAMΔCTE and POLRMT.
POLRMT and TFB2M do not form a stable complex in the absence of promoter DNA
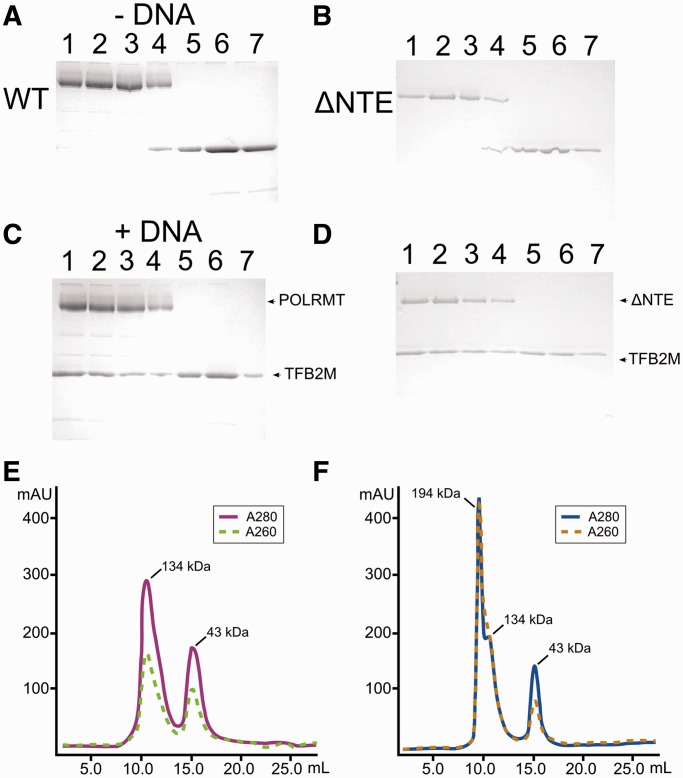
Given that POLRMT and TFB2M can be co-purified (22), we hypothesized that stable association of the initiation complex with the promoter would require an interaction between POLRMT and TFB2M. Surprisingly, we were not able to observe any evidence of an interaction between these proteins in pull-down experiments (Figure 2). However, this approach might not be sufficiently sensitive for low-affinity interactions. Thus, we decided to analyze the putative association between POLRMT and TFB2M using isothermal calorimetry. Titration of POLRMT with TFB2M resulted in no appreciable thermal effect (Supplementary Figure S4). Although it cannot be excluded that the thermal effect is too small to be detected, together with the results of the pull-down experiments these results suggest that the two proteins do not directly bind to one another. Given that POLRMT and TFB2M can mediate TFAM-independent initiation and both proteins can be co-purified under some conditions, we speculated that a stable complex might nevertheless form in the presence of promoter DNA. We used gel-filtration chromatography to test this hypothesis and found that the peaks corresponding to POLRMT and TFB2M can be fully resolved on a Sephadex 200 10/300 gel-filtration column (Figure 4A and Supplementary Figure S5). In contrast, a similar experiment performed in the presence of a linear promoter DNA substrate (LSP27, see ‘Materials and Methods’ section) demonstrated that the initial fractions (corresponding to the high molecular weight species) now contained both POLRMT and TFB2M (Figure 4C). Our results suggest that whereas POLRMT and TFB2M do not interact strongly with each other in the absence of promoter DNA, in the presence of the promoter they can form a stable POLRMT:TFB2M:DNA ternary complex. Interestingly, in this case the NTE of POLRMT is dispensable for the interaction (Figure 4B and D), suggesting that the observed ternary complex only depends on the presence of the C-terminal region of POLRMT. It is important to note that this approach does not allow us to determine whether this interaction is specific and to which degree it is relevant to transcription initiation.
Figure 4.
POLRMT and TFB2M interact in a DNA-dependent manner. Gel-filtration experiment demonstrating that both proteins could be resolved as separate peaks on a Sephadex 200 10/300 gel filtration column in the absence of promoter DNA (A) and (B). However, when linear promoter DNA was included in the experiment, both POLRMT and POLRMTΔNTE co-eluted with TFB2M, suggesting that a stable POLRMT:TFB2M complex is formed in the presence of promoter DNA (C) and (D). Actual size-exclusion chromatograms for experiments without the promoter DNA (E) and with promoter DNA (F).
Single-particle reconstruction of POLRMT
We next decided to generate a docking model of the initiation complex based on our EM map. The N-terminal domain of POLRMT was not resolved in the electron density of the recent crystal structure of the enzyme, presumably due to significant flexibility or disorder (10). Thus, in an attempt to obtain a full-length structural characterization of POLRMT and facilitate docking into our EM envelope, we decided to perform a model-independent EM single-particle reconstruction of POLRMT. Furthermore, we reasoned that this determination might allow us to obtain a confident assignment of the position of POLRMT within the complex. We analyzed 3764 images of POLRMT preserved in 1% uranyl acetate stain and reconstructed its 3D structure using reference-free alignment and common-line Euler angle assignment (see ‘Materials and Methods’ section), followed by iterative projection matching refinement of the initial model as implemented in the EMAN software package. The structure we obtained after 12 cycles of refinement has an elongated globular shape and displays a robust fit (cross-correlation coefficient 0.86) with the existing X-Ray structure of POLRMT (Figure 5A). Manual and computer-aided (SITUS and our in-house FFT-based search software; see ‘Materials and Methods’ section) docking of POLRMT (PDB: 3SPA) resulted in identical docking orientations. Consistent with the previously published crystallographic results, it is clear that the N-terminal domain of POLRMT is also disordered in the EM structure, providing further evidence for its flexibility, at least in the absence of promoter DNA.
Figure 5.
Localization of the components of the transcription initiation complex. (A) Surface representation of the EM map for POLRMT apoenzyme. The flexibly fitted crystal structure (PDB ID: 3SPA) is shown superimposed (cross-correlation coefficient 0.86) in cartoon representation with a 90° rotation. (B) SAXS/WAXS model of TFB2M. Surface representation (gray) of a filtered average bead model that describes the data (χ2 = 1.59). Shown inside the surface is the X-ray crystallographic structure of TFB1M (PDB: 4GC5). (C) The experimental and theoretical scattering profiles of TFB2M and TFB1M. The observed scattering profile for TFB2M is well fit by the calculated profile derived from the TBF1M crystal structure (χ2 = 1.68). (D) EM and SAXS data reveal the relative positions of POLRMT and TB2M within the complex. The EM envelope for the full complex is shown as a gray mesh. The POLRMT EM map and TFB2M SAXS model have been positioned within the EM envelope and are shown in green and orange, respectively. (E) Relative orientation of the three protein components of the initiation complex (PDB IDs: 3SPA, 4GC5 and 3TQ6). The EM envelope for the full complex is shown as grey mesh, whereas POLRMT, TFAM and TFB2M are shown as ribbons colored green, purple and orange, respectively. The asterisk indicates additional volume corresponding to the N-terminal region of POLRMT that was not resolved in the crystal structure.
TFB2M is structurally homologous to TFB1M
The only component of the initiation complex for which there is no known structure is the TFB2M transcription factor. However, a structure of its mouse paralog TFB1M, (18% identity) has recently been solved (47). TFB2M and TFB1M are thought to share a common methyltransferase core domain, but TFB2M contains a larger N-terminal tail whose function is not yet known, but that appears to be dispensable for transcription initiation (21). Thus, we decided to assess whether the TFB1M structure would be an appropriate model for TFB2M to be used for docking into our EM envelope. Given the relatively small size of TFB2M (45 kD), we decided to carry out small- and wide-angle X-ray scattering experiments in order to obtain its overall structure and further characterize the shape of this protein.
The SAXS/WAXS data show that human TFB2M has a slightly larger maximal dimension (Dmax) compared to mouse TFB1M, at 109 ± 3 Å versus 90 ± 5 Å, respectively (Supplementary Figure S6; for mmTFB1M, the Dmax was determined from the crystal structure), which is consistent with the presence of a larger N-terminal tail in hsTFB2M. Consistently, the experimental pairwise-distance distribution [P(r)] function for human TFB2M is relatively skewed, suggesting an elongated asymmetric shape with the bulk mass of the protein having dimensions ∼75 × 35 Å2.
The SAXS/WAXS data were used to generate low-resolution ab initio models of 3D arrangements of scattering centers, or beads that provide the shape of the molecular envelope for human TFB2M (see ‘Materials and Methods’ section). A molecular surface representation of the resulting filtered average bead model for human TFB2M is shown in Figure 5B, and the overall shape is a deformed prolate ellipsoid with a narrower extended tail. The wider section has dimensions that closely resemble those of mouse TFB1M (∼75 × 35 Å2). The remaining volume is consistent with the larger N-terminal tail of TFB2M. A theoretical scattering profile was calculated using the mouse TFB1M crystal structure (47,48) and compared directly with the experimental scattering data for human TFB2M, resulting in a robust fit that further supports the structural similarity of these two proteins and conservation of the methyltransferase fold (Figure 5C).
Docking model of the transcription initiation complex
The volume of the EM envelope for the initiation complex is consistent with the expected volume from the individual structures of each component. Comparison of the EM map for POLRMT and the TFB2M SAXS/WAXS envelope with the EM map of the full initiation complex allows for preliminary localization of the individual components. The majority of volume in the larger lobe corresponds to POLRMT, whereas the remaining space accommodates TFB2M (Figure 5D). The size and shape of the smaller lobe corresponds to the expected volume for TFAM (Figure 5E).
Importantly, a wealth of biochemical information exists pertaining to certain aspects of the organization of the complex, including details of specific protein–protein and protein–DNA interactions (49,50). In addition, both the initiation site and TFAM-binding sites have been clearly defined, (51) and together, these details provide an important set of biochemical constraints that should be recapitulated by a valid docking model. We thus set out to reconcile all existing information, including our EM results, into a final model for the quaternary complex. We proceeded by docking each individual component into their corresponding segments, while retaining multiple suboptimal solutions as described in methods (see ‘Materials and Methods’ section). We then selected the best scoring model that satisfied all structural and biochemical constraints, while avoiding significant inter-component clashes.
The final model (Figure 6) suggests that the organization of the initiation complex is more compact than initially expected (21,50). Both POLRMT and TFB2M occupy the larger lobe in the EM envelope, but they associate with opposite sides of the open promoter around the initiation site. Consistent with the absence of a strong interaction, they do not establish extensive protein–protein contacts. TFAM is bound to its binding site upstream of the initiation site and does not appear to establish extensive contacts with any of the other two proteins, but its C-terminal tail is located in close proximity to the N-terminal end of POLRMT, which is in agreement with our biochemical observations. Interestingly, although our EM structure of POLRMT exhibits the same disorder in the N-terminus seen in the POLRMT crystal structure, in the quaternary complex additional density in the map is apparent (indicated with an asterisk in Figures 5E and 6). The additional density appears to correspond to the N-terminus of POLRMT, implying that this domain becomes well ordered in the context of a fully assembled initiation complex. Additional density was also observed adjacent to the expected location of the upstream duplex between POLRMT/TFB2M and TFAM. This density likely corresponds to the large N-terminal tail of TFB2M that is also observed in the SAXS/WAXS analysis. Based on structural predictions, this tail might contact the upstream duplex, consistent with previous observations (21). It is important to note that DNA and proteins interact differently with uranyl ions; such that DNA, unlike proteins, is poorly visible on negatively stained grids at ∼25 Å resolution. Consistently, our EM map displays a clear groove in the density that correlates well with the path of DNA in our docking model (Supplementary Figure S7). Moreover, superposing the POLRMT elongation complex on the POLRMT molecule in our docking model (Supplementary Figure S7B) reveals the exit point for the DNA downstream of the active site, which is also consistent with the presence of a second groove in the density in our EM map (Supplementary Figure S7C). Thus, the features in our map appear to be consistent with the expected location of the DNA according to our model.
Figure 6.
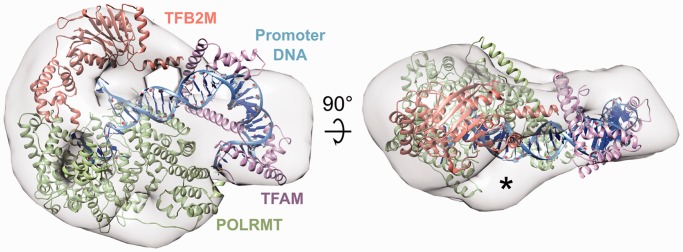
Model of the intact mitochondrial transcription initiation machinery. The EM envelope for the full complex is shown as a grey mesh, while POLRMT, TFAM, TFB2M and the promoter DNA are shown as ribbons colored green, purple, orange and blue, respectively. The asterisk indicates additional volume corresponding to the N-terminal region of POLRMT that was not resolved in the crystal structure.
DISCUSSION
In this study, we have performed the first structural characterization of the human mitochondrial transcription initiation machinery by combining biochemical, biophysical and structural data. Using negative staining EM, we determined the overall shape of the transcription initiation complex, and, taking into account existing and newly obtained biochemical data about the interactions between different initiation components, we established their placement within the EM envelope.
Our results suggest that the structural architecture of the mitochondrial transcription initiation complex is compact and does not follow a linear arrangement. Instead, the initiation proteins are organized around the promoter DNA forming a bi-lobed shape and are thus likely to establish at least transient physical contacts with one another. To establish which of these contacts are important for stabilization of the initiation complex, we examined all possible pairwise interactions between the initiation proteins and we found that under our experimental conditions, only the interaction between POLRMT and TFAM is strong enough to be detected and characterized by a highly sensitive method such as ITC. We further demonstrate that the non-conserved N-terminus of POLRMT and the C-terminal tail of TFAM mediate this interaction, which is consistent with the fact that both regions are essential for promoter-dependent initiation (10,26). Moreover, our results are indirectly corroborated by the fact that the yeast TFAM homolog, Abf2, which does not interact with the polymerase, lacks the C-terminal domain that we have identified to be responsible for the interaction of human TFAM with POLRMT (52)
Interestingly, although TFB2M is known to be indispensable for initiation of transcription (53), was shown to co-purify with POLRMT (22) and was suggested to interact with the C-terminal tail of TFAM (54), our data suggest that no biologically relevant amounts of pre-assembled initiation complex or binary TFAM–TFB2M or POLRMT–TFB2M complexes can exist in the absence of the promoter DNA. Thus, the promoter DNA segment likely serves as the main scaffold holding all components of the initiation complex together, whereas the interaction between TFAM and POLRMT acts as an additional stabilizing factor. In our opinion, this hypothesis reconciles our findings with earlier observations (23,52). It is important to note that in order to facilitate our structural studies, our purification protocols aimed to quantitatively eliminate any nucleic acid contaminants from the protein sample. It is possible that small traces of DNA could have facilitated co-purification or immunocoprecipitation of the initiation factors. It is also possible that, in the presence of DNA, those interactions might be bona fide protein–protein interactions, perhaps as a consequence of DNA-induced or other conformational changes. In this respect, the C-terminus of TFAM has been shown to be a site of post-translational modifications (55) and any such modifications are likely to affect the nature of the interactions between the components of the initiation complex.
The idea that TFAM physically associates with other components of the initiation machinery had been proposed earlier (50,52), but that model was based on the assumption that POLRMT and TFB2M associate to the promoter via an interaction between TFB2M and TFAM. It is important to note, however, that the relatively low affinity of the POLRMT–TFAM interaction strongly suggests that a POLRMT–TFAM complex does not exist outside of the context of initiation.
We were able to generate an EM-based model of the transcription initiation complex, where previously determined X-ray crystallographic structures of the constituent components were docked into the bi-lobed EM envelope and their positions further refined by taking into account their binding sites on the LSP-containing DNA. According to the model we obtained, the larger lobe is occupied by POLMRT and TFB2M, which bracket the promoter DNA on opposite sides, whereas TFAM occupies the smaller lobe upstream of the initiation site. Previously, TFAM was shown to substantially bend the promoter DNA and its interaction with DNA was thoroughly characterized using X-ray crystallography (24,56). Both the ability of TFAM to bend DNA and its interaction with the N-terminus of POLRMT seem to play a critical role in establishing the compact organization of the initiation complex. In fact, in our EM complex, the N-terminus of POLRMT appears to be at least partially ordered, further supporting its role in mediating an interaction during initiation. Deletion of the N-terminus of POLRMT decouples it from TFAM, explaining why this mutant is unable to initiate transcription (10).
The recent structure of the elongation complex of POLRMT provides the first glimpse of the interaction between the RNA polymerase and its DNA and RNA substrates (46). An overlay of this structure with our model of the transcription initiation complex further supports the notion that TFB2M is in a position to bind the DNA substrate near the active site [Supplementary Figure S7 (21)]. It is important to note, however, that it is not possible to ascertain whether the conformations observed in the elongation complex are relevant in the context of initiation, where TFAM and TFB2M likely impose additional structural constraints. In fact, our EM model is consistent with a shift in the position of the upstream duplex [similar to that observed for T7 RNA polymerase (57)] suggesting that a distinct elongation mode is adopted upon dissociation of TFAM and TFB2M.
The observed arrangement of initiation complex is fully consistent with earlier cross-linking studies that revealed that both POLRMT and TFB2M interact with the promoter DNA (21). Our structure implies that the methyltransferase fold of TFB2M interacts predominantly with the single-stranded non-transcribed strand, consistent with its expected binding mode (47). Moreover, our model also suggests that the large N-terminal tail of TFB2M interacts with the upstream duplex, which is again consistent with cross-linking studies (21). Importantly, one aspect that provides additional validation of our EM structure is the fact that the docking is in good agreement with the known position of TFAM-binding relatively to the initiation site (21).
Although our study clarifies the mechanism by which TFAM participates in transcription initiation, it does not explain why, unlike in yeast, TFAM is necessary for this process in mammalian mitochondria. This might be related to the fact that mammalian mitochondrial promoters have a more variable consensus sequence than yeast (58,59), and thus TFAM might provide additional selectivity to the DNA recognition process. Moreover, TFAM has been shown to differentially regulate the human mitochondrial promoters (60), implying that relying on TFAM for initiation might provide an additional opportunity for transcriptional regulation. This also suggests that promoter-specific differences might exist in the organization of the mitochondrial initiation complex. Clearly, TFAM is needed for efficient transcription from LSP and HSP1, however, that fact does not imply that a two-component initiation complex (TFB2M–POLRMT) is not capable of transcription initiation (29). Moreover, transcription from HSP2 appears to take place in the absence of TFAM (60,61).
Furthermore, even the participation of TFB2M is not absolutely necessary for transcription initiation in vivo, because POLRMT alone is capable of transcription initiation for lagging DNA strand replication (33). These, as well as other observations imply that the TFAM–TFB2–POLRMT ensemble represents a flexible and multipurpose machine that, depending on the circumstances, is capable of transcription initiation from diverse sites. Such flexibility may facilitate regulation of mitochondrial replication and transcription. Nevertheless, at least in the context of LSP initiation, the data in this manuscript strongly indicate that the POLRMT–TFAM interaction is critical for assembly of the initiation complex at LSP and likely important for its stabilization. Based on these results, we favor a model where TFAM helps recruit POLRMT to LSP through this interaction. Furthermore, considering our structural model, it is likely that the POLRMT–TFAM interaction introduces an additional structural restraint that modulates the position of the upstream duplex with respect to POLRMT. Thus, release of the POLRMT–TFAM interaction after polymerization by POLRMT might be the event that determines the transition to elongation mode.
In summary, we have presented the first structural characterization of the complete human mitochondrial initiation complex and provided evidence that formation of this complex depends critically on the promoter DNA but is aided by a low-affinity interaction between POLRMT and TFAM. Our EM model for transcription initiation is consistent with previous experimental evidence, and reveals the compact arrangement of POLRMT, TFB2M and TFAM around the LSP. In addition to shedding light on the nature of mitochondrial transcription complexes, this will serve as the basis for elucidation of the many structural and functional questions that still remain unanswered.
ACCESSION NUMBERS
The EM density map for the mitochondrial transcription initiation complex can be found at the Electron Microscopy Data Bank under accession number EMD-2529.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors wish to thank all members of the Garcia-Diaz, Kozakov and Beglov laboratories for helpful discussions and support as well as Marc Allaire and Lin Yang at NSLS beamline X9 for assistance with SAXS/WAXS experiments.
FUNDING
NYSTAR and Research facilities Improvement Program [C06 RR017528-01-CEM from the National Center for Research Resources, National Institutes of Health] for use of the EM facility at NYSBC; the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences [DE-AC02-98CH10886 for the use of the National Synchrotron Light Source, Brookhaven National Laboratory]; National Institutes of Health [NIH F30-ES022930 to K.E.G, NIH R00-ES015421 to M.G.-D., NIH R01-GM100021 to M.G.-D., NIH R01-GM93147 to D.K., NIH R01- GM61687 to D.B., NSF DBI1047082 to D.K.]; Russian Ministry of Education and Science [14.A18.21.1973 to D.K.]; United Mitochondrial Disease Foundation [59042 to M.G.-D.]. Funding for open access charge: NIH [R01-GM100021].
Conflict of interest statement. None declared.
REFERENCES
- 1.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Ann. Rev.Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kroemer G, Reed JC. Mitochondrial control of cell death. Nat. Med. 2000;6:513–519. doi: 10.1038/74994. [DOI] [PubMed] [Google Scholar]
- 4.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 5.Mancuso M, Orsucci D, LoGerfo A, Calsolaro V, Siciliano G. Clinical features and pathogenesis of Alzheimer's disease: involvement of mitochondria and mitochondrial DNA. Adv. Exp. Med. Biol. 2010;685:34–44. doi: 10.1007/978-1-4419-6448-9_4. [DOI] [PubMed] [Google Scholar]
- 6.Shutt TE, Shadel GS. A compendium of human mitochondrial gene expression machinery with links to disease. Environ. Mol. Mutagen. 2010;51:360–379. doi: 10.1002/em.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saini G, Jensen DS, Wiest LA, Vail MA, Dadson A, Lee ML, Shutthanandan V, Linford MR. Core-shell diamond as a support for solid-phase extraction and high-performance liquid chromatography. Anal Chem. 2010;82:4448–4456. doi: 10.1021/ac1002068. [DOI] [PubMed] [Google Scholar]
- 8.Filee J, Forterre P, Sen-Lin T, Laurent J. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 2002;54:763–773. doi: 10.1007/s00239-001-0078-x. [DOI] [PubMed] [Google Scholar]
- 9.McGeoch AT, Bell SD. Extra-chromosomal elements and the evolution of cellular DNA replication machineries. Nat. Rev. Mol. Cell Biol. 2008;9:569–574. doi: 10.1038/nrm2426. [DOI] [PubMed] [Google Scholar]
- 10.Ringel R, Sologub M, Morozov YI, Litonin D, Cramer P, Temiakov D. Structure of human mitochondrial RNA polymerase. Nature. 2011;478:269–273. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- 11.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 12.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 13.Xu F, Ackerley C, Maj MC, Addis JBL, Levandovskiy V, Lee J, MacKay N, Cameron JM, Robinson BH. Disruption of a mitochondrial RNA-binding protein gene results in decreased cytochrome b expression and a marked reduction in ubiquinol–cytochrome c reductase activity in mouse heart mitochondria. Biochem. J. 2008;416:15–26. doi: 10.1042/BJ20080847. [DOI] [PubMed] [Google Scholar]
- 14.Lightowlers RN, Chrzanowska-Lightowlers ZM. PPR (pentatricopeptide repeat) proteins in mammals: important aids to mitochondrial gene expression. Biochem. J. 2008;416:e5–6. doi: 10.1042/BJ20081942. [DOI] [PubMed] [Google Scholar]
- 15.Paratkar S, Deshpande AP, Tang GQ, Patel SS. The N-terminal domain of the yeast mitochondrial RNA polymerase regulates multiple steps of transcription. J. Biol. Chem. 2011;286:16109–16120. doi: 10.1074/jbc.M111.228023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mercer,T.R., Neph,S., Dinger,M.E., Crawford,J., Smith,Martin A., Shearwood,A.-Marie J., Haugen,E., Bracken,Cameron P., Rackham,O., Stamatoyannopoulos,John A. et al. (2011) The human mitochondrial transcriptome. Cell, 146, 645–658. [DOI] [PMC free article] [PubMed]
- 17.Paratkar S, Patel SS. Mitochondrial transcription factor Mtf1 traps the unwound non-template strand to facilitate open complex formation. J. Biol. Chem. 2010;285:3949–3956. doi: 10.1074/jbc.M109.050732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savkina M, Temiakov D, McAllister WT, Anikin M. Multiple functions of yeast mitochondrial transcription factor Mtf1p during initiation. J. Biol. Chem. 2010;285:3957–3964. doi: 10.1074/jbc.M109.051003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotney J, McKay SE, Shadel GS. Elucidation of separate, but collaborative functions of the rRNA methyltransferase-related human mitochondrial transcription factors B1 and B2 in mitochondrial biogenesis reveals new insight into maternally inherited deafness. Hum. Mol. Genet. 2009;18:2670–2682. doi: 10.1093/hmg/ddp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seidel-Rogol BL, McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat. Genet. 2003;33:23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]
- 21.Sologub M, Litonin D, Anikin M, Mustaev A, Temiakov D. TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 2009;139:934–944. doi: 10.1016/j.cell.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 23.Kang D, Kim SH, Hamasaki N. Mitochondrial transcription factor A (TFAM): roles in maintenance of mtDNA and cellular functions. Mitochondrion. 2007;7:39–44. doi: 10.1016/j.mito.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 24.Ngo HB, Kaiser JT, Chan DC. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat. Struct. Mol. Biol. 2011;18:1290–1296. doi: 10.1038/nsmb.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubio-Cosials A, Sidow JF, Jimenez-Menendez N, Fernandez-Millan P, Montoya J, Jacobs HT, Coll M, Bernado P, Sola M. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat. Struct. Mol. Biol. 2011;18:1281–1289. doi: 10.1038/nsmb.2160. [DOI] [PubMed] [Google Scholar]
- 26.Dairaghi DJ, Shadel GS, Clayton DA. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- 27.Shi Y, Dierckx A, Wanrooij PH, Wanrooij S, Larsson NG, Wilhelmsson LM, Falkenberg M, Gustafsson CM. Mammalian transcription factor A is a core component of the mitochondrial transcription machinery. Proc. Natl Acad. Sci. USA. 2012;109:16510–16515. doi: 10.1073/pnas.1119738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohgaki K, Kanki T, Fukuoh A, Kurisaki H, Aoki Y, Ikeuchi M, Kim SH, Hamasaki N, Kang D. The C-terminal tail of mitochondrial transcription factor a markedly strengthens its general binding to DNA. J. Biochem. 2007;141:201–211. doi: 10.1093/jb/mvm020. [DOI] [PubMed] [Google Scholar]
- 29.Shutt TE, Lodeiro MF, Cotney J, Cameron CE, Shadel GS. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc. Natl Acad. Sci. USA. 2010;107:12133–12138. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brewer LR, Friddle R, Noy A, Baldwin E, Martin SS, Corzett M, Balhorn R, Baskin RJ. Packaging of single DNA molecules by the yeast mitochondrial protein Abf2p. Biophys. J. 2003;85:2519–2524. doi: 10.1016/s0006-3495(03)74674-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shutt TE, Bestwick M, Shadel GS. The core human mitochondrial transcription initiation complex: It only takes two to tango. Transcription. 2011;2:55–59. doi: 10.4161/trns.2.2.14296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuste JM, Wanrooij S, Jemt E, Granycome CE, Cluett TJ, Shi Y, Atanassova N, Holt IJ, Gustafsson CM, Falkenberg M. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol. Cell. 2010;37:67–78. doi: 10.1016/j.molcel.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 33.Yakubovskaya E, Mejia E, Byrnes J, Hambardjieva E, Garcia-Diaz M. Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell. 2010;141:982–993. doi: 10.1016/j.cell.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 35.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 36.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 37.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 38.Guinier A. La diffraction des rayons X aux tres petits angles; application a l'etude de phenomenes ultramicroscopiques. Annals Physics. 1939;12:161–237. [Google Scholar]
- 39.Semenyuk AV, Svergun DI. GNOM - a program package for small-angle scattering data processing. J. Appl. Crystallogr. 1991;24:537–540. [Google Scholar]
- 40.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 41.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 44.Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys. Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins. 2006;65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 46.Schwinghammer K, Cheung AC, Morozov YI, Agaronyan K, Temiakov D, Cramer P. Structure of human mitochondrial RNA polymerase elongation complex. Nat. Struct. Mol. Biol. 2013;20:1298–1303. doi: 10.1038/nsmb.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guja KE, Venkataraman K, Yakubovskaya E, Shi H, Mejia E, Hambardjieva E, Karzai AW, Garcia-Diaz M. Structural basis for S-adenosylmethionine binding and methyltransferase activity by mitochondrial transcription factor B1. Nucleic Acids Res. 2013;41:7947–7959. doi: 10.1093/nar/gkt547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Svergun D, Barberato C, Koch MHJ. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 49.Sologub M, Kochetkov SN, Temiakov DE. Transcription and its regulation in mammalian and human mitochondria. Mol. Biol. 2009;43:215–229. [PubMed] [Google Scholar]
- 50.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 51.Chang DD, Clayton DA. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell. 1984;36:635–643. doi: 10.1016/0092-8674(84)90343-x. [DOI] [PubMed] [Google Scholar]
- 52.Hallberg BM, Larsson NG. TFAM forces mtDNA to make a U-turn. Nat. Struct. Mol. Biol. 2011;18:1179–1181. doi: 10.1038/nsmb.2167. [DOI] [PubMed] [Google Scholar]
- 53.Litonin D, Sologub M, Shi Y, Savkina M, Anikin M, Falkenberg M, Gustafsson CM, Temiakov D. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem. 2010;285:18129–18133. doi: 10.1074/jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol. Cell. Biol. 2003;23:5816–5824. doi: 10.1128/MCB.23.16.5816-5824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dinardo MM, Musicco C, Fracasso F, Milella F, Gadaleta MN, Gadaleta G, Cantatore P. Acetylation and level of mitochondrial transcription factor A in several organs of young and old rats. Biochem. Biophys. Res. Commun. 2003;301:187–191. doi: 10.1016/s0006-291x(02)03008-5. [DOI] [PubMed] [Google Scholar]
- 56.Rubio-Cosials A, Sidow JF, Jimenez-Menendez N, Fernandez-Millan P, Montoya J, Jacobs HT, Coll M, Bernado P, Sola M. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat. Struct. Mol. Biol. 2011;18:1281–1289. doi: 10.1038/nsmb.2160. [DOI] [PubMed] [Google Scholar]
- 57.Steitz TA. The structural changes of T7 RNA polymerase from transcription initiation to elongation. Curr. Opin. Struct. Biol. 2009;19:683–690. doi: 10.1016/j.sbi.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foury F, Roganti T, Lecrenier N, Purnelle B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 1998;440:325–331. doi: 10.1016/s0014-5793(98)01467-7. [DOI] [PubMed] [Google Scholar]
- 59.Osinga KA, De Vries E, Van der Horst GT, Tabak HF. Initiation of transcription in yeast mitochondria: analysis of origins of replication and of genes coding for a messenger RNA and a transfer RNA. Nucleic Acids Res. 1984;12:1889–1900. doi: 10.1093/nar/12.4.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lodeiro MF, Uchida A, Bestwick M, Moustafa IM, Arnold JJ, Shadel GS, Cameron CE. Transcription from the second heavy-strand promoter of human mtDNA is repressed by transcription factor A in vitro. Proc. Natl Acad. Sci. USA. 2012;109:6513–6518. doi: 10.1073/pnas.1118710109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zollo O, Tiranti V, Sondheimer N. Transcriptional requirements of the distal heavy-strand promoter of mtDNA. Proc. Natl Acad. Sci. USA. 2012;109:6508–6512. doi: 10.1073/pnas.1118594109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.